Abstract
Prostate cancer is the second leading cause of cancer-related death among the American male population, and society is in dire need of new approaches to treat this disease. Here we report the design, synthesis, and biological evaluation of a class of bifunctional small molecules, called antibody-recruiting molecules targeting prostate-cancer (ARM-Ps), that enhance the recognition of prostate cancer cells by the human immune system. ARM-P derivatives were designed rationally via the computational analysis of crystallographic data, and we demonstrate here that these materials are able to: (1) bind PSMA with high affinity (high pM to low nM), (2) template the formation of ternary complexes between anti-DNP antibodies, ARM-P, and LNCaP human prostate cancer cells, and (3) mediate the antibody-dependent killing of LNCaP cells in the presence of human effector cells. This manuscript describes the application of fundamental chemical principles to the design of a novel class of molecules with high therapeutic potential. We believe that this general small-molecule-based strategy could give rise to novel directions in treating cancer and other diseases.
Prostate cancer is the second leading cause of cancer-related death among the American male population, and it has been predicted that one out of every six American men will develop prostate cancer during their lifetime.1 Available treatment options, including chemical/surgical castration, radiation therapy, and chemotherapy, are often ineffective against advanced disease, and are also associated with severe side effects.2 Thus, new approaches to treat prostate cancer are highly desirable. To this end, monoclonal antibody therapies have shown promise;2 however no such agent has yet successfully obtained FDA approval for treating prostate cancer. Further, antibody drugs are limited by severe side effects, lack of oral bioavailabiliy, and high cost.3 Here we describe a novel technology for prostate cancer treatment that we believe could address many of the limitations of currently available therapies, and combines advantages of both small-molecule-based and antibody-based strategies.
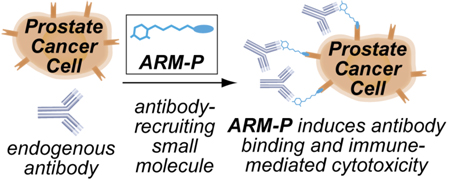
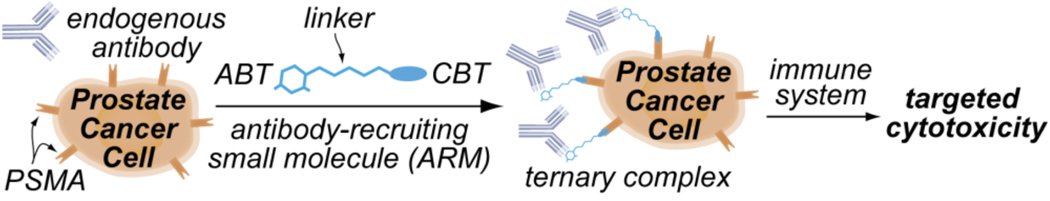
The key component of our approach is what we call antibody-recruiting small molecules targeting prostate cancer (ARM-Ps). These are bifunctional materials capable of redirecting antibodies already present in the human bloodstream to prostate cancer cell surfaces, and increasing their destruction by effector cells of the immune system (Figure 1). As shown, ARMs are composed of an antibody-binding terminus (ABT), a cell surface binding terminus (CBT), and a linker region. In this manuscript, it is demonstrated that ternary complexes formed between ARM-Ps, human prostate cancer cells (LNCaP cells), and antibodies recognizing the 2,4-dinitrophenyl (DNP) group lead to targeted cell-mediated cytotoxicity of LNCaP cells. The power of this approach derives from the observation that anti-DNP antibodies are already found in the human bloodstream in a high percentage of the human population,4 and are competent to mediate target cell killing.5,6 Several approaches have appeared that utilize bifunctional materials to recruit antibodies to human pathogens,7 but ARM-Ps are the first class of antibody-recruiting small molecules that target prostate cancer. The general strategy reported herein has the potential to initiate novel directions in treating cancer and other diseases.
Figure 1.
Schematic depiction of the reported approach to prostate cancer targeting. An antibody-recruiting small molecule (ARM) binds the cell-surface prostate cancer marker prostate-specific membrane antigen (PSMA), thus recruiting antibodies to these cells for recognition and targeted killing by the immune system. Bifunctional ARMs are composed of an antibody binding terminus (ABT), a linker region, and a cell-binding terminus (CBT).
