Abstract
Amastin is a transmembrane glycoprotein found on the cell surfaces of trypanosomatid parasites. Encoded by a large, diverse gene family, amastin was initially described from the intracellular, amastigote stage of Trypanosoma cruzi and Leishmania donovani. Genome sequences have subsequently shown that the amastin repertoire is much larger in Leishmania relative to Trypanosoma. However, it is not known when this expansion occurred, whether it is associated with the origins of Leishmania and vertebrate parasitism itself, or prior to this. To examine the timing of amastin diversification, as well as the evolutionary mechanisms regulating gene repertoire and sequence diversity, this study sequenced the genomic regions containing amastin loci from two related insect parasites (Leptomonas seymouri and Crithidia sp.) and estimated a phylogeny for these and other amastin sequences. The phylogeny shows that amastin includes four subfamilies with distinct genomic positions, secondary structures, and evolution, which were already differentiated in the ancestral trypanosomatid. Diversification in Leishmania was initiated from a single ancestral locus on chromosome 34, with rapid derivation of novel loci through transposition and accelerated sequence divergence. This is absent from related organisms showing that diversification occurred after the origin of Leishmania. These results describe a substantial elaboration of amastin repertoire directly associated with the origin of Leishmania, suggesting that some amastin genes evolved novel functions crucial to cell function in leishmanial parasites after the acquisition of a vertebrate host.
Keywords: amastin, Leishmania, evolution, phylogeny, comparative genomics
Introduction
The Trypanosomatidae are unicellular, eukaryotic parasites of the phylum Kinetoplastida. They cause various vector-borne diseases in humans and other vertebrates and include Trypanosoma brucei (African sleeping sickness), Trypanosoma cruzi (Chagas disease), and Leishmania spp. (leishmaniases). These diseases are largely endemic to developing countries, where they have significant negative effects on public health and, in the case of T. brucei which also causes “Nagana” in cattle, on economic development. In each case, there is no vaccine and drug treatment is frequently inadequate due to a combination of hazardous side effects and parasite resistance (Barrett 2006). In 2005, the draft genome sequences of T. brucei (Berriman et al. 2005), T. cruzi (El-Sayed et al. 2005), and Leishmania major (Ivens et al. 2005) were completed, providing a basis for understanding their basic biology, pathology, and developing novel therapies. Four years on and after the publication of further Leishmania genome sequences (Peacock et al. 2007), considerable value is still to be extracted regarding the relative genetic repertoires of the different trypanosomatid genomes, and we have barely begun to place each disease in its evolutionary context by explaining how these organisms came have their distinct parasitic strategies. This study addresses one principal gene family in trypanosomatids: amastin.
Amastin genes became prominent in screens for vaccine candidates in T. cruzi (Teixeira et al. 1994) and Leishmania donovani (Wu et al. 2000), which identified transcripts that were developmentally restricted to the parasite life stage in humans (the amastigote). Amastin protein sequences are among the most immunogenic of all leishmanial surface antigens in mice (Stober et al. 2005) and solicit strong immune responses in humans, particularly in association with visceral leishmaniasis (Rafati et al. 2006). So, amastin proteins appear to operate at the host–parasite interface and are implicated in severe disease. The structure of amastin in both T. cruzi and Leishmania comprises four hydrophobic transmembrane domains, interspersed with serine–threonine rich, extracellular domains and probable glycosylation sites (Teixeira et al. 1994; Rochette et al. 2005). The sequences of these regions, as well as the C-terminus, vary substantially among gene family members (Rochette et al. 2005). Amastin proteins also have a predicted 24 amino acid signal peptide and are expressed across the plasma membrane, including the flagellum (Teixeira et al. 1994; Wu et al. 2000). This structural model is a starting point for understanding function but is based on only a sample of all amastin sequences; in this study, sequence variation is placed within its structural and phylogenetic contexts.
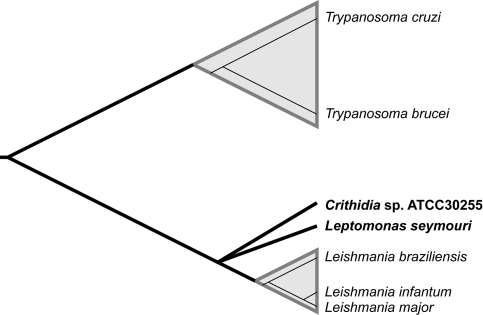
Existing genome sequences clearly show that amastin is more abundant in Leishmania spp. than either T. brucei or T. cruzi (Ivens et al. 2005). Amastin loci are found on multiple chromosomes and initial phylogenetic analyses indicated that genes cluster by genomic position and might display orthology across the family (Rochette et al. 2005). Amastin also occurs in large tandem gene arrays in both T. cruzi (Teixeira et al. 1995; El-Sayed et al. 2005) and Leishmania spp. (Wu et al. 2000; Ivens et al. 2005). At a proximate level, this explains the disparity between Leishmania, in which amastin is the most numerous gene family of all, and Trypanosoma, where it is rare. It strongly suggests an evolutionary expansion in Leishmania and a functional shift relative to its trypanosomatid ancestor. Hence, this study addresses whether the leishmanial expansion precedes or coincides with the origins of Leishmania itself by examining two insect parasitic trypanosomatids, Leptomonas seymouri and a species of Crithidia. These organisms are related most closely to Leishmania, as shown in figure 1, and their precise phylogenetic positions mean that the timing of the expansion in amastin repertoire depends on whether these species shared the expansion or not.
FIG. 1.
A consensus of the trypanosomatid phylogeny based on published studies, showing the phylogenetic relationships of the insect trypanosomatid parasites Leptomonas seymouri and Crithidia sp.
Considerable work has also gone into understanding the regulation of amastin expression as a paradigm of stage-specific, differential expression in trypanosomatids. Trypanosomatid genes typically lack individual promoters (VanHamme and Pays 1995) and are transcribed as long polycistrons (Imboden et al. 1987; Flinn and Smith 1992; Wong et al. 1993). Indeed, our knowledge of posttranscriptional gene regulation is derived partly from amastin (Stiles et al. 1999; Campbell et al. 2003). As suggested above, amastin transcripts and proteins are greatly more abundant in the amastigote than the epimastigote from which it differentiates in both T. cruzi (Teixeira et al. 1994, 1995) and L. donovani (Wu et al. 2000). However, there is no difference in transcription rate between life stages in either genus (Teixeira et al. 1994; Nozaki and Cross 1995; Wu et al. 2000). Observations of conserved noncoding sequences within the 3′untranslated region (UTR) (Teixeira et al. 1994; Boucher et al. 2002; McNicoll et al. 2005) and significant differences in mRNA half-life (Nozaki and Cross 1995; Coughlin et al. 2000; Wu et al. 2000) led to the hypothesis that amastin expression is upregulated in amastigotes through two mechanisms: 1) enhanced transcript stability stimulated by acidic conditions (Coughlin et al. 2000) and 2) enhanced translational efficiency in response to increased temperature (Boucher et al. 2002; McNicoll et al. 2005). Other processes ensure that amastin is suppressed in other life stages, such as mRNA degradation (Haile et al. 2008). These processes are mediated by conserved motifs within the 3′UTR, some of which coincide with short interspersed degenerated retroposons elements (SIDER) retroposons, which may mediate amastigote-specific expression generally (Bringaud et al. 2007; Smith et al. 2009). Given that 3′UTR structure is crucial to regulation of expression, this study analyzed 3′UTR structural diversity to make predictions about functional redundancy and differentiation.
