Abstract
Polycystic ovary syndrome (PCOS) is one of the most common endocrine disorders in women. It is characterized by chronic anovulation, hyperandrogenism, obesity and a predisposition to type 2 diabetes mellitus (T2DM). Since obesity plays an important role in the etiology of PCOS, we sought to determine if variants in the perilipin gene (PLIN), a gene previously implicated in the development of obesity, were also associated with PCOS. We typed six single nucleotide polymorphisms (haplotype tagging and/or previously associated with obesity or related metabolic traits) in PLIN in 305 unrelated non-Hispanic white women (185 with PCOS and 120 without PCOS). None of the variants was associated with PCOS (P < 0.05). However, the variant rs1052700*A was associated with increased risk for glucose intolerance (impaired glucose tolerance or T2DM) in both non-PCOS (OR = 1.75 [1.02-3.01], P = 0.044) and PCOS subjects (OR = 1.67 [1.08-2.59], P = 0.022). It was also associated with increased LDL (P = 0.007) and total cholesterol levels (P = 0.042). These results suggest that genetic variation in PLIN may affect glucose and lipid metabolism in women both with and without PCOS.
Keywords: Perilipin, polycystic ovary syndrome (PCOS), type 2 diabetes mellitus, glucose, lipid
INTRODUCTION
The clinical manifestations of polycystic ovary syndrome (PCOS), one of the most common endocrine disorders in women include chronic anovulation, hyperandrogenism, obesity, and a predisposition to type 2 diabetes mellitus (T2DM) [1-4]. Insulin resistance is a key contributor to the phenotypic manifestations of PCOS and appears to be a heritable component of the disorder [4-7]. Thus, efforts to identify genes contributing to the etiology of PCOS have focused on those affecting metabolic pathways related to obesity and insulin action [8-10].
Perilipins are hormonally-regulated phosphoproteins found on the surface of lipid storage droplets in adipocytes and steroidogenic cells of the adrenal cortex, testes and ovaries [11]. They modulate deposition and mobilization of triglycerides in the adipocyte by inhibiting hormone sensitive lipase (HSL)-mediated lipolysis [12,13]. Targeted knockout of the perilipin gene in mice results in constitutive activation of adipocyte HSL and resistance to diet-induced and genetic obesity [12,13]. Variants in the perilipin gene (PLIN) has been previously associated with measures of obesity and lipid levels in humans [14-23]. We therefore hypothesized that variants in PLIN may be associated with the phenotype of PCOS. To test this hypothesis, we examined variants in PLIN for association with PCOS as well as metabolic abnormalities characteristic of PCOS.
RESEARCH DESIGN AND METHODS
Non-Hispanic white women with PCOS and without PCOS living in Chicago and St. Louis were recruited for this study. Subjects with PCOS were recruited without regard to personal or family history of glucose tolerance. All were at least 2 years post-menarche and <40 years of age. A diagnosis of PCOS required the presence of oligo/amenorrhea, hyperandrogenemia (plasma free testosterone level ≥ 34.7 pmol/L), hyperandrogenism as evidenced by infertility, hirsutism, acne or androgenetic alopecia, and exclusion of nonclassic 21-hydroxylase deficiency congenital adrenal hyperplasia, hypothyroidism, or significant elevations in serum prolactin (when screened for clinically indicated Cushing's syndrome). The non-PCOS group was comprised of healthy post-pubertal girls and women. The study was approved by the Institutional Review Boards of the University of Chicago and Washington University, St. Louis and written informed consent was obtained from each subject.
All individuals, with the exception of those with known T2DM, had an oral glucose tolerance test (OGTT). After an overnight fast, blood samples were obtained at times −15 and 0 min. 75 g of dextrose was then administered orally and blood samples were obtained at 30, 60, 90 and 120 min for measurement of glucose and insulin concentrations. Glucose tolerance status was based upon the plasma glucose concentration at 2 h using criteria of the American Diabetes Association [24]. A diagnosis of normal glucose tolerance (NGT), impaired glucose tolerance (IGT), or T2DM was assigned if the glucose level at 2 h was <7.8 mmol/L, between 7.8 and 11.1 mmol/L, or > 11.1 mmol/L, respectively.
