Abstract
Peroxisome proliferator chemicals (PPC) are thought to mediate their effects in rodents on hepatocyte growth and liver cancer through the nuclear receptor peroxisome proliferator–activated receptor (PPAR) α. Recent studies indicate that the plasticizer di-(2-ethylhexyl) phthalate (DEHP) increased the incidence of liver tumors in PPARα-null mice. We hypothesized that some PPC, including DEHP, induce transcriptional changes independent of PPARα but dependent on other nuclear receptors, including the constitutive-activated receptor (CAR) that mediates phenobarbital (PB) effects on hepatocyte growth and liver tumor induction. To determine the potential role of CAR in mediating effects of PPC, a meta-analysis was performed on transcript profiles from published studies in which rats and mice were exposed to PPC and compared the profiles to those produced by exposure to PB. Valproic acid, clofibrate, and DEHP in rat liver and DEHP in mouse liver induced genes, including Cyp2b family members that are known to be regulated by CAR. Examination of transcript changes by Affymetrix ST 1.0 arrays and reverse transcription-PCR in the livers of DEHP-treated wild-type, PPARα-null, and CAR-null mice demonstrated that (1) most (∼94%) of the transcriptional changes induced by DEHP were PPARα-dependent, (2) many PPARα-independent genes overlapped with those regulated by PB, (3) induction of genes Cyp2b10, Cyp3a11, and metallothionine-1 by DEHP was CAR dependent but PPARα-independent, and (4) induction of a number of genes (Cyp8b1, Gstm4, and Gstm7) was independent of both CAR and PPARα. Our results indicate that exposure to PPARα activators including DEHP leads to activation of multiple nuclear receptors in the rodent liver.
Keywords: peroxisome proliferator–activated receptor α, transcript profiling, liver cancer, di-(2-ethylhexyl) phthalate, constitutive-activated receptor, pregnane X receptor
Phthalates, diesters of benzenedicarboxylic acid (phthalic acids), are high production volume chemicals used in numerous industrial applications (polyvinyl chloride [PVC] products, personal care products, automotive industries, and residential construction). Phthalate compounds have become ubiquitous environmental contaminants as they migrate out from PVC-containing items into food, air, dust, water, and soils (Clark et al., 2003). Diet, particularly fatty food (e.g., fish, oils), is the main source of phthalate exposure in the general public (Clark et al., 2003; Meek and Chan, 1994; Wormuth et al., 2006). Other sources include blood storage bags (Labow et al., 1986; Shintani, 1985) and kidney dialysis and blood transfusion equipment (Nassberger et al., 1987; Sjoberg et al., 1985). An increasing number of studies sampling human urine reveal the ubiquitous phthalate exposure of consumers in industrialized countries (Wormuth et al., 2006). Concern over human exposure comes from the toxicological effects of phthalates, which include an increased incidence of liver tumors in mice and rats (Klaunig et al., 2003) and irreversible changes in the development of the male reproductive tract (Corton and Lapinskas, 2005; Foster et al., 2001).
Many phthalates belong to a large class of structurally diverse chemicals, including herbicides, industrial solvents, and pharmaceutical agents known as peroxisome proliferator chemicals (PPC). PPC refers to any chemical that when administered to rats and mice elicits a classic pleiotropic response in the liver, including a dramatic increase in the size and number of peroxisomes, induction of peroxisomal and microsomal fatty acid–oxidizing enzymes, and an increase in the size and number of hepatocytes (Moody et al., 1991; Rao and Reddy, 1987). Sustained oral administration of most PPC at doses sufficient to induce moderate to marked peroxisome proliferation in rats and mice results in liver tumor formation (Reddy and Lalwani, 1983), most likely through a nongenotoxic mode of action (Klaunig et al., 2003).
Many of the effects of PPC are mediated by three members of the nuclear receptor superfamily called the peroxisome proliferator–activated receptors ([PPAR] α, β, and γ). The PPARα subtype plays a dominant role in mediating the effects of hypolipidemic and xenobiotic PPC in the liver (Corton et al., 2000; Klaunig et al., 2003). Definitive evidence that PPARα is involved in the short-term effects of PPC exposure comes from experiments performed in PPARα-null mice (Lee et al., 1995). PPARα-null mice do not exhibit the classic short-term effects of PPC exposure, including peroxisome proliferation, hepatomegaly, induction of lipid-metabolizing enzymes such as Cyp4a family members, and increases in hepatocyte proliferation (summarized in Corton, 2009). The hypolipidemic agents WY-14,643 (WY) and bezafibrate increased liver tumors in wild-type but not in PPARα-null mice (Hays et al., 2005; Peters et al., 1997). In contrast, exposure to one of the most commonly used phthalates in industry, di-(2-ethylhexyl) phthalate (DEHP), resulted in a low level of liver tumors in PPARα-null but not in wild-type mice (Ito et al., 2007). The lack of statistically significant increases in liver tumors in the wild-type mice may be due to the relatively low levels of DEHP in the diet fed to the mice in the Ito et al. study compared to other studies (Klaunig et al., 2003). Additionally, PPARα-null but not wild-type mice exhibited sustained increases in markers of oxidative stress after DEHP exposure (Ito et al., 2007; Takashima et al., 2008). These data demonstrate that PPARα-independent biological events underlie the DEHP-induced mouse liver tumors in PPARα-null mice.
PPC including phthalates may induce transcriptional effects in the liver through other nuclear receptors. These candidate nuclear receptors include constitutive activated androstane receptor (CAR) and pregnane X receptor (PXR), which regulate the expression of subsets of xenobiotic-metabolizing enzymes (XME) in response to exposure to drugs and environmental chemicals (Stanley et al., 2006; Timsit and Negishi, 2007). Activators of CAR and PXR, such as phenobarbital (PB) and pregnenolone-16-alpha-carbonitrile (PCN), respectively, regulate an overlapping set of XMEs, including members of the CYP2B and CYP3A families and growth regulatory genes in the rat and mouse liver (Nelson et al., 2006). Like PPC, exposure to CAR or PXR activators can lead to increases in liver weight and hepatocyte hyperplasia that are abolished in mice nullizygous for the individual receptors (Chen et al., 2003; Huang et al., 2005; Staudinger et al., 2001; Yamamoto et al., 2004). Chronic exposure to the CAR activators PB or 1,4-bis-[2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) leads to increases in liver cancer in wild-type but not in CAR-null mice (Huang et al., 2005; Yamamoto et al., 2004).
Evidence is accumulating that phthalate ester plasticizers activate CAR and PXR. In transactivation assays, monoethylhexyl phthalate (MEHP), the primary metabolite of DEHP, activated mouse and human PXR (Hurst and Waxman, 2004) and mouse CAR (Baldwin and Roling, 2009). DEHP activated human PXR (Cooper et al., 2008; Takeshita et al., 2001) and a splicing variant of human CAR (CAR2) (DeKeyser et al., 2009). DEHP also increased the protein expression of Cyp3a11, a PXR target in livers of both wild-type and PPARα-null mice (Fan et al., 2004). Di-n-butyl phthalate (DBP) activated CAR and PXR in transactivation assays (Wyde et al., 2005). DBP increased the expression of CYP2B1 and CYP3A1 messenger RNA (mRNA) or protein in male and female rats (Fan et al., 2004) as well as dams and gestation day 19 fetuses (Wyde et al., 2005). In addition to phthalates, other PPC likely activate CAR or PXR. Exposure to PPARα-null mice to perfluorooctanoic acid (PFOA) results in a transcript profile similar to that of PB (Rosen et al., 2008a,b). Perfluorodecanoic acid was shown to regulate the CAR target gene and protein Cyp2b10 in wild-type but not in CAR-null mice (Cheng and Klaassen, 2008). The ability of PPC including DEHP to activate gene expression through nuclear receptors other than PPARα has not been comprehensively evaluated.
Toxicogenomic analyses in which global gene expression profiles of chemicals with different modes of action are directly compared can be invaluable to make predictions as to the pathway basis for toxicity. Given the evidence that DEHP increases liver tumors in the absence of PPARα (Ito et al., 2007) and indications that DEHP and other PPC can activate CAR and PXR, we compared the transcript profiles produced by structurally diverse PPC to known activators of CAR and PXR in rats and mice. We tested the role of PPARα and CAR in mediating the effects of DEHP directly by examining responses in wild-type, PPARα-null mice, and CAR-null mice. Overall, the data demonstrate that a number of PPC including DEHP exhibit transcriptional responses independent of PPARα but dependent on CAR.
MATERIALS AND METHODS
Animal studies.
