Abstract
Background: dementia risk conferred by apolipoprotein-E (APOE) and angiotensin-1-converting enzyme (ACE) polymorphisms have been reported for the MRC Cognitive Function and Ageing Study (CFAS) at 6-year follow-up. We concentrate on incident dementia risk over 10 years.
Methods: participants come from MRC CFAS, a multi-centre longitudinal population-based study of ageing in England and Wales. Three follow-up waves of data collection were used: 2, 6 and 10 years. Logistic regressions were undertaken to investigate associations between APOE (n = 955) and ACE (n = 856) alleles/genotypes and incident dementia. Two types of control groups were used: non-demented and highly functioning non-demented. Results were back-weighted.
Results: compared to APOE ε3, ε2 conferred protection of odds ratio (OR) = 0.3 (95% confidence interval, CI = 0.1–0.6) and ε4 risk of OR = 2.9 (95% CI = 1.7–4.9) for incident dementia. Compared to ε3/ε3, the ε3/ε4 and ε4/ε4 genotypes conferred risks of OR = 3.6 (95% CI = 1.8–7.3) and OR = 7.9 (95% CI = 1.6–39.2), respectively. The ε3/ε2 genotype protected against dementia (OR = 0.2, 95% CI = 0.1–0.7), and ε2/ε2 had a similar protective effect but with wide CIs (OR = 0.3, 95% CI = 0.1–1.7). Restricting the control group accentuated these differentials. The effects of ACE alleles/genotypes on incident dementia risk were small.
Conclusions: APOE but not ACE is associated with late-onset incident dementia in the population. Using longer term follow-up with proper adjustment for attrition and incident cases increases estimates of risk.
Keywords: apolipoprotein-E, angiotensin-1-converting enzyme, population, dementia, old
Introduction
Apolipoprotein-E (APOE) and angiotensin-1-converting enzyme (ACE) polymorphisms have received a great deal of attention in relation to dementia risk (e.g. [1, 2]). APOE, found on chromosome 19, is involved in lipid transport in the body [3]. There are three alleles—ε2, ε3, ε4—with ε3 being the most common [1]. APOE is associated with many neuropathological features of Alzheimer’s disease (AD), the most common form of dementia, including plaque and tangle formation, β-amyloid deposition and cholinergic dysfunction (e.g. [4]). Two meta-analyses have reported that in population-based studies those with the ε4/ε4 genotype as compared to those with the ε3/ε3 are 12–13 times more likely to develop AD [1, 5]. Results are less consistent regarding individual alleles, with not all population-based studies reporting that the ε4 allele confers a risk of AD/dementia or the ε2 allele a protective effect [6–16].
ACE appears to be involved in blood pressure regulation and electrolyte balance [17]. It is found on chromosome 17 and there are two allele types, D (a deletion) and I (an insertion), relating to intron 16. Neuropathological studies have been inconsistent as to whether ACE is associated with neuropathological features of AD (e.g. [18, 19]). One recent meta-analysis of clinical/necropsy (i.e. non-population-based) samples reported that the D/D genotype conferred a protective effect on AD (odds ratio, OR = 0.8; 95% confidence interval, CI = 0.8–0.9), the I/D genotype a risk (OR = 1.1, 95% CI = 1.0–1.2) while the I/I had had no effect on AD—with referents being the combination of remaining genotypes [2]. Another meta-analysis on clinical/necropsy samples (i.e. not population-based) reported that the I allele conferred a 1.1 risk (95% CI = 1.0–1.2) for AD compared to the D allele; however, this association was non-significant when adjusting for Hardy–Weinberg equilibrium deviations and non-Caucasian ancestry [1]. There have been few population-based studies of ACE and of these they have generally reported no or small associations between the ACE polymorphism and AD [19–21].
As stated above, there is a great deal of evidence for APOE being associated with AD or dementia, and the evidence concerning ACE is not so consistent. Whether the effects and/or the sizes of such effects reported previously are relevant in a population context is uncertain. This is because the majority of previous research has been conducted on selected clinic/necropsy samples which do not represent the population most at risk of dementia. Accordingly, the current study aimed to assess incident late-onset dementia risk as conferred by APOE and ACE polymorphisms in a population-based sample with a long follow-up (10 years).