The function of amastin is not known. As an abundant surface antigen with stage-specific expression, amastin could be a transporter crucial to life inside the vertebrate cell or a signal transducer allowing the parasite the sense or manipulate the host environment beyond. This study provides a basis for functional characterization by defining the evolutionary dynamics of amastin variation, which will have considerable functional implications.
Materials and Methods
Selection and Preparation of Comparator Species
Two insect parasitic trypanosomatids were selected for comparative analysis. Leptomonas seymouri American Type Culture Collection (ATCC) 30220 (host: Dysdercus suturellus) is among the closest relatives of Leishmania that still lacks a vertebrate host. Crithidia sp. ATCC 30255 was previously recorded as Crithidia deanei but was recently shown to be more related to Crithidia luciliae or Leptomonas bifurcata (Yurchenko et al. 2009). As trypanosomatid lineages close to the base of the Leishmania clade, these two species are effective comparators for events that postdate the divergence of Trypanosoma and Leishmania but predate the origin of Leishmania itself. Leptomonas seymouri and Crithidia sp. cells were obtained from the ATCC and cultured at 25 °C for 48 h in crithidia culture medium (ATCC medium 355). Cells were harvested by centrifugation, and the pellet was then resuspended overnight at 37 °C in 500 μl of extraction buffer comprising 50 mM Tris–HCl, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid, with an additional 50 μl of 10% sodium dodecyl sulfate and 3 μl proteinase K. Whole genomic DNA was prepared through phenol–chloroform extraction.
Library Construction
For each species, genomic DNA was sheared and end repaired. Fragments in the range of 25–40 kbp were ligated into CopyControl pCC1FOS fosmids (Epicenter Biotechnologies) according to the supplier's instructions. Ligated fosmids were combined with a phage-packaging mix (Gigapack XL2; Stratagene) and used to transform XL2-Blue MRF ultracompetent cells (Stratagene). Each genomic library consisted of 4,608 clones and approximately 160 Mbp of genomic sequence; assuming a typical trypanosomatid haploid genome size of 35 Mbp, this is sufficient to provide at least 4× coverage of both L. seymouri and Crithidia sp. genomes. Each genomic library was immobilized on Nytran Supercharge nylon membranes (Schleicher and Schuell Bioscience).
Library Probing and DNA Sequencing of Leptomonad and Crithidial Amastin Loci
The L. seymouri genomic library was screened for amastin using 32P radiolabeled probes derived from amastin polymerase chain reaction (PCR) products, or, to confirm the absence of amastin at particular loci, those of conserved flanking genes. Initially, degenerate oligonucleotide primers were designed against conserved regions of all leishmanial amastin sequences to amplify any potential amastin sequence in L. seymouri (or Crithidia sp.). Three primer pairs were successful: 1) amaF1 (TACGCCGMTCGACATGTTTCG)/amaR1 (GATGTTDAGCGYCAGGCA) (320 bp product); 2) amaF2 (TCGGTACGGGGGTCGACATGT)/amaR1 (GATGTTDAGCGYCAGGCA) (330 bp product); and 3) amaF4 (ATGGGGTTCGAGGCTCTCCGC)/amaR2 (ACCAGGCCAATCACCTGGGTG) (570 bp product). When applied together, these probes identified three clones that were confirmed as containing amastin. The leptomonad amastin sequences subsequently obtained from these fosmids were used to design specific oligonucleotide probes, but these did not identify any further positive clones.
The Crithidia sp. genomic library was fully end sequenced using a Sanger method, producing 10,092 sequence reads that are available from the Wellcome Trust Sanger Institute Crithidia project page (http://www.sanger.ac.uk/sequencing/Crithidia/ATCC30255/). From the several positive matches to amastin in this sequence library, nine fosmids were selected for complete sequencing. The Crithidia sp. fosmid library was subsequently probed using the same three pairs of L. major–derived degenerate probes used successfully against L. seymouri and specific Crithidia amastin probes, but these did not identify any positive clones beyond those already seen in the end-sequence library.
Fosmid inserts from positive clones, selected either from library probing or end sequencing, were sequenced to between 2 and 10× read coverage using a whole shotgun approach and capillary (Sanger) sequencing method. Fosmid inserts were assembled using Phrap (http://www.phrap.org/phredphrapconsed.html) and visualized using Gap4 (http://staden.sourceforge.net/manual/gap4_unix_2.html). PCR products were generated to close residual gaps between finished contigs. Finished sequence was annotated using Artemis (Rutherford et al. 2000; Berriman and Rutherford 2003), and coding sequences were initially defined by eye. Whole sequences were compared with EMBL sequence databases using both BlastN and BlastP algorithms. Coding sequences were scrutinized for possible transmembrane helices and signal peptides using TMHMM (Krogh et al. 2001) and SignalP (Emanuelsson et al. 2007), respectively.
Table 1 describes all ama loci defined in this study using the nomenclature described below. Collectively, fosmid inserts from the L. seymouri or Crithidia sp. genomic libraries represented all leishmanial loci (plus a novel crithidial locus), except ama24 and ama34C. So to inspect these positions, degenerate oligonucleotide primers were designed against Leishmania and Trypanosoma orthologs of conserved flanking genes (LmjF24.1240/Tb11.02.3670 and LmjF34.1090/Tb927.4.3160, respectively) to generate probes from each species. The primer combination AMA24F (GCGCAGATGTTTCGMAAGTTGAA)/AMA24R2 (CCGCGGTGCACHTCGTACA) produced an ∼800-bp product homologous to LmjF24.1240/Tb11.02.3670 in Crithidia sp., that was applied to both fosmid libraries; a single positive clone was identified from L. seymouri, but none was seen in Crithidia sp. Although a probe was successfully generated for the ama34C flanking gene in both species, it did not identify a positive clone in either fosmid library.
Table 1.
Nomenclature and Genomic Positions of Amastin Loci Used in This Study, Each Defined by Conserved Flanking Genes in Leishmania major.
