Abstract
Gamma-radiation exposure of humans is a major public health concern as the threat of terrorism and potential hostile use of radiological devices increases worldwide. We report here the effects of sublethal γ-radiation exposure on the mouse urinary metabolome determined using ultra-performance liquid chromatography-coupled time-of-flight mass spectrometry-based metabolomics. Five urinary biomarkers of sublethal radiation exposure that were statistically significantly elevated during the first 24 h after exposure to doses ranging from 1 to 3 Gy were unequivocally identified by tandem mass spectrometry. These are deaminated purine and pyrimidine derivatives, namely, thymidine, 2′-deoxyuridine, 2′-deoxyxanthosine, xanthine and xanthosine. Furthermore, the aminopyrimidine 2′-deoxycytidine appeared to display reduced urinary excretion at 2 and 3 Gy. The elevated biomarkers displayed a time-dependent excretion, peaking in urine at 8–12 h but returning to baseline by 36 h after exposure. It is proposed that 2′-deoxyuridine and 2′-deoxyxanthosine arise as a result of γ irradiation by nitrosative deamination of 2′-deoxycytidine and 2′-deoxyguanosine, respectively, and that this further leads to increased synthesis of thymidine, xanthine and xanthosine. The urinary excretion of deaminated purines and pyrimidines, at the expense of aminopurines and aminopyrimidines, appears to form the core of the urinary radiation metabolomic signature of mice exposed to sublethal doses of ionizing radiation.
INTRODUCTION
Scenarios involving accidental and intentional exposure of human populations to ionizing radiation have been described (1, 2). Among these has emerged the increasing threat of terrorist attacks involving radiological or nuclear devices with potential for causing mass casualties for which available countermeasures and strategies for dosimetry are of limited use (2, 3). To address this deficiency, the U.S. Homeland Security Council and the Office of Science and Technology Policy created the Weapons of Mass Destruction Medical Countermeasures Subcommittee to oversee the research and development of improved countermeasures (3). The Subcommittee lists the development of biomarkers and devices for biodosimetry among its highest priority areas of research (3).
Metabolomics is an area of research properly suited for the development and application of rapid, high-throughput, minimally invasive radiation biodosimetry (4). Metabolomics is the global, quantitative characterization of the metabolic phenotype, i.e. small molecule complement, of biofluids, cells, tissues, organs and organisms under specific sets of conditions such as genotypic differences or environmental exposures (5). The importance of metabolomics as a priority research area is reflected in the National Institutes of Health Roadmap for Medical Research wherein it is named as a new pathway of discovery initiative (6). This powerful platform has been used in drug development, screening and metabolism (7) and to identify novel biomarkers of exposure (8), effect (9) and disease (10).
We set out to develop a method for rapid, noninvasive radiation biodosimetry using metabolomics on an ultra-performance liquid chromatography-time-of-flight mass spectrometry (UPLC-TOFMS) platform and recently reported the successful detection of urine biomarkers of γ radiation in the mouse (4). Among the elevated biomarkers identified in urine collected over the first 24 h after exposure to 3 and 8 Gy, thymidine (dT) exhibits the lowest estimates of variance and the highest estimates of exposure-dependent mean concentration differences. In this report, these findings are extended by identifying, in addition to dT, one additional pyrimidine nucleoside (2′-deoxyuridine) and three purine biomarkers (2′-deoxyxanthosine, xanthine and xanthosine) that are elevated in urine of the exposed mouse in the first 24 h after exposure to 0, 1, 2 or 3 Gy γ radiation. Clear dose dependence and temporal characteristics of these candidate radiation biomarkers were established. These findings represent an important step in addressing priorities within the call for countermeasures against radiation and will help guide the development of rapid, non-invasive radiation biodosimetry in humans.
MATERIALS AND METHODS
Compounds
The following compounds were obtained from Sigma-Aldrich Co., St. Louis, MO: 2′-deoxycytidine (dC), 2′-deoxyuridine (dU), 4-nitrobenzoic acid, creatinine, debrisoquine hemisulfate, thymidine (dT), xanthine, and xanthosine (X). We received 2′-deoxyxanthosine (dX) as a kind gift from Dr. Peter Dedon, Department of Biological Engineering, MIT. All inorganic reagents and solvents were of the highest purity obtainable.
Animals
Male C57BL/6-N mice at 8 and 12 weeks of age, obtained from Charles River Laboratories, Inc. (Frederick, MD), were used for this study. Mice were fed NIH-31 rodent chow (Zeigler, Gardners, PA) ad libitum with free access to fresh drinking water and were housed three to five per cage under a standard 12-h light, 12-h dark cycle. During urine collection, mice were housed individually in metabolic cages for up to 24 h at a time. After urine collection, mice were returned to their home cages together with up to four littermates. Mice were monitored daily for outward signs of distress or adverse health effects. All animal handling and experimental protocols were designed for maximum possible well-being, conformed to the guidelines stipulated by the NIH Office of Animal Care and Use, and were approved prior to the initiation of this study by the NIH Animal Care and Use Committee.
Radiation Exposure
Individual body weights were recorded, and mice were placed into sterile, dry RadDisk™ Rodent Microisolation Irradiator Disks (Braintree Scientific, Inc., Braintree, MA) exposed to single doses of 0 (sham), 1, 2, or 3 Gy (n = 6 per dose) γ radiation emitted from a 137Cs source in a Mark I Model 68 small animal irradiator (J. L. Shepherd & Associates, San Fernando, CA) operating at 2.57 Gy/min as described previously (4). Physical exposures within the irradiator chamber were assessed using models AT-742 (0–2 Gy) and AT-746 (0–6 Gy) direct reading dosimeters (Arrow-Tech, Inc., Rolla, ND) placed directly on the disks. Mice were placed individually into metabolic cages immediately after radiation exposure for urine collection.
Urine Collection
Urine samples from mice housed individually in Nalgene metabolic cages (Tecniplast USA, Inc., Exton, PA) were collected over continuous 24-h periods with alternating 24-h rest intervals as described previously (4). Briefly, mice were housed in groups of up to five during 24-h rest periods and individually in metabolic cages during 24-h urine collection periods. Dose was assigned randomly, and treatment groups were mixed rather than segregated after exposure. Three 24-h urine samples per mouse were obtained at 6, 4 and 2 days before exposure. The mice were exposed and immediately placed into metabolic cages for urine collection over 24 h (day 1 after exposure). Subsequent 24-h samples were collected 3, 5, 7 and 9 days after exposure. In a subsequent experiment, mice were housed individually in metabolic cages before and for 20 h immediately after radiation exposure while urine was collected every 4 h (4, 8, 12, 16 and 20 h after exposure). The animals were placed back with their cage mates until housed again in metabolic cages for a 4-h urine collection at 36 h after exposure. A final 24-h urine collection was taken at day 4 after exposure. In both experiments, body weights were recorded immediately prior to placement in the metabolic cages, and urine sample volumes were recorded and used to calculate the rates of urine formation. All urine samples were stored at −80°C until analyzed.
UPLC-TOFMS Analyses
Urine samples were analyzed by UPLC-TOFMS in order by time and by mouse as described (4) with slight modifications. Urine aliquots (20 µl) were diluted 1:5 with deionized water (80 µl) and centrifuged at 13,000 g for 20 min at 4°C to remove particulates. For analysis of urine in electrospray negative-ionization (ESI−) mode, supernatants were transferred to auto-sampler vials containing 4-nitrobenzoic acid at a final concentration of 40 µM for internal standard (IS). For analysis by electrospray positive-ionization (ESI+) mode, 4-nitrobenzoic acid was replaced by 1 µM debrisoquine hemisulfate as IS. The samples (5 µl injection) were resolved on a reverse-phase 2.1 × 50-mm ACQUITY UPLC® BEH C18 1.7-µm column (Waters Corp, Milford, MA) using an ACQUITY UPLC® system (Waters) with a gradient mobile phase comprising 0.1% formic acid solution (A) and acetonitrile containing 0.1% formic acid solution (B). Each sample was resolved for 10 min at a flow rate of 0.5 ml/min. The gradient consisted of 100% A for 0.5 min, 80% A/20% B for 3.5 min, 5% A/95% B for 4 min, 100% B for 1 min, and finally 100% A for 1 min. The column eluent was introduced directly into the mass spectrometer by electrospray.
Time-of-flight mass spectrometry was performed on a Q-TOF Premier® (Waters) operating in either negative-ion or positive-ion electrospray ionization mode with a capillary voltage of 3 kV and a sampling cone voltage of 30 V. The desolvation gas flow was 720 liter/h and the temperature was set to 350°C. The cone gas flow was 50 liter/h, and the source temperature was 120°C. Quadrupole settings were 4.7 for LM resolution, 15 for HM resolution, 1.0 for ion energy, and 2.0 pre-filter. Collision cell settings were 0.4 ml/min, 5.0 collision energy, 2.0 cell entrance, and −15 cell exit. The TOF detector was set to 1650 with automatic mass range. Acquisition was made in V mode with extended dynamic range, scan time was 0.3 s, and inter-scan delay was 0.08 s. Accurate mass was maintained by the LockSpray® interface (Waters) with the introduction of sulfadimethoxine (310.0736 Da) in 50% aqueous acetonitrile (250 pg/µl) at a rate of 30 µl/min. The LockMass was measured as an average of five scans with a mass window of 0.5 Da, a scan time of 0.3 s at a frequency of every 10 s, sampling cone set to 60 V, and collision energy set to 5 V. Data were acquired in centroid mode from 50 to 850 Da in MS scanning. Tandem MS (MSMS) collision energy was ramped from 5 to 35 V.