These analyses update previous analyses completed on the Medical Research Council Cognitive Function and Ageing Study (CFAS) sample in relation to APOE [22] and ACE [23]. This paper extends these analyses in that it includes another follow-up wave of data collection, increasing the follow-up time from 6 to 10 years which increases the number of incident dementia cases by around 60%. Results were weighted back to the original MRC CFAS sample, to fully take account of drop-out, which has recently become a standard technique. Notably, these previous papers on the MRC CFAS sample reported that the ε4 APOE allele conferred a small risk for all dementia [22] whilst the ACE polymorphism did not confer any dementia risk [23].
Methods
Sample
MRC CFAS has been fully described in Brayne et al. [24] and will only be briefly described here. It is a large longitudinal population-based multi-centre study on ageing and dementia in England and Wales, with participants aged ≥65 at baseline. Sampling was based on geographical areas using general practitioner registration details. The initial response rate was 82%, and there has been a drop-out rate of 13–29% between follow-up waves due to death, moving away or refusal [25, 26]. Four of the six study centres were used in these analyses: East Cambridgeshire, Gwynedd, Newcastle and Nottingham. The remaining two centres, Oxford and Liverpool, collected and analysed blood samples at different times and according to a different study protocol and are thus not included here.
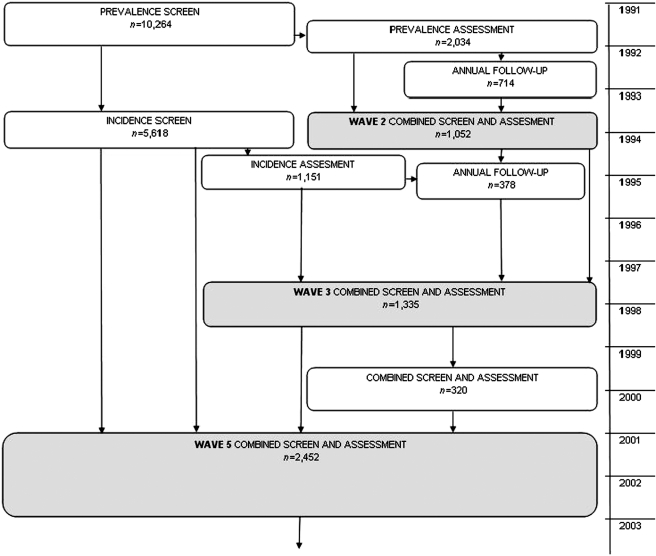
In relation to the four centres included, there were 10,264 participants aged ≥65 years (stratified by equal numbers aged 65–74 and >75) at baseline (‘prevalence screen’) beginning in 1991. The study employed a two-phase design, with a more comprehensive assessment given to a subsample (n = 2,034) at baseline (‘prevalence assessment’) stratified by age and cognitive function (biased toward those older and those with worse cognitive function). There have been multiple re-screens and re-assessments to detect incident dementia, of which the current study employs those at 2 (wave 2), 6 (wave 3) and 10 (wave 5) years. For this analysis, the 2-year follow-up included only those in the prevalence assessment subsample (n = 1,052), the 6-year follow-up included those in the prevalence assessment subsample and another subsample of those part of the incidence screen (n = 1,335), and the 10-year follow-up included all study participants still alive (n = 2,452). The MRC CFAS study design is illustrated in Figure 1.
Figure 1.
An illustration of the MRC CFAS study design and the number of participants seen at each screen/assessment relative to the four centres used in the current study. The grey shading indicates assessments that were used to define cases of incident dementia.
Blood or a buccal swab was collected at wave 3, 6 years into the study, with 62% (n = 1,070 or n = 945 excluding prevalent dementia at baseline) of participants who were included in the wave 3 interview consenting. It is these participants who consented to give genetic material that are included in these analyses. They had a mean age of 73.8 (SD = 6.5) at baseline; 60% women. Ethnic background was asked in a subset of the sample, of which 99% reported being of white British background.
Measures
A computer-automated version of the Geriatric Mental State known as the Automated Geriatric Examination for Computer Assisted Taxonomy (AGECAT) was used to assess the presence of dementia at interview by trained interviewers from professions allied to health. Those with an AGECAT organicity rating of O3 and above (score range 0–5) were classified as demented.