The first study was carried out at U.S. Environmental Protection Agency (EPA), Research Triangle Park, NC, and utilized wild-type and PPARα-null male mice 7.5 months ± 2.5 weeks of age. PPARα-null mice (129S4/SvJae-Pparatm1Gonz/J, stock #003580) and wild-type mice (129S1/SvlmJ, stock #002448) were originally purchased from Jackson Laboratory (Bar Harbor, ME) and maintained as an inbred colony on the 129/Sv background at the U.S. EPA. PPARα-null and wild-type male mice used in the study were housed one per cage and were allowed to acclimate for a period of 1 week prior to the start of the study. Food (LabDiet 5001; PMI Nutrition International, St Louis, MO) and municipal tap water were provided ad libitum. Municipal tap water was filtered through sand and charcoal to reduce organic material, remove chlorine, and most of the chloramine. Chlorine was added back to the filtered water to maintain bacteria-free drinking water for the animals. Animal facilities were controlled for temperature (20°C–24°C) and relative humidity (40–60%) and kept under a 12-h light-dark cycle. The mice were a kind gift from Dr. Barbara Abbott. Exposure studies were similar to those published earlier (Currie et al., 2005) in which 1150 mg/kg gave robust transcriptional responses in wild-type mice. A dose of 200 mg/kg was selected as it was more similar to that used earlier (Ito et al., 2007). Wild-type and PPARα-null mice were given one daily gavage dose of DEHP (200 or 1150 mg/kg/body weight per day) in corn oil or corn oil alone each day for 4 days.
The second study was carried out at the University of Kansas Medical Center (Kansas City, KS). Eight-week-old adult C57BL/6 mice were purchased from Jackson Laboratory to establish a breeding colony. Breeding pairs of CAR-null mice on the C57BL/6 background were obtained from Dr. Ivan Rusyn (University of North Carolina, Chapel Hill, NC), which were engineered by Tularik, Inc. (South San Francisco, CA), as described previously (Ueda et al., 2002). CAR-null and C57BL/6 mice were administered DEHP identical to study 1.
Livers were removed 24 h after the last dose. Portions of the livers were rapidly snap frozen in liquid nitrogen and stored at −70°C until analysis. All animal studies were conducted under federal guidelines for the use and care of laboratory animals and were approved by Institutional Animal Care and Use Committees.
RNA isolation.
Total RNA was isolated and purified from mouse livers according to the mirVana miRNA Isolation Kit (Ambion, Austin, TX), which isolates total RNA including microRNAs. The integrity of each RNA sample was determined using an Agilent 2100 Bioanalyzer (Agilent, Foster City, CA), and RNA quantity was determined using a Nanodrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE).
Microarray hybridization and analysis.
Liver gene expression analysis was performed according to the Affymetrix-recommended protocol using Affymetrix Mouse ST v1.0 GeneChips containing probes for over 28,000 well-annotated genes. Total RNA (300 ng per sample) was labeled using the Affymetrix GeneChip WT cDNA Synthesis and Amplification kit protocol and hybridized to the arrays as described by the manufacturer (Affymetrix, Santa Clara, CA). The complementary RNA hybridization cocktail was incubated overnight at 45°C while rotating in a hybridization oven. After 16 h of hybridization, the cocktail was removed and the arrays were washed and stained in an Affymetrix GeneChip fluidics station 450, according to the Affymetrix-recommended protocol. Arrays were scanned on an Affymetrix GeneChip scanner. Four mice per dose group were examined. A detailed description of the microarray experiment is available through Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov/geo/, as accession number GSE18564. Data (.cel files) were analyzed and statistically filtered using X-Ray version 3.9934 software (www.biotiquesystems.com/Products-Solutions/XRAY/). Input files were normalized with full quantile normalization (Irizarry et al., 2003). Statistically significant genes were identified using mixed model analysis of variance with a false discovery rate (Benjamini-Hochberg test) of p ≤ 0.05. Fold-change values < |± 1.3| were removed.
Reanalysis of published microarray data.
A summary of the microarray studies reanalyzed is shown in Table 1. The doses selected in these studies would be expected to elicit close to a maximal transcriptional response. The raw data files analyzed in this project (.cel files from Affymetrix DNA chips) were either downloaded from GEO or communicated through the original authors. All the Affymetrix .cel files were first analyzed by Bioconductor SimpleAffy to assess data quality (Wilson and Miller, 2005). All .cel files passed this QC step. Data (.cel files) were background corrected and statistically filtered using Rosetta Resolver version 7.1 software (Rosetta Inpharmatics, Kirkland, WA). The background correction was done by Resolver’s specific data processing pipeline (Affymetrix Rosetta-Intensity Profile Builder). Statistically significant genes were identified using one-way ANOVA with a false discovery rate (Benjamini-Hochberg test) of ≤ 0.05, followed by a post hoc test (Scheffe) for significance. Fold-change values < |± 1.5| were removed. As most of the experiments in rats used the RG-U34A array, we compared profiles in the RG-U34A annotation file from Affymetrix (http://www.affymetrix.com/analysis/index.affx). We identified probe set IDs (a total of 8799) on the U34A chip that exhibited sequence similarity with those on the RAE230_2 chip using the “good match” comparison and then built fold-change values for those genes from the RAE230_2 chip, which were altered significantly. Signature genes were identified in the rat liver as those that were expressed in at least two of the three PB and PCN treatment groups (common genes), expressed in at least two of three PB groups but in only one or less of the PCN groups (CAR signature genes), or expressed in at least two of three PCN groups but in only one or less of the PB groups (PXR signature genes). Pearson's correlation coefficients were generated in Excel (2003). CAR signature genes in the mouse liver were defined as those that were altered by both PB and TC in wild-type mice, and the fold changes were of greater magnitude than that in similarly treated CAR-null mice. Heat maps were generated using Eisen Lab Cluster and Treeview software (http://rana.lbl.gov/EisenSoftware.htm). A detailed description of each experiment is available through GEO at the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov/geo/, as accession numbers indicated in Table 1.
TABLE 1.
Characteristics of the Studies Used in the Rat and Mouse Liver Microarray Analysis
| Species | Reference | GEO accession number | Strain | Dose frequency | Chemical (dose)a | Time of treatment | Vehicle | Array Type | Replicates | Number of .cel files |
| Rat | Nelson et al. (2006) | GSE14712 | SD | Once daily | PB (100) or PCN (100) | 6 h, 1 day, 5 days | 1% Tween 80 in 0.5% aqueous methylcellulose | RGU34A | 5 | 80 |
| Rat | Ellinger-Ziegelbauer et al. (2005) | GSE14712 | SD (Wistar) | Once daily | WY (60) | 1 day, 3 days, 7 days | Carboxymethyl-cellulose | RAE230A | 3 | 12 |
| Rat | Jolly et al. (2005) | GSE2303 | SD | Once | DEHP (20,000) or VPA (2000) | 4, 24, 48 h | Distilled water | U34A | 3–5 | 16 (DEHP) and 15 (VPA) |
| Rat | Jolly et al. (2005) | GSE2303 | SD | Once | CLO (1000) | 4, 24, 48 h | 0.9% saline | U34A | 3–5 | 16 |
| Mouse | www.nursa.org/10.1621/datasets.01003 | Wild type and CAR- null | Once | TCPOBOP (3) | 3 days | Corn oil | 430_2 | 2 | 8 | |
| Mouse | www.nursa.org/10.1621/datasets.01003 | Wild type and CAR- null | Once daily | PB (100) | 3 days | Corn oil | 430_2 | 2 | 8 | |
| Mouse | Sanderson et al. (2008) | GSE8292, GSE8295 | SV129 and PPAR null | Once daily | WYb | 6 h, 5 days | 430_2 | 3–5 (6 h) and 4 (5 days) | 17 (6 h) and 16 (5 days) | |
| Mouse | Currie et al. (2005) | C57Bl/6 | Once daily | DEHP (1150) | 2, 8, 24 h, 3 days | Corn oil | 430_2 | 3 | 24 |
Note. All exposure experiments were conducted on male rats or mice by gavage. SD, Sprague-Dawley.
All doses are in milligrams per kilogram per day.
400 μl of a 10 mg/ml solution/day.
Evaluation of selected genes by real-time reverse transcription-PCR.
The levels of expression of selected genes were quantified using real-time reverse transcription-PCR (RT-PCR) analysis. Briefly, total RNA was reverse transcribed with murine leukemia virus reverse transcriptase and oligo(dT) primers. The forward and reverse primers for selected genes (Supplementary file 1) were designed using Primer Express software, version 2.0 (Applied Biosystems, Foster City, CA). The SYBR green DNA PCR kit (Applied Biosystems) was used for real-time PCR analysis. The relative differences in expression between groups were expressed using cycle threshold (Ct) values as follows. The Ct values of the genes were first normalized with β-actin and glyceraldehyde 3-phosphate dehydrogenase of the same sample. Assuming that the Ct value is reflective of the initial starting copy and that there is 100% efficiency, a difference of one cycle is equivalent to a twofold difference in starting copy. Means and SE (n = 4) for RT-PCR data were calculated by Student’s t-test. The level of significance was set at p ≤ 0.05.