| Range (kbp) |
Upstream Locus |
Downstream Locus |
|||||
| Locus | Chromosome | Identifier | Description | Identifier | Description | ||
| ama8Aa | 8 | 94 | 101 | LmjF08.0260 | Hypothetical protein (1) | LmjF08.0280 | Ribosomal protein L2 |
| ama8B | 8 | 286 | 386 | LmjF08.0660 | Serine/threonine protein kinase | LmjF08.0890 | Mitochondrial DNA polymerase beta (3) |
| ama17a | 17 | — | — | — | — | LmjF17.1030 | Hypothetical protein |
| ama18a | 18 | 190 | 194 | LmjF18.0450 | Serine carboxypeptidase | LmjF18.0460 | Tubulin-specific chaperone |
| ama24A | 24 | 444 | 454 | LmjF24.1240 | Hypothetical protein | LmjF24.1300 | DNAJ-domain protein (1) |
| ama24Ba | 24 | 780 | 769 | LmjF24.2130 | Ubiquitin-conjugating enzyme E2 | LmjF24.2100 | Hypothetical protein |
| ama28 | 28 | 516 | 521 | LmjF28.1380 | Haloacid dehalogenase-like protein | LmjF28.1410 | DNA polymerase kappa (1) |
| ama30 | 30 | 274 | 280 | LmjF30.0840 | Hypothetical protein | LmjF30.0880 | Adenosine kinase |
| ama31 | 31 | 153 | 154 | LmjF31.0440 | Cytoskeleton-associated protein CAP5.5 | LmjF31.0460 | Calpain-like cysteine peptidase |
| ama34A | 34 | 201 | 202 | LmjF34.0495 | Splicing factor ptsr1 interacting protein | LmjF34.0510 | Phosphoglycan beta 1,2 arabinosyltransferase |
| ama34B | 34 | 416 | 427 | LmjF34.0940 | Serine/threonine protein kinase (1) | LmjF34.1000 | Myosin IB heavy chain (1) |
| ama34Ca | 34 | 368 | 374 | LmjF34.0820 | Elongation factor 1-beta | LmjF34.0840 | Elongation factor 1-beta |
| ama34D | 34 | 484 | 483 | LmjF34.1090 | Dihydroxyacetonephosphate acetyltransferase | LmjF34.1070 | Hypothetical protein |
| ama34E | 34 | 867 | 710 | LmjF34.2000 | Pyroglutamyl-peptidase I | LmjF34.1555 | Ubiquitin-conjugating enzyme |
| ama34F | 34 | 1,285 | 1,282 | LmjF34.2780 | 40S ribosomal protein S19 protein (1) | LmjF34.2810 | Zinc carboxypeptidase |
| ama36 | 36 | 468 | 473 | LmjF36.1260 | Fructose-1,6-bisphosphate aldolase (1) | LmjF36.1290 | Hypothetical protein |
NOTE.—Where a flanking gene is followed by a number in parentheses, this signifies the number of nonconserved genes between the amastin and the flanking gene named. Ama17 was only found in Crithidia sp., and no upstream information was available in this instance.
Those loci were not present in L. major, but each locus was still defined by conserved flanking genes that were present in L. major.
Data Collection and Nomenclature
Amastin sequences were collected from published genome sequences of L. major MON-103, Leishmania infantum JPCM5, Leishmania braziliensis M2904, T. cruzi CL Brener, and T. brucei TREU927 hosted by the GeneDB web site (http://www.genedb.org/). Searching these genomes for amastin using TBlastN identified all known amastin genes, except for Tb11.01.1000, which has been previously noted as unannotated (Rochette et al. 2005), and a gene relic at a conserved position in L. major (downstream of LmjF34.2800). When added to the sequences obtained from L. seymouri and Crithidia sp., the data set comprised 170 gene sequences from seven species, four of which were partial sequences.
To effectively compare amastin repertoire between species, a nomenclature is required that defines genes by genomic position and by phylogenetic lineage. These properties are reliable because gene order tends to be better conserved among trypanosomatids than gene repertoire, and the amastin family itself predates the origins of the individual genomes. By convention, genes are classified by their chromosome and position relative to L. major. The flanking genes up- and downstream in L. major that define each position have been selected because they are conserved in Trypanosoma and therefore should be conserved in any given trypanosomatid. For example, ama31 is on chromosome 31 and flanked by calpain-like cysteine peptidases. Where there are several loci on a single chromosome, they are labeled sequentially in a 5′ to 3′ direction, for example, ama34A to F. Similarly, where there is a tandem gene array, genes are numbered from the 5′-most copy, for example, ama34E.1 to 15. This system will allow future amastin gene sequences from any trypanosomatid to be integrated into the existing scheme easily and for their phylogenetic and genomic affinities to be immediately apparent.
Multiple Sequence Alignment
Translated nucleotide sequences for all amastin coding sequences were initially orientated using ClustalW (Larkin et al. 2007), and the alignment was completed manually and back translated for analysis. The 5′-most half of ama28 genes, which is unique to this position and has no homologous domain in other amastin genes, was removed; the 3′ half of ama28 aligned unambiguously. After inserting gaps for other minor indels, the final alignment was 269 amino acids in length and provided 808 characters when back translated.
Phylogenetic Estimation
The amastin phylogeny was estimated from both nucleotide and protein sequence alignments with two methods: maximum likelihood (ML) using PHYML v3.0 (Guindon and Gascuel 2003) and Bayesian inference (BI) using MrBayes v3.1.2. (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). The ML nucleotide tree was estimated using a general time reversible + G base substitution model, with a gamma distribution of rate heterogeneity estimated from the data. Robustness was evaluated with 500 nonparametric bootstrap replicates, whereas the approximate likelihood ratio test (aLRT) function within PHYML (Anisimova and Gascuel 2006) was used to test the accuracy of each branch using log-likelihood ratio tests. The ML protein tree was estimated using a WAG model (Whelan and Goldman 2001), as prescribed by Protest (Abascal et al. 2005), with an additional rate heterogeneity parameter. The BI nucleotide and protein trees were estimated using the gamma rates function in MrBayes and two Markov chain Monte Carlo chains run in parallel over 5,000,000 generations, sampling every 100 generations. A burn-in of 1,000 trees was sufficient to ensure convergence of all parameters, according to the potential scale reduction factor within MrBayes. All other parameters were set to default. The accuracy of the BI trees was assessed using the posterior probabilities of each node. A final tree was estimated using a Neighbor-Joining algorithm and logdet genetic distances, which can correct for base composition bias (Lockhart et al. 1994), to confirm that base composition was not introducing in phylogenetic error.
Structural Analysis of Sequence Variation
The genomic regions generated for L. seymouri and Crithidia sp. included untranscribed and intergenic regions as well as amastin coding sequences. 3′UTR sequences were defined for all L. major amastin genes using the patterns described by Benz et al. (2005) and compared using BlastN. In addition, corresponding nucleotide sequences were collected from L. infantum, L. braziliensis, L. seymouri, and Crithidia sp. for each L. major locus and compared using BlastN. 3′UTRs were considered “conserved” if they displayed >40% identity over >300 bp. This established the degree of sequence conservation both between gene copies and across species boundaries.
Sequence alignments for each subfamily were analyzed for evidence of recombination, that is, situations where a single sequence displays affinities to multiple, other sequences at different points along its length. The pairwise homoplasy index (PHI) provides a single significance value for any such phylogenetic incompatibility among sequences, even in the presence of rate heterogeneity (Bruen et al. 2006). Each sequence alignment was analyzed in Splitstree v4 (Huson and Bryant 2006) using the PHI menu option. Beyond simply identifying multiple evolutionary signals within the alignment, the LRT tool within TOPALi (Milne et al. 2009) was also used to predict the location of recombination breakpoints.
Finally, sequence alignments for each subfamily were analyzed for positive selection using five different methods: PAML (Yang 2007) within the TOPALi package, as well as the single likelihood ancestor counting, fixed-effects likelihood (FEL), random-effects likelihood methods (Pond and Frost 2005a), and the internal IFEL method (Pond et al. 2006), all employed by the adaptive evolution server (Pond and Frost 2005a, 2005b). Each method scanned the nucleotide alignments on a per-codon basis for positions where the ratio of nonsynonymous substitutions per site to synonymous substitutions per site (dN/dS) was greater than 1 (ω > 1). These methods approach the calculation of ω in subtly different ways, and their relative performance is still debated (Suzuki and Nei 2001, 2004; Zhang 2004; Pond and Frost 2005a; Zeng et al. 2007), so only those codons with ω > 1 in all five tests are reported here.
Results
The Amastin Repertoire of the Insect Parasites L. seymouri and Crithidia sp.