The Mini-Mental State Examination (MMSE) was used to obtain further evidence of cognitive impairment (score range from 0 to 30). The MMSE was employed to create two different control groups: ‘non-demented’ and ‘highly functioning (HF) non-demented’. The non-demented group included participants not classified as demented (i.e. an AGECAT organicity rating of ≤O2). The HF non-demented group was not demented and also displayed no cognitive impairment, as defined as an MMSE of ≥26. By excluding those with an MMSE <26, cases with mild cognitive impairment (MCI) or low baseline cognitive ability are mostly excluded. MCI is a prodrome of dementia that has been shown to predict conversion to dementia [27] and appears to relate to APOE status [28]. Thus, by excluding those with MCI, those cases most likely to convert to dementia are dropped. However, those with naturally low baseline cognitive abilities are also excluded when using this MMSE cut-off, which reduces the representativeness of the sample to the population. Control groups similar to the HF non-demented group are commonly employed in genetic studies, including those using the MRC CFAS sample [22, 23]. However, the complete non-demented group most closely resembles the general population of individuals without dementia.
APOE and ACE genotyping was carried out in line with Wenham et al. [29] and Evans et al. [30], respectively, blind to clinical status. Ambiguous results were re-run up to three times, after which they were recorded as ‘unknown’. APOE genotype was determined in 955 participants and ACE in 856 participants (excluding those with prevalent dementia at baseline).
Ethics statement
MRC CFAS has multi-centre research ethics committee’s approval and ethical approval from the relevant local research ethics committees.
Analysis
Logistic regression analyses were undertaken to examine the associations between APOE/ACE polymorphisms and incident dementia (with dementia as the outcome). Prevalent dementia cases at baseline were excluded from analyses as these participants who went on to consent to DNA collection 6 years later are atypical dementia cases in terms of length of illness without death. Incident dementia was assessed at 2, 6 and 10 years (corresponding to waves 2, 3 and 5, respectively). Analyses were adjusted for wave, age group at interview (65–74, 75–84, 85–94 and 95+), education (low education ≤9 and high education >9 years) and social class. If education information was missing, it was coded as ‘low education’. If social class information was missing, it was coded as ‘social class missing’. ACE analyses were also adjusted for APOE genotype. Analyses were run relative to APOE and ACE genotype and allele status. The ε3 allele and ε3/ε3 genotype were employed as the reference groups in APOE analyses, and the D allele and I/D genotype were employed as the reference groups for ACE. Allelic analyses were undertaken with the assumption that the two alleles from each case were independent.
Each analysis was repeated with both control groups (non-demented and HF non-demented controls); however, analyses relating to the non-demented control group were focused on. Adjusted as well as adjusted and weighted results are presented. Back weighting was employed to provide a population estimate which takes into account the MRC CFAS sampling procedure and those who dropped out prior to the respective case finding interview. Those selected for the more comprehensive prevalence assessment were older and more likely to be cognitively impaired, though all cognitive abilities were represented. Further, those who dropped out were more likely to be cognitively impaired and older than those that did not [25]. Thus, both the sampling procedure and drop-outs influence the age of the sample. Given genetic associations with dementia, particularly with APOE, are stronger in the young as compared to the old [31]; not weighting back would most likely incorrectly estimate genetic associations.
Results
Table 1 displays the distribution of APOE and ACE genotypes and allele frequencies for cases and controls (both non-demented and HF non-demented) relative to 2-, 6- and 10-year follow-ups.
Table 1.