Plasma glucose was measured immediately using an automated glucose analyzer (YSI Model 2300 STAT, Yellow Springs Instruments Co., Yellow Springs, OH). Serum insulin was measured using a double-antibody technique in the Ligand Assay Core laboratories of the University of Chicago and Washington University Diabetes Research and Training Centers.
Genomic DNA was isolated from peripheral blood lymphocytes. We genotyped seven single nucleotide polymorphisms (SNPs) (rs4578621, rs2289487, rs6496589, rs894160, rs894162, rs2304795 and rs1052700) using Taqman-based assays on an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA); however, the assay for rs894160 failed. These seven SNPs were selected because they tagged (r2 = 0.8) all SNPs within 2 kb upstream and downstream of PLIN with minor allele frequency (MAF) > 0.01 in the HapMap phase II Utah residents with ancestry from northern and western Europe (CEU), or showed association with obesity or metabolic traits in previous studies [14-23]. All polymorphisms (e.g. rs4578621 C>T) are labeled by their dbSNP rs# with the common allele followed by the rare allele in the minus strand orientation (forward transcription orientation).
Hardy-Weinberg equilibrium (HWE) was assessed for each SNP in PCOS and non-PCOS groups separately. Estimated pairwise linkage disequilibrium (LD) was performed using JLIN (http://www.genepi.com.au/projects/jlin; [25]). Allele frequencies differences between PCOS and non-PCOS groups were compared using a χ2 test or Fisher's exact test as appropriate and presented with odds ratio (OR) and 95% confidence interval (CI). We compared the haplotype frequency distribution in subjects with and without PCOS using PHASE (v. 2.0.2, http://www.stat.washington.edu/stephens/software.html [26,27]. Multivariate linear regression was used to assess the association between genetic variants at PLIN and phenotypic traits with or without adjustment for age and BMI as covariates. Subjects with impaired glucose tolerance or T2DM were removed from consideration in this portion of the analysis as their values for many of the metabolic phenotypes could be affected by hyperglycemia and/or hyperinsulinemia. Continuous variables that were not normally distributed were logarithmically transformed and expressed as mean ± standard deviation (SD) or geometric mean (95% CI). Additive genetic models were used except for rs4578621 in the non-PCOS group in which case a dominant genetic model for the minor allele was used due to the small number (N=2) of individuals homozygous for the minor allele. All statistical tests were performed with SPSS (SPSS for Windows, v. 11.5; SPSS, Chicago, IL, USA) unless specified otherwise.
RESULTS
We studied 185 and 120 women with and without PCOS respectively. All were of European descent and classified as non-Hispanic white. Compared to the non-PCOS group, those with PCOS were significantly younger (Table 1). Within the PCOS group, 102 subjects (55%) had NGT, 72 (39%) had IGT and 11 (6%) had T2DM, based on the results of an OGTT. In contrast, 77 (64%) of non-PCOS women had NGT, 40 (33%) had IGT and 3 (3%) had T2DM.
Table 1.