RESULTS
Expression profiles were compared after exposure of rodents to a number of xenobiotics and drugs that activate well-characterized pathways of gene expression. The list of studies is shown in Table 1. To address whether CAR and/or PXR were activated upon exposure to DEHP and other PPC, the expression profiles induced by classical activators of CAR and PXR (PB and PCN, respectively) in the rat liver and activators of CAR in mouse liver (PB and TCPOBOP) were compared. PB and PCN transcript profiles in rat livers were compared to known activators of PPARα, including two hypolipidemic compounds (WY and clofibrate [CLO]), an anti-epilepsy drug (valproic acid [VPA]) as well as DEHP. In mouse livers, PB, and TCPOBOP transcript profiles were compared to those induced by DEHP in wild-type mice and WY in both wild-type and PPARα-null mice. WY is selective for the PPARα subtype in liver; almost all genes regulated by WY were dependent on PPARα in mice (Anderson et al., 2004a,b; Rosen et al., 2008a,b).
Similarities in the Transcriptional Profiles of PPARα, CAR, and PXR Activators in the Rat Liver
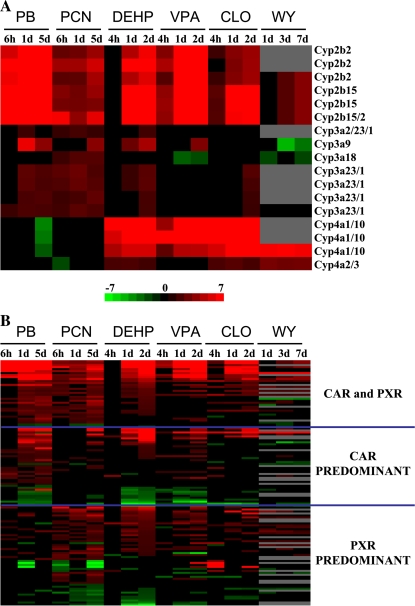
The expression of CYP family members is often used as an indicator of xenobiotic activation of nuclear receptors. The expression of all CYP family members on the Affymetrix RU-U34A array that exhibited significant change(s) in at least two of the 18 treatment comparisons were compared (Supplementary file 2). Figure 1A shows the expression of CYP family members best known to be regulated by PPARα, CAR, and PXR (CYP4A, CYP2B, and CYP3A, respectively). As expected, the PPC, DEHP, VPA, CLO, and WY but not PB and PCN exposure led to increased expression in CYP4A family members. DEHP, VPA, and CLO activated CYP2B family members that were also activated by PB and to a lesser extent by PCN. WY was a weak inducer of only CYP2B15. DEHP also activated CYP3A family members that were activated by both PB and PCN. CLO activated CYP3A23/1 only at 2 days, and WY decreased the expression of CYP3A9, CYP3A18, and CYP3A23/1. Overall, the expression profiles indicate that DEHP, VPA, and CLO exhibit the ability to alter the expression of CYP genes in multiple families that are regulated by PPARα and CAR.
FIG. 1.
Gene expression in rat liver after exposure to activators of CAR, PXR, and PPARα. (A) Expression of CYP genes. CYP2B, CYP3A, and CYP4A family members significantly altered after chemical treatment in one or more of the comparisons were grouped by family. The profiles show that DEHP, VPA, and CLO exhibit increased expression of a number of CYP genes that overlap with PB and PCN (CYP2B and CYP3A family members) and WY (CYP4A family members). Genes indicated as pairs (e.g., CYP4A1/10) are interrogated by one Affymetrix probe set. (B) Comparison of genes regulated exclusively by PB, PCN, or both PB and PCN. Genes that were regulated by PB and/or PCN were identified, and their expression pattern was examined across all the treatment groups. The intensity scale indicates fold change due to chemical exposure relative to controls for A and B.
The expression of CAR and/or PXR signature genes including CYP genes as detailed in the “Materials and Methods” section were compared to the expression of these genes after exposure to various PPARα activators (Fig. 1B). Correlation coefficients were generated for these comparisons between 1-day treatment groups. DEHP, VPA, and CLO but not the WY treatment groups exhibited strong correlations with the PB and to a lesser extent the PCN groups (Table 2). DEHP, VPA, and CLO when compared to the PB group gave higher correlation coefficients than when PB was compared to the PCN group. These data indicate that under these exposure conditions, DEHP, VPA, and CLO but not WY treatments exhibit transcriptional similarities to classical CAR and PXR inducers.
TABLE 2.
Correlation Coefficients between Transcriptional Changes after Exposure to the Indicated Chemicals in the Rat Liver
| DEHP | VPA | CLO | WY | PCN | |
| PB | 0.66 | 0.72 | 0.51 | 0.04 | 0.41 |
| PCN | 0.46 | 0.42 | 0.41 | 0.15 |
Note. Pearson's correlation coefficients were generated as described in the “Materials and Methods” section.
Overlap in Genes Regulated by DEHP and Activators of CAR in Mouse Liver
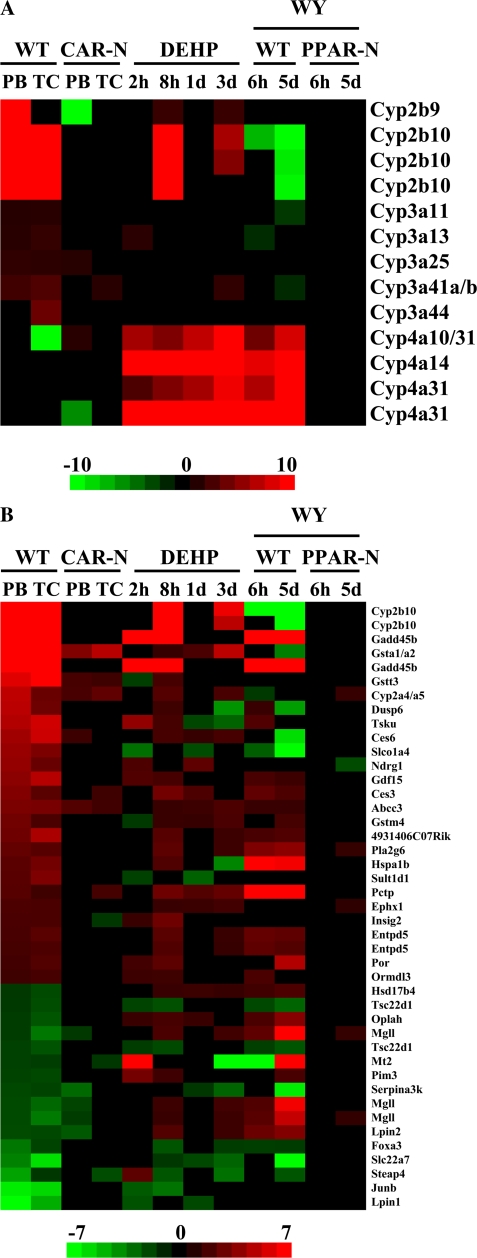
To investigate the possibility that DEHP regulates gene expression similar to activators of CAR in mouse liver, the profiles from wild-type mice treated with DEHP were compared to those from livers of mice exposed to CAR activators. Wild-type and CAR-null mice were administered the CAR activators PB or TCPOBOP. As expected, DEHP and WY treatment in wild-type mice resulted in strong induction of Cyp4a family members (Fig. 2A). DEHP-treated wild-type mice also induced expression of Cyp2b9 and Cyp2b10 at 8 h and 3 days but not at 1 day. CAR and regulated genes such as Cyp2b10 exhibit circadian regulation; CAR expression peaks near the time of the 8-h sacrifice (Zhang et al., 2009). We speculate that the relatively weak induction of Cyp2b10 by DEHP in wild-type mice was enhanced at 8 h due to the increased expression of CAR. DEHP exposure led to inconsistent induction of Cyp3a family members. In contrast, WY treatment downregulated expression of Cyp2b and Cyp3a family members.
FIG. 2.
Gene expression in mouse liver after exposure to activators of CAR, PXR, and PPARα. (A) Expression of Cyp genes. Cyp2b, Cyp3a, and Cyp4a family members that were significantly altered after chemical treatment in one or more of the comparisons were grouped by family. Genes regulated by both PB and TCPOBOP were identified and their expression was compared to the same genes after four different times of DEHP exposure in wild-type mice or WY treatment for 6 h or 5 days in wild-type and PPARα-null mice. The profiles show that DEHP exhibits increased expression of a number of Cyp genes that overlap with PB and PCN (Cyp2b and Cyp3a family members) and WY (Cyp4a family members). The changes due to WY were PPARα dependent. (B) Comparison of CAR genes to those regulated by DEHP and WY. The intensity scale indicates fold change due to chemical exposure relative to controls.