Data Processing and Multivariate Data Analysis
Centroided and integrated mass spectrometric data from the UPLC-TOFMS were processed to generate a multivariate data matrix using MarkerLynx® software (Waters) (11, 12). For each sample, relative concentration data for each urine ion (generated by MarkerLynx processing) were normalized by the corresponding relative creatinine concentration ([Normalized Ion]s = [Ion]s /[creatinine] s, where s = sample number). Normalization by creatinine concentration was chosen as a means to adjust for individual differences in glomerular filtration rates (13, 14). Normalized relative concentration data were Pareto-scaled (data set divided by the square root of its standard deviation) (15) and analyzed by principal components analysis (PCA) (16) and orthogonal projections to latent structures (OPLS) (17) using SIMCA-P+ software v12.0 (Umetrics, Kinnelon, NJ). PCA was used as an initial investigation for differences among the urine samples and to identify potential outlier samples. A PCA score in the component 2 compared to component 1 space that falls outside the Hotelling’s T2 ellipse [a generalization of Student’s t test for multivariate data (18)] may be considered for removal. However, we did not remove any outliers in this study because there were no obvious outliers. OPLS analyses were used to determine which metabolites contribute most to the separation in the scores space. Samples were classified as either from control (y = 0) or irradiated (y = 1) mice for supervised OPLS analyses.
Selection of Candidate Markers
First, candidates were selected by examining the OPLS component 1 loadings S-plots of ion confidence (a measure of model correlation expressed as p(corr)[1]P) as a function of ion contribution (a measure of covariance expressed as p[1]P). In loadings S-plots, the ions positioned most distant from the origin in the upper right and lower left quadrants were interrogated for consideration as candidate biomarkers. Ions that appeared to exhibit differential excretion according to exposure status were then compared as a function of dose using the appropriate statistical tests, which are described below. Data acquired from UPLC-TOFMS often include urine ions detected in only one or a few samples. These ions can appear spuriously as candidate biomarkers in OPLS loadings S-plots. Therefore, the t test and Mann-Whitney tests were used to validate the OPLS confidence values (p(corr)[1]P). Candidates that exhibited P values for dose-dependent differences below 0.05 were pursued. Third, random forests analysis, described below, was used in an attempt to validate the OPLS-driven candidate selection.
Exact masses of candidates were checked against online databases, including Madison Metabolomics Consortium Database (20) and ChemSpider (21, 22), for identity matches. The list was examined and curated to identify artifacts such as isotopes, in-source fragments, and sodium adducts of other candidates. Authentic standards at 20–60 µM in 0.1% formic acid were then used to confirm the identities of the markers with UPLC-TOFMSMS, which fragments molecules in a consistent manner. Therefore, correctly identified urine metabolites provide a TOFMSMS fragmentation spectrum identical to the fragmentation spectrum of the authentic standards. In addition to dT and N-hexanoylglycine, biomarkers of radiation exposure identified previously (4), a total of 21 ions were chosen based on S-plot coordinates and lowest P values derived from both two-tailed t tests (parametric) of the mean normalized relative concentrations and Mann-Whitney U tests (nonparametric) of the normalized data (19). In this report, we refer to ions that satisfied the statistical tests for exposure-dependent differences that we could unequivocally identify as urine biomarkers of radiation exposure. The ions that satisfied statistical tests for difference that could not be unequivocally identified are referred to as candidate urine biomarkers and are listed by their respective m/z and Tret.
Random Forests Data Analysis
To address concerns of over-fitting the data, a common concern with large datasets with comparatively few observations, we implemented the machine learning algorithm random forests (RF) (23) into the biomarker discovery effort. We chose the RF algorithm because it has been demonstrated to handle large data sets independent of data scale, to be less susceptible to over-fitting the data, and to provide convenient estimates of the most important variables in sample classification. Here, RF was used to compare sham and 1 Gy, sham and 2 Gy, sham and 3 Gy, or sham and all doses, and important variables were identified as those highly ranked on the variable importance list. RF was run in the R software environment using the following parameters: ntree = 10000, importance = TRUE. The value for mtry, or the number of variables used at each split, was the default value (the square root of the total number of variables). The variable importance measure was used to assign RF rankings. Due to the inherent variability in random forests analysis, 25 independent random forests models were constructed and the variable importance ranks were averaged across all 25 models. Bootstrapping of the results from the 25 independent random forests was used to determine the 95% confidence intervals of the variable importance ranks. The common significant variables to all three doses defined by the variable importance list of the random forests algorithm were defined in a Venn diagram using the ABarray package (Applied Biosystems, Foster City, CA).
Quantification of Urine Biomarkers
Absolute urine creatinine concentrations were determined so that urinary biomarker levels could be normalized as an adjustment for differences in glomerular filtration. Briefly, urine samples were diluted 1:1000 in 10 mMammonium formate buffer (pH 3.5) and separated on a Phenomenex Synergi Polar-RP column (Torrance, CA). Analysis was carried out using a high-performance LC system consisting of a PerkinElmer Series 200 quaternary pump, vacuum degasser and autosampler (PerkinElmer Life and Analytical Sciences, Boston, MA) with a 100-µl loop interfaced to an API2000 SCIEX triple-quadrupole tandem mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). The multiple reaction monitoring transitions were monitored in positive-ionization mode for creatinine [114.0→86.1 mass/charge (m/z)] and debrisoquine hemisulfate IS (176.1→134.2 m/z).
Urine radiation biomarkers were quantified by UPLC-TOFMS and QuanLynx software (Waters) using extracted ion chromatographic peak areas as described previously (4). Briefly, authentic standards of dT ([M-H]− 241.0824), dU ([M-H]− 227.0668), dX ([M-H]− 267.0729), xanthine ([M-H]− 151.0256), and X ([M-H]− 283.0679) in aqueous solution at concentrations ranging from 0.19 to 100 µM were analyzed in duplicate by UPLC-TOFMS. For IS, 4-nitrobenzoic acid ([M-H]− 166.0141) was included in each vial at a final concentration of 40 µM. The standard mixtures were used to calculate observed standard peak area:IS peak area ratios (dependent variable), which were plotted as a function of expected concentrations (independent variable). Linear regression with a forced y-intercept of zero was used to create standard curves with Prism (GraphPad Software, Inc., San Diego, CA). The slopes obtained by linear regression for each authentic standard (astd) and the observed analyte peak area/IS peak area ratios (Ranalyte) for each biomarker were used to calculate normalized biomarker concentrations ([B]) according to the equation [B] = [5(astd × Ranalyte)]/(mmol creatinine).
Statistical Analyses
All observations were tested for normal distribution by the skewness and kurtosis test (19). Data that were not normally distributed were compared by the Mann-Whitney U test when comparing exposed to unexposed groups. Means were calculated from normally distributed data and compared with appropriate parametric tests for difference as described below. Mean body weight, mean urine sample volume, and mean urine rate of formation plotted over time were stratified by dose and analyzed by linear regression to determine whether any changes were associated with radiation exposure and were therefore potential confounding factors. The regression lines of the means stratified by dose were compared. Variances were tested for equality using the F test (19). At each time, mean urine sample volumes, mean body weights, and mean [B] were tested for difference according to dose by a t test assuming equal or unequal variances (depending on F-test outcome) when comparing exposed to unexposed, or one-way analysis of variance (ANOVA) (α = 0.05) with Bonferroni correction for multiple comparisons when comparing within exposed groups (19). We reasoned that the comparison of mean [B] from exposed animals to control animals can be accomplished in pairs by a t test with an α of 0.05 without an increased risk of type I error [falsely rejecting the null hypothesis which states that the two means are equal (19)] because of the multiple comparison nature of comparing each exposed group to the control group according to dose, and we determined that ANOVA with Bonferroni correction is unnecessarily stringent for these comparisons (24). We did, however, use ANOVA with Bonferroni correction for comparing means of exposed mouse urines with each other. Corrected P < 0.05 was interpreted to indicate significant differences in the means compared. All statistical analyses were performed using STATA (Stata Corp LP, College Station, TX). Graphical presentations of mean [B] comparisons were prepared using Prism.
RESULTS
Animal Performance
No adverse health effects or changes in behavior were detected in any of the mice through 30 days after exposure. Measures of overall health (body weight and urine sample volume) were recorded and compared to identify potential health-related confounding. No differences in mean body weights, mean urine sample volumes, and mean rates of urine formation of the groups were fond at any time (data not shown). Furthermore, linear regression analyses revealed no significant changes in any of these variables (data not shown). Finally, we found no exposure- or time-dependent differences in urine creatinine concentrations (data not shown).