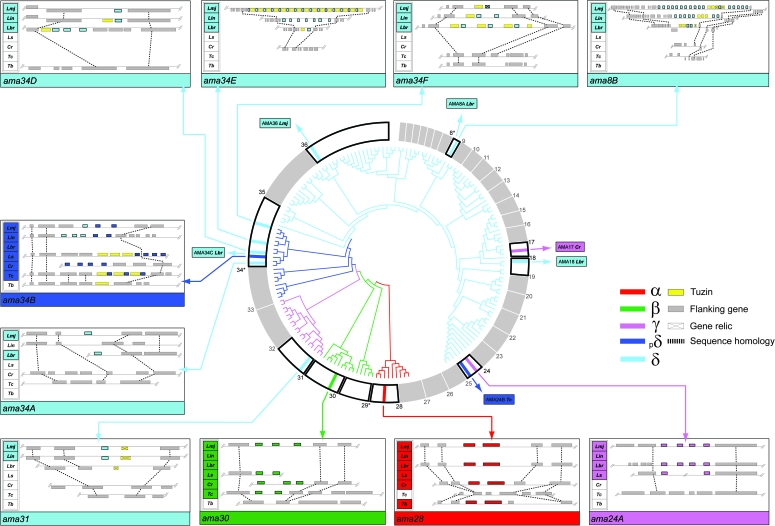
Various amastin genes, along with their surrounding genomic regions, were recovered from L. seymouri and Crithidia sp. genomic libraries based on the similarity of Crithidia fosmid end sequences to amastin or by probing libraries with amastin PCR products or those derived from conserved flanking genes. Novel genomic sequences are listed in table 2 with their GenBank accession numbers. Leptomonas seymouri amastin loci correspond to ama30 (LmjF30.0870), ama24A (LmjF24.1250), and ama34B (LmjF34.0970) in L. major. Despite specifically probing for suspected amastin loci, no further loci were identified. Crithidia sp. amastin genes correspond to ama28 (LmjF28.1400), ama30, and ama34B in L. major; additionally, a novel Crithidia-specific locus was discovered corresponding to a position on chromosome 17 in L. major (closest to LmjF17.1030 and designated ama17). Amastin genes were not found elsewhere; regions corresponding to ama8, ama31, ama34A, and ama34E in L. major were sequenced in Crithidia sp. and shown to lack amastin. Amastin repertoire across seven trypanosomatid species is presented within a L. major context in figure 2, and this clearly shows that insect parasites related to Leishmania spp. have a smaller amastin complement.
Table 2.
Genomic Regions from Leptomonas seymouri and Crithidia sp. Sequenced in This Study.
| Species | Clone | Method | Locus | Sequence Assembly | Total Length (bp) | GenBank Accession Number | |
| Read Number | Contig Number | ||||||
| L. seymouri | 9c15 | Library screening with upstream flanking gene fragment | ama24 | 757 | 3 | 26,722 | GQ153671 |
| 1n19 | Library screening with mixed amastin fragments | ama30 | 1,118 | 1 | 38,992 | GQ153670 | |
| 1j06 | Library screening with mixed amastin fragments | ama34B | 1,243 | 1 | 39,593 | GQ153669 | |
| Crithidia sp. | 5g03 | End-sequence library: Blast match to downstream flanking gene | ama8a | 538 | 1 | 6,053 | GQ153662 |
| 6b24 | End-sequence library: Blast match to amastin | ama17 | 532 | 3 | 38,272 | GQ153665 | |
| 9h15 | End-sequence library: Blast match to downstream flanking gene | ama28 | 689 | 1 | 28,616 | GQ153667 | |
| 7m16 | End-sequence library: Blast match to amastin | ama30 | 695 | 1 | 37,238 | GQ153666 | |
| 12a17 | End-sequence library: Blast match to downstream flanking gene | ama31a | 569 | 3 | 43,87* | GQ153668 | |
| 5p16 | End-sequence library: Blast match to downstream flanking gene | ama34Aa | 628 | 2 | 39,331 | GQ153663 | |
| 2 e16 | End-sequence library: Blast match to amastin | ama34B | 624 | 1 | 37,400 | GQ153661 | |
| 1m01 | End-sequence library: Blast match to upstream flanking gene | ama34Ea | 389 | 2 | 37,943 | GQ153660 | |
| 6b12 | End-sequence library: Blast match to upstream flanking gene | ama36a | 696 | 1 | 31,933 | GQ153664 | |
NOTE.—The total sequence length of Crithidia sp. clone 12a17 exceeds the theoretical maximum of 40 kbp because it contained repetitive material that was assembled multiple times; this did not affect evaluation of the amastin locus because this was nonrepetitive. Blast, basic alignment search tool.
These loci lacked amastin in L. seymouri or Crithidia sp.
FIG. 2.
Amastin repertoire across the Trypanosomatidae. The amastin repertoire across seven species of trypanosomatid (Lm, Leishmania major; Lin, Leishmania infantum; Lbr, Leishmania braziliensis; Ls, Leptomonas seymouri; Cr, Crithidia sp.; Tc, Trypanosoma cruzi; Tb, Trypanosoma brucei) is mapped on to the chromosomes of L. major in a circular arrangement with amastin loci marked by colored bars; shading denotes distinct subfamilies, as defined by the phylogeny. Loci are named according to the L. major chromosome bearing that position and sequentially where multiple, syntenic loci exist, for example, ama34A to F (see Materials and methods). A ML cladogram is shown within the circle, again shaded by subfamily and rooted with α-amastin sequences. For each locus, a cartoon describes the comparative gene order in selected species; the name of the species is shaded where amastin is present.
Amastin Diversity Comprises four Subfamilies that Are Structurally and Positionally Distinct
The combined data set of five completed genome sequences and genomic sequences from L. seymouri (three fosmid inserts) and Crithidia sp. (nine fosmid inserts) produced 170 genes occupying 16 unique loci on nine different L. major chromosomes. Some of these loci were conserved across trypanosomatids and contained distinct amastin subtypes as noted previously (Rochette et al. 2005), whereas six loci were specific to a single species. The classification of amastin genes based on L. major genomic position (see fig. 2) proved to be consistent with phylogenetic relationships and secondary structure (see below). The four subfamilies are termed α, β, γ, and δ, respectively:
α-Amastin: conserved across trypanosomatids as a tandem pair of structurally distinct isoforms on chromosome 28 in L. major (ama28).
β-Amastin: conserved across trypanosomatids as a tandem gene array on chromosome 30 in L. major (ama30), comprising multiple copies of two structurally distinct isoforms.
γ-Amastin: conserved across Leishmania, but not Trypanosoma, as a tandem gene array on chromosome 24 in L. major (ama24A). The tandem duplicates comprise multiple copies of two structurally distinct isoforms. Related genes are found in Crithidia sp. at a position corresponding to chromosome 17 in L. major.
δ-Amastin: a highly diverse clade found at several loci across various chromosomes (ama8A-B, ama18, ama31, ama34A-F, and ama36) in Leishmania, but not other genera. Only one locus (ama34B) is conserved across the Trypanosomatidae as a tandem gene array; due to its basal branching position in the amastin phylogeny (see below), this is referred to as proto-δ-amastin hereafter.