Genes attributed to CAR activation were identified (i.e., genes coordinately regulated by PB and TCPOBOP in a CAR-dependent manner), and of those 244 genes, the correlation between PB and DEHP or WY treatment groups was determined. DEHP treatment for 2 h, 8 h, 1, day, or 3 days gave correlation coefficients of 0.11, 0.83, 0.03, and 0.21, respectively, with genes altered by PB treatment. WY treatments at 6 h or 5 days gave coefficients of 0.05 and −0.19, respectively, with genes altered by PB treatment. The expression of 43 genes that were expressed in at least two of the four DEHP treatment groups showed a high degree of concordance to expression of CAR signature genes especially the 8-h DEHP treatment group (Fig. 2B). WY but not DEHP treatment altered the expression of a number of genes opposite to that of PB and TCPOBOP, including Cyp2b10, Gsta1/a2, and Ces6. Overall, these results indicate that DEHP treatment induces a gene expression pattern that overlaps with CAR activators.
PPARα Is Required for the Majority of Transcriptional Changes after DEHP Exposure
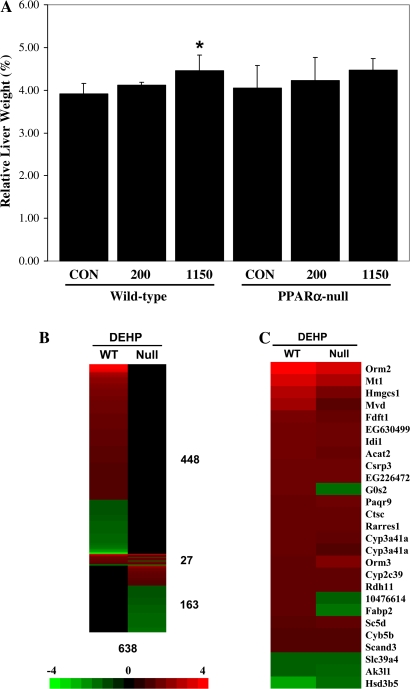
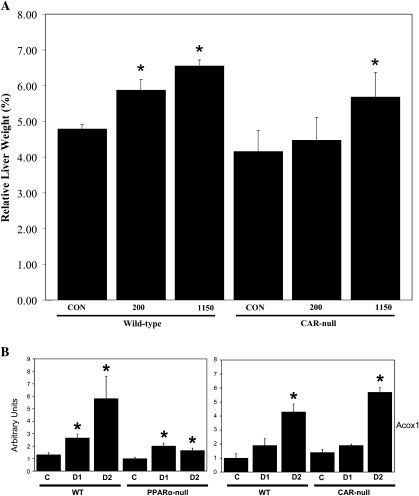
To uncover the gene regulatory effects that may contribute to liver tumor induction by DEHP, we exposed wild-type and PPARα-null mice to 200 or 1150 mg/kg of DEHP once each day for 4 days. Liver to body weights increased only in the wild-type mice exposed to 1150 mg/kg/day but not to 200 mg/kg/day (Fig. 3A). Global gene expression changes were assessed using Affymetrix ST 1.0 mouse chips containing ∼28,000 genes. There were 475 genes that exhibited altered expression by DEHP in wild-type mice of which only 27 (∼6% of 475) were also differentially expressed in PPARα-null mice after DEHP exposure (Fig. 3B, Supplementary file 3). All but 3 of the 27 genes exhibited similar regulation in the wild-type and PPARα-null mice (Fig. 3C). The 27 genes included those involved in xenobiotic metabolism and cholesterol biosynthesis. In the absence of PPARα, DEHP exposure altered an additional 163 genes (Fig. 3B). Thus, this analysis revealed that PPARα controls the expression of most (∼94%) of the genes regulated by DEHP in wild-type mice. The absence of PPARα leads to the altered regulation of additional genes.
FIG. 3.
Identification of PPARα-dependent and -independent genes regulated by DEHP in the mouse liver. (A) Liver to body weights in wild-type and PPARα-null mice after exposure to DEHP. (B) Genes regulated by DEHP in wild-type and PPARα-null mice. Significantly altered genes were identified as described in the “Materials and Methods” section. The genes were rank ordered based on their fold change after exposure to DEHP in wild-type or PPARα-null mice. There were a total of 475 and 190 genes that were significantly altered by DEHP in wild-type (WT) or PPARα-null (Null) mice, respectively. Of these, 448 and 163 genes were uniquely altered in wild-type or PPARα-null mice, respectively. Twenty-seven genes exhibited altered expression in both mouse strains. (C) Genes regulated by DEHP in both wild-type and PPARα-null mice. The intensity scale in B indicates fold change due to DEHP exposure relative to controls in B and C.
Characterization of the PPARα-Independent Genes Regulated by DEHP
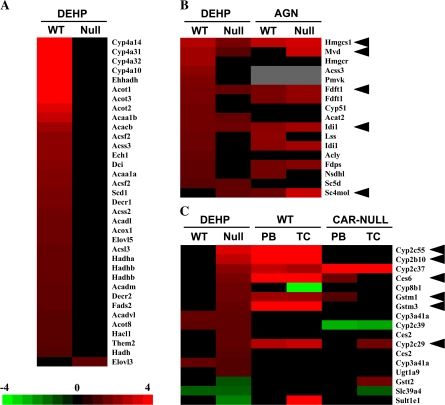
The functional categories of DEHP-regulated genes between wild-type and PPARα-null mice were compared. Ingenuity Pathways Analysis showed statistical significance of altered changes in groups of genes involved in fatty acid metabolism, tryptophan metabolism, valine, leucine and isoleucine degradation, butanoate metabolism, propanoate metabolism, and β-alanine metabolism in wild-type mice and to a lesser extent in PPARα-null mice, whereas xenobiotic metabolism signaling, metabolism of xenobiotics by cytochrome P450, and biosynthesis of steroids were unaffected by genotype. The previous analysis of genes regulated by PFOA in PPARα-null mice demonstrated that many lipid-metabolizing genes, especially those involved in fatty acid β-oxidation, were similarly regulated in PFOA-treated wild-type and PPARα-null mice (Rosen et al., 2008a,b). In contrast, the consistent upregulation of these genes by DEHP was PPARα dependent, with the exception of Elovl3 that was altered only in PPARα-null mice (Fig. 4A).
FIG. 4.
Functional analysis of DEHP-regulated genes. (A) Fatty acid catabolism and synthesis genes. All but Elovl3 were regulated by DEHP in a PPARα-dependent manner. (B) Cholesterol biosynthetic genes. Genes involved in cholesterol biosynthesis altered by DEHP in wild-type or PPARα-null mice were compared to those regulated by the RXR pan-agonist AGN194,204 (AGN) in wild-type and PPARα-null mice. Many of the genes were regulated by DEHP or AGN independently of PPARα (arrowheads). (C) Xenobiotic metabolism genes. XME genes that exhibited altered regulation in PPARα-null mice were rank ordered and compared to the regulation by PB or TCPOBOP (TC) in wild-type and CAR-null mice. Arrowheads indicate genes that exhibit increased expression by DEHP in PPARα-null mice and by both PB and TC in wild-type but not in CAR-null mice. Genes represented more than once were identified through the analysis of different probe sets. The intensity scale in A indicates fold change due to DEHP exposure relative to controls in A–C.
Many genes involved in cholesterol biosynthesis were uniformly upregulated by DEHP in PPARα-null mice (Fig. 4B). Given the observation of a similar induction of cholesterol biosynthetic genes after exposure to a RXR pan-agonist (AGN194,204; AGN) (Anderson et al., 2004a), the expression of these genes was compared between DEHP and AGN. Many of these cholesterol biosynthetic genes were regulated by DEHP and AGN in a PPARα-independent manner, including 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (Hmgcs1), mevalonate (diphospho) decarboxylase (Mvd), farnesyl diphosphate farnesyl transferase 1 (Fdft1), isopentenyl-diphosphate delta isomerase (Idi1), and sterol-C4-methyl oxidase-like (Sc4mol). These results indicate that DEHP regulates many of the cholesterol biosynthetic genes in a manner similar to an RXR agonist.
As shown previously for PFOA, DEHP altered the expression of many XMEs independently of PPARα. Most of these XMEs were regulated exclusively in PPARα-null mice (Fig. 4C). The expression of all DEHP-regulated PPARα-independent genes were compared to those regulated by the two CAR activators, PB or TCPOBOP (TC), from a previous study (www.nursa.org/10.1621/datasets.01003). Very little concordance was observed except for a number of XMEs that were almost uniformly upregulated (Fig. 4C, arrowheads). Additional non-XME genes also exhibited concordance including hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5), NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 (Ndufb2), and programmed cell death 6 (Pdcd6), which were downregulated by DEHP and either PB or TCPOBOP (data not shown). It cannot be ruled out that the lack of concordance is due to the comparison of results between two microarray platforms, i.e., mouse 430_2 versus mouse ST v1.0. Many of the XMEs were upregulated by DEHP in PPARα-null but not in wild-type mice and by PB and TC in a CAR-dependent manner, indicating that at least a subset of the XMEs regulated by DEHP are CAR dependent.