Distribution of APOE and ACE genotypes and allele frequencies for cases and controls (both non-demented and HF non-demented), relative to years 2, 6 and 10 (corresponding to waves 2, 3 and 5, respectively)
| 2 years/wave 2 |
6 years/wave 3 |
10 years/wave 5 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incident demented |
Non-demented |
HF non-demented |
Incident demented |
Non-demented |
HF non-demented |
Incident demented |
Non-demented |
HF non-demented |
||||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| APOE | ||||||||||||||||||
| Allele | ||||||||||||||||||
| ε2 | 4 | 10 | 166 | 9 | 70 | 9 | 5 | 3 | 161 | 9 | 70 | 9 | 8 | 7 | 77 | 10 | 34 | 9 |
| ε3 | 28 | 70 | 1,471 | 79 | 639 | 78 | 114 | 78 | 1,357 | 79 | 595 | 77 | 77 | 71 | 616 | 79 | 290 | 80 |
| ε4 | 8 | 20 | 233 | 12 | 109 | 13 | 27 | 18 | 206 | 12 | 103 | 13 | 23 | 21 | 83 | 11 | 40 | 11 |
| Total | 40 | 1,870 | 818 | 146 | 1,724 | 768 | 108 | 776 | 364 | |||||||||
| Genotype | ||||||||||||||||||
| ε2/ε2 | 0 | 0 | 7 | 1 | 3 | 1 | 1 | 1 | 6 | 1 | 4 | 1 | 0 | 0 | 3 | 1 | 2 | 1 |
| ε2/ε3 | 3 | 15 | 128 | 14 | 55 | 13 | 2 | 3 | 126 | 15 | 55 | 14 | 7 | 13 | 62 | 16 | 27 | 15 |
| ε2/ε4 | 1 | 5 | 24 | 3 | 9 | 2 | 1 | 1 | 23 | 3 | 7 | 2 | 1 | 2 | 9 | 2 | 3 | 2 |
| ε3/ε3 | 11 | 55 | 583 | 62 | 251 | 61 | 46 | 63 | 537 | 62 | 229 | 60 | 26 | 48 | 244 | 63 | 114 | 63 |
| ε3/ε4 | 3 | 15 | 177 | 19 | 82 | 20 | 20 | 27 | 157 | 18 | 82 | 21 | 18 | 33 | 66 | 17 | 35 | 19 |
| ε4/ε4 | 2 | 10 | 16 | 2 | 9 | 2 | 3 | 4 | 13 | 2 | 7 | 2 | 2 | 4 | 4 | 1 | 1 | 1 |
| Total | 20 | 935 | 409 | 73 | 862 | 384 | 54 | 388 | 182 | |||||||||
| ACE | ||||||||||||||||||
| Allele | ||||||||||||||||||
| I | 20 | 50 | 838 | 50 | 365 | 49 | 64 | 53 | 773 | 50 | 341 | 48 | 51 | 53 | 349 | 50 | 165 | 48 |
| D | 20 | 50 | 834 | 50 | 385 | 51 | 56 | 47 | 779 | 50 | 363 | 52 | 45 | 47 | 349 | 50 | 177 | 52 |
| Total | 40 | 1,672 | 750 | 120 | 1,552 | 704 | 96 | 698 | 342 | |||||||||
| Genotype | ||||||||||||||||||
| D/D | 7 | 35 | 227 | 27 | 107 | 29 | 15 | 25 | 213 | 27 | 104 | 30 | 11 | 23 | 98 | 28 | 49 | 29 |
| I/D | 6 | 30 | 380 | 45 | 171 | 46 | 26 | 43 | 353 | 45 | 155 | 44 | 23 | 48 | 153 | 44 | 79 | 46 |
| I/I | 7 | 35 | 229 | 27 | 97 | 26 | 19 | 32 | 210 | 27 | 93 | 26 | 14 | 29 | 98 | 28 | 43 | 25 |
| Total | 20 | 836 | 375 | 60 | 776 | 352 | 48 | 349 | 171 | |||||||||
Table 2 displays the ORs and CIs—adjusted (for age group, sex, education and social class) as well as adjusted and weighted—for APOE genotype and allele status. It can be seen that the ε4 allele conferred a significant risk of dementia (OR = 2.9, 95% CI = 1.7–4.9) and the ε2 allele a significant protective effect (OR = 0.3, 95% CI = 0.1–0.6) relative to the non-demented control group (ε3 referent). Regarding APOE genotypes, the ε3/ε4 and ε4/ε4 genotypes conferred significant risks for dementia (OR = 3.6, 95% CI = 1.8–7.3 and OR = 7.9, 95% CI = 1.6–39.2, respectively) relative to the non-demented control group. The ε2/ε3 was associated with a decreased OR = 0.3 (95% CI = 0.1–0.7), with the ε2/ε2 genotype conferring a similar protective effect of OR = 0.3 with a wide 95% CI (0.1–1.7), though only one individual with ε2/ε2 had dementia. Allele and genotype effects were generally more extreme for the HF non-demented control group comparisons.