Clinical characteristics of non-Hispanic white female PCOS and non-PCOS subjects classified by glucose tolerance status
| NGT |
IGT and T2DM |
|||||
|---|---|---|---|---|---|---|
| non-PCOS | PCOS | P | non-PCOS | PCOS | P | |
| N | 77 | 102 | 43 | 83 | ||
| Age (yr) | 32.3 ± 10.9 | 28.4 ± 6.1 | 0.005 | 41.6 ± 10.6 | 30.8 ± 6.3 | <0.001 |
| Body mass index (kg/m2) | 26.8 ± 6.8 | 36.3 ± 8.0 | <0.001 | 31.6 ± 8.4 | 38.2 ± 7.3 | <0.001 |
| Glycohemoglobin (%) | 5.3 (5.2-5.4) | 5.3 (5.1-5.4) | 0.676 | 5.6 (5.5-5.8) | 5.5 (5.3-5.7) | 0.298 |
| Fasting glucose (mmol/l) | 5.0 (4.9-5.0) | 5.0 (4.9-5.1) | 0.792 | 5.3 (5.0-5.5) | 5.4 (5.3-5.6) | 0.269 |
| 2-h glucose (mmol/l) | 6.5 (6.4-6.7) | 6.3 (6.2-6.5) | 0.151 | 9.2 (8.8-9.6) | 9.3 (9.0-9.6) | 0.577 |
| Fasting insulin (pmol/l) | 30.2 (25.6-35.5) | 120.2 (105.2-137.3) | <0.001 | 54.6 (42.6-70) | 162.0 (138.0-190.2) | <0.001 |
| 2-h insulin (pmol/l) | 197.5 (174.3-223.8) | 603.4 (525.5-692.9) | <0.001 | 406.5 (336.6-490.8) | 1186.2 (1034-1360.7) | <0.001 |
| HOMA-IR | 1.1 (0.9-1.3) | 4.4 (3.8-5.1) | <0.001 | 2.1 (1.6-2.8) | 6.5 (5.5-7.7) | <0.001 |
Mean ± SD or geometric mean (95% CI) are shown.
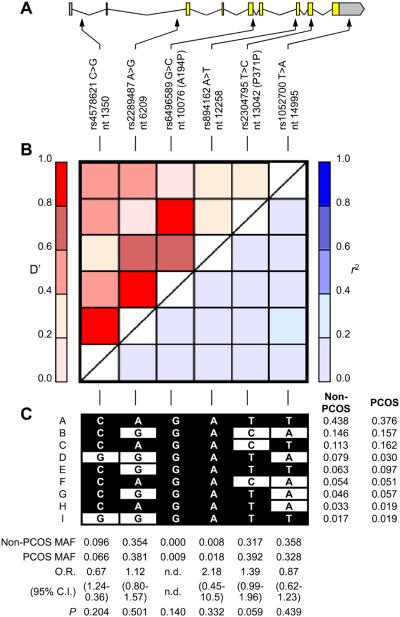
We studied six single nucleotide polymorphisms (SNPs) including two coding and four non-coding SNPs that were successfully genotyped (Figure 1). All SNPs were in Hardy-Weinberg equilibrium in both PCOS and non-PCOS groups. These SNPs covered 85% of HapMap phase II SNPs in this region at r2 of 0.8 in the CEU samples, and/or were associated with metabolic or obesity related phenotypes in previous studies [14-23]. There were no significant differences in allele frequencies between PCOS and non-PCOS subjects (Figure 1) although the minor C allele of rs2304795 (rs2304795*C) approached but did not reach statistical significance with PCOS (OR = 1.39 (95% CI = 0.99-1.96), P = 0.059). PCOS and non-PCOS groups had significantly different six-SNP haplotype frequencies (P = 0.04), but after the two rare polymorphisms (rs6496589 and rs894162; minor allele frequency (MAF) < 0.02) were removed from consideration, the four-SNP haplotype frequencies were no longer significantly different (P = 0.08). These two SNPs were not included in subsequent analyses.
Fig. 1.
PLIN structure, linkage disequilibrium, and genotype and haplotype frequencies. A: Intron/exon structure of PLIN and the location of SNPs studied. The sequence and intron/exon boundary of PLIN was based on GenBank accession number AB005293. rs4578621 C>T, etc., denote the polymorphism and refer to the common and rare alleles, respectively. Nucleotide (nt) positions are indicated (the “A” of the initiator Methionine ATG codon at NT_010274 position 5188832 is nt position 1), and synonymous and nonsynonymous coding region polymorphisms follow in brackets. B: Pairwise linkage disequilibrium of PLIN SNPs. D' is shown in upper left triangle and r2 in lower right triangle. C: Common haplotypes. Haplotypes occurring at frequency > 0.01 are shown with their frequencies in PCOS and non-PCOS groups on the right. Common alleles are indicated by black squares, and rare alleles in white squares. Beneath each SNP is the minor allele frequency (MAF) in the PCOS and non-PCOS groups and the results of allelic association tests.