CAR-Dependent Transcriptional Changes Induced by DEHP
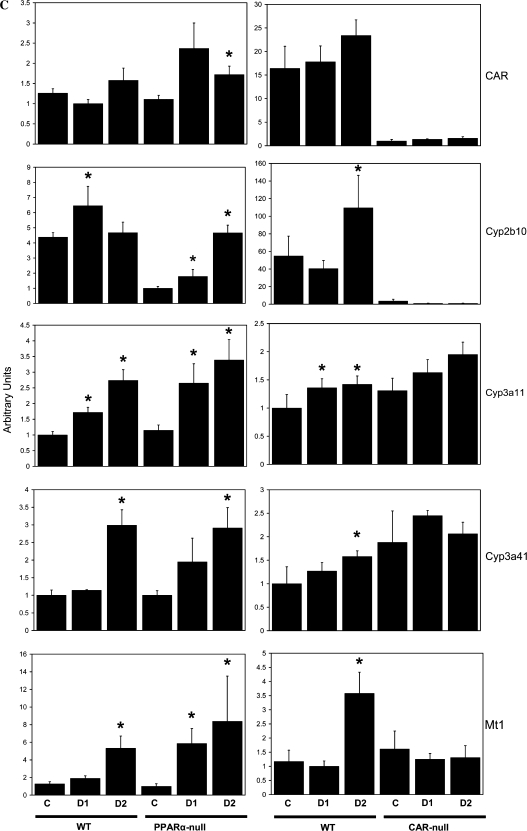
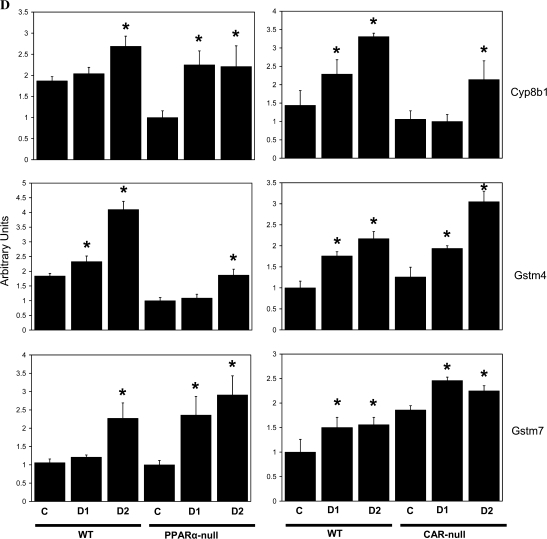
To directly test the hypothesis that a subset of DEHP-regulated genes are mediated by CAR, we exposed wild-type and CAR-null mice once each day to 200 or 1150 mg/kg/day DEHP for 4 days. Induction of liver weights was observed in the wild-type mice at both doses and the CAR-null mice at the highest dose (Fig. 5A). RT-PCR analysis of gene expression of PPARα and CAR target genes revealed genes that can be divided into three classes, based on PPARα- and/or CAR-dependent induction of expression. DEHP induction of acyl-Coenzyme A oxidase 1, palmitoyl (Acox1) exhibited partial dependence of induction on PPARα. Induction in wild-type mice at the highest dose was approximately fivefold compared to approximately twofold in PPARα-null mice. Partial induction of Acox1 and other genes involved in fatty acid catabolism was observed in PPARα-null mice by PFOA (Rosen et al., 2008a,b) and was attributed to minor activation of other PPAR subtypes. Acox1 induction was not affected by CAR genotype (Fig. 5B). A number of genes including Cyp2b10, Cyp3a11, Cyp3a41a, and metallothionein 1 (Mt1) were induced by DEHP that was dependent on CAR but not on PPARα (Fig. 5C). Cyp2b10 and Cyp3a11 are known targets of CAR from previous studies (Maglich et al., 2002). Metallothionein protein levels were increased by DEHP but not by PB in mouse liver (Waalkes and Ward, 1989). The expression of the CAR gene itself was increased by DEHP in PPARα-null but not in wild-type mice. A number of putative CAR and PXR targets exhibited PPARα- and CAR-independent induction, including Cyp8b1, Gstm4, and Gstm7 (Fig. 5D). These results indicate that DEHP requires CAR for induction of a subset of genes.
FIG. 5.
DEHP responses in wild-type and CAR-null mice. Wild-type and CAR-null mice were treated with DEHP (200 [day 1] or 1150 mg/kg [day 2]) by gavage for 4 days. (A) Liver to body weights. (B–D) Gene expression in the livers of DEHP-treated wild-type, PPARα-null, and CAR-null mice determined by RT-PCR. (B) Induction of Acox1 by DEHP is partially dependent on PPARα but not on CAR. (C) Induction of Cyp2b10, Cyp3a11, Cyp3a41a, and Mt1 is dependent on CAR but not on PPARα. The expression of the CAR gene is also shown. (D) PPARα- and CAR-independent induction of Cyp8b1, Gstm4, and Gstm7 by DEHP. There were four animals per treatment group, and each sample was analyzed in duplicate. Variability is expressed as SEM. Means and SE (n = 4) for RT-PCR data were calculated by Student’s t-test. The level of significance was set at p ≤ 0.05. *Indicates statistically significant change relative to control animals of the same genotype.
DISCUSSION
DEHP Induces Gene Expression through Multiple Nuclear Receptors
A feature of PPARα activators is their structural heterogeneity, leading to the hypothesis that a subset of these compounds could interact with other nuclear receptors. Consistent with this, the present study identified PPARα and CAR as targets of DEHP indirectly through meta-analysis of transcript profiles of livers from rats treated with nuclear receptor activators and directly through transcriptional analysis in wild-type mice and mice nullizygous for these nuclear receptors. Microarray analysis of PPC-treated rats showed an overlap in the profiles of DEHP-, VPA-, and CLO-treated rats with classical activators of CAR and to a lesser extent an activator of PXR. The overlapping genes included CYP gene families that are often considered signature genes for nuclear receptor activation. Consistent with these findings, VPA was recently shown to activate CAR and PXR in transactivation assays (Cerveny et al., 2007). In contrast to the other PPC, WY treatment in rats resulted in little or no overlap with genes induced by CAR and PXR activators, providing evidence that WY is more PPARα-specific. A comparison of DEHP-treated wild-type and PPARα-null mice revealed that PPARα is required for ∼94% of all transcriptional changes in wild-type mice. The PPARα-dependent genes were the typical target genes of PPC identified in many other studies and included those involved in fatty acid β-oxidation (Anderson et al., 2004a,b; Wong and Gill, 2002; Woods et al., 2007). The remaining 6% of the genes were dominated by those involved in xenobiotic metabolism, which are known target genes of CAR or PXR (Stanley et al., 2006; Timsit and Negishi, 2007), and cholesterol biosynthesis, which are regulated by a number of transcription factors, including RXR (Anderson et al., 2004a). XMEs, including Cyp2b10, Cyp3a11, and Cyp3a41a as well as Mt1, were induced by DEHP that was partially or completely dependent on CAR but not on PPARα. Cyp2b10 is primarily a CAR target gene but can be induced by PXR activation (Maglich et al., 2002). In the present study, DEHP induced Cyp2b10 in CAR-null mice to low levels possibly through PXR activation. The results of the present study are consistent with a recent study in which an array of 320 nuclear receptor–targeted genes was used to show that in mouse liver, most DEHP-regulated genes were PPARα-dependent and more specifically the induction of Cyp2b10 was PPARα-independent (Eveillard et al., 2009). The identification of Cyp8b1, Gstm4, and Gstm7 as genes regulated by DEHP independent of PPARα and CAR indicates the existence of a class of XMEs possibly regulated by PXR or the oxidant-activated transcription factor nuclear factor erythroid 2-related factor 2.
Other environmentally relevant PPC, such as PFOA (Rosen et al., 2008a,b) and PFOS (Rosen, Corton, Scmid, Zehr, Das, Abbott, and Lau, in preparation), exhibit similar transcriptional features as DEHP. PPARα is required for 85 and 92% of the genes regulated by PFOA and PFOS, respectively. WY, originally developed as a hypolipidemic drug, requires PPARα for > 99% of the transcriptional changes in wild-type mice (Rosen et al., 2008a,b). The activation of CAR and/or PXR in vivo by DEHP was not unexpected because DEHP or its proximate metabolite MEHP trans-activate CAR or PXR in vitro (Baldwin and Roling, 2009; Cooper et al., 2008; DeKeyser et al., 2009; Hurst and Waxman, 2004; Takeshita et al., 2001). Taken together, the present results provide direct evidence that DEHP, and likely other PPC, regulate gene expression in liver through PPARα and CAR. Similar experiments in wild-type and PXR-null mice will be useful to determine the extent of involvement of PXR.