Metabolomics of 24-h Mouse Urine Samples before and after Exposure to 0, 1, 2 or 3 Gy γ Radiation
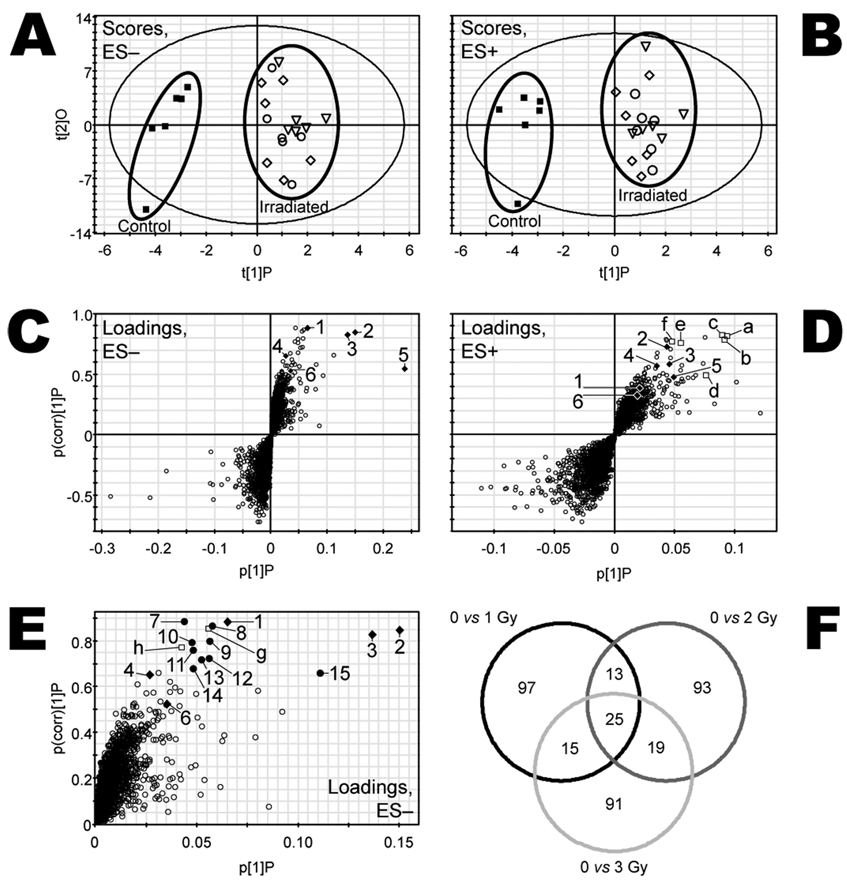
Urine samples were collected over 24 h from 24 mice housed individually in metabolic cages 6, 4 and 2 days prior to irradiation with 0 (sham), 1, 2 or 3 Gy (n = 6 mice per dose) and subsequently at days 1, 3, 5, 7 and 9 after exposure. Deconvolution of data collected from UPLC-TOFMS analysis of the urine samples yielded a large data matrix consisting of m/z, UPLC column retention time (Tret), and peak areas for approximately 4000 ESI− and 4000 ESI+ urinary ions. These data were normalized by the respective peak areas of urinary creatinine to control for differences in glomerular filtration, urine concentration and sample collection. Unsupervised PCA analyses with either the ESI− or ESI+ data sets did not show appreciable exposure-dependent differences in the scores space before or after exposure, nor were outliers detected (data not shown). Therefore, OPLS supervised multivariate data analysis and the machine learning algorithm random forests (below) were used to detect exposure-dependent differences in metabolite profiles. No meaningful differences in the OPLS scores space were observed before exposure, whereas exposure-dependent differences were apparent in the urine samples collected over the first 24 h at day 1 after exposure. Figure 1 shows the OPLS scores of the data from samples at this time for both ESI− (panel A) and ESI+ (panel B) data sets. Clear separation by exposure status (exposed and unexposed) was evident. Meaningful OPLS scores separation was not observed at subsequent times after exposure. Therefore, attention was focused on the differences in metabolite profiles observed in the first 24 h after exposure.
FIG. 1.
Multivariate data analyses of UPLC-TOFMS data. Urine was collected from each mouse for 24 h immediately after exposure. OPLS scores for the samples are shown as component 2 (t[2]O) compared to component 1 (t[1]P) for samples from mice exposed to 0 (■), 1 (■), 2 (◊) or 3 Gy (○)γ radiation analyzed by both ESI− (panel A) and ESI+ (panel B) modes. The spatial separation of scores according to exposure was used to find candidate biomarkers. Respective OPLS loadings plots are shown as p(corr)[1]P compared to p[1]P (panels C and D). Each urine metabolite of unique m/z-Tret pair is represented by an open circle (○) and plotted according to its relative abundance (abscissa axes) and association with radiation exposure (ordinate axes). Ions plotted in the upper right quadrant correlate positively with irradiation. Identified urine markers of exposure (♦) are labeled 1–6 in panel C and are listed in Table 1. The ions labeled a–f (□) in panel D are in-source fragments of or, in two cases, sodiated forms of markers 1–6 (also labeled). Panel E is the upper right quadrant of panel C, expanded for better view of candidate markers 7–15 (●),which are also listed in Table 2 by their respective m/z-Tret pairs and ions g and h (□) determined to be isotopes of no. 2 and no. 15, respectively. Random Forests analyses were conducted with combined ESI− and ESI+ data comparing 0 and 1, 0 and 2, and 0 and 3 Gy. The Venn diagram (panel F), in which each pairwise comparison is represented by a circle, was constructed using the 150 most important ions found by Random Forests in this pooled data set. The numbers represent the ions that are unshared or shared among each of the three comparisons. There are 25 ions that are important to all three comparisons. Radiation biomarkers 1–4 and 6 and their associated in-source fragments and Na+ adducts are among these 25 ions, as are candidate markers 7–14.
The observed separation in OPLS scores was used to identify the ions in the urine that correlated most with exposure status. Figure 1C and D shows plots of the urine ions as measure of correlation to the OPLS model (p(corr)[1]P, ordinate) as a function of the measure of relative abundance (p[1]P, abscissa) for ESI− and ESI+ TOFMS data, respectively. Figure 1E is an enlargement of the upper right quadrant of the plot in Fig. 1C. Ions that have higher relative abundance in the urine samples are plotted along the abscissa more distant from the origin. The ions that are different according to exposure status are plotted on the ordinate according to the correlation value (range −1 to 1), where larger distance from the origin correlates with a larger interclass difference. Together these two measures place higher-abundance ions with larger class-dependent differences further away from the origin in both dimensions. The ions that are elevated in samples from irradiated mice are plotted in the upper right quadrants, and the radiation-attenuated ions are plotted in the lower left quadrants. While these measures alone are not reliable indicators of statistically significant differences in ion concentrations in urine of exposed and unexposed mice, they are useful for identifying candidates for further scrutiny. Numbered 1–15 and lettered a–h in these S-plots are the ions we have identified as urine biomarkers of radiation exposure (numbers 1–6), candidate biomarkers (numbers 7–15), or isotopes, sodium adducts, or in-source fragments thereof (letters a–h). These ions were selected for chemical identification after inspection of the means and variance estimates. Authentic biomarkers that are elevated rather than attenuated in the irradiated urine samples were more reliably observed. Additionally, the ESI− data set contained more reliable, authentic ions suitable for biomarker candidacy. The selected ions that could be unequivocally identified and their observed m/z, predicted m/z, ppm mass errors, UPLC column retention times, and empirical formulae are listed in Table 1.
TABLE 1.
Identification of Mouse Urinary Biomarkers of γ-Radiation Exposure
| Ion no.a | Tret(min) | Observed m/z | Predicted m/z | Error (ppm) | Formula |
|---|---|---|---|---|---|
| 1 | 1.80 | 267.0746 | 267.0729 | 6.4 | C10H12N4O5 |
| 2 | 1.12 | 227.0668 | 227.0668 | 0.0 | C9H12N2O5 |
| 3 | 1.91 | 241.0816 | 241.0824 | 3.3 | C10H14N2O5 |
| 4 | 0.64 | 151.0266 | 151.0256 | 6.6 | C5H4N4O2 |
| 5 | 3.66 | 172.0983 | 172.0974 | 5.2 | C8H15NO3 |
| 6 | 1.80 | 283.0694 | 283.0679 | 5.3 | C10H12N4O6 |
| a | 1.89 | 265.0863 | 265.0800 | 23 | C10H13N2O5Na+ |
| b | 1.12 | 251.0659 | 251.0644 | 6.0 | C9H12N2O5Na+ |
| c | 1.07 | 113.0335 | 113.0351 | 14 | C4H4N2O2 |
| d | 1.91 | 127.0463 | 127.0508 | 35 | C5H6N2O2 |
| e | 1.80 | 153.0457 | 153.0413 | 29 | C5H4N4O2 |
| f | 1.08 | 117.0560 | 117.0552 | 6.8 | C5H10O3 |
| g | 1.13 | 228.0709 | 228.0746 | 16 | C9H12N2O5 |
| h | 0.52 | 265.9845 | — | — | Unknown |
| Random Forests | ||||||
|---|---|---|---|---|---|---|
| OPLS | 95% CI | |||||
| Identity | p(1)P | p(corr)(1)P | MI | Lower | Upper | Rank |
| 2′-Deoxyxanthosine | 0.065 | 0.882 | 1.84 | 1.48 | 2.20 | 1 |
| 2′-Deoxyuridine | 0.150 | 0.846 | 3.52 | 3.20 | 3.80 | 4 |
| Thymidine | 0.137 | 0.828 | 2.60 | 2.12 | 3.04 | 3 |
| Xanthine | 0.027 | 0.651 | 11.3 | 10.9 | 11.7 | 12 |
| N-Hexanoylglycine | 0.237 | 0.541 | 2506 | 1903 | 3095 | 1146 |
| Xanthosine | 0.035 | 0.523 | 2063 | 1370 | 2736 | 550 |
| Na+ adduct of no. 3 | 0.094 | 0.814 | — | — | — | — |
| Na+ adduct of no. 2 | 0.091 | 0.795 | — | — | — | — |
| Fragment of no. 10b | 0.089 | 0.825 | — | — | — | — |
| Fragment of no. 3 | 0.076 | 0.498 | — | — | — | — |
| Fragment of no. 1 and/or no. 6 | 0.055 | 0.758 | — | — | — | — |
| Fragment of no. 10b | 0.048 | 0.774 | 2262 | 1802 | 2705 | 745 |
| Isotope of no. 2 | 0.055 | 0.859 | 5.48 | 5.20 | 5.76 | 5 |
| Isotope of no. 15b | 0.042 | 0.776 | 19.6 | 18.6 | 20.6 | 20 |
Notes. Tret, retention time; OPLS, orthogonal projections to latent structures; p(1)P, contribution; p(corr)(1)P, correlation; MI, mean importance; CI, confidence interval.
As shown in Fig. 1C and D. These are assignments of ion number, not ranks. Ions 1– 6 are sorted in descending order of p(corr)(1)P, however. Ions a–h are unsorted.
See Table 2.
Random forests analysis yielded ion rankings that mostly agreed with the OPLS loadings S-plots coordinates of the ions highlighted in this report. However, there were a few notable exceptions. N-Hexanoylglycine (Table 1, ion no. 5) and X (Table 1, ion no. 6) were ranked considerably lower by RF (mean importance 2506 and 2063, respectively), even though these two ions correlated with the OPLS model as indicated by their respective p(corr)[1]P values of 0.5411 and 0.5233. Similarly, candidate ion no. 15, m/z = 264.9886 (Table 2), was also assigned a relatively high p(corr)[1]P value of 0.6631, but its mean importance rank by RF was only 101. This discrepancy demonstrates the utility of using more than one multivariate data analysis approach for identifying candidate biomarkers in UPLC-TOFMS data matrices. The Venn diagram (Fig. 1F) was exceptionally useful for defining a subset of ions that was common and important to all three doses as well as for validating the results obtained with OPLS analyses. While conservative, this approach provided an unbiased selection of the most important ions for class discrimination, as opposed to only selecting ions from the OPLS loadings S-plots based on their distances from the origin.