Our first goal in constructing ARM-Ps was to design an appropriate cell-binding terminus (CBT), and to this end, we chose to target the prostate-specific membrane antigen (PSMA). PSMA is a cell surface protein that is highly overexpressed on prostate cancer cells versus normal cells of the prostate, and its expression increases with clinical stage.8 This protein has been exploited as a target both in prostate cancer imaging9 and in monoclonal antibody therapy for the disease.10
Several small molecule ligands have been developed that bind PSMA selectively and with high affinity, including 2-PMPA (1)12 and the glutamate ureas (2, Figure 2C).13 These compounds competitively inhibit PSMA’s enzymatic activity, and have been successfully modified for imaging, and targeted drug delivery applications.14 At the outset of our studies, we were intrigued by observations that 2 could accommodate a wide range of R-groups at C2, including various alkyl heterocycle substituents, with minimal loss of inhibitory potency.13 We therefore reasoned that we might be able to incorporate a linker to join the ABT and CBT at this position.
Figure 2.
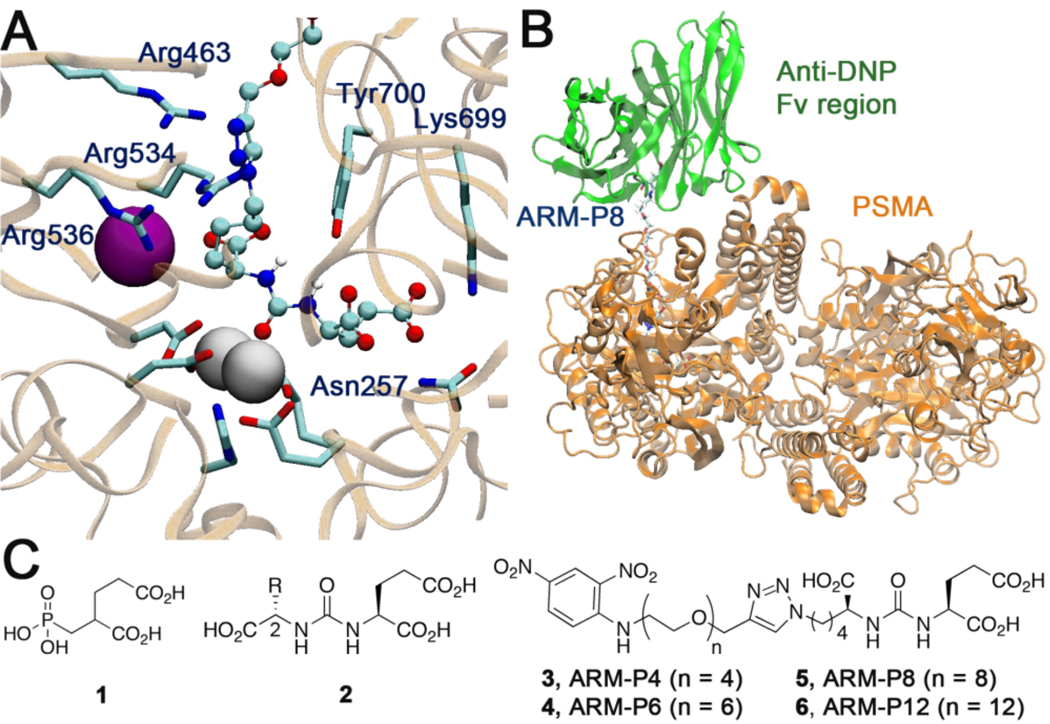
Structure-based design studies. (A) Modeled complex illustrating the design of a CBT for use in ARM-Ps. (B) Structural model of the ternary complex between the Fv region of an anti-DNP antibody, ARM-P, and the PSMA dimer. (C) Known PSMA-binding small molecules and structures of ARM-P derivatives utilized in this study. Figures 2A and B were created with the program VMD.11
Thus, starting from a crystal structure for the complex of PSMA with 1,15 the corresponding complex with 2 (R=H) was modeled using the program BOMB (biochemical and organic model builder).16 Stabilizing interactions are indicated with active site zinc ions, as well as hydrogen bonding and salt-bridge interactions with Tyr700, Lys699, Arg534, Arg536, and Asn257 (Figure 2A). This model was found subsequently to be consistent with the recently published co-crystal structure of PSMA in complex with urea-based ligands.17 Next, BOMB was used to construct complexes of 2 with alternative DNP linking groups. Among plausible designs, 1-butyl-4-alkyl-1,2,3-triazole analogues (e.g., 3–6) were judged promising owing to favorable electrostatic interactions with Arg463, π-stacking interactions with Tyr700, the orientation of the linker towards solvent, and ease of synthesis.