Since glucose intolerance (IGT and T2DM) is common in PCOS, we tested the hypothesis that genetic variation in PLIN may modulate the glucose and insulin responses to an oral glucose challenge in these subjects. The rs1052700*A allele was associated with glucose intolerance in PCOS (OR = 1.67 (1.08-2.59), P = 0.022; Table 2). Similar effects were observed in subjects without PCOS (OR = 1.75 (1.02-3.01), P = 0.044). The rs4578621*G allele was also associated with glucose intolerance in PCOS subjects (OR = 2.61 (1.05-6.46), P = 0.039) but not in non-PCOS subjects (OR = 1.17 (0.48-2.82), P = 0.729). The associations observed with these two SNPs represent independent observations as there is no substantial linkage disequilibrium (LD) between them (D' = 0.52, r2 = 0.05, Figure 1).
Table 2.
Association of PLIN polymorphisms with glucose tolerance in non-Hispanic white women
| rs4578621 C/G * | rs2289487 A/G * | rs2304795 T/C * | rs1052700 T/A * | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comparison | N | MAF | OR† (95% CI) |
P | MAF | OR† (95% CI) |
P | MAF | OR† (95% CI) |
P | MAF | OR† (95% CI) |
P |
| non-PCOS NGT vs. | 77 | 0.091 | 1 | 0.318 | 1 | 0.299 | 1 | 0.312 | 1 | ||||
| non-PCOS IGT/T2DM | 43 | 0.105 | 1.17 (0.48-2.82) |
0.729 | 0.419 | 1.54 (0.89-2.66) |
0.119 | 0.349 | 1.26 (0.72-2.20) |
0.423 | 0.442 | 1.75 (1.02-3.01) |
0.044 |
| PCOS NGT vs. | 102 | 0.040 | 1 | 0.358 | 1 | 0.363 | 1 | 0.277 | 1 | ||||
| PCOS IGT/T2DM | 83 | 0.099 | 2.61 (1.05-6.46) |
0.039 | 0.410 | 1.25 (0.82-1.90) |
0.308 | 0.428 | 1.31 (0.86-2.00) |
0.203 | 0.390 | 1.67 (1.08-2.59) |
0.022 |
The first and second alleles represent the major and minor alleles, respectively.
for minor allele as risk allele.
MAF, minor allele frequency
The effect of PLIN upon metabolic measures in normoglycemic non-PCOS subjects were assessed by examining the genotypic associations with BMI, lipids, glucose and insulin levels obtained during the OGTT (Table 3). The variant rs1052700*A was associated with low density lipoprotein cholesterol (LDL-C) and total cholesterol levels even after adjustment for age and BMI in the case of LDL-C (P = 0.007 and Padj = 0.014 for LDL-C; P = 0.042 and Padj = 0.073 for total cholesterol). The variant rs2289487*G was also associated with LDL-C level (P = 0.036 and Padj = 0.046) and fasting glucose level (P = 0.045 and Padj = 0.039) in normoglycemic non-PCOS subjects. However, none of the metabolic phenotypes including BMI, sex hormone concentrations, glucose and insulin levels at basal and during OGTT showed significant association with PLIN in the normoglycemic PCOS women (data not shown).
Table 3.