The Ancestral Amastin Repertoire Was Diverse and Has Been Modified by Frequent Gene Gain and Loss
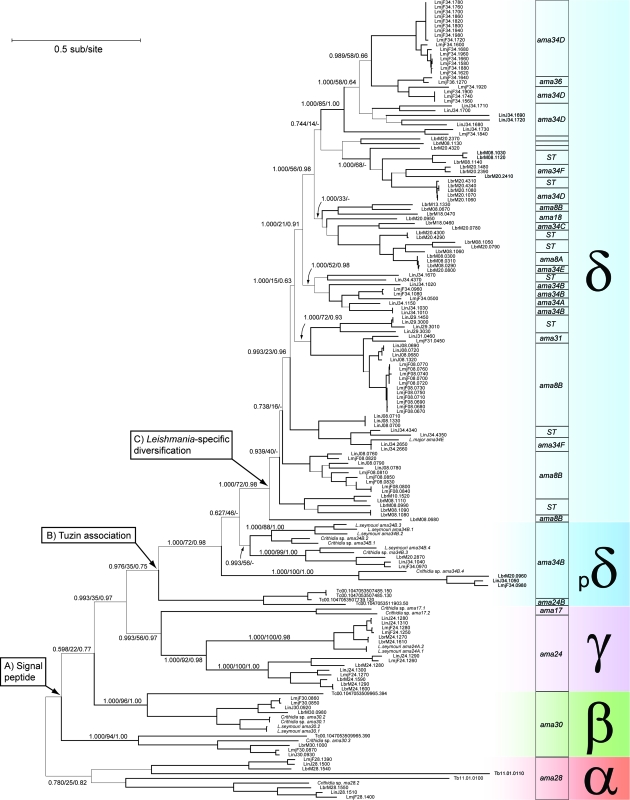
A ML phylogeny was estimated from an 808-bp multiple alignment of all amastin nucleotide sequences. The phylogeny is shown in figure 3, where it is subdivided by subfamily and shown alongside genomic position. It was well resolved and robust in all but the most basal nodes. The ML topology was largely concordant with Bayesian trees estimated using both nucleotide and amino acid alignments and with a Neighbor-Joining tree estimated using logdet distances, indicating that base composition bias had no significant effect on topology. Three subfamilies (α, γ, and δ) are monophyletic, whereas β-amastin splits into two closely related lineages corresponding to the distinct isoforms already noted and is paraphyletic with δ-amastin; this suggests that δ-amastin originated from one β-amastin isoform and does not negate a common origin of both β-amastin isoforms at ama30. Thus, the phylogeny reinforces the four-subfamily classification that was based on taxonomic distribution and conserved genomic position. γ-Amastin is not found in Trypanosoma, but its phylogenetic position, intermediate between β- and δ-amastin, demands that it once existed in Trypanosoma and has been lost.
FIG. 3.
A ML phylogeny for all amastin gene copies in seven trypanosomatid species. Branch lengths are drawn proportion to evolutionary change. Basal nodes are labeled with three branch support values: Bayesian posterior probability/aLRT statistic/nonparametric bootstrap. All branches supported by bootstrap values greater than 90 are drawn in bold. GeneDB identifiers are used for taxon names when derived from published genome projects. Clades are shaded by subfamily. The genomic position of individual sequences or clades is indicated on the right. Genes derived from subtelomeric regions are labeled as “ST.” Nodes relating to three evolutionary events are noted: (A) the acquisition of a signal peptide; (B) linkage of amastin with tuzin; and (C) the diversification of δ-amastin in Leishmania only.
When gene repertoire is interpreted in a phylogenetic context, it is clear that gene loss has occurred in several lineages: 1) γ-amastin is absent from T. cruzi and T. brucei; 2) α-amastin is absent from T. cruzi but not T. brucei; 3) β-amastin is absent from T. brucei but not T. cruzi; 4) ama34A is missing from L. infantum; and v) ama34F is present in L. major only as a pseudogene. Within subfamilies, especially δ-amastin for which there are so many loci, there are likely to be more frequent gene losses and gains; in two cases, ama31 in L. braziliensis and ama34B in T. brucei, no amastin was observed, but a tuzin gene persists, strongly suggesting that amastin was present formerly. Gene gains are also required to explain the differences in amastin complement, for example, ama17 is a γ-amastin locus found only in Crithidia sp.; its basal phylogenetic position within the subfamily suggests that γ-amastin may have occupied more positions previously. ama8A, 18, and 34C are all unique expansions of δ-amastin in L. braziliensis, whereas ama24B only exists in T. cruzi. But the single largest incidence of gene gain is the evolution of δ-amastin itself in the lineage leading to Leishmania.
δ-Amastin Is a Unique Elaboration of the Ancestral Gene Repertoire in Leishmania spp.
From the comparative genomics and phylogenetic analyses shown in figures 2 and 3, the expansion of Leishmania repertoire clearly derives from a single subfamily: δ-amastin. The proto-δ-amastin locus (ama34B) branches basally and represents the original position from which all δ-amastin were subsequently derived; this is consistent with the presence of ama34B in other trypanosomatids and the common ancestor. ama34B positional orthologs cluster together, and the ama34B clade bisects δ-amastin and the other subfamilies. Because the long tandem gene arrays of δ-amastin are absent from Crithidia sp. and L. seymouri, but present in all Leishmania genome sequences currently available, δ-amastin must have evolved with, or shortly after, the origin of Leishmania. From ama34B, δ-amastin loci have been established through multiple transposition events on chromosomes 34 and 8, as well as more species-specific transpositions to chromosomes 18, 31, and 36. It is also notable that ama34B, although containing the proto-δ-amastin genes described above, also includes derived δ-amastin (more closely related to genes at ama34E and ama8B), indicating that δ-amastin has secondarily transposed back to ama34B from elsewhere. δ-Amastin sequences are apparently highly mobile. When genomic position is mapped onto the phylogeny, δ-amastin genes sharing a chromosome are not always monophyletic, indicating frequent, independent movements of δ-amastin around the genomes of different Leishmania species. In fact, L. braziliensis δ-amastin sequences largely cluster together and away from L. major/L. infantum genes, irrespective of shared genomic position, suggesting that some form of concerted evolution or rapid replacement has occurred since their speciation to abolish any sequence orthology.
Tuzin Is Associated with δ-Amastin Only
Amastin genes are often contiguous with tuzin, a conserved transmembrane protein with unknown function (Teixeira et al. 1995, 1999). With a renewed definition of amastin diversity, it is now clear that tuzin is only found associated with δ-amastin, that is, it is never found at ama28, ama30, or ama24. It is present at ama34B in L. seymouri, T. cruzi, and T. brucei (in the latter, it indicates that amastin has been lost), but not in Leishmania spp. Therefore, one can infer that tuzin became associated with amastin in a common trypanosomatid ancestor at the proto-δ-amastin locus. It has subsequently spread to new positions in Leishmania spp. together with δ-amastin. In some cases and species, tuzin has been deleted, for example, at ama8 in Leishmania spp., ama31 in L. braziliensis, and ama34E in L. infantum. However, in general, the association has persisted during the diversification of δ-amastin, suggesting that there is a strong, if flexible, functional link between the two gene families.
The Roles of Positive Selection and Recombination in δ-Amastin Microevolution
A Leishmania-specific radiation of δ-amastin loci represents a major macroevolutionary transition, which presages a functional change that should be reflected in microevolutionary changes to δ-amastin gene sequences. A multiple alignment of δ-amastin sequences from L. major was scrutinized for the signatures of molecular adaptation and recombination to assess the role of functional differentiation in the expansion of δ-amastin. Analysis for positive selection was carried out on a per-codon basis for each amastin subfamily separately. No codons were predicted to be under positive selection in α, β, or γ-amastin alignments. For δ-amastin, 13 codons were identified, where ω > 1 in two or more of the five tests carried out. These are shown in their structural context in supplementary figure 1 (Supplementary Material online). δ-Amastin shows the same substitution dynamic as other subfamilies, that is, sequence conservation is greatest around the putative transmembrane domains, whereas the C-terminal domain and the two extracellular domains are most variable. Six sites predicted to be evolving adaptively are in the C-terminal domain, but even discounting this region (because some sites may be nonhomologous due to very rapid evolutionary change), six codons in the two extracellular domains are also predicted to be under positive selection.