To estimate viable linker lengths, ternary complexes (PSMA, ARM-Ps, the Fv region of an anti-DNP antibody18) were constructed using the program FIRST (Figure 2B).19 Constrained geometric simulations20 were then performed to assemble the complex with the ABT and CBT binding sites in close proximity. The modeling suggested that at least 6 oxyethylene units would be necessary to prevent steric clashes between the two proteins, and that longer linkers might be preferable to prevent excessive dehydration of the protein-protein interface.
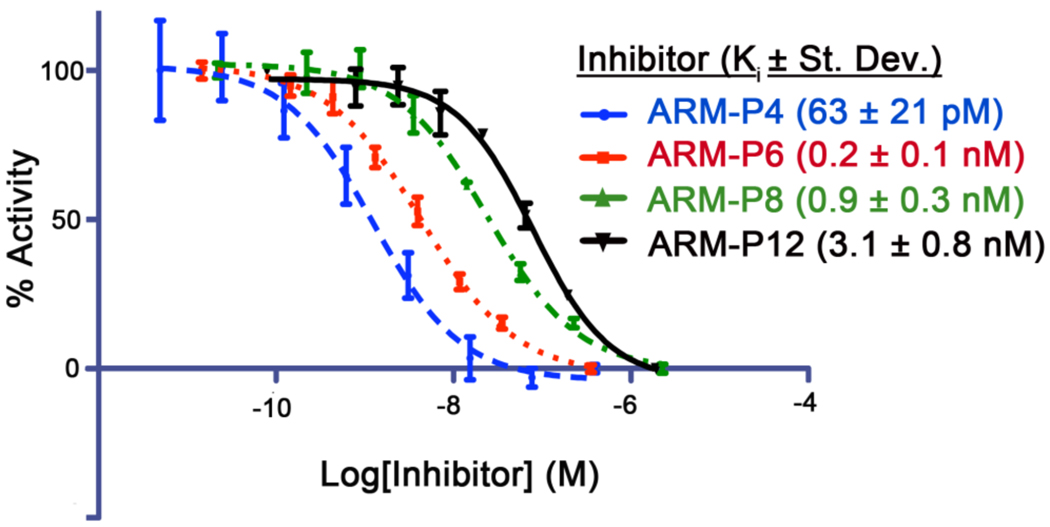
Thus, ARM-Ps 3–6 were synthesized (Figure 2C, called ARM-P4 through ARM-P12, for the number of oxyethylene units in the linker) and tested for binding to PSMA through a standard enzymatic inhibition assay.22 This assay measures the ability of designed small molecules to inhibit PSMA-catalyzed cleavage of the peptide substrate N-acetyl-aspartyl-glutamate (NAAG). As depicted in Figure 3, compound 3 (ARM-P4) inhibits PSMA with a Ki of 63 pM, a value similar to the most potent PSMA-binding small molecule developed to date.17 Although we anticipated that the flexible PEG linker in 3 could lead to entropic penalties in binding PSMA, perhaps stabilizing interactions with the triazole, DNP, and/or PEG portions of ARM-P4 are compensating. Interestingly, compounds 4–6, which contain 6, 8, and 12 oxyethylene units, exhibited decreased inhibitory potency versus ARM-P4 (3). The origin of the trend is currently under investigation.
Figure 3.
Representative PSMA inhibition curves for ARM-Ps. Ki values were calculated from measured IC50 and KM values through the Cheng-Prusoff equation21 and are reported as the average of 3 runs ± standard deviation.