Association of PLIN polymorphisms with metabolic traits in normoglycemic non-Hispanic white women without PCOS
| rs4578621 |
rs2289487 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | P* | Padj† | AA | AG | GG | P* | Padj† | ||
| N | 63 | 14 | 0 | N | 36 | 33 | 8 | ||||
| Age (yr) | 31.8 ± 10.7 | 34.6 ± 11.7 | 0.374 | Age (yr) | 33.1 ± 11.6 | 32.2 ± 10.6 | 28.8 ± 8.9 | 0.363 | |||
| BMI (kg/m2) | 26.9 ± 7.2 | 26.4 ± 5.3 | 0.771 | BMI (kg/m2) | 26 ± 5.7 | 26.8 ± 6.3 | 30.8 ± 11.8 | 0.117 | |||
| Total cholesterol (mg/dL) | 165.8 ± 31 | 172.6 ± 14.2 | 0.480 | 0.555 | Total cholesterol (mg/dL) | 159.7 ± 23.7 | 174 ± 31.8 | 162.4 ± 26.2 | 0.262 | 0.276 | |
| Triglycerides (mg/dL) | 70.4 (58.6-84.7) | 60.3 (44.8-81.1) | 0.440 | 0.550 | Triglycerides (mg/dL) | 64.5 (48.7- 85.3) |
77 (63.3-93.7) | 45.6 (22-94.5) | 0.832 | 0.543 | |
| HDL (mg/dL) | 54.4 ± 12.4 | 53.5 ± 10.4 | 0.836 | 0.574 | HDL (mg/dL) | 56.6 ± 12.5 | 52.9 ± 11.4 | 50.6 ± 12.5 | 0.186 | 0.383 | |
| LDL (mg/dL) | 94.3 ± 26.7 | 106 ± 17.7 | 0.173 | 0.178 | LDL (mg/dL) | 86.9 ± 20.3 | 103.7 ± 27.6 | 101.2 ± 26 | 0.036 | 0.046 | |
| Glycohemoglobin (%) | 5.3 (5.2-5.4) | 5.3 (5.1-5.4) | 0.883 | 0.757 | Glycohemoglobin (%) | 5.3 (5.1-5.4) | 5.3 (5.2-5.4) | 5.4 (5.1-5.7) | 0.263 | 0.317 | |
| Fasting glucose (mmol/l) | 4.9 (4.9-5) | 5 (4.7-5.3) | 0.678 | 0.763 | Fasting glucose (mmol/l) | 4.9 (4.8-4.9) | 5 (4.9-5.2) | 5 (4.7-5.4) | 0.045 | 0.039 | |
| 2-h glucose (mmol/l) | 6.5 (6.3-6.7) | 6.5 (6.2-6.8) | 0.904 | 0.920 | 2-h glucose (mmol/l) | 6.5 (6.2-6.8) | 6.5 (6.3-6.8) | 6.8 (6.2-7.4) | 0.511 | 0.438 | |
| Fasting insulin(pmol/l) | 29.7 (24.6-35.9) | 32.2 (22.5-46) | 0.712 | 0.263 | Fasting insulin(pmol/l) | 26.9 (21.5- 33.8) |
30.8 (24-39.6) | 46.3 (21.5- 99.4) |
0.071 | 0.537 | |
| 2-h insulin (pmol/l) | 195.2 (169.5- 224.9) |
207.9 (154.2- 280.4) |
0.699 | 0.343 | 2-h insulin (pmol/l) | 189.1 (160.4- 222.9) |
200.3 (162.1- 247.4) |
227.5 (129.1- 401) |
0.398 | 0.923 | |
| HOMA-IR | 1.1 (0.9-1.3) | 1.2 (0.8-1.7) | 0.693 | 0.271 | HOMA-IR | 1 (0.8-1.2) | 1.1 (0.9-1.5) | 1.7 (0.8-3.9) | 0.053 | 0.418 | |
| rs2304795 |
rs1052700 |
||||||||||
| TT | TC | CC | P * | Padj † | TT | TA | AA | P * | Padj † | ||
| N | 40 | 28 | 9 | N | 36 | 34 | 7 | ||||
| Age (yr) | 32.6 ± 10.9 | 31.4 ± 10.8 | 33.8 ± 12.4 | 0.978 | Age (yr) | 31.4 ± 10.8 | 33.7 ± 11.2 | 29.6 ± 10.8 | 0.856 | ||