Interactions between PPARα and CAR
Functional antagonism between transcription factors involved in opposing biological functions is a common molecular mechanism for regulation of gene expression. There is a growing body of evidence that PPARα and CAR have antagonist properties. In the absence of PPARα, CAR expression levels were increased (Martin et al., 2007) and PB-induced hyperplasia was greater when compared to that in wild-type mice (Columbano et al., 2001). The transcript profiles of DEHP-treated mice in the present study showed that the mRNA of a number of XMEs that are known CAR target genes were induced to a greater extent by DEHP in PPARα-null mice than in wild-type mice. Using RT-PCR Cyp2b10 and Mt1 as well as the CAR gene itself were induced more in PPARα-null mice than in wild-type mice. An increase in CAR regulation of gene expression in the absence of PPARα was also observed in a microarray comparison between wild-type and PPARα-null mice after PFOA (Rosen et al., 2008a,b) and PFOS (Rosen, Corton, Scmid, Zehr, Das, Abbott and Lau, in preparation) treatments. In the reverse situation, absence of CAR may lead to increased induction of PPARα-dependent responses. CAR-null mice exhibited increased hepatic fatty acid β-oxidation typically controlled by PPARα; treatment of wild-type mice with TCPOBOP decreased expression of PPARα mRNA as well as PPARα target genes in liver (Maglich et al., 2009).
Functional antagonism between PPARα and CAR can occur at a number of levels. PPARα agonists may act as CAR antagonists, much like the CAR reverse agonist androstanol; exposure to WY or ciprofibrate leads to CAR nuclear localization but not target gene induction (Guo et al., 2007). PPARα and CAR may also compete for a number of shared coactivators, including PPAR-binding protein, PRIC320, and PGC1α (Corton and Brown-Borg, 2005; Jia et al., 2005; Mäkinen et al., 2002; Surapureddi et al., 2006), but apparently not the shared heterodimer partner, RXR (Guo et al., 2007). Furthermore, CAR-RXR heterodimers from mice and humans can bind and trans-activate at a PPAR-RXR DNA–binding site, and in humans, this transactivation does not require the CAR DNA–binding domain (Guo et al., 2007; Stoner et al., 2007). Further work is required to clarify the functional significance of PPARα and CAR antagonistic effects.
Mode of Action of Liver Tumor Induction by DEHP
Chronic exposure to DEHP resulted in a low level of liver tumors in PPARα-null mice but in not wild-type mice (Ito et al., 2007). DEHP induction of tumors in PPARα-null mice is in part the basis for an argument against a PPARα mode of action in wild-type mice (Guyton et al., 2009). However, there are a number of findings that argue against this view. First, the transcript profile comparison in the present study showed that the vast majority of genes altered by DEHP in wild-type mice were not similarly altered in PPARα-null mice. Thus, the DEHP-induced tumors in wild-type mice could only be PPARα independent if the ∼6% of the PPARα-independent gene changes were responsible for the tumors, an unlikely scenario given the magnitude of the PPARα-dependent effects. Second, wild-type and PPARα-null mice exhibited differences in DEHP-induced carcinogenesis. DEHP did not induce equivalent levels of tumors in the wild-type and PPARα-null mice; there were no statistically significant increases in liver tumors in the wild-type mice under these exposure conditions (200 ppm). DEHP increased the expression of growth control genes in PPARα-null mice but not in wild-type mice at equivalent doses (Ito et al., 2007). Transcript profiling and RT-PCR showed highly dissimilar changes in gene expression in the liver tumors from the wild-type and PPARα-null mice, indicating different molecular mechanisms of their origins (Takashima et al., 2008). Lastly, in the absence of PPARα, DEHP altered a unique set of genes not similarly altered in wild-type mice (Fig. 3B). Some of these genes are known targets of CAR (Fig. 4C). Taken together, these data indicate that although DEHP can induce marginal increases in liver tumors in PPARα-null mice, the mode of action is different from that in wild-type mice. The transcriptional responses induced by DEHP in wild-type and nullizygous mouse strains indicates that CAR is important in the induction of tumors in PPARα-null mice.
CAR and Endocrine Modulation by DEHP
Activation of CAR may lead to a number of extrahepatic effects due to CAR-dependent regulation of genes that metabolize thyroid and steroid hormones. CAR controls phase II genes uridine 5′-diphosphate-glucuronosyltransferases (UGT) and sulfotransferases (SULT) involved in the metabolism of thyroid hormones (Klaassen and Hood, 2001; Kretschmer and Baldwin, 2005). PB increases metabolism of thyroid hormones and leads to a decrease in thyroid hormone levels followed by compensatory increases in thyroid stimulating hormone, thyroid epithelial hyperplasia, and tumors (Klaassen and Hood, 2001; McClain et al., 1988; Qatanani et al., 2005). CAR activation by DEHP is consistent with increases in thyroid hyperplasia and hypertrophy observed in rats (Elcombe et al., 2002; Price et al., 1988). The indication that increased exposure to DEHP metabolites was associated with decreases in thyroid hormone in men (Meeker et al., 2007) indicates that the relationships between DEHP exposure, CAR activation, and alteration in thyroid hormone metabolism should be further characterized.
A growing number of endocrine disrupting chemicals including pesticides activate PXR and sometimes CAR resulting in increases in the expression of steroid hydroxylases, including CYP2B and CYP3A family members, potentially perturbing normal steroid metabolism (Kretschmer and Baldwin, 2005). DEHP activates the CAR-regulated gene CYP2B6 in human primary hepatocytes (Eveillard et al., 2009; Goyak et al., 2008). PB exhibits classical antiandrogen effects in developing rats, including reduced ano-genital distance and delayed testicular descent (Gupta et al., 1980) overlapping with the male reproductive tract defects induced by active phthalates (summarized in Corton and Lapinskas, 2005). Whether there are increases in fetal liver steroid hydroxylases after exposure to CAR inducers has not been adequately explored for linkage to decreases in testosterone levels. In humans, an association has been noted between treatment of pregnant mothers with CAR activators, PB or phenytoin, and a higher risk for developmental abnormalities, such as undescended testes and genital malformations in male offspring (Dessens et al., 2001). Workers exposed to high levels of DEHP and DBP had associated decreases in serum-free testosterone (Pan et al., 2006). Additional studies are required to determine the significance of endocrine disruption by DEHP through different nuclear receptors, including CAR.
Conclusions
The present analysis of gene expression in wild-type and PPARα-null mice demonstrate that like other PPARα activators, including WY, PFOA, and PFOS, DEHP transcriptional responses are overwhelmingly dependent on PPARα. A minor number of genes in wild-type mice including Cyp2b10 are dependent on CAR for activation. In the absence of PPARα, CAR-dependent effects are likely important. The chronic activation of the CAR mode of action in the PPARα-null mice likely underlies the low level of liver tumor induced by DEHP in these mice.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
U.S. Environmental Protection Agency; National Institutes of Health (ES013714 to C.D.K.).