TABLE 2.
Additional Candidate Mouse Urinary Biomarkers of γ-Radiation Exposure
| Random forests | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 95% CI | P valueb | ||||||||||
| Ion no.a | Tret (min) | Observed [M-H]− m/z |
OPLS p[1]P | p(corr)[1]P | MI | Lower | Upper | Rank | 0 vs. 1 Gy | 0 vs. 2 Gy | 0 vs. 3 Gy |
| 7 | 1.13 | 402.1510 | 0.043 | 0.888 | 2.04 | 1.72 | 2.36 | 2 | 0.003 | 0.001 | <0.001 |
| 8 | 1.85 | 417.1173 | 0.057 | 0.870 | 6.00 | 5.72 | 6.28 | 6 | 0.007 | 0.002 | 0.002 |
| 9 | 1.01 | 325.0300 | 0.056 | 0.801 | 6.84 | 6.44 | 7.28 | 7 | 0.011 | <0.001 | <0.001 |
| 10 | 1.09 | 265.0401 | 0.047 | 0.797 | 10.2 | 9.68 | 10.7 | 10 | 0.022 | <0.001 | <0.001 |
| 11 | 1.17 | 273.0716 | 0.048 | 0.764 | 7.92 | 7.68 | 8.16 | 8 | 0.025 | <0.001 | <0.001 |
| 12 | 1.90 | 277.0744 | 0.056 | 0.726 | 11.0 | 10.5 | 11.4 | 11 | <0.001 | 0.021 | <0.001 |
| 13 | 1.90 | 430.1804 | 0.052 | 0.723 | 25.0 | 23.2 | 27.1 | 24 | 0.008 | 0.010 | <0.001 |
| 14 | 0.50 | 433.0170 | 0.048 | 0.682 | 16.1 | 15.2 | 17.0 | 16 | 0.117 | 0.002 | <0.001 |
| 15 | 0.50 | 264.9886 | 0.111 | 0.663 | 418 | 123 | 800 | 101 | 0.141 | 0.010 | 0.002 |
Notes. Tret, retention time; OPLS, orthogonal progjections to latent structures; p[1]P, contribution; p(corr)[1]P, correlation; MI, mean importance; CI, confidence interval.
As shown in Fig. 1E. These are assignments of ion number, not ranks. Ions 7–15 are sorted in descending order of p(corr)[1]P.
According to comparisons of mean relative normalized concentrations.
Identification and Quantification of Mouse Urinary Biomarkers in the First 24 h after Exposure to 1, 2 and 3 Gy Gamma Radiation
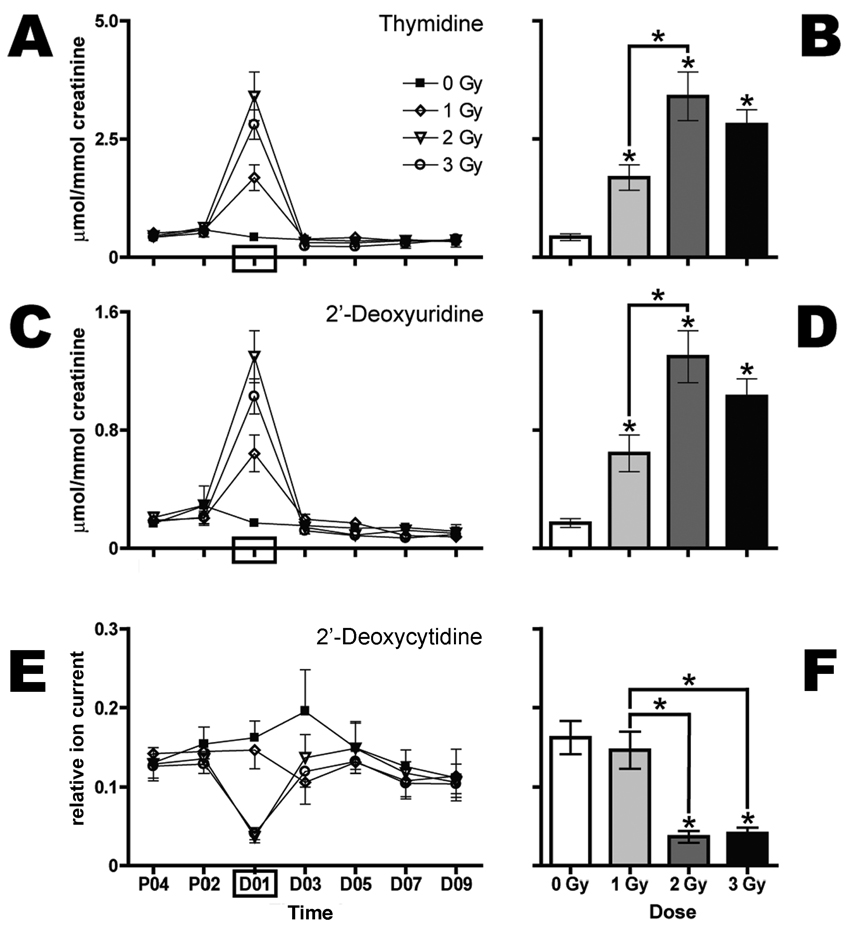
Consistent with a previous report of biomarkers of exposure to 3 Gy and above (4), this study also revealed elevations in dT (Table 1, no. 3) in the urine from mice irradiated with 1, 2 or 3 Gy collected over the first 24 h after exposure. Here these findings are extended and show a dose–response relationship for elevated dT (Fig. 2A). The pairwise comparison of mean concentrations at day 1 after exposure is shown in Fig. 2B. In addition, we observed dose-dependent elevated concentrations of dU (Fig. 2C and D) and dX (Fig. 3A and B). These ions correspond to no. 2 and no. 1 in Table 1, respectively. Together these three ions represent the most dramatic metabolic response observed. Table 1 also lists an in-source fragment (no. d) and a sodium adduct (no. a) of dT, a M+1 isotope (no. g) and a sodium adduct (no. b) of dU, and a possible in-source fragment (no. e) of dX.
FIG. 2.
Gamma-radiation exposure elicits dose-dependent increases in urinary excretion of dT and dU during the first 24 h after exposure and a decrease in urinary excretion of dC. Urine samples were collected prior to (P04, P02) and after (D01, D03, D05, D07, D09) exposure to 0 (■), 1 (◊), 2 (▽) or 3 Gy (○) Gy (n = 6 per dose) and analyzed by UPLC-TOFMS. Thymidine (panels A and B) and dU (panels C and D), expressed as mean µmol/mmol creatinine and stratified by dose, are shown over time (panels A and C). 2′-Deoxycytidine (panels E and F), expressed as relative ion current (normalized against creatinine), is shown over time (panel E). Mean normalized concentrations of dT (panel B), dU (panel D) and the relative concentration of dC (panel F) in urine from mice exposed to 1, 2 or 3 Gy were compared to the corresponding means of sham controls (0 Gy) at day 1 after exposure (D01) by a two-tailed t test with unequal variances and α = 05. Comparisons of mean normalized concentrations of dT, dU and dC among the three groups of exposed mice were made by one-way ANOVA with Bonferroni correction and α = 0.05. Error bars represent ± SEM, and * indicates P < 0.05.
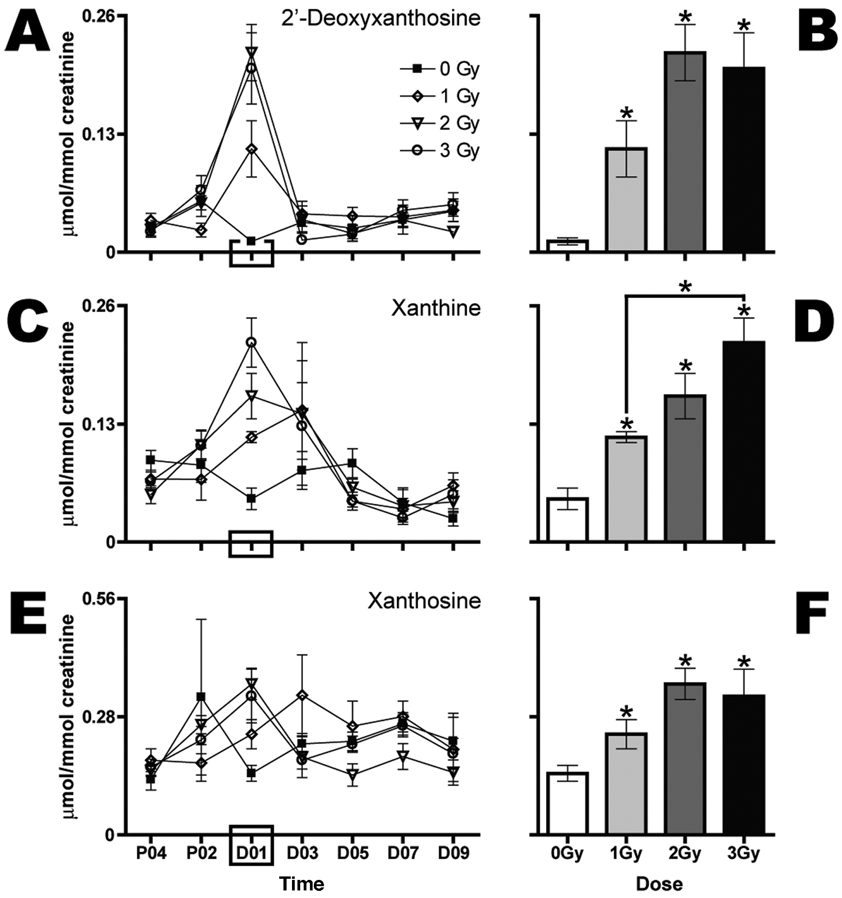
FIG. 3.