PHI failed to detect any incompatible phylogenetic signals in the α (P = 0.442), β (P = 0.779), and γ-amastin (P = 0.738) alignments, indicating that recombination does not occur between their gene copies. However, significant incompatibility was detected for δ-amastin (P = 4.3 × 10−5), but only when the C-terminal region was included. Closer inspection of this region showed that some tandem gene copies on chromosome 34 or 8 in L. major resembled recombinant products of other copies, for instance, LmjF34.1760 was a product of the LmjF34.1720 (at its 5′ end) and LmjF34.1920 (at its C-terminal domain). Individual sequence reads spanned the entire lengths of each gene, precluding misassembly. Further analysis of L. major δ-amastin sequences using the LRT tool in TOPALi predicted two recombination breakpoints, around position 41 (i.e., first extracellular domain) and position 158 (i.e., second extracellular domain, just upstream of the fourth transmembrane domain).
3′UTR Structural Variation Supports the Functional Differentiation of Amastin Genes
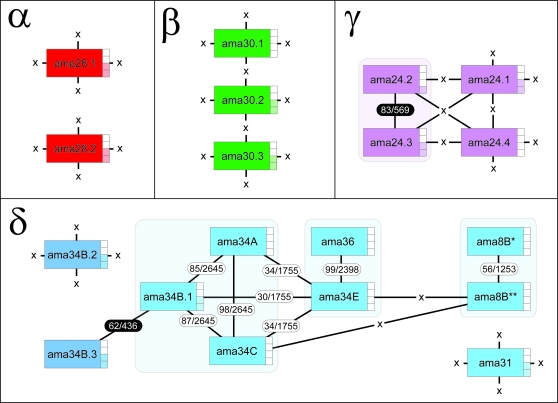
Given their role in posttranscriptional control of gene expression, comparison of 3′UTR sequences could provide evidence for functional differentiation or redundancy. Figure 4 shows the level of sequence similarity between 3′UTRs of different subfamilies in L. major, which are known to be dissimilar (Rochette et al. 2005). Yet, even within the α, β, and γ subfamilies, no similarity was found among the structurally distinct tandem duplicates at these loci. One exception was a 569-bp region showing 83% identity between the γ-amastin copies ama24.2 and ama24.3; however, this coincided with a SIDER retroposon found in both 3′UTRs (Rochette et al. 2005; Smith et al. 2009), but they were otherwise dissimilar. Similarly, no homology was found between 3′UTRs of the proto-δ-amastin genes and any other δ-amastin gene, other than for one weak match coinciding with SIDER sequence (Smith et al. 2009). In contrast, sequence homology was conserved among the 3′UTRs of various δ-amastin loci. The amastin phylogeny shows that ama36 is very closely related and probably derived from the large tandem gene array at ama34E. In accordance with this, the 3′UTRs of these genes are virtually identical, as they are among the tandem duplicates of the array. Very good homology (85–98% nucleotide identity over 2,645 bp) exists between ama34A, ama34B.1, and ama34C. ama8 Genes form two arrays either side of a tuzin gene cluster (see fig. 2); 3′UTRs are identical within each array but are poorly conserved between the two (56% over 1,253 bp). When 3′UTRs were compared interspecifically for orthologous loci, they were generally conserved in L. infantum and L. braziliensis, but not L. seymouri or Crithidia sp. There was no evidence for SIDER retroposons within the 3′UTRs of L. seymouri or Crithidia sp. However, δ-amastin 3′UTRs were generally not conserved within Leishmania; only 3′UTRs at ama34B and ama31 are conserved across the genus.
FIG. 4.
Comparison of 3′UTR sequences across all Leishmania major amastin loci. All L. major 3′UTRs were compared with each other using BlastN to produce a percentage of sequence similarity. Genes are represented as boxes, color coded by subfamily. Lines are drawn between genes with sequence affinity, and each is labeled with the percentage of sequence similarity and the length of the sequence match in base pairs. In two cases, sequence similarity was due solely to inclusion of a SIDER retroposon; these similarity values are shaded black. Where no affinity was detected, an X is shown; these loci were unique in their 3′UTR sequences. Each 3′UTR was also compared with sequences found in corresponding positions in four other species. Where the sequence was conserved, this is indicated by a shaded box to the right of the gene label: (boxes from top to bottom) Crithidia sp., Leptomonas seymouri, Leishmania braziliensis, and Leishmania infantum.
Discussion
This comparative analysis indicates that the amastin gene family is both diverse and ancient. It comprises four subfamilies with distinct phylogenetic identities and genomic locations, each containing gene duplicates with distinct molecular structures in both coding and noncoding regions. The elaboration of the family predates the diversification of the various parasite species, proving that amastin is an ancient feature of all trypanosomatid genomes and that the obvious disparity in copy number between Trypanosoma and Leishmania is due to the expansion of one particular subfamily (δ-amastin) in the ancestral Leishmania sp. from one particular genomic location (ama34B). It is now possible to reconstruct the evolution of the family and to make predictions about gene function.
The ancestral amastin gene, present in the common ancestor of all Trypanosomatidae, is most likely to resemble α-amastin. Relative to other amastin genes, α-amastin is between 35% and 64% larger, with 3–10 additional transmembrane helices at the 5′ end. Both α-amastin gene copies lack recognizable signal peptides and appear to be expressed constitutively. Importantly, α-amastin is the only family member in T. brucei, which lacks an amastigote life cycle stage. The process of diversification began with the origin of β-amastin from the α locus (or from a progenitor of both) through transposition to a new location (ama30) and deletion of coding sequence from the 5′ end. The β-amastin locus includes two divergent sequence types that are consistently paraphyletic in the phylogeny, indicating that the ancestor of γ- and δ-amastin arose through transpositive gene duplication from the upstream β-amastin isoform in particular (i.e., the ortholog of LmjF30.0860). Subsequent duplication events must be inferred to explain the origin of dimorphic tandem gene arrays of γ-amastin at ama24 and proto-δ-amastin at ama34B.
So far, these loci are conserved throughout the trypanosomatids at consistent genomic positions. Where they are absent, the shape of the phylogeny determines that we interpret absence as losses, rather than species-specific acquisitions. The absence of α-amastin from T. cruzi is due to deletion because it is otherwise present in T. brucei, Crithidia sp., and Leishmania. Similarly, the absence of β-amastin from T. brucei is due to loss because other trypanosomes possess it and because there is evidence that T. brucei had proto-δ-amastin (see below), which originated after β-amastin. The absence of γ-amastin from trypanosomes and Crithidia sp. is most likely to be evolutionary loss because these organisms possess β- and δ-amastin, which, in temporal terms, bracket the episode when γ-amastin originated. Finally, the absence of proto-δ-amastin from T. brucei looks like deletion because a tuzin gene (which is usually associated with δ-amastin) is still present. In summary, subsequent deletion events notwithstanding the amastin gene family was already differentiated into its four principal subfamilies (each arranged as a dimorphic tandem gene array) in the ancestral trypanosomatid genome.