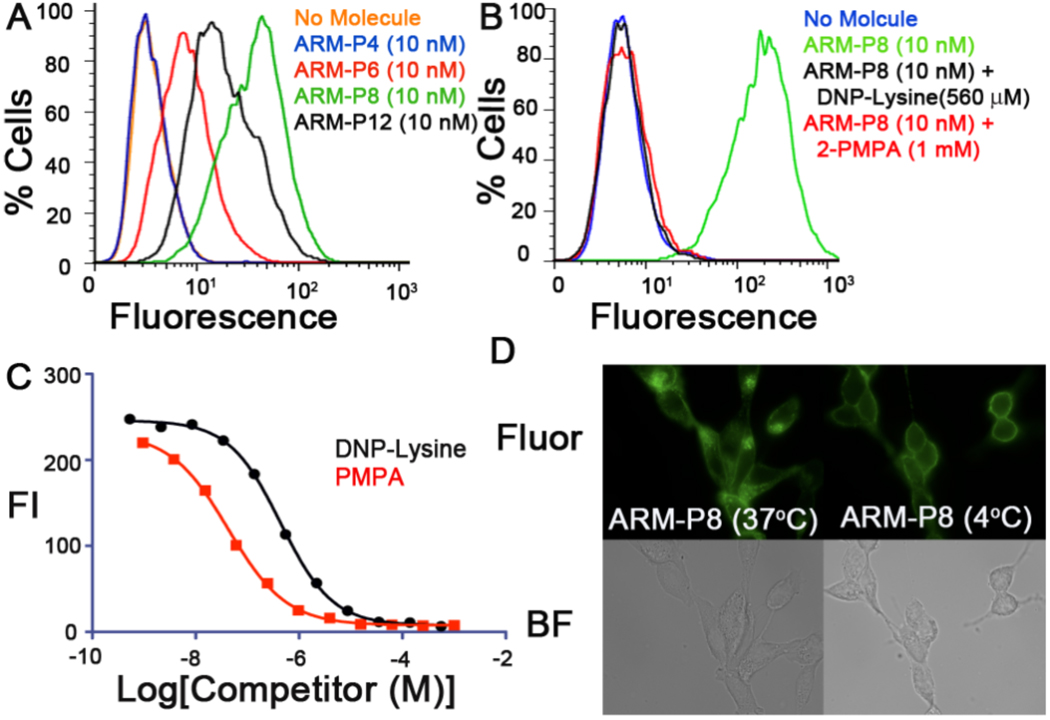
To evaluate the capacity of ARM-P derivatives to template ternary complex formation in a cellular environment, we performed live-cell flow cytometry assays with PSMA-expressing LNCaP cells and Alexafluor488 conjugated anti-DNP antibodies. Since the anti-DNP antibody represented the fluorescent component in these studies, rightward shifts of flow cytometry histograms indicate increased levels of ternary complex formation. These experiments revealed an intriguing trend (Figure 4A): although ARM-P4 (3) possessed the highest affinity in PSMA-binding assays, maximal amounts of ternary complex were formed in the presence of ARM-P8 (5). These results are consistent with predictions from computational modeling studies (Figure 2B, see above), which suggest that linker lengths of n=4 and 6 could lead to unfavorable steric interactions between antibody and PSMA. The relative decrease in ternary complex formation for ARM-P12 versus ARM-P8 may simply result from the decreased affinity of ARM-P12 for PSMA, consistent with results in Figure 3.
Figure 4.
Evaluating ternary complex formation. (A) and (B) Representative traces from flow cytometry experiments. (C) Dose dependence of competitor concentration on ternary complex. (D) Epifluorescence (Fluor) and brightfield (BF) microscopy experiments performed in the presence of ARM-P8 at 37 °C and 4 °C.