Supplementary Material
Acknowledgments
We wish to thank Dr Barbara Abbott for the wild-type and PPARα-null mice; Drs Lois Lehman-McKeeman, Heidrun Ellinger-Ziegelbauer, and Richard Currie for Affymetrix data from their published studies; Kathleen Wallace and Tanya Moore for their assistance with the animal studies; Dr Jie Liu for assistance in the RT-PCR experiments; and Drs Susan Hester and Sheau-Fung Thai for their critical review of this manuscript. It has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. Author contributions: H.R. and B.V. analyzed the microarray data and C.W. generated and analyzed the RT-PCR data. L.A., M.G., and C.K. performed animal experiments. J.C.C. conceived of the study, participated in study design and animal studies, analyzed microarray data, and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- Anderson SP, Dunn C, Laughter A, Yoon L, Swanson C, Stulnig TM, Steffensen KR, Chandraratna RA, Gustafsson JA, Corton JC. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor alpha, retinoid X receptor, and liver X receptor in mouse liver. Mol. Pharmacol. 2004a;66:1440–1452. doi: 10.1124/mol.104.005496. [DOI] [PubMed] [Google Scholar]
- Anderson SP, Howroyd P, Liu J, Qian X, Bahnemann R, Swanson C, Kwak MK, Kensler TW, Corton JC. The transcriptional response to a peroxisome proliferator-activated receptor alpha agonist includes increased expression of proteome maintenance genes. J. Biol. Chem. 2004b;279:52390–52398. doi: 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- Baldwin WS, Roling JA. A concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol. Sci. 2009;107:93–105. doi: 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny L, Svecova L, Anzenbacherova E, Vrzal R, Staud F, Dvorak Z, Ulrichova J, Anzenbacher P, Pavek P. Valproic acid induces CYP3A4 and MDR1 gene expression by activation of constitutive androstane receptor and pregnane X receptor pathways. Drug Metab. Dispos. 2007;35:1032–1041. doi: 10.1124/dmd.106.014456. [DOI] [PubMed] [Google Scholar]
- Chen C, Staudinger JL, Klaassen CD. Nuclear receptor, pregname X receptor, is required for induction of UDP-glucuronosyltranferases in mouse liver by pregnenolone-16 alpha-carbonitrile. Drug Metab. Dispos. 2003;31:908–915. doi: 10.1124/dmd.31.7.908. [DOI] [PubMed] [Google Scholar]
- Cheng X, Klaassen CD. Critical role of PPAR-alpha in perfluorooctanoic acid- and perfluorodecanoic acid-induced downregulation of Oatp uptake transporters in mouse livers. Toxicol. Sci. 2008;106:37–45. doi: 10.1093/toxsci/kfn161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Cousins I, Mac Kay D. Assessment of critical exposure pathways. In: Staples C, editor. The Handbook of Environmental Chemistry, 3Q. Phthalate Esters. New York, NY: Springer; 2003. pp. 227–262. [Google Scholar]
- Columbano A, Ledda-Columbano GM, Pibiri M, Concas D, Reddy JK, Rao MS. Peroxisome proliferator-activated receptor-alpha mice show enhanced hepatocyte proliferation in response to the hepatomitogen 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene, a ligand of constitutive androstane receptor. Hepatology. 2001;34:262–266. doi: 10.1053/jhep.2001.26172. [DOI] [PubMed] [Google Scholar]
- Cooper BW, Cho TM, Thompson PM, Wallace AD. Phthalate induction of CYP3A4 is dependent on glucocorticoid regulation of PXR expression. Toxicol. Sci. 2008;103:268–277. doi: 10.1093/toxsci/kfn047. [DOI] [PubMed] [Google Scholar]
- Corton JC. Mode of action analysis and human relevance of liver tumors induced by PPARα activation. In: Hsu C-H, Stedeford T, editors. Cancer Risk Assessment: Chemical Carcinogenesis from Biology to Standards Quantification. Hoboken, NJ: John Wiley & Sons, Inc.; 2009. [Google Scholar]
- Corton JC, Anderson SP, Stauber A. Central role of peroxisome proliferator-activated receptors in the actions of peroxisome proliferators. Annu. Rev. Pharmacol. Toxicol. 2000;40:491–518. doi: 10.1146/annurev.pharmtox.40.1.491. [DOI] [PubMed] [Google Scholar]
- Corton JC, Brown-Borg HM. Peroxisome proliferator-activated receptor gamma coactivator 1 in caloric restriction and other models of longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:1494–1509. doi: 10.1093/gerona/60.12.1494. [DOI] [PubMed] [Google Scholar]
- Corton JC, Lapinskas PJ. Peroxisome proliferator-activated receptors: mediators of phthalate ester-induced effects in the male reproductive tract? Toxicol. Sci. 2005;83:4–17. doi: 10.1093/toxsci/kfi011. [DOI] [PubMed] [Google Scholar]
- Currie RA, Bombail V, Oliver JD, Moore DJ, Lim FL, Gwilliam V, Kimber I, Chipman K, Moggs JG, Orphanides G. Gene ontology mapping as an unbiased method for identifying molecular pathways and processes affected by toxicant exposure: application to acute effects caused by the rodent non-genotoxic carcinogen diethylhexylphthalate. Toxicol. Sci. 2005;86:453–469. doi: 10.1093/toxsci/kfi207. [DOI] [PubMed] [Google Scholar]
- DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol. Pharmacol. 2009;75:1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessens AB, Cohen-Kettenis PT, Mellenbergh GJ, Koppe JG, Poll NE, Boer K. Association of prenatal phenobarbital and phenytoin exposure with genital anomalies and menstrual disorders. Teratology. 2001;64:181–188. doi: 10.1002/tera.1063. [DOI] [PubMed] [Google Scholar]
- Elcombe CR, Odum J, Foster JR, Stone S, Hasmall S, Soames AR, Kimber I, Ashby J. Prediction of rodent nongenotoxic carcinogenesis: evaluation of biochemical and tissue changes in rodents following exposure to nine nongenotoxic NTP carcinogens. Environ. Health Perspect. 2002;110:363–375. doi: 10.1289/ehp.02110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Stuart B, Wahle B, Bomann W, Ahr HJ. Comparison of the expression profiles induced by genotoxic and nongenotoxic carcinogens in rat liver. Mutat. Res. 2005;575:61–84. doi: 10.1016/j.mrfmmm.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Eveillard A, Mselli-Lakhal L, Mogha A, Lasserre F, Polizzi A, Pascussi JM, Guillou H, Martin PG, Pineau T. Di-(2-ethylhexyl)-phthalate (DEHP) activates the constitutive androstane receptor (CAR): a novel signalling pathway sensitive to phthalates. Biochem. Pharmacol. 2009;77:1735–1746. doi: 10.1016/j.bcp.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Fan LQ, You L, Brown-Borg H, Brown S, Edwards RJ, Corton JC. Regulation of phase I and phase II steroid metabolism enzymes by PPAR alpha activators. Toxicology. 2004;204:109–121. doi: 10.1016/j.tox.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Foster PM, Mylchreest E, Gaido KW, Sar M. Effects of phthalate esters on the developing reproductive tract of male rats. Hum. Reprod. Update. 2001;7:231–235. doi: 10.1093/humupd/7.3.231. [DOI] [PubMed] [Google Scholar]
- Goyak KM, Johnson MC, Strom SC, Omiecinski CJ. Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol. Appl. Pharmacol. 2008;231:216–224. doi: 10.1016/j.taap.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Sarkar J, Suino-Powell K, Xu Y, Matsumoto K, Jia Y, Yu S, Khare S, Haldar K, Rao MS, et al. Induction of nuclear translocation of constitutive androstane receptor by peroxisome proliferator-activated receptor alpha synthetic ligands in mouse liver. J. Biol. Chem. 2007;282:36766–36776. doi: 10.1074/jbc.M707183200. [DOI] [PubMed] [Google Scholar]
- Gupta C, Shapiro BH, Yaffe SJ. Reproductive dysfunction in male rats following prenatal exposure to phenobarbital. Pediatr. Pharmacol. (New York) 1980;1:55–62. [PubMed] [Google Scholar]
- Guyton KZ, Chiu WA, Bateson TF, Jinot J, Scott CS, Brown RC, Caldwell JC. A reexamination of the PPAR-α activation mode of action as a basis for assessing human cancer risks of environmental contaminants. Environ. Health Perspect. 2009;117:1664–1672. doi: 10.1289/ehp.0900758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays T, Rusyn I, Burns AM, Kennett MJ, Ward JM, Gonzalez FJ, Peters JM. Role of peroxisome proliferator-activated receptor-alpha (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis. 2005;26:219–227. doi: 10.1093/carcin/bgh285. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Hurst CH, Waxman DJ. Environmental phthalate monoesters activate pregnane X receptor-mediated transcription. Toxicol. Appl. Pharmacol. 2004;199:266–274. doi: 10.1016/j.taap.2003.11.028. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yamanoshita O, Asaeda N, Tagawa Y, Lee CH, Aoyama T, Ichihara G, Furuhashi K, Kamijima M, Gonzalez FJ, et al. Di(2-ethylhexyl)phthalate induces hepatic tumorigenesis through a peroxisome proliferator-activated receptor alpha-independent pathway. J. Occup. Health. 2007;49:172–182. doi: 10.1539/joh.49.172. [DOI] [PubMed] [Google Scholar]