Gamma-radiation exposure elicits dose-dependent increases in urinary excretion of three purine-containing metabolites during the first 24 h after exposure. Urine samples were collected prior to (P04, P02) and after (D01, D03, D05, D07, D09) exposure to 0 (■), 1 (◊), 2 (▽) or 3 Gy (○) Gy (n = 6 per dose). 2′-Deoxyxanthosine (panels A and B), xanthine (panels C and D), and X (panels E and F) expressed as mean µmol/mmol creatinine stratified by dose are shown over time (panels A, C and E). Mean normalized concentrations of dX (panel B), xanthine (panel D) and X (panel F) in urine from mice exposed to 1, 2 or 3 Gy were compared to the corresponding mean normalized concentrations of the sham control group (0 Gy) at day 1 after exposure (D01) by a two-tailed t test assuming unequal variances with α = 0.05. Comparisons of mean normalized concentrations among the three groups of exposed mice were made by one-way ANOVA with Bonferroni correction and α = 0.05. Error bars represent ± SEM, and * indicates P < 0.05.
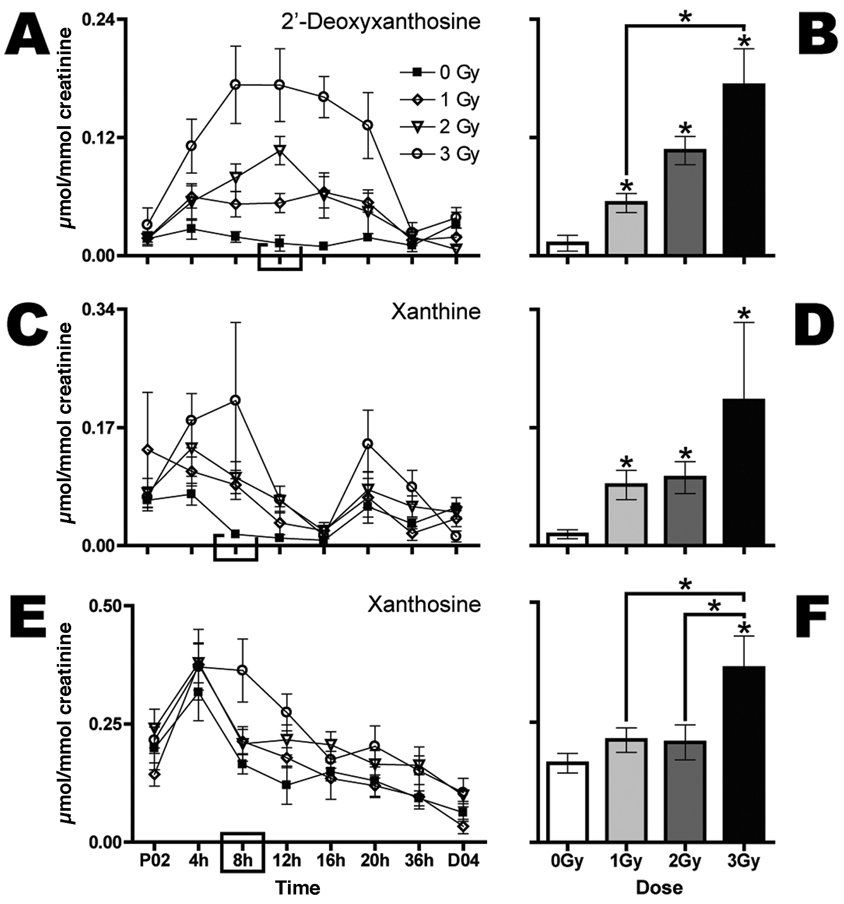
Two other new elevated biomarkers and one new potentially attenuated biomarker of radiation exposure were observed in the first 24-h urine samples after exposure. Both xanthine (Fig. 3C and D) and X (Fig. 3E and F) are elevated in the urine collected immediately after exposure. However, the elevations are modest; different dose groups are significantly different from 0 Gy and not from each other. Additionally, the variance estimates and baseline fluctuations of these two ions are higher compared with the ions shown in Fig. 2A–D and Fig. 3A and B. The only ion that has been identified as potentially attenuated after exposure thus far is dC. Based upon relative ion abundance, this ion is significantly attenuated in the first 24-h urine samples from mice irradiated with 2 and 3 Gy but not 1 Gy (Fig. 2E, F).
Also listed in Table 1 is an ion that was identified as a urinary biomarker of radiation exposure in a previous report (4), N-hexanoylglycine (no. 5). As can be seen from Table 1 and Fig. 1C, the relative abundance measure of this ion (p[1]P = 0.24) is high in the loadings space and yet its correlation to the model (p(corr)[1]P = 0.54) is lower than the ions we focus our attention on in this report. However, N-hexanoylglycine represents a “biochemically interesting compound” as opposed to one that is merely highly correlated to the OPLS model because of its consistent outlying position in the OPLS loadings S-plots (25). Random forests analysis, however, which does not embody a concentration-dependent component, ranks this ion as 1146. It is thus not included in the 25 consistent biomarkers shown in the Venn diagram (Fig. 1F), and we did not quantify N-hexanoylglycine for presentation in this study. One further biomarker is worthy of mention, and that is the anonymous ion no. 8 in Table 2 with [M-H]− = 417.1173 m/z and Tret = 1.9 min. An earlier study reported a similar ion of m/z = 417.1143 and Tret = 1.82 min and tentatively assigned it as “putative thymidine 5′-β-d-glucuronide” (4). Its abundance was too low to be studied by tandem mass spectrometry. A glucuronide of dT has hitherto never been reported.
Temporal Dynamics of Urinary Biomarkers during the First 96 h after Exposure to 1, 2 or 3 Gy Gamma Radiation
Having established that the core 0–24-h urinary biomarkers of 1–3 Gy γ-radiation exposure in mice were dT, dU, 2-deoxyxanthosine, xanthine and X, a more detailed study of the time dependence of their urinary elimination was mounted. It must be borne in mind that the utility in the field of human biomarkers of γ-radiation exposure will be related to their rate of appearance and disappearance in the sampled biofluid. Accordingly, the five core biomarkers were investigated further at 48 h before irradiation and at 4, 8, 12, 16, 20, 36 and 96 h postirradiation.
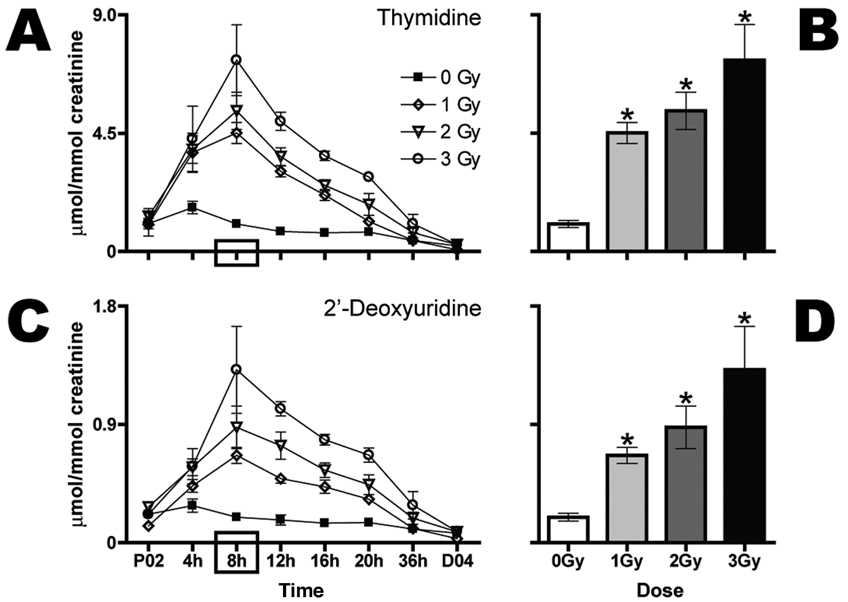
Figure 4A shows the time course of urinary dT excretion postirradiation. At 1, 2 and 3 Gy, the maximum concentrations (µmol/mmol creatinine) were observed at 8 h. The parallel declines in urinary dT had a half-life of approximately 8 h. Consequently, values had returned to baseline by 36 h. At all three doses, the elevation in urinary dT above basal levels was statistically significant at 8 h (Fig. 4B). Similarly, dU urinary excretion peaked at 8 h postirradiation (Fig. 4C) for all three doses and also declined with a half-life of approximately 8 h to basal levels at 36 h. At all three doses, the elevation in urinary dU above basal levels was statistically significant at 8 h (Fig. 4D).
FIG. 4.
Gamma-radiation exposure elicits dose-dependent increases in urinary excretion of dT and dU as early as 4 h after exposure. Urine samples were collected prior to (P02) and every 4 h after (4, 8, 12, 16 and 20 h) exposure to 0 (■), 1 (◊), 2 (▽) or 3 Gy (○) Gy (n = 6 per dose) radiation. Urine was then collected over 16 h (36 h) and later for a full 24 h at day 4 after exposure (D04). Thymidine (panels A and B) and dU (panels C and D) expressed as mean µmol/mmol creatinine stratified by dose are shown over time (panels A and C). Mean normalized concentrations of dT (panel B) and dU (panel D) for each dose at 8 h were compared to the respective sham control by a two-tailed t test assuming unequal variances with α = 05. Comparisons of mean normalized concentrations among the three groups of exposed mice were made by one-way ANOVA with Bonferroni correction and α = 0.05. Error bars represent ± SEM, and * indicates P < 0.05.