ARM-P8 was therefore chosen for evaluation in subsequent studies. Flow cytometry experiments performed in the excess of the competing ligands 2-PMPA or bis-DNP lysine (Figure 4B), reveal baseline levels of ternary complex. Further, no ternary complex formation was observed under these conditions using DU145 prostate cancer cells, which lack PSMA.23 Together, these data confirm that small-molecule-mediated antibody recruitment is dependent upon binding of ARM-P8 to both PSMA and anti-DNP antibodies. Further, competition with 2-PMPA and bis-DNP lysine was found to be concentration-dependent (Figure 4C). The Ki value determined for 2-PMPA in these studies was found to be 5.0 nM, which is almost identical to that determined in enzymatic assays (2.3 nM).23 This result implies that ARM-P binding to PSMA does not benefit from increased affinity due to multivalent presentation.7c,g Fluorescent microscopy experiments (Figure 4D) further confirm flow cytometry data, and demonstrate localization of the ternary complex to the cell membrane. No fluorescence was observed in the absence of ARM-P8.23 Endocytosis of fluorescent features at 37 °C, but not 4 °C, is consistent with reported behavior of PSMA.24
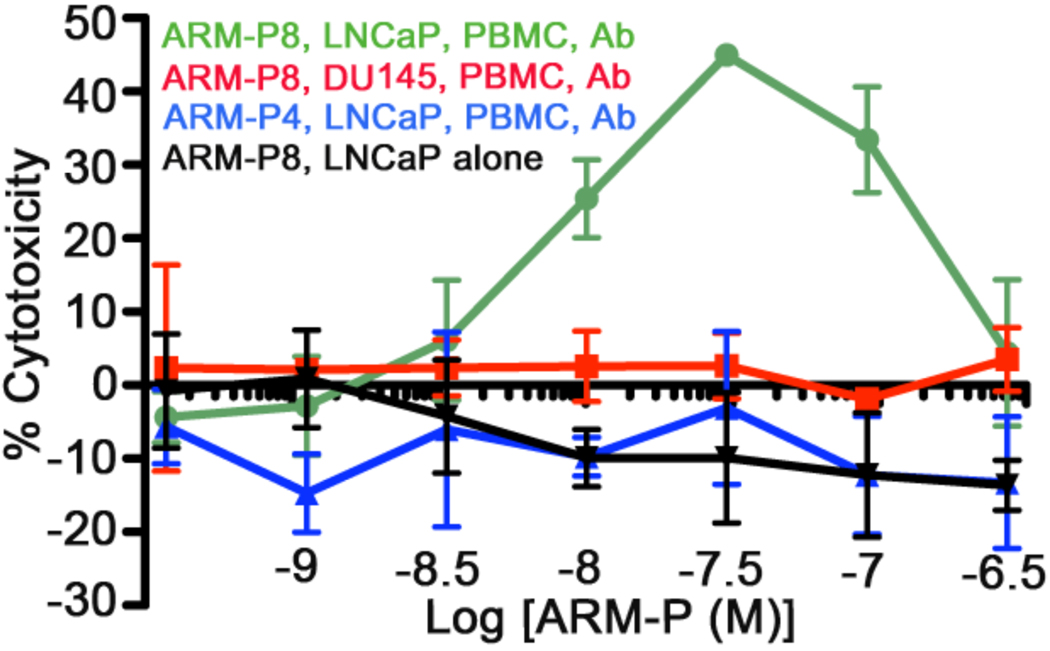
Having established that ARM-P8 possessed optimal linker length in forming ternary complexes, we tested its ability to induce cell-mediated cytotoxicity of LNCaP prostate cancer cells. This process is known to take place by way of interactions between Fc-receptors on cytotoxic effector cells contained in peripheral blood (such as NK cells, macrophages, and dendritic cells), and the Fc (constant) regions of antibodies.6 Thus, LNCaP cells were combined with peripheral blood mononuclear cells (PBMCs), anti-DNP antibodies, and ARM-P8, and cell death was measured using a commercially available calcein-release-assay (Figure 5).25 As expected (vide infra), ARM-P8 at concentrations up to 30 nM led to enhanced cell killing, while treatment with ARM-P4 (3) led to no changes in cell viability. The antibody concentration employed in these experiments is slightly below that found in human serum,4 and lower concentrations were also found to be efficacious (Figure S5). DU145 cells were not susceptible to ARM-P8 mediated cell killing, and no cytotoxicity was observed in either LNCaP or DU145 cell lines after treatment with ARM-P8 in the absence of effector PBMCs, indicating that this compound is not itself cytotoxic (Figure S5).
Figure 5.