Amastin diversity remained unchanged until the origin of Leishmania. Leishmania differs from closely related insect parasites such as Leptomonas spp. in that the amastigote stage takes place inside a vertebrate macrophage. The expansion of δ-amastin should be viewed in the context of the adaptation of the amastigote to this novel life stage after the acquisition of vertebrate parasitism. By mapping chromosomal position on to the phylogeny, we can see that δ-amastin was elaborated through a sequence of transpositions increasing the number of loci and tandem duplications increasing copy number at each locus. This has not simply increased gene dosage; positive selection has contributed to substantial divergence of variable protein domains among δ-amastin, whereas downstream regulatory regions have evolved rapidly, such that they are not conserved between Leishmania spp. This indicates that a diverse range of δ-amastin proteins are expressed in various situations. When amastin expression was observed specifically in the amastigote, those genes were recognizably δ-amastin (Teixeira et al. 1994; Wu et al. 2000). Recent studies have confirmed this on a genome scale (Akopyants et al. 2004; Holzer et al. 2006; Rochette et al. 2009), although Alcolea et al. (2009) present data suggesting that some δ-amastin isoforms originating from ama34C and ama8B may be expressed in the metacyclic stage. However, other amastin subfamilies are not restricted to the amastigote, and this has consequences for the evolution of amastin function.
Transcriptomic and proteomic surveys (Saxena et al. 2003; Akopyants et al. 2004; Almeida et al. 2004; Dea-Ayuela et al. 2006; Holzer et al. 2006; Leifso et al. 2007; Bridges et al. 2008) have consistently failed to show that α-amastin is developmentally regulated in any trypanosomatid, suggesting that it is constitutively expressed. Almeida et al. (2004) compared procyclic, metacyclic, and amastigote life stages of L. major using cDNA microarrays and showed that β-amastin (accession number AA060767) was only expressed in procyclics. A 2-fold upregulation of β-amastin was also observed in L. mexicana procyclics (Holzer et al. 2006). Recent RNA-profiling studies found that γ-amastin is upregulated in L. infantum axenic amastigotes, but not in intracellular forms (Rochette et al. 2008, 2009). When γ-amastin was observed to be upregulated in L. major intracellular amastigotes, the enhancement was modest (1.9-fold), indeed less than any δ-amastin transcript (average 9.4-fold increase; Rochette et al. 2008). Taken together, the amastigote-specific function of amastin may possibly be limited to δ-amastin and is certainly derived within the phylogeny of the family. If one considers that the diversification of amastin in Leishmania is not shared with Crithidia sp. or L. seymouri, that it concerns amastigote-specific forms only, and can be traced back to ama34B, which is also the site of amastigote-specific genes in T. cruzi (Minning et al. 2003), these data strongly support the hypothesis that δ-amastin is part of the evolutionary modification of the leishmanial amastigote to a new host environment. The derivation of novel protein sequences under selection and the distinct regulatory regions associated with certain loci further suggests that δ-amastin has undergone an adaptive radiation associated with the acquisition of vertebrate parasitism.
At present, our best guess at the generic function of amastin is of a membrane-bound signal transducer, binding ligands at the host–amastigote interface, with various downstream effectors within the cell. However, with an eye to future functional genetic studies of the various amastin genes, we can make some predictions based on the evolutionary dynamics seen here. Amastin subfamilies are consistently structurally divergent and they have distinct regulatory regions and do not recombine; therefore, they are likely to be functionally differentiated. Similarly, the distinct tandem duplicates found at α, β, and γ loci are likely to have different functions because they also maintain their distinctiveness across evolutionary time, presumably due to purifying selection. Functional differentiation has been observed in other tandem gene duplicates in trypanosomatids, for example, glucose transporters in T. brucei (Bringaud and Baltz 1994), phosphoglycerate kinase in Trypanosoma congolense (Parker et al. 1995), and adenylate cyclases in L. donovani (Sanchez et al. 1995). So we can predict functional differentiation among δ-amastin wherever distinct coding and regulatory sequences have evolved and are maintained across species boundaries. Thus, ama31 and ama34C, as well as ama8B and ama34E, could have different functions, but tandem duplicates at these loci will be functionally redundant, as will ama34E and ama36. The biology of amastin genes remains obscure, but with a better understanding of when sequence variation evolved, how it can be described, and where it is to be found in trypanosomatid genomes, we can begin to explain the explosive innovation of this gene family in Leishmania.
Supplementary Material
Supplementary figure 1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
All novel sequence data were produced by the Pathogen Sequencing Unit of the Wellcome Trust Sanger Institute. This work was funded by the Wellcome Trust through a Wellcome Trust Sanger Institute Postdoctoral Fellowship.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Akopyants NS, Matlib RS, Bukanova EN, Smeds MR, Brownstein BH, Stormo GD, Beverley SM. Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol Biochem Parasitol. 2004;136:71–86. doi: 10.1016/j.molbiopara.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Alcolea PJ, Alonso A, Sánchez-Gorostiaga A, Moreno-Paz M, Gómez MJ, Ramos I, Parro V, Larraga V. Genome-wide analysis reveals increased levels of transcripts related with infectivity in peanut lectin non-agglutinated promastigotes of Leishmania infantum. Genomics. 2009;93:551–564. doi: 10.1016/j.ygeno.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Almeida R, Gilmartin BJ, McCann SH, et al. (15 co-authors) Expression profiling of the Leishmania life cycle: cDNA arrays identify developmentally regulated genes present but not annotated in the genome. Mol Biochem Parasitol. 2004;136:87–100. doi: 10.1016/j.molbiopara.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Barrett MP. The rise and fall of sleeping sickness. Lancet. 2006;367:1377–1378. doi: 10.1016/S0140-6736(06)68591-7. [DOI] [PubMed] [Google Scholar]
- Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. Messenger RNA processing sites in Trypanosoma brucei. Mol Biochem Parasitol. 2005;143:125–134. doi: 10.1016/j.molbiopara.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, et al. (102 co-authors) The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Berriman M, Rutherford K. Viewing and annotating sequence data with Artemis. Brief Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- Boucher N, Wu Y, Dumas C, Dube M, Sereno D, Breton M, Papadopoulou B. A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3′-untranslated region element. J Biol Chem. 2002;277:19511–19520. doi: 10.1074/jbc.M200500200. [DOI] [PubMed] [Google Scholar]
- Bridges DJ, Pitt AR, Hanrahan O, Brennan K, Voorheis HP, Herzyk P, de Koning HP, Burchmore RJ. Characterisation of the plasma membrane subproteome of bloodstream form Trypanosoma brucei. Proteomics. 2008;8:83–99. doi: 10.1002/pmic.200700607. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Baltz T. African trypanosome glucose transporter genes: organization and evolution of a multigene family. Mol Biol Evol. 1994;11:220–230. doi: 10.1093/oxfordjournals.molbev.a040104. [DOI] [PubMed] [Google Scholar]