The purine derivatives, dX, xanthine and X behaved differently. For example, dX displayed more of a parabolic rise, with broad peak values for 3 Gy between 8 and 16 h (Fig. 5A). The 2-Gy dose peaked at 12 h and the 1-Gy dose exhibited a shallow rise between 4 and 20 h. Biomarker excretion returned to baseline values by 36 h. Figure 5B shows dose-specific mean concentrations of urinary dX at 12 h. At all three doses this biomarker was elevated statistically significantly above baseline values, and the 3-Gy value was significantly greater than the 1-Gy value, even by the stringent Bonferroni correction for multiple comparisons. The purine base xanthine displayed variable baseline values over the 96 h of the investigation (Fig. 5C). There were two apparent peaks in its urinary excretion, one at 8 h and the other at 20 h. This second peak appeared to coincide with the rapid decline in dX urinary excretion (Fig. 5A) and may represent conversion of one to the other by hydrolytic removal of 2′-deoxyribose. Despite the variable basal excretion profile, xanthine excretion was statistically significantly elevated above sham control values at all three doses of γ radiation (Fig. 5D). Moreover, xanthine is reliably elevated in urine from irradiated compared to control mice at day 1 after exposure in independent experiments. The purine nucleoside X displayed an early peak at 4–8 h in its elevated excretion above baseline, after which there was a decline in urinary excretion to basal levels at 16 h (Fig. 5E). Only at 3 Gy was the peak excretion (8 h) significantly elevated above baseline, but it was also elevated above the 1- and 2-Gy excretion values (Fig. 5F). As with xanthine, X is consistently elevated at day 1 after exposure in independent experiments, despite the wide baseline fluctuation and variance.
FIG. 5.
Gamma-radiation exposure elicits dose-dependent increases in urinary excretion of dX, xanthine and X as early as 4 h after exposure. Urine samples were collected prior to (P02) and every 4 h after (4, 8, 12, 16 and 20 h) exposure to 0 (■), 1 (◊), 2 (▽) or 3 Gy (○) Gy (n = 6 per dose) radiation. Urine was then collected over 16 h (36 h) and later for a full 24 h at day 4 after exposure (D04). 2′-Deoxyxanthosine (panels A and B), xanthine (panels C and D) and X (panels E and F) expressed as mean µmol/mmol creatinine stratified by dose are shown over time (panels A, C and E). Mean normalized concentrations of dX (panel B), xanthine (panel D) and X (panel F) for each dose at 12 h (panel B) or 8 h (panels D and F) were compared to the respective sham control by a two-tailed t test assuming unequal variances with α = 0.05. Comparisons of mean normalized concentrations among the three groups of exposed mice were made by one-way ANOVA with Bonferroni correction and α = 0.05. Error bars represent ± SEM, and * indicates P < 0.05.
Thus two pyrimidine 2′-deoxynucleosides (dT and dU), one purine 2′-deoxynucleoside (dX), one purine nucleoside (X), and one purine base (xanthine) were statistically significantly elevated in urine after γ irradiation. In general, these excretion profiles followed patterns that would be expected for rapid production after γ irradiation and with a similar rate of decay.
Biochemical Interrelationship of the Biomarkers
From a mechanistic standpoint it is important to understand the origins of and interrelationships between the two elevated and the one potentially attenuated pyrimidine biomarker, together with the three elevated purine biomarkers. While dC was attenuated in urine from mice irradiated with 2 and 3 Gy compared to controls at day 1 after exposure, this observation was made with relative ion abundance data and must be regarded as preliminary.
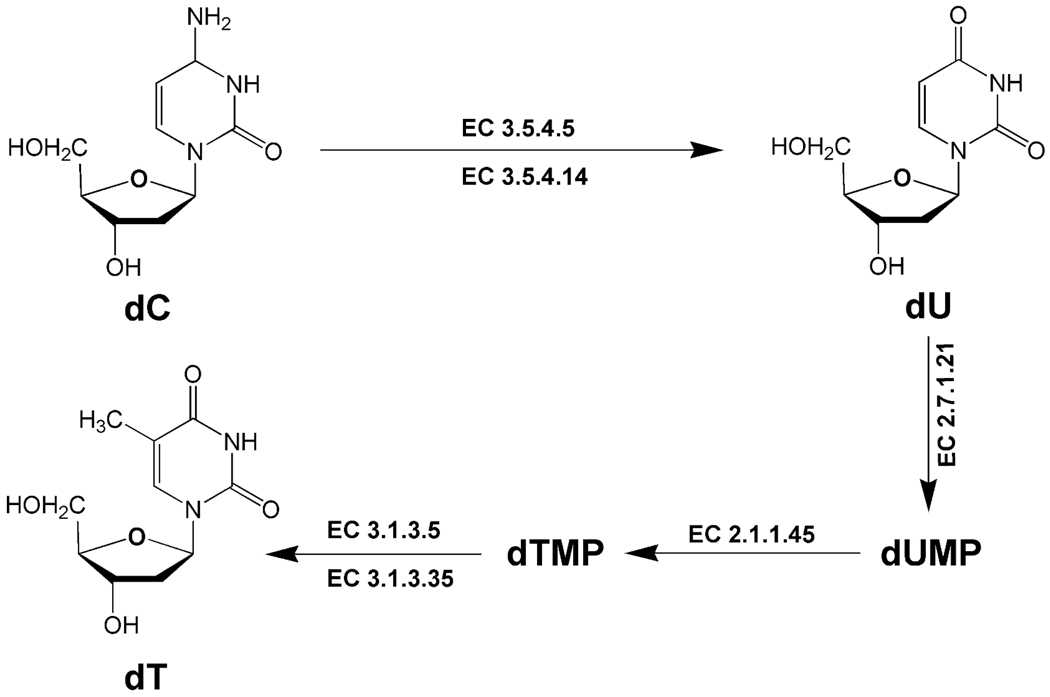
Do these purines and pyrimidines originate from DNA, from RNA or from both, or are they excreted as a result of the effect of γ radiation on cellular pools of pyrimidines and purines? The pyrimidine biomarkers dT, dU and dC should be derived from DNA since they are all deoxynucleosides. Their origin may be better understood if we can determine why the excretion of dC goes down concomitantly with a rise in excretion of dU and dT. Figure 6 shows that dC is deaminated to dU by the enzymes cytidine deaminase (EC 3.5.4.5) and deoxycytidine deaminase (EC 3.5.4.14). Under normal conditions, mouse plasma has undetectable levels (<0.1 µM) of dC and mouse liver, kidney and lung have deoxycytidine deaminase activity (26). Moreover, dU can be converted to dT via dUMP and its conversion to dTMP by thymidylate synthase (EC 2.1.1.45). Interestingly, hepatic activity of thymidylate synthase has been reported to increase significantly in mice that received whole-body γ radiation at doses of 2–7 Gy (27). Therefore, superficially at least, an elevated urinary excretion of dT and dU, concurrent with an attenuated excretion of dC, might be consistent with an increased turnover of DNA and the interconversions of deoxynucleosides presented in Fig. 6.
FIG. 6.
Enzymic deamination of pyrimidine nucleosides. 2′-Deoxycytidine (dC) is converted to dU and dT. Enzymes listed are cytidine deaminase (EC 3.5.4.5), deoxycytidine deaminase (EC 3.5.4.14), thymidine kinase (EC 2.7.1.21), thymidylate synthase (EC 2.1.1.45), 5′-nucleotidase (EC 3.1.3.5) and thymidylate 5′-phosphatase (EC 3.1.3.35).
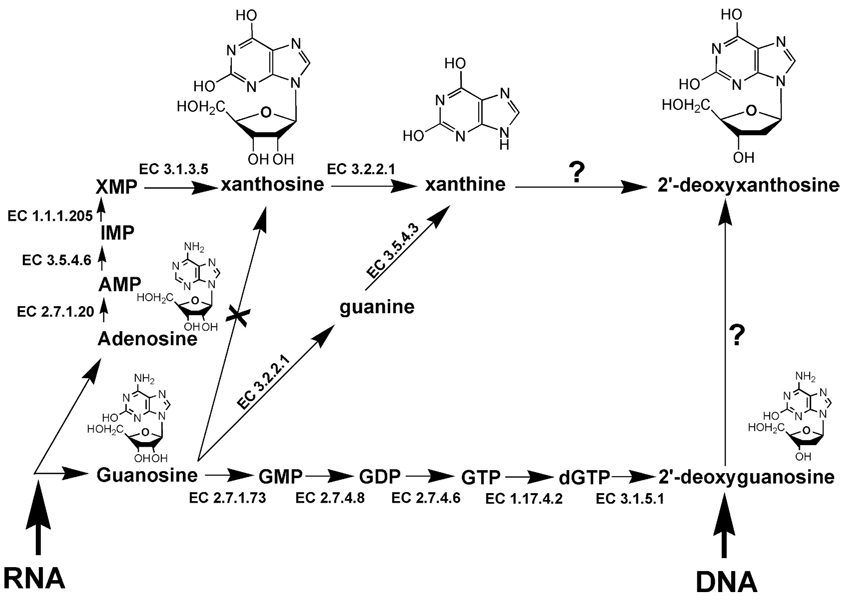
Three different types of purine comprise the other observed biomarker group, a purine nucleoside (X), a purine base (xanthine), and a purine 2′-deoxynucleoside (dX). Their potential biochemical relationships are shown in Fig. 7. It is clear that X is derived from RNA but only via the adenosine (A) pool. Guanosine (G) is not converted to X. However, xanthine can arise either directly by the action of purine nucleosidase (EC 3.2.2.1) on X or from the action of this enzyme on G to give guanine, followed by its conversion to xanthine by guanine deaminase (EC 3.5.4.3). Thus xanthine may arise from the A or G pool. Although G may be converted to dG by a series of steps, dG is not reported to be converted enzymically to dX, nor has xanthine been reported to be converted to dX. An important question therefore arises as to the origin of dX in the urine of γ-irradiated mice.
FIG. 7.