Antibody-Dependent Cellular Cytotoxicity (ADCC) Assays. LNCaP (PSMA-positive) and DU145 (PSMA-negative) cells were treated with the ARM-P derivatives at the indicated concentrations, and cell death was measured with and without exposure to anti-DNP antibody (Ab, 24 µg/mL) and peripheral blood mononuclear cells (PBMC). Points represent the average of 4 measurements ± standard deviation. All depicted trends were observed on at least three separate occasions.
Notably, the intriguing bell-shaped pattern observed in cell killing measurements with ARM-P8 has been observed in various situations in which bifunctional ligands template ternary linkages.26 Such behavior results from the binding dynamics of these systems – at large total concentrations of bifunctional small molecule, unbound material competes with the ternary complex, driving the system toward formation of binary complexes. We view this self-antagonistic trend in ARM-P8-mediated cytotoxicity experiments as evidence that cell killing proceeds via reversible formation of a ternary complex. Furthermore, from a clinical standpoint, such a model reveals a unique advantage associated with this novel class of bifunctional therapeutics: they are auto-inhibitory, and could serve clinically as the antidote for their own overdose.
Here we report the structure-based design of a class of prostate cancer-targeted antibody-recruiting small molecules (ARM-Ps) capable of binding to prostate-specific membrane antigen with high affinity (pM to nM), and recruiting antibodies to PSMA-expressing cells. We have also demonstrated that one member of this class, ARM-P8, is capable of inducing antibody- and PBMC-dependent cytotoxicity at concentrations in the nanomolar range. This ARM-based strategy could have profound advantages in the treatment of human cancers. Its auto-inhibitory pharmacology (See Figure 5, above) represents a unique regulatory mechanism worthy of further study. Also, it exploits pre-existing immune mechanisms, not cytotoxic compounds, in cell killing and thus could lead to safer cancer therapies.
Supplementary Material
Acknowledgement
The authors thank John Hines, Thomas Gniadek, and Jacob Appelbaum for helpful suggestions and experimental assistance. This work was funded by the National Institutes of Health through the NIH Director’s New Innovator Award Program (DP22OD002913, DAS), and the National Foundation for Cancer Research (WLJ). JM acknowledges support from a Marie Curie International Fellowship from the European Commission (FP7-PEOPLE-2008-4-1-IOF, 234796-PPIdesign).
Footnotes
Supporting Information Available: Detailed experimental procedures and compound characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.American Cancer Society. Cancer Facts and Figures 2009. 2009 Available online.
- 2.Olson WC, Heston WD, Rajasekaran AK. Rev. Recent Clin. Trials. 2007;2:182–190. doi: 10.2174/157488707781662724. [DOI] [PubMed] [Google Scholar]
- 3.Allen TM. Nat. Rev. Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 4.Antibodies recognizing the 2,4-dinitrophenyl (DNP) epitope have been estimated to constitute 1% of circulating IgM (approx. 10 µg/mL in human serum) and 0.8% of circulating IgG (approx. 40–120 µg/mL in human serum). See Karjalainen K, Makela O. Eur. J. Immunol. 1976;6:88–93. doi: 10.1002/eji.1830060204. Farah FS. Immunology. 1973;25:217–226. and Rowe DS, Anderson SG, Skegg J. In: Immunoglobulins. Merler E, editor. National Academy of Sciences Press; 1970. p. 361. The prevalence of anti-DNP antibodies has been estimated at between 18–90% of humans (see Ortega E, Kostovetzky M, Larralde C. Mol. Immunol. 1984;21:883–888. doi: 10.1016/0161-5890(84)90143-3. and Jormalainen S, Makela O. Eur. J. Immunol. 1971;1:471–478. doi: 10.1002/eji.1830010613. )
- 5.Muller-Eberhard HJ. Annu. Rev. Biochem. 1988;57:321–347. doi: 10.1146/annurev.bi.57.070188.001541. [DOI] [PubMed] [Google Scholar]
- 6.Hale G, Clark M, Waldmann H. J. Immunol. 1985;134:3056–3061. [PubMed] [Google Scholar]
- 7.For antibacterial approaches, see: Bertozzi CR, Bednarski MD. J. Am. Chem. Soc. 1992;114:5543–5546. Bertozzi CR, Bednarski MD. J. Am. Chem. Soc. 1992;114:2242–2245. Krishnamurthy V, Quinton L, Estroff L, Metallo S, Isaacs J, Mizgerd J, Whitesides G. Biomaterials. 2006:12. doi: 10.1016/j.biomaterials.2006.02.006. For anti-HIV approaches, see: Shokat KM, Schultz PG. J. Am. Chem. Soc. 1991;113:1861–1862. Naicker KP, Li H, Heredia A, Song H, Wang L. Org. and Biomolec. Chem. 2004;2:660–664. doi: 10.1039/b313844e. Perdomo MF, Levi M, llberg MS, Vahlne A.Proc. Natl. Acad. Sci. U.S.A 2008619118201 For anti-cancer approaches, see: Carlson C, Mowery P, Owen R, Dykhuizen EC, Kiessling L. ACS Chem. Biol. 2007;2:119–127. doi: 10.1021/cb6003788. Popkov M, Gonzalez B, Sinha S, Barbas C. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4378–4383. doi: 10.1073/pnas.0900147106. and Lu Y, You F, Vlahov I, Westrick E, Fan M, Low PS, Leamon CP. Mol. Pharm. 2007;4:695–706. doi: 10.1021/mp070050b.