- Bringaud F, Müller M, Cerqueira GC, Smith M, Rochette A, El-Sayed NM, Papadopoulou B, Ghedin E. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–1307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Coughlin BC, Teixeira SM, Kirchhoff LV, Donelson JE. Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3′-untranslated region position-dependent cis-element and an untranslated region-binding protein. J Biol Chem. 2000;275:12051–12060. doi: 10.1074/jbc.275.16.12051. [DOI] [PubMed] [Google Scholar]
- Dea-Ayuela MA, Rama-Iñiguez S, Bolás-Fernández F. Proteomic analysis of antigens from Leishmania infantum promastigotes. Proteomics. 2006;6:4187–4194. doi: 10.1002/pmic.200600101. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Bartholomeu DC, et al. (82 co-authors) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Flinn HM, Smith DF. Genomic organisation and expression of a differentially-regulated gene family from Leishmania major. Nucleic Acids Res. 1992;20:755–762. doi: 10.1093/nar/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haile S, Dupé A, Papadopoulou B. Deadenylation-independent stage-specific mRNA degradation in Leishmania. Nucleic Acids Res. 2008;36:1634–1644. doi: 10.1093/nar/gkn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Imboden MA, Laird PW, Affolter M, Seebeck T. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res. 1987;15:7357–7368. doi: 10.1093/nar/15.18.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, et al. (102 co-authors) The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (12 co-authors) ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Lockhart PJ, Steel MA, Hendy MD, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- McNicoll F, Müller M, Cloutier S, Boilard N, Rochette A, Dubé M, Papadopoulou B. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J Biol Chem. 2005;280:35238–35246. doi: 10.1074/jbc.M507511200. [DOI] [PubMed] [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minning TA, Bua J, Garcia GA, McGraw RA, Tarleton RL. Microarray profiling of gene expression during trypomastigote to amastigote transition in Trypanosoma cruzi. Mol Biochem Parasitol. 2003;131:55–64. doi: 10.1016/s0166-6851(03)00189-0. [DOI] [PubMed] [Google Scholar]
- Nozaki T, Cross GA. Effects of 3’ untranslated and intergenic regions on gene expression in Trypanosoma cruzi. Mol Biochem Parasitol. 1995;75:55–67. doi: 10.1016/0166-6851(95)02512-x. [DOI] [PubMed] [Google Scholar]
- Parker HL, Hill T, Alexander K, Murphy NB, Fish WR, Parsons M. Three genes and two isozymes: gene conversion and the compartmentalization and expression of the phosphoglycerate kinases of Trypanosoma (Nannomonas) congolense. Mol Biochem Parasitol. 1995;69:269–279. doi: 10.1016/0166-6851(94)00208-5. [DOI] [PubMed] [Google Scholar]
- Peacock CS, Seeger K, Harris D, et al. (42 co-authors) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol. 2005a;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005b;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Grossman Z, Gravenor MB, Richman DD, Brown AJ. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafati S, Hassani N, Taslimi Y, Movassagh H, Rochette A, Papadopoulou B. Amastin peptide-binding antibodies as biomarkers of active human visceral leishmaniasis. Clin Vaccine Immunol. 2006;13:1104–1110. doi: 10.1128/CVI.00188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette A, McNicoll F, Girard J, Breton M, Leblanc E, Bergeron MG, Papadopoulou B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol Biochem Parasitol. 2005;140:205–220. doi: 10.1016/j.molbiopara.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Rochette A, Raymond F, Corbeil J, Ouellette M, Papadopoulou B. Whole-genome comparative RNA expression profiling of axenic and intracellular amastigote forms of Leishmania infantum. Mol Biochem Parasitol. 2009;165:32–47. doi: 10.1016/j.molbiopara.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Rochette A, Raymond F, Ubeda JM, Smith M, Messier N, Boisvert S, Rigault P, Corbeil J, Ouellette M, Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sanchez MA, Zeoli D, Klamo EM, Kavanaugh MP, Landfear SM. A family of putative receptor-adenylate cyclases from Leishmania donovani. J Biol Chem. 1995;270:17551–17558. doi: 10.1074/jbc.270.29.17551. [DOI] [PubMed] [Google Scholar]
- Saxena A, Worthey EA, Yan S, Leland A, Stuart KD, Myler PJ. Evaluation of differential gene expression in Leishmania major Friedlin procyclics and metacyclics using DNA microarray analysis. Mol Biochem Parasitol. 2003;129:103–114. doi: 10.1016/s0166-6851(03)00100-2. [DOI] [PubMed] [Google Scholar]
- Smith M, Bringaud F, Papadopoulou B. Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics. 2009;10:240. doi: 10.1186/1471-2164-10-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles JK, Hicock PI, Shah PH, Meade JC. Genomic organization, transcription, splicing and gene regulation in Leishmania. Ann Trop Med Parasitol. 1999;93:781–807. doi: 10.1080/00034989957781. [DOI] [PubMed] [Google Scholar]
- Stober CB, Lange UG, Roberts MT, Gilmartin B, Francis R, Almeida R, Peacock CS, McCann S, Blackwell JM. From genome to vaccines for leishmaniasis: screening 100 novel vaccine candidates against murine Leishmania major infection. Vaccine. 2005;24:2602–2616. doi: 10.1016/j.vaccine.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nei M. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol Biol Evol. 2001;18:2179–2185. doi: 10.1093/oxfordjournals.molbev.a003764. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nei M. False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol. 2004;21:914–921. doi: 10.1093/molbev/msh098. [DOI] [PubMed] [Google Scholar]
- Teixeira SM, Kirchhoff LV, Donelson JE. Post-transcriptional elements regulating expression of mRNAs from the amastin/tuzin gene cluster of Trypanosoma cruzi. J Biol Chem. 1995;270:22586–22594. doi: 10.1074/jbc.270.38.22586. [DOI] [PubMed] [Google Scholar]
- Teixeira SM, Kirchhoff LV, Donelson JE. Trypanosoma cruzi: suppression of tuzin gene expression by its 5′-UTR and spliced leader addition site. Exp Parasitol. 1999;93:143–151. doi: 10.1006/expr.1999.4446. [DOI] [PubMed] [Google Scholar]
- Teixeira SM, Russell DG, Kirchhoff LV, Donelson JE. A differentially expressed gene family encoding “amastin,” a surface protein of Trypanosoma cruzi amastigotes. J Biol Chem. 1994;1994(269):20509–20516. [PubMed] [Google Scholar]
- Vanhamme L, Pays E. Control of gene expression in trypanosomes. Microbiol Rev. 1995;59:223–240. doi: 10.1128/mr.59.2.223-240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wong S, Morales TH, Neigel JE, Campbell DA. Genomic and transcriptional linkage of the genes for calmodulin, EF-hand 5 protein, and ubiquitin extension protein 52 in Trypanosoma brucei. Mol Cell Biol. 1993;13:207–216. doi: 10.1128/mcb.13.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, El Fakhry Y, Sereno D, Tamar S, Papadopoulou B. A new developmentally regulated gene family in Leishmania amastigotes encoding a homolog of amastin surface proteins. Mol Biochem Parasitol. 2000;110:345–357. doi: 10.1016/s0166-6851(00)00290-5. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yurchenko VY, Lukes J, Jirku M, Maslov D. Selective recovery of the cultivation-prone components from mixed trypanosomatid infections: a case of several novel species isolated from Neotropical Heteroptera. Int J Syst Evol Microbiol. 2009;59:893–909. doi: 10.1099/ijs.0.001149-0. [DOI] [PubMed] [Google Scholar]
- Zeng K, Mano S, Shi S, Wu CI. Comparisons of site- and haplotype-frequency methods for detecting positive selection. Mol Biol Evol. 2007;24:1562–1574. doi: 10.1093/molbev/msm078. [DOI] [PubMed] [Google Scholar]
- Zhang J. Frequent false detection of positive selection by the likelihood method with branch-site models. Mol Biol Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.