| BMI (kg/m2) | 27.1 ± 6.6 | 27.4 ± 7.9 | 24 ± 3.3 | 0.399 | BMI (kg/m2) | 25.6 ± 6 | 28.2 ± 7.9 | 26.7 ± 4.5 | 0.260 | ||
| Total cholesterol (mg/dL) | 170.6 ± 31.3 | 164.6 ± 25 | 159.3 ± 28.6 | 0.291 | 0.315 | Total cholesterol (mg/dL) | 159.3 ± 24.8 | 171.3 ± 31.6 | 185.3 ± 15.9 | 0.042 | 0.073 |
| Triglycerides (mg/dL) | 79.9 (63.6- 100.2) |
57.8 (44.5-75.1) | 58.2 (36.9- 91.9) |
0.066 | 0.119 | Triglycerides (mg/dL) | 67.2 (54.5-83) | 69.1 (52.9- 90.3) |
70.2 (46.3- 106.4) |
0.851 | 0.998 |
| HDL (mg/dL) | 55.1 ± 12.7 | 52.9 ± 12.2 | 54.7 ± 8.4 | 0.732 | 0.436 | HDL (mg/dL) | 56.5 ± 13.3 | 52.4 ± 10.4 | 53.3 ± 13.9 | 0.279 | 0.326 |
| LDL (mg/dL) | 96.4 ± 25.6 | 98.2 ± 28.1 | 91.9 ± 19.6 | 0.835 | 0.955 | LDL (mg/dL) | 87.4 ± 21.7 | 101.4 ± 27.2 | 117.8 ± 14.2 | 0.007 | 0.014 |
| Glycohemoglobin (%) | 5.3 (5.2-5.4) | 5.3 (5.2-5.4) | 5.3 (5.1-5.5) | 0.976 | 0.768 | Glycohemoglobin (%) | 5.3 (5.2-5.4) | 5.3 (5.2-5.4) | 5.3 (5-5.5) | 0.919 | 0.765 |
| Fasting glucose (mmol/l) | 4.9 (4.8-5) | 5 (4.8-5.2) | 5.1 (4.8-5.4) | 0.112 | 0.059 | Fasting glucose (mmol/l) | 4.9 (4.8-5) | 5.1 (4.9-5.2) | 4.9 (4.6-5.2) | 0.147 | 0.248 |
| 2-h glucose (mmol/l) | 6.4 (6.1-6.6) | 6.8 (6.5-7) | 6.5 (5.9-7.2) | 0.150 | 0.162 | 2-h glucose (mmol/l) | 6.4 (6.2-6.6) | 6.7 (6.3-7) | 6.6 (6.2-7) | 0.279 | 0.224 |
| Fasting insulin(pmol/l) | 27.8 (22.7-34.1) | 37.3 (27-51.7) | 21.4 (15.3- 29.9) |
0.906 | 0.454 | Fasting insulin(pmol/l) | 26.3 (21.1- 32.7) |
34.1 (25.6- 45.5) |
34.1 (21.3- 54.5) |
0.152 | 0.335 |
| 2-h insulin (pmol/l) | 188.1 (159.9- 221.2) |
232 (185.8- 289.7) |
143.6 (93.3- 220.9) |
0.853 | 0.874 | 2-h insulin (pmol/l) | 181.3 (154.8- 212.3) |
213.8 (172.4- 265.1) |
211.3 (116.4- 383.7) |
0.252 | 0.366 |
| HOMA-IR | 1 (0.8-1.2) | 1.4 (1-1.9) | 0.8 (0.6-1.2) | 0.791 | 0.359 | HOMA-IR | 0.9 (0.8-1.2) | 1.3 (0.9-1.7) | 1.2 (0.8-2) | 0.129 | 0.295 |
Mean ± SD or geometric mean (95% CI) are shown.
Not adjusted for age and BMI.
Adjusted for age and BMI.
DISCUSSION
Our results are consistent with earlier studies indicating that variants in the PLIN gene are associated with metabolic abnormalities related to obesity, insulin resistance and glucose intolerance in women [14-23]. Our results further suggest that genetic variation in PLIN is associated with glucose intolerance in non-Hispanic white women. These effects, particularly for the variant rs1052700, were seen in women both with and without PCOS suggesting they are not related to PCOS per se.