Table 2.
ORs and 95% CIs for associations between APOE allele/genotype and dementia
| Comparison using non-demented controls |
Comparison using HF non-demented controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| Adjusteda | Adjusteda and weightedb | Adjusteda | Adjusteda and weightedb | |||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| Allele | ||||||||
| ε2 | 0.6 | 0.3–0.9 | 0.3 | 0.1–0.6 | 0.6 | 0.3–1.1 | 0.2 | 0.1–0.5 |
| ε3 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| ε4 | 2.2 | 1.6–3.0 | 2.9 | 1.7–4.9 | 2.4 | 1.6–3.5 | 3.2 | 1.8–5.6 |
| Genotype | ||||||||
| ε2/ε2 | 0.5 | 0.0–5.7 | 0.3 | 0.1–1.7 | 0.1 | 0.0–6.0 | 0.1 | 0.0–0.4 |
| ε2/ε3 | 0.6 | 0.3–1.1 | 0.2 | 0.1–0.7 | 0.7 | 0.3–1.4 | 0.3 | 0.1–0.8 |
| ε2/ε4 | 0.6 | 0.2–2.3 | 0.2 | 0.0–1.3 | 1.2 | 0.3–5.4 | 0.6 | 0.2–2.1 |
| ε3/ε3 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| ε3/ε4 | 2.3 | 1.5–3.6 | 3.6 | 1.8–7.3 | 2.1 | 1.3–3.5 | 3.1 | 1.4–6.5 |
| ε4/ε4 | 5.0 | 1.9–13.0 | 7.9 | 1.6–39.2 | 9.1 | 3.0–27.2 | 18.1 | 4.9–67.0 |
For age group, sex, education and social class.
For study design and drop-out.
Table 3 displays the ORs and 95% CIs—adjusted (for age group, sex, education and social class) as well as adjusted and weighted—for ACE genotype and allele status. From Table 3, it can be seen that the ACE I allele conferred a small dementia risk and the I/I and D/D genotypes a small degree of protection; however, all estimates are consistent with no effect.
Table 3.
ORs and 95% CIs for associations between ACE allele/genotype and dementia
| Comparison using non-demented controls |
Comparison using HF non-demented controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| Adjusteda | Adjusteda and weightedb | Adjusteda | Adjusteda and weightedb | |||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| Allele | ||||||||
| I | 0.9 | 0.7–1.2 | 1.1 | 0.5–2.3 | 1.2 | 0.9–1.6 | 1.3 | 0.8–2.2 |
| D | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| Genotype | ||||||||
| I/I | 0.9 | 0.6–1.4 | 0.5 | 0.3–1.0 | 0.9 | 0.6–1.4 | 0.5 | 0.3–1.0 |
| I/D | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| D/D | 1.2 | 0.8–1.8 | 0.8 | 0.5–1.4 | 1.2 | 0.8–1.8 | 0.8 | 0.5–1.4 |
For age group, sex, education, social class and APOE genotype.
For study design and drop-out.
Discussion
Dementia risk in the population is associated with APOE but not ACE. Effects were generally larger when employing the high functioning non-demented as compared to the total population of non-demented individuals. This is likely to be due to the exclusion of those with MCI who are at high risk of converting to dementia [27]. This finding suggests that highly selected control groups which are typically employed in genetic association studies are likely to lead to the overestimation of effect sizes, and their relevance to the population must be interpreted with caution. However, although effects were generally more extreme, 95% CIs from analyses employing either type of control group overlapped, which suggests a consistency in the direction of effects.
As expected, effects were also larger when they were weighted back to the original population sample, which accounted for the sampling procedure and drop-outs in the study, both of which influence the age of the sample. As introduced previously, APOE genotype affects age at dementia onset [31] and thus not weighting is likely to lead to incorrect estimates. This finding has important implications for future population-based genetic studies.