Enzymic deamination of purine nucleosides. Guanosine (G) from RNA is converted through multiple steps to 2′-deoxyguanosine (dG). Adenosine (A) from RNA is converted through adenosine 5′-monophosphate (AMP), inosine 5′-monophosphate (IMP), xanthosine 5′-monophosphate (XMP) to X, which can be converted to xanthine, but not further to dX. Xanthine may also arise from G via guanine. The enzymic conversion of dG to dX has not been reported. The enzymes listed are inosine-guanosine kinase (EC 2.7.1.73), guanylate kinase (EC 2.7.4.8), nucleoside-diphosphate kinase (EC 2.7.4.6), ribonucleoside-triphosphate reductase (EC 1.17.4.2), dGTPase (EC 3.1.5.1), purine nucleosidase (EC 3.2.2.1), guanine deaminase (EC 3.5.4.3), adenosine kinase (EC 2.7.1.20), AMP deaminase (EC 3.5.4.6), IMP dehydrogenase (EC 1.1.1.205) and 5′-nucleotidase (EC 3.1.3.5).
The elevated purine and pyrimidine radiation biomarkers have a common thread, in that they are all deaminated nucleosides or their derivatives. It is tempting to invoke a role for deoxycytidine deaminase, guanine deaminase and AMP deaminase (EC 3.5.4.6 that converts AMP to IMP; Fig. 7). However, none of these deaminases can explain the presence of dX in urine. The alternative explanation for the elevated excretion of hydroxylated purine and pyrimidine derivatives (and the decreased excretion of the aminopyrimidine dC) is the action of reactive oxygen species (ROS) and reactive nitrogen species (RNS). Both dU and dX have been reported to be formed from dC and dG in DNA in situ by so-called nitrosative deamination (28, 29). These possibilities will be discussed in more detail.
DISCUSSION
The effects on the mouse urinary metabolome of sublethal γ irradiation were investigated. Five urinary biomarkers of radiation exposure, statistically significantly elevated between 1 and 3 Gy, were unequivocally identified by tandem mass spectrometry. These are all deaminated purine and pyrimidine derivatives: dT, dU, dX, xanthine and X. All are elevated above baseline in urine at the three doses studied, 1, 2 and 3 Gy. Furthermore, we observed reduced urinary excretion of the aminopyrimidine dC at 2 and 3 Gy. However, this ion was only partially quantified. All of the biomarkers displayed a time-dependent excretion, peaking in urine at 8–12 h but returning to baseline by 36 h. The enhanced excretion of another nine compounds in urine was also observed, based upon detection of their negative ions by UPLC-TOFMS, but the chemical identities of these metabolites remain to be elucidated.
This study involved the dimensions of both dose and time. With respect to time, it was first observed that differential excretion of the biomarkers according to exposure status occurred within the first 24 h after exposure, consistent with earlier findings (4). The response within the first 24 h was then characterized by collecting urine over 4-h periods up to 20 h. In both experiments, where 24-h and 4-h urine samples were collected, the irradiations were conducted in the late afternoon, at 5:00 p.m. ± 1 h. Late afternoon was chosen in the second experiment to maximize the chances of having 4-h urine samples from every mouse since the mice are active and produce more urine during the dark cycle. However, the possibility cannot be ruled out that diurnal variation in urine metabolite profiles confounds the observations (30, 31). On the other hand, if there is confounding by diurnal variation, then it can be argued that it is probably minimal given that the maximum differences (exposed compared to control) observed for each dose within the 4-h samples are similar to those seen in the corresponding 24-h samples.
The loadings S-plots shown in Fig. 1C–E suggest that there are rich pools of both ESI− and ESI+ urine ions from which to select candidate biomarkers of radiation exposure. In fact, however, the loadings that correlate well with the model classification of exposure do not always present in urine in a manner that is useful for biodosimetry. An ideal biomarker is consistently elevated or attenuated in urine from all exposed individuals, and the individual variation is reasonably small. In a matrix of UPLC-TOFMS data, there are often a substantial number of ions detected in one or only a few samples but not in all of the samples of a given class or sample set. The variance estimates of the means of these types of ions are very large. The SIMCA software nevertheless calculates a correlation score for these ions without regard to the variance estimates of the means, and if they were elevated or attenuated in only one or two samples from exposed mice, they will nonetheless correlate reasonably well in an OPLS model in such a case. A test for exposure-specific differences in means will produce a null result because the variance estimates are relatively large. We focused our attention on ions that may be more useful for biodosimetry and less so for modeling by OPLS alone and eliminated those that were not significantly different from control concentrations. In fact, more of these ions that show promise for biodosimetry applications were found in the ESI− data set than in the ESI+ data set.
Our observations raise a number of important questions regarding the effect of γ radiation on the mouse. First, the excretion of N-hexanoylglycine: We had reported in a previous study that this urinary biomarker was elevated at doses of both 3 and 8 Gy (4). Multivariate data analysis using OPLS (Fig. 1C) shows that N-hexanoylglycine is by far the most abundant murine urinary biomarker of γ radiation but that it is comparatively less well correlated to the OPLS model (p(corr)[1]P = 0.54). Thus it is presumed that this biomarker, but no other metabolite of fatty acid metabolism, results from a perturbation of hexanoyl-CoA β-oxidation, resulting in its conjugation with glycine in an alternative pathway. This biomarker was not investigated further in the present study because it was significantly elevated only at doses of 3 Gy and above, and we focused here on the effects of doses of 3 Gy and below.
A dose-dependent elevated excretion of dT, dU, dX, X and xanthine was observed in the first 24 h postirradiation, together with a putative dose-related attenuated excretion of dC. It has already been noted (see Fig. 6) that the pattern of excretion of the three pyrimidine derivatives dT, dU and dC might be explained by deamination of dC to dU by cytidine deaminase and/or deoxycytidine deaminase, followed by ultimate synthesis of dT by thymidylate synthase, an enzyme reported to be induced in mouse liver by γ radiation (27). It would appear, therefore, that production of deaminated pyrimidines is due to γ-radiation exposure. The same pattern holds true for the purine derivatives that were found to be elevated in urine. Metabolomic analysis did not reveal an enhanced urinary excretion of guanosine (G), deoxyguanosine (dG), adenosine (A) or deoxyadenosine (dA). All of these nucleosides are aminopurines. In contrast, dX, X and xanthine were all elevated in urine after γ irradiation in a dose- and time-related manner (Fig. 3 and Fig. 5). These purines are all deaminated and their formation from G,A, dG or dA in response to γ radiation is not readily explained by simple enzyme-mediated reactions (Fig. 7). Thus an alternative explanation must be sought. It was reported that γ irradiation of mice results in a statistically significant two- to threefold increase in serum nitrate concentration 2.5–3.0 h postirradiation that returns to baseline after 12 h (32). This nitrate increase is consistent with the effect in mice of sublethal γ -radiation exposure on nitric oxide (NO) synthesis in the liver, intestine, lung, kidney, brain, spleen and heart (33), on increased hepatic nitrite concentration and peroxidative damage (34), and on attenuated hepatic glutathione concentration (34, 35). There is abundant evidence therefore that γ irradiation of mice increases hepatic NO synthesis.
One important property of cellular NO is that it may autooxidize to form nitrous anhydride (N2O3), which then can participate in so-called nitrosative deamination of both purines and pyrimidines in DNA in situ (29). Dedon and his colleagues have reported that dC can undergo such nitrosative deamination to dU, dA to 2′-deoxyinosine (dI) and dG to dX and 2′-deoxyoxanosine (dO) (28, 29). Although we did not detect elevated urinary dI (C10H12N4O4; [M-H]− = 251.0780) or dO (C10H12N4O5; [M-H]− = 267.0729) in our metabolomic analyses (see Table 2), all other observations regarding the urinary excretion of elevated concentrations of deaminated pyrimidines and purines are consistent with the hypothesis that sublethal γ irradiation of mice leads to in vivo nitrosative deamination by N2O3 of DNA bases in situ. Clearly, further investigations with agents that can ameliorate NO are warranted. Such a strategy may provide new avenues for the development of radioprotective drugs.
A relationship between ionizing radiation and the urinary excretion of dT in rats was reported in the Russian literature over 40 years ago (36, 37) and subsequently between ionizing radiation and dU and the pyrimidine metabolite β-aminoisobutyric acid (38, 39). The elevated excretion of dT in rats after subcutaneous injection of 90Sr was somewhat modest (38–64%) and did not appear until the 5th to 9th day after injection (36). This is clearly different from the present findings. In addition, an increased excretion of β-aminoisobutyric acid was not observed. However, in the historical Russian literature, it was reported that whole-body X irradiation with 6.5 Gy produced a transient eightfold increase in urinary dT excretion at day 1 in rats (37), which is remarkably similar to the sevenfold increase in urinary dT reported herein after 3 Gy γ irradiation and in a previous study (4) in the mouse. Thus dT and dU may be biomarkers of ionizing radiation exposure not only in mouse and rat but also in humans. This hypothesis awaits rigorous testing.
The classical urinary biomarkers of repaired oxidative DNA damage are 8-hydroxy-2′-deoxyguanosine (8-OH-dG) (40) together with thymine glycol and thymidine glycol (41). No evidence of elevated 8-OH-dG (C10H13N5O5; [M-H]− = 282.0838) or of thymine glycol (C5H8N2O4; [M-H]− = 159.0406) was uncovered (see Table 2). However, thymidine glycol (C10H14N2O7; [M-H]− = 273.0723) may possibly correspond to unidentified ion no. 11 (Table 2). These findings suggest that 1–3 Gy of γ radiation does not simply cause oxidation of dG by ROS, leading to increased urinary excretion of 8-OH-dG, and further helps substantiate the hypothesis that the effects of these doses of ionizing radiation on the mouse occur through a specific mechanism, such as nitrosative deamination of dC and dG. Further investigations are required to provide a more detailed mechanism of DNA damage from exposure to sublethal ionizing radiation.
In this report attention is focused mainly on urinary anions that were elevated in irradiated mice. In fact, the loadings scatter plots (both ESI− and ESI+) were interrogated to determine whether any attenuated species reliably indicated radiation exposure. However, little emphasis is placed on this pool of ions for two reasons. First, eventual human biomarkers of radiation exposure are more useful if they exhibit up-regulation in association with exposure. Given that initial positive results with urine biodosimetry were validated with follow-up methodologies, it can be argued that false negatives in radiation exposure assessment are more detrimental to the objective of biodosimetry than are false positives. Second, a lower success in observing meaningful, consistent and verifiable attenuated biomarkers of radiation exposure has been attained, despite what the loadings scatter plots suggest, dC being a notable exception.