The molecular mechanism(s) by which perilipins affect glucose and lipid metabolism in vivo remain unclear. Perilipins increase triacylglycerol storage in adipocytes by forming a physical barrier that reduces the action of soluble lipases on stored lipids, thus inhibiting triacylglycerol hydrolysis in the basal state [12,13,28]. Phosphorylation of perilpins allows HSL to hydrolyze triacylglycerol [12,13,29]. In addition, tumor necrosis factor–α, one of the key adipokines associated with insulin resistance, has been reported to affect perilipin function [30]. Thus, it seems reasonable to assume that perilipin might affect glucose intolerance via the modulation of lipid metabolism. We also observed that rs2289487*G and rs1052700*A are associated with LDL-C levels. A previous study showed that two other SNPs in PLIN, rs2289487 and rs894160, were associated with LDL-C levels [15]. The variant rs2289487 is in moderate LD with rs1052700 (r2 = 0.349) and rs894160 (r2=0.733) in the CEU HapMap sample suggesting that the SNP cluster rs1052700, rs894160 and rs2289487 are associated with LDL-C levels.
In summary, our studies suggest that genetic variation in PLIN affects glucose and lipid metabolism in women and perhaps risk of PCOS. However, our sample size is small and as a consequence, it is important to determine if these results can be replicated in studies of larger groups of women, both non-Hispanic white and other racial groups.
Acknowledgments
This study was supported by U.S. Public Health Service grants DK-20595 (University of Chicago DRTC), DK-20579 (Washington University DRTC), DK-31842, DK-47486, DK-55889, RR-00055 (University of Chicago CRC) and RR-24992 (Washington University CTSA), a Clinical Research Award to D.A.E., an American Diabetes Association Mentored Postdoctoral Fellowship Award (to N.J.C., M.G.H.), and a gift from the Kovler Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors report no conflict of interest.
REFERENCES
- 1.Knochenhauer E, Key T, Kahsar-Miller M, Waggoner W, Boots L, Azziz R. Prevalence of the polycystic ovary syndrome in unselected black and white women of the southeastern United States: a prospective study. J Clin Endocrinol Metab. 1998;83:3078–3082. doi: 10.1210/jcem.83.9.5090. [DOI] [PubMed] [Google Scholar]
- 2.Solomon CG. The epidemiology of polycystic ovary syndrome. Prevalence and associated disease risks. Endocrinol Metab Clin North Am. 1999;28:247–263. doi: 10.1016/s0889-8529(05)70069-4. [DOI] [PubMed] [Google Scholar]
- 3.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–2749. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 4.Ehrmann DA. Polycystic ovary syndrome. N Engl J Med. 2005;352:1223–1236. doi: 10.1056/NEJMra041536. [DOI] [PubMed] [Google Scholar]
- 5.Legro R, Driscoll D, Strauss J, 3rd, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci U S A. 1998;95:14956–14960. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legro RS, Bentley-Lewis R, Driscoll D, Wang SC, Dunaif A. Insulin resistance in the sisters of women with polycystic ovary syndrome: association with hyperandrogenemia rather than menstrual irregularity. J Clin Endocrinol Metab. 2002;87:2128–2133. doi: 10.1210/jcem.87.5.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sam S, Legro RS, Essah PA, Apridonidze T, Dunaif A. Evidence for metabolic and reproductive phenotypes in mothers of women with polycystic ovary syndrome. Proc Natl Acad Sci U S A. 2006;103:7030–7035. doi: 10.1073/pnas.0602025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara M, Alcoser SY, Qaadir A, Beiswenger KK, Cox NJ, Ehrmann DA. Insulin resistance is attenuated in women with polycystic ovary syndrome with the Pro(12)Ala polymorphism in the PPARgamma gene. J Clin Endocrinol Metab. 2002;87:772–775. doi: 10.1210/jcem.87.2.8255. [DOI] [PubMed] [Google Scholar]
- 9.Ehrmann DA, Tang X, Yoshiuchi I, Cox NJ, Bell GI. Relationship of insulin receptor substrate-1 and -2 genotypes to phenotypic features of polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:4297–4300. doi: 10.1210/jc.2002-020216. [DOI] [PubMed] [Google Scholar]