The study is not without limitations. Longitudinal follow-up studies have drop out between waves. However, the statistical method of back weighting accounted for any bias this along with attrition (due to drop-out) within the centres included may have introduced [25]. The AGECAT diagnostic algorithm was used to classify participants as demented or non-demented. Although it would have been preferential to have participants individually assessed by a clinician, this was not possible for such a large sample, and AGECAT classifications are closely related to diagnoses made by psychiatrists with the Diagnostic and Statistical Manual IIIR [32, 33]. The study also assessed dementia in general rather than specific clinical diagnoses of AD, which adds weight to the relevance of findings to the population but does not provide information regarding risk relative to specific subtypes of dementia such as AD.
Results reinforce the importance of APOE alleles in terms of dementia risk in the population, with previous population-based studies being somewhat inconsistent [6–10, 12], perhaps due to smaller sample sizes, the inclusion of prevalent dementia and/or short follow-up times. Regarding APOE genotype, the ε3/ε4 and ε4/ε4 genotypes were consistently associated with incident dementia risk—conferring four to eight times the risk (relative to the non-demented control group). This result is similar to two meta-analyses on population studies that reported the ε4/ε4 genotype was associated with 12–13 times greater AD risk [1, 5]. There was a protective effect of the ε2/ε2 genotype which is similar to a meta-analysis by Farrer et al. [5] who reported the AD risk conferred by ε2/ε2 to be OR = 0.9 (95% CI = 0.3–2.8).
It is possible that those with an APOE ε2 allele died before being eligible for entry into the study, given their increased mortality at younger ages [34], and thus the protective effect found could be an artefact of survival. However, the impact of these effects is concerned with those who survive into the age when dementia becomes most prevalent, in old age. APOE results from this study are more extreme than those previously reported on the MRC CFAS sample [22] most likely due to the longer follow-up time and use of back weighting.
Previous studies have reported that the ACE I allele confers a risk for incident dementia [1] and that the I/I and D/D genotypes protect against dementia relative to the I/D genotype [2, 19]. Our results were consistent, with small effects seen in these directions. This result is in line with small effect sizes or null results from population-based studies [19–21, 23], suggesting that the ACE effect is at best weak.
Results from the current study reiterate the importance of APOE in relation to incident dementia risk in the population. The current study was large and addressed many of the methodological issues in previous population-based genetic association studies: long follow-up time, exclusion of prevalent dementia at baseline and weighting back to the original population. Differences between non-demented and HF non-demented control group comparisons also highlighted how control selection affects genetic association estimates.
It should be noted that despite the large sample size and long follow-up time it would still be desirable for these to be increased in future studies. Only around 20% of cases of incident dementia in the MRC CFAS sample displayed an ε4 allele (as compared to around 12% of the non-demented regardless of control group), so it remains neither necessary nor sufficient, supporting suggestions that many other environmental and biological (including genetic) factors are involved in the clinical manifestation of dementia. From these results, it does not appear that ACE substantially raises the risk of incident dementia.
Key points
APOE associated with incident dementia in the old.
ACE does not substantially raise the risk of incident dementia.
Control selection affects genetic association estimates.
Acknowledgments
The authors would like to thank the respondents for their time and continued support. The MRC CFAS is supported by the Medical Research Council (G990140) and the Health Department. H.A.D.K. was supported by a BUPA Foundation grant (RHAG/094) and a Marie Curie International Incoming Fellowship. F.E.M. and L.G. are funded by UK.MRC.U.1052.00.0013. D.C.R. is a Wellcome Trust Senior Fellow in Clinical Science. MRC CFAS is part of the Peterborough and Cambridge NHS CLAHRC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of interest
No conflict interest.