The need for noninvasive high-throughput radiation biodosimetry tools is ever more pressing and prescient. A recent report from a U.S. Congressional Commission (December 2, 2008) states in the first paragraph of its executive summary, “The Commission believes that unless the world community acts decisively and with great urgency, it is more likely than not that a weapon of mass destruction will be used in a terrorist attack somewhere in the world by the end of 2013” (42). Even if preparedness for a nuclear or radiological terrorist attack are high, the tools available for the mass screening of at-risk individuals for radiation exposure are pitifully few. Metabolomics offers a means of screening large populations in relatively short periods, providing that (1) a robust radiation metabolomic signature with unambiguous dose–response characteristics can be developed, and (2) a portable device suitable for use in the field by first responders can be designed, built, tested and validated. These are ambitious aims but ones that must be pursued aggressively. This report represents a second step in the characterization of a radiation metabolomic signature, with emphasis on both the dose response and the time course of the response. In addition, some novel insights into the mechanisms of radiation effects at sublethal doses are beginning to emerge. These studies may also ultimately have a bearing on the development of new strategies for prophylaxis and treatment of radiation sickness.
ACKNOWLEDGMENTS
We wish to thank Dr. Peter Dedon of MIT for his generous gift of 2′-deoxyxanthosine. We also wish to thank Dr. Gabriel Eichler, Genomics and Bioinformatics Group, Laboratory of Molecular Pharmacology, NCI, for his advice with random forests analysis and statistics. This work was performed as part of the Columbia University Center for Medical Countermeasures against Radiation (P.I. David Brenner) and funded by NIH (NIAID) grant U19 AI067773-02 and also supported by the National Cancer Institute Intramural Research Program. JBT was supported in part by the Cancer Prevention Fellowship Program, Office of Preventive Oncology, National Cancer Institute. ADP is supported by the Pharmacology Research Associate in Training program, National Institute of General Medical Sciences. JRI is grateful to U.S. Smokeless Tobacco Company for a grant for collaborative research.
REFERENCES
- 1.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 2.Chao NJ. Accidental or intentional exposure to ionizing radiation: biodosimetry and treatment options. Exp. Hematol. 2007;35:24–27. doi: 10.1016/j.exphem.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Pellmar TC, Rockwell S. Priority list of research areas for radiological nuclear threat countermeasures. Radiat. Res. 2005;163:115–123. doi: 10.1667/rr3283. [DOI] [PubMed] [Google Scholar]
- 4.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat. Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 6.Zerhouni E. Medicine. The NIH Roadmap. Science. 2003;302:63–72. doi: 10.1126/science.1091867. [DOI] [PubMed] [Google Scholar]
- 7.Zhen Y, Slanar O, Krausz KW, Chen C, Slavik J, McPhail KL, Zabriskie TM, Perlik F, Gonzalez FJ, Idle JR. 3,4-Dehydrodebrisoquine, a novel debrisoquine metabolite formed from 4-hydroxydebrisoquine that affects the CYP2D6 metabolic ratio. Drug Metab. Dispos. 2006;34:1563–1574. doi: 10.1124/dmd.105.008920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giri S, Krausz KW, Idle JR, Gonzalez FJ. The metabolomics of (+/−)-arecoline 1-oxide in the mouse and its formation by human flavin-containing monooxygenases. Biochem. Pharmacol. 2007;73:561–573. doi: 10.1016/j.bcp.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Zhen Y, Krausz KW, Chen C, Idle JR, Gonzalez FJ. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol. Endocrinol. 2007;21:2136–2151. doi: 10.1210/me.2007-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Berriz GF, Roth FP, Gerszten RE. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 11.Castro-Perez J, Plumb R, Granger JH, Beattie I, Joncour K, Wright A. Increasing throughput and information content for in vitro drug metabolism experiments using ultra-performance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2005;19:843–848. doi: 10.1002/rcm.1859. [DOI] [PubMed] [Google Scholar]
- 12.Plumb R, Castro-Perez J, Granger J, Beattie I, Joncour K, Wright A. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:2331–2337. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 13.Dunn SR, Qi Z, Bottinger EP, Breyer MD, Sharma K. Utility of endogenous creatinine clearance as a measure of renal function in mice. Kidney Int. 2004;65:1959–1967. doi: 10.1111/j.1523-1755.2004.00600.x. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi N, Boysen G, Li F, Li Y, Swenberg JA. Tandem mass spectrometry measurements of creatinine in mouse plasma and urine for determining glomerular filtration rate. Kidney Int. 2007;71:266–271. doi: 10.1038/sj.ki.5002033. [DOI] [PubMed] [Google Scholar]
- 15.Noda I. Scaling techniques to enhance two-dimensional correlation spectra. J. Mol. Struct. 2008;883–884:216–227. [Google Scholar]
- 16.Pearson K. On lines and planes of closest fit to systems of points in space. Philos. Mag. 1901;2:559–572. [Google Scholar]
- 17.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J. Chemometrics. 2002;16:119–128. [Google Scholar]
- 18.Hotelling H. The generalization of Student’s ratio. Ann. Math. Stat. 1931;2:360–378. [Google Scholar]
- 19.Rosner B. Fundamentals of Biostatistics. 5th ed. Duxbury, Pacific Grove, CA: 2000. [Google Scholar]
- 20.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 21.Williams AJ. Internet-based tools for communication and collaboration in chemistry. Drug Discov. Today. 2008;13:502–506. doi: 10.1016/j.drudis.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Williams AJ. Public chemical compound databases. Curr. Opin. Drug Discov. Dev. 2008;11:393–404. [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 24.Browning BL. PRESTO: rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies. BMC Bioinformat. 2008;9:309. doi: 10.1186/1471-2105-9-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 26.Chan TS, Lakhchaura BD, Hsu TF. Differences in deoxycytidine metabolism in mouse and rat. Biochem. J. 1983;210:367–371. doi: 10.1042/bj2100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batra V, Kesavan V, Mishra KP. Modulation of enzymes involved in folate dependent one-carbon metabolism by gamma-radiation stress in mice. J. Radiat. Res. (Tokyo) 2004;45:527–533. doi: 10.1269/jrr.45.527. [DOI] [PubMed] [Google Scholar]
- 28.Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat. Protoc. 2008;3:1287–1298. doi: 10.1038/nprot.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 30.Plumb RS, Granger JH, Stumpf CL, Johnson KA, Smith BW, Gaulitz S, Wilson ID, Castro-Perez J. A rapid screening approach to metabonomics using UPLC and oa-TOF mass spectrometry: application to age, gender and diurnal variation in normal/Zucker obese rats and black, white and nude mice. Analyst. 2005;130:844–849. doi: 10.1039/b501767j. [DOI] [PubMed] [Google Scholar]
- 31.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Marrie TJ. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 32.Ohta S, Matsuda S, Gunji M, Kamogawa A. The role of nitric oxide in radiation damage. Biol. Pharm. Bull. 2007;30:1102–1107. doi: 10.1248/bpb.30.1102. [DOI] [PubMed] [Google Scholar]
- 33.Voevodskaya NV, Vanin AF. Gamma-irradiation potentiates l-arginine-dependent nitric oxide formation in mice. Biochem. Biophys. Res. Commun. 1992;186:1423–1428. doi: 10.1016/s0006-291x(05)81565-7. [DOI] [PubMed] [Google Scholar]
- 34.Agrawal A, Chandra D, Kale RK. Radiation induced oxidative stress: II Studies in liver as a distant organ of tumor bearing mice. Mol. Cell Biochem. 2001;224:9–17. doi: 10.1023/a:1011990704575. [DOI] [PubMed] [Google Scholar]
- 35.Baliga MS, Jagetia GC, Venkatesh P, Reddy R, Ulloor JN. Radioprotective effect of abana, a polyherbal drug following total body irradiation. Br. J. Radiol. 2004;77:1027–1035. doi: 10.1259/bjr/83720350. [DOI] [PubMed] [Google Scholar]
- 36.Uspenskaia MS, Rabinkova EV. [The excretion of thymidine in the urine in Sr90-injured rats] Med. Radiol. (Mosk.) 1965;10:19–22. [PubMed] [Google Scholar]
- 37.Zharkov Iu A, Fedorova TA, Mikhailova LF. [The excretion of thymidine in the urine of rats after total body x-ray irradiation in various doses] Radiobiologiia. 1965;5:675–680. [PubMed] [Google Scholar]
- 38.Mazurik VK, Bryksina LE, Saprygin DB, Iarilin AA. [A study of the role of lymphoid tissue in post-radiation hyperexcretion of deoxycytidine, deoxyuridine and thymidine by use of specific antisera] Radiobiologiia. 1970;10:346–349. [PubMed] [Google Scholar]
- 39.Mazurik VK, Bryksina LE, Bibikhin LN. [The relationship between the excretion of deoxyuridine, thymidine and beta-aminoisobutyric acid by rats and the radiation dose and length of time following total irradiation] Radiobiologiia. 1970;10:43–48. [PubMed] [Google Scholar]
- 40.Loft S, Fischer-Nielsen A, Jeding IB, Vistisen K, Poulsen HE. 8-Hydroxydeoxyguanosine as a urinary biomarker of oxidative DNA damage. J. Toxicol. Environ. Health. 1993;40:391–404. doi: 10.1080/15287399309531806. [DOI] [PubMed] [Google Scholar]
- 41.Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc. Natl. Acad. Sci. USA. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham B, Talent J, Allison G, Cleveland R, Rademaker S, Roemer T, Sherman W, Sokolski H, Verma R. World at Risk. The Report of the Commission on the Prevention of WMD Proliferation and Terrorism. New York: Vintage Books; 2008. [Google Scholar]