- 10.Barber TM, Bennett AJ, Groves CJ, Sovio U, Ruokonen A, Martikainen H, et al. Association of variants in the fat mass and obesity associated (FT0) gene with polycystic ovary syndrome. Diabetologia. 2008;51:1153–1158. doi: 10.1007/s00125-008-1028-6. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg A, Egan J, Wek S, Garty N, Blanchette-Mackie E, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 12.Martinez-Botas J, Anderson J, Tessier D, Lapillonne A, Chang B, Quast M, et al. Absence of perilipin results in leanness and reverses obesity in Leprdb/db mice. Nat Genet. 2000;26:474–479. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- 13.Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan W, Chen S, Huang J, Shen Y, Qiang B, Gu D. Polymorphisms in PLIN and hypertension combined with obesity and lipid profiles in Han Chinese. Obes Res. 2004;12:1733–1737. doi: 10.1038/oby.2004.214. [DOI] [PubMed] [Google Scholar]
- 15.Qi L, Shen H, Larson I, Schaefer EJ, Greenberg AS, Tregouet DA, et al. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res. 2004;12:1758–1765. doi: 10.1038/oby.2004.218. [DOI] [PubMed] [Google Scholar]
- 16.Qi L, Corella D, Sorli JV, Portoles O, Shen H, Coltell O, et al. Genetic variation at the perilipin (PLIN) locus is associated with obesity-related phenotypes in white women. Clin Genet. 2004;66:299–310. doi: 10.1111/j.1399-0004.2004.00309.x. [DOI] [PubMed] [Google Scholar]
- 17.Qi L, Tai ES, Tan CE, Shen H, Chew SK, Greenberg AS, et al. Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med. 2005;83:448–456. doi: 10.1007/s00109-004-0630-4. [DOI] [PubMed] [Google Scholar]
- 18.Corella D, Qi L, Sorli JV, Godoy D, Portoles O, Coltell O, et al. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J Clin Endocrinol Metab. 2005;90:5121–5126. doi: 10.1210/jc.2005-0576. [DOI] [PubMed] [Google Scholar]
- 19.Jang Y, Kim OY, Lee JH, Koh SJ, Chae JS, Kim JY, et al. Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 2006;30:1601–1608. doi: 10.1038/sj.ijo.0803312. [DOI] [PubMed] [Google Scholar]
- 20.Corella D, Qi L, Tai ES, Deurenberg-Yap M, Tan CE, Chew SK, et al. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high-saturated fat, low-carbohydrate diet. Diabetes Care. 2006;29:1313–1319. doi: 10.2337/dc06-0045. [DOI] [PubMed] [Google Scholar]
- 21.Kang ES, Cha BS, Kim HJ, Kim HJ, Kim SH, Hur KY, et al. The 11482G>A polymorphism in the perilipin gene is associated with weight gain with rosiglitazone treatment in type 2 diabetes. Diabetes Care. 2006;29:1320–1324. doi: 10.2337/dc05-2466. [DOI] [PubMed] [Google Scholar]
- 22.Qi L, Zhang C, Greenberg A, Hu FB. Common variations in perilipin gene, central obesity, and risk of type 2 diabetes in US women. Obesity. 2008;16:1061–1065. doi: 10.1038/oby.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deram S, Nicolau CY, Perez-Martinez P, Guazzelli I, Halpern A, Wajchenberg BL, et al. Effects of Perilipin (PLIN) gene variation on metabolic syndrome risk and weight loss in obese children and adolescents. J Clin Endocrinol Metab. 2008;93:4933–4940. doi: 10.1210/jc.2008-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2008;31(Suppl 1):S55–S60. doi: 10.2337/dc08-S055. [DOI] [PubMed] [Google Scholar]
- 25.Carter KW, McCaskie PA, Palmer LJ. JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics. 2006;7:60. doi: 10.1186/1471-2105-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C. Perilipin A increases triacylglycerol strage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–38493. doi: 10.1074/jbc.M007322200. [DOI] [PubMed] [Google Scholar]
- 29.Sztalryd C, Xu G, Dorward H, Tansey J, Contreras J, Kimmel A, et al. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161:1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souza SC, de Vargas LM, Yamamoto MT, Lien P, Franciosa MD, Moss LG, et al. Overexpression of perilipin A and B blocks the ability of tumor necrosis factor alpha to increase lipolysis in 3T3-L1 adipocytes. J Biol Chem. 1998;273:24665–24669. doi: 10.1074/jbc.273.38.24665. [DOI] [PubMed] [Google Scholar]