References
- 1.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann DJ, Cortina-Borja M, Warden DR, Smith AD, Sleegers K, et al. Large meta-analysis establishes the ACE insertion–deletion polymorphism as a marker of Alzheimer's disease. Am J Epidemiol. 2005;162:305–17. doi: 10.1093/aje/kwi202. [DOI] [PubMed] [Google Scholar]
- 3.Cedazo-Minguez A. Apolipoprotein E and Alzheimer's disease: molecular mechanisms and therapeutic opportunities. J Cell Mol Med. 2007;11:1227–38. doi: 10.1111/j.1582-4934.2007.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beffert U, Poirier J. Apolipoprotein E, plaques, tangles and cholinergic dysfunction in alzheimer's disease. Ann NY Acad Sci. 1996;777:166–74. doi: 10.1111/j.1749-6632.1996.tb34415.x. [DOI] [PubMed] [Google Scholar]
- 5.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–56. [PubMed] [Google Scholar]
- 6.Kuusisto J, Koivisto K, Kervinen K, Mykkanen L, Helkala EL, et al. Association of apolipoprotein E phenotypes with late onset Alzheimer's disease: population based study. BMJ. 1994;309:636–38. doi: 10.1136/bmj.309.6955.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brayne C, Harrington CR, Wischik CM, Huppert FA, Chi LY, et al. Apolipoprotein E genotype in the prediction of cognitive decline and dementia in a prospectively studied elderly population. Dementia. 1996;7:169–74. doi: 10.1159/000106873. [DOI] [PubMed] [Google Scholar]
- 8.Qiu C, Kivipelto M, Aguero-Torres H, Winblad B, Fratiglioni L. Risk and protective effects of the APOE gene towards Alzheimer's disease in the Kungsholmen project: variation by age and sex. J Neurol Neurosurg Psychiatry. 2004;75:828–33. doi: 10.1136/jnnp.2003.021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–68. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 10.Myers RH, Schaefer EJ, Wilson PWF, D'Agostino R, Ordovas JM, et al. Apolipoprotein E element 4 association with dementia in a population-based study: The Framingham Study. Neurology. 1996;46:673–77. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- 11.Juva K, Verkkoniemi A, Viramo P, Polvikoski T, Kainulainen K, et al. APOE epsilon4 does not predict mortality, cognitive decline, or dementia in the oldest old. Neurology. 2000;54:412–15. doi: 10.1212/wnl.54.2.412. [DOI] [PubMed] [Google Scholar]
- 12.Skoog I, Hesse C, Aevarsson O, Landahl S, Wahlstrom J, et al. A population study of apoE genotype at the age of 85: relation to dementia, cerebrovascular disease, and mortality. J Neurol Neurosurg Psychiatry. 1998;64:37–43. doi: 10.1136/jnnp.64.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M-X, Stern Y, Marder K, Bell K, Gurland B, et al. The APOE-{epsilon}4 allele and the risk of Alzheimer disease among African Americans, Whites, and Hispanics. JAMA. 1998;279:751–55. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Fratiglioni L, Guo Z, Basun H, Corder EH, et al. Incidence of dementia in relation to stroke and the apolipoprotein E {epsilon}4 allele in the very old : findings from a population-based longitudinal study. Stroke. 2000;31:53–60. doi: 10.1161/01.str.31.1.53. [DOI] [PubMed] [Google Scholar]
- 15.Havlik RJ, Izmirlian G, Petrovitch H, Ross GW, Masaki K, et al. APOE-epsilon4 predicts incident AD in Japanese-American men: The Honolulu-Asia Aging Study. Neurology. 2000;54:1526–9. doi: 10.1212/wnl.54.7.1526. [DOI] [PubMed] [Google Scholar]
- 16.Kivipelto M, Helkala EL, Laakso MP, Hanninen T, Hallikainen M, et al. Apolipoprotein E epsilon4 allele, elevated midlife total cholesterol level, and high midlife systolic blood pressure are independent risk factors for late-life Alzheimer disease. Ann Intern Med. 2002;137:149–55. doi: 10.7326/0003-4819-137-3-200208060-00006. [DOI] [PubMed] [Google Scholar]
- 17.Catto A, Carter AM, Barrett JH, Stickland M, Bamford J, et al. Angiotensin-converting enzyme insertion/deletion polymorphism and cerebrovascular disease. Stroke. 1996;27:435–40. [PubMed] [Google Scholar]
- 18.Chalmers KA, Culpan D, Kehoe PG, Wilcock GK, Hughes A, et al. APOE promoter, ACE1 and CYP46 polymorphisms and beta-amyloid in Alzheimer's disease. Neuroreport. 2004;15:95–8. doi: 10.1097/00001756-200401190-00019. [DOI] [PubMed] [Google Scholar]