- 8.Holmes EH, Greene TG, Tino WT, Boynton AL, Aldape HC, Misrock SL, Murphy GP. Prostate Suppl. 1996;7:25–29. [PubMed] [Google Scholar]
- 9.Mohammed AA, Shergill IS, Vandal MT, Gujral SS. Expert Rev. Mol. Diagn. 2007;7:345–349. doi: 10.1586/14737159.7.4.345. [DOI] [PubMed] [Google Scholar]
- 10.Slovin SF. Expert Opin. Ther. Targets. 2005;9:561–570. doi: 10.1517/14728222.9.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 12.Slusher BS, et al. Nat. Med. 1999;5:1396–1402. doi: 10.1038/70971. [DOI] [PubMed] [Google Scholar]
- 13.Kozikowski AP, Zhang J, Nan F, Petukhov PA, Grajkowska E, Wroblewski JT, Yamamoto T, Bzdega T, Wroblewska B, Neale JH. J. Med. Chem. 2004;47:1729–1738. doi: 10.1021/jm0306226. [DOI] [PubMed] [Google Scholar]
- 14.Humblet V, Misra P, Bhushan KR, Nasr K, Ko Y, Tsukamoto T, Pannier N, Frangioni JV, Maison W. J. Med. Chem. 2009;52:544–550. doi: 10.1021/jm801033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barinka C, Rovenska M, Mlcochova P, Hlouchova K, Plechanovova A, Majer P, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. J. Med. Chem. 2007;50:3267–3273. doi: 10.1021/jm070133w. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen WL. Acc. Chem. Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barinka C, Byun Y, Dusich CL, Banerjee SR, Chen Y, Castanares M, Kozikowski AP, Mease RC, Pomper MG, Lubkowski J. J. Med. Chem. 2008;51:7737–7743. doi: 10.1021/jm800765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James LC, Roversi P, Tawfik DS. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs DJ, Rader AJ, Kuhn LA, Thorpe MF. Proteins. 2001;44:150–165. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 20.Wells S, Menor S, Hespenheide B, Thorpe MF. Phys. Biol. 2005;2:S127–S136. doi: 10.1088/1478-3975/2/4/S07. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 22.Natarajan A, Du W, Xiong C-Y, DeNardo GL, DeNardo SJ, Gervay-Hague J. Chem. Commun. 2007:695–697. doi: 10.1039/b611636a. [DOI] [PubMed] [Google Scholar]
- 23.Details of these experiments can be found in the Supporting Information.
- 24.Anilkumar G, Barwe SP, Christiansen JJ, Rajasekaran SA, Kohn DB, Rajasekaran AK. Microvasc. Res. 2006;72:54–61. doi: 10.1016/j.mvr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Neri S, Mariani E, Meneghetti A, Cattini L, Facchini A. Clin. Diagn. Lab. Immunol. 2001;8:1131–1135. doi: 10.1128/CDLI.8.6.1131-1135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mack ET, Perez-Castillejos R, Suo Z, Whitesides GM. Anal. Chem. 2008;80:5550–5555. doi: 10.1021/ac800578w. and references contained therein.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.