- Jia Y, Guo GL, Surapureddi S, Sarkar J, Qi C, Guo D, Xia J, Kashireddi P, Yu S, Cho YW, et al. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly RA, Goldstein KM, Wei T, Gao H, Chen P, Huang S, Colet JM, Ryan TP, Thomas CE, Estrem ST. Pooling samples within microarray studies: a comparative analysis of rat liver transcription response to prototypical toxicants. Physiol. Genomics. 2005;22:346–355. doi: 10.1152/physiolgenomics.00260.2004. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Hood AM. Effects of microsomal enzyme inducers on thyroid follicular cell proliferation and thyroid hormone metabolism. Toxicol. Pathol. 2001;29:34–40. doi: 10.1080/019262301301418838. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Babich MA, Baetcke KP, Cook JC, Corton JC, David RM, DeLuca JG, Lai DY, McKee RH, Peters JM, et al. PPARalpha agonist-induced rodent tumors: modes of action and human relevance. Crit. Rev. Toxicol. 2003;33:655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: xenosensors of endocrine disrupters? Chem. Biol. Interact. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Labow RS, Tocchi M, Rock G. Contamination of platelet storage bags by phthalate esters. J. Toxicol. Environ. Health. 1986;19:591–598. doi: 10.1080/15287398609530955. [DOI] [PubMed] [Google Scholar]
- Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Lobe DC, Moore JT. The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J. Lipid Res. 2009;50:439–445. doi: 10.1194/jlr.M800226-JLR200. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Mäkinen J, Frank C, Jyrkkärinne J, Gynther J, Carlberg C, Honkakoski P. Modulation of mouse and human phenobarbital-responsive enhancer module by nuclear receptors. Mol. Pharmacol. 2002;62:366–378. doi: 10.1124/mol.62.2.366. [DOI] [PubMed] [Google Scholar]
- Martin PG, Guillou H, Lasserre F, Déjean S, Lan A, Pascussi JM, Sancristobal M, Legrand P, Besse P, Pineau T. Novel aspects of PPARalpha-mediated regulation of lipid and xenobiotic metabolism revealed through a nutrigenomic study. Hepatology. 2007;45:767–777. doi: 10.1002/hep.21510. [DOI] [PubMed] [Google Scholar]
- McClain RM, Posch RC, Bosakowski T, Armstrong JM. Studies on the mode of action for thyroid gland tumor promotion in rats by phenobarbital. Toxicol. Appl. Pharmacol. 1988;94:254–265. doi: 10.1016/0041-008x(88)90267-0. [DOI] [PubMed] [Google Scholar]
- Meek ME, Chan PKL. Bis(2-ethylhexyl)phthalate—evaluation of risks to health from environmental exposure in Canada. J. Environ. Sci. Health Part C. 1994;12:179–194. [Google Scholar]
- Meeker JD, Calafat AM, Hauser R. Di(2-ethylhexyl) phthalate metabolites may alter thyroid hormone levels in men. Environ. Health Perspect. 2007;115:1029–1034. doi: 10.1289/ehp.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DE, Reddy JK, Lake BG, Popp JA, Reese DH. Peroxisome proliferation and nongenotoxic carcinogenesis: commentary on a symposium. Fundam. Appl. Toxicol. 1991;16:233–248. doi: 10.1016/0272-0590(91)90108-g. [DOI] [PubMed] [Google Scholar]
- Nassberger L, Abin A, Ostelius J. Exposure of patients to phthalates from polyvinyl chloride tubes and bags during dialysis. Nephron. 1987;45:286–290. doi: 10.1159/000184165. [DOI] [PubMed] [Google Scholar]
- Nelson DM, Bhaskaran V, Foster WR, Lehman-McKeeman LD. p53-independent induction of rat hepatic Mdm2 following administration of phenobarbital and pregnenolone 16alpha-carbonitrile. Toxicol. Sci. 2006;94:272–280. doi: 10.1093/toxsci/kfl115. [DOI] [PubMed] [Google Scholar]
- Pan G, Hanaoka T, Yoshimura M, Zhang S, Wang P, Tsukino H, Inoue K, Nakazawa H, Tsugane S, Takahashi K. Decreased serum free testosterone in workers exposed to high levels of di-n-butyl phthalate (DBP) and di-2-ethylhexyl phthalate (DEHP): a cross-sectional study in China. Environ. Health Perspect. 2006;114:1643–1648. doi: 10.1289/ehp.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Cattley RC, Gonzalez FJ. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029–2033. doi: 10.1093/carcin/18.11.2029. [DOI] [PubMed] [Google Scholar]
- Price SC, Chescoe D, Grasso P, Wright M, Hinton RH. Alterations in the thyroids of rats treated for long periods with di-(2-ethylhexyl) phthalate or with hypolipidaemic agents. Toxicol. Lett. 1988;40:37–46. doi: 10.1016/0378-4274(88)90181-6. [DOI] [PubMed] [Google Scholar]
- Qatanani M, Zhang J, Moore DD. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005;146:995–1002. doi: 10.1210/en.2004-1350. [DOI] [PubMed] [Google Scholar]
- Rao MS, Reddy JK. Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis. 1987;8:631–636. doi: 10.1093/carcin/8.5.631. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Lalwani ND. Carcinogenesis by hepatic peroxisome proliferators: evaluation of the risk of hypolipidemic drugs and industrial plasticizers to man. Crit. Rev. Toxicol. 1983;12:1–58. doi: 10.3109/10408448309029317. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Abbott BD, Wolf DC, Corton JC, Wood CR, Schmid JE, Das KP, Zehr RD, Blair ET, Lau C. Gene profiling in the livers of wild-type and PPARalpha-null mice exposed to perfluorooctanoic acid. Toxicol. Pathol. 2008a;36:592–607. doi: 10.1177/0192623308318208. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Lee JS, Ren H, Vallanat B, Liu J, Waalkes MP, Abbott BD, Lau C, Corton JC. Toxicogenomic dissection of the perfluorooctanoic acid transcript profile in mouse liver: evidence for the involvement of nuclear receptors PPAR alpha and CAR. Toxicol. Sci. 2008b;103:46–56. doi: 10.1093/toxsci/kfn025. [DOI] [PubMed] [Google Scholar]
- Sanderson LM, de Groot PJ, Hooiveld GJ, Koppen A, Kalkhoven E, Müller M, Kersten S. Effect of synthetic dietary triglycerides: a novel research paradigm for nutrigenomics. PLoS ONE. 2008;3:e1681. doi: 10.1371/journal.pone.0001681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani H. Determination of phthalic acid, mono-(2-ethylhexyl) phthalate and di-(2-ethylhexyl) phthalate in human plasma and blood products. J. Chromatogr. 1985;33:279–290. doi: 10.1016/0378-4347(85)80041-4. [DOI] [PubMed] [Google Scholar]
- Sjoberg PO, Bondesson UG, Sedin EG, Gustafsson JP. Exposure of newborn infants to plasticizers. Plasma levels of di-(2-ethylhexyl) phthalate and mono-(2-ethylhexyl) phthalate during exchange transfusion. Transfusion. 1985;25:424–428. doi: 10.1046/j.1537-2995.1985.25586020115.x. [DOI] [PubMed] [Google Scholar]
- Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. PXR and CAR: Nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab. Rev. 2006;38:515–597. doi: 10.1080/03602530600786232. [DOI] [PubMed] [Google Scholar]
- Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab. Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- Stoner MA, Auerbach SS, Zamule SM, Strom SC, Omiecinski CJ. Transactivation of a DR-1 PPRE by a human constitutive androstane receptor variant expressed from internal protein translation start sites. Nucleic Acids Res. 2007;35:2177–2190. doi: 10.1093/nar/gkm090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochem. Biophys. Res. Commun. 2006;343:535–543. doi: 10.1016/j.bbrc.2006.02.160. [DOI] [PubMed] [Google Scholar]
- Takashima K, Ito Y, Gonzalez FJ, Nakajima T. Different mechanisms of DEHP-induced hepatocellular adenoma tumorigenesis in wild-type and Ppar alpha-null mice. J. Occup. Health. 2008;50:169–180. doi: 10.1539/joh.l7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita A, Koibuchi N, Oka J, Taguchi M, Shishiba Y, Ozawa Y. Bisphenol-A, an environmental estrogen, activates the human orphan nuclear receptor, steroid and xenobiotic receptor-mediated transcription. Eur. J. Endocrinol. 2001;145:513–517. doi: 10.1530/eje.0.1450513. [DOI] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. CAR and PXR: The xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Waalkes MP, Ward JM. Induction of hepatic metallothionein in male B6C3F1 mice exposed to hepatic tumor promoters: effects of phenobarbital, acetaminophen, sodium barbital, and di(2-ethylhexyl) phthalate. Toxicol. Appl. Pharmacol. 1989;100:217–226. doi: 10.1016/0041-008x(89)90308-6. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: a BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- Wong JS, Gill SS. Gene expression changes induced in mouse liver by di(2-ethylhexyl) phthalate. Toxicol. Appl. Pharmacol. 2002;185:180–196. doi: 10.1006/taap.2002.9540. [DOI] [PubMed] [Google Scholar]
- Woods CG, Kosyk O, Bradford BU, Ross PK, Burns AM, Cunningham ML, Qu P, Ibrahim JG, Rusyn I. Time course investigation of PPARalpha- and Kupffer cell-dependent effects of WY-14,643 in mouse liver using microarray gene expression. Toxicol. Appl. Pharmacol. 2007;225:267–277. doi: 10.1016/j.taap.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormuth M, Scheringer M, Vollenweider M, Hungerbuhler K. What are the sources of exposure to eight frequently used phthalic acid esters in Europeans? Risk Anal. 2006;26:803–824. doi: 10.1111/j.1539-6924.2006.00770.x. [DOI] [PubMed] [Google Scholar]
- Wyde ME, Kirwan SE, Zhang F, Laughter A, Hoffman HB, Bartolucci-Page E, Gaido KW, Yan B, You L. Di-n-butyl phthalate activates constitutive androstane receptor and pregnane X receptor and enhances the expression of steroid-metabolizing enzymes in the liver of rat fetuses. Toxicol. Sci. 2005;86:281–290. doi: 10.1093/toxsci/kfi204. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Zhang YK, Yeager RL, Klaassen CD. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab. Dispos. 2009;37:106–115. doi: 10.1124/dmd.108.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.