- 19.Sleegers K, den Heijer T, van Dijk EJ, Hofman A, Bertoli-Avella AM, et al. ACE gene is associated with Alzheimer's disease and atrophy of hippocampus and amygdala. Neurobiol Aging. 2005;26:1153–9. doi: 10.1016/j.neurobiolaging.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Myllykangas L, Polvikoski T, Sulkava R, Verkkoniemi A, Tienari P, et al. Cardiovascular risk factors and Alzheimer's disease: a genetic association study in a population aged 85 or over. Neurosci Lett. 2000;292:195–8. doi: 10.1016/s0304-3940(00)01467-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JW, Li XQ, Zhang ZX, Chen D, Zhao HL, et al. Association between angiotensin-converting enzyme gene polymorphism and Alzheimer's disease in a Chinese population. Dement Geriatr Cogn Disord. 2005;20:52–6. doi: 10.1159/000085075. [DOI] [PubMed] [Google Scholar]
- 22.Yip AG, Brayne C, Easton D, Rubinsztein DC. Apolipoprotein E4 is only a weak predictor of dementia and cognitive decline in the general population. J Med Genet. 2002;39:639–43. doi: 10.1136/jmg.39.9.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip AG, Brayne C, Easton D, Rubinsztein DC. An investigation of ACE as a risk factor for dementia and cognitive decline in the general population. J Med Genet. 2002;39:403–6. doi: 10.1136/jmg.39.6.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brayne C, McCracken C, Matthews FE. Cohort profile: the Medical Research Council Cognitive Function and Ageing Study (CFAS) Int J Epidemiol. 2006;35:1140–5. doi: 10.1093/ije/dyl199. [DOI] [PubMed] [Google Scholar]
- 25.Matthews FE, Chatfield M, Freeman C, McCracken C, Brayne C. Attrition and bias in the MRC cognitive function and ageing study: an epidemiological investigation. BMC Public Health. 2004;4:12. doi: 10.1186/1471-2458-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews FE, Chatfield M, Brayne C. An investigation of whether factors associated with short-term attrition change or persist over ten years: data from the Medical Research Council Cognitive Function and Ageing Study (MRC CFAS) BMC Public Health. 2006;6:185. doi: 10.1186/1471-2458-6-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruscoli M, Lovestone S. Is MCI really just early dementia? A systematic review of conversion studies. Int Psychogeriatr. 2004;16:129–40. doi: 10.1017/s1041610204000092. [DOI] [PubMed] [Google Scholar]
- 28.Caselli RJ, Reiman EM, Osborne D, Hentz JG, Baxter LC, et al. Longitudinal changes in cognition and behavior in asymptomatic carriers of the APOE e4 allele. Neurology. 2004;62:1990–5. doi: 10.1212/01.wnl.0000129533.26544.bf. [DOI] [PubMed] [Google Scholar]
- 29.Wenham PR, Price WH, Blandell G. Apolipoprotein E genotyping by one-stage PCR. Lancet. 1991;337:1158–9. doi: 10.1016/0140-6736(91)92823-k. [DOI] [PubMed] [Google Scholar]
- 30.Evans AE, Poirier O, Kee F, Lecerf L, McCrum E, et al. Polymorphisms of the angiotensin-converting-enzyme gene in subjects who die from coronary heart disease. Q J Med. 1994;87:211–4. [PubMed] [Google Scholar]
- 31.Blacker D, Haines JL, Rodes L, Terwedow H, Go RCP, et al. ApoE-4 and age at onset of Alzheimer's disease: The NIMH Genetics Initiative. Neurology. 1997;48:139–47. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Copeland JR, Dewey ME, Griffiths-Jones HM. A computerized psychiatric diagnostic system and case nomenclature for elderly subjects: GMS and AGECAT. Psychol Med. 1986;16:89–99. doi: 10.1017/s0033291700057779. [DOI] [PubMed] [Google Scholar]
- 33.Schaub RT, Linden M, Copeland JR. A comparison of GMS-A/AGECAT, DSM-III-R for dementia and depression, including subthreshold depression (SD)—results from the Berlin Aging Study (BASE) Int J Geriatr Psychiatry. 2003;18:109–17. doi: 10.1002/gps.799. [DOI] [PubMed] [Google Scholar]
- 34.Ewbank DC. The APOE gene and differences in life expectancy in Europe. J Gerontol A Biol Sci Med Sci. 2004;59:B16–20. doi: 10.1093/gerona/59.1.b16. [DOI] [PubMed] [Google Scholar]