Abstract
Fenofibrate has been widely used for the treatment of dyslipidaemia with a long history. Species differences of its metabolism were reported, but its metabolites in rodent have not been fully investigated.
Urine and plasma samples were collected before and after oral dosages of fenofibrate in Sprague–Dawley rats. Urine samples were subjected to ultra-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOF-MS) analysis, and projection to latent structures discriminant analysis was used for the identification of metabolites.
New metabolites in urine and plasma were also studied by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The metabolism pathway was studied in rat hepatocytes. Synthesized and purchased authentic compounds were used for metabolite identification by LC-MS/MS.
Five ever-reported metabolites were identified and another four new ones were found. Among these new metabolites, fenofibric acid taurine and reduced fenofibric acid taurine indicate new phase II conjugation pathway of fenofibrate.
Keywords: Metabolomics, metabolites, metabolism pathway, fenofibrate, rat
Introduction
Fibrates form a group of agonists of the peroxisome proliferator-activated receptor α (PPARα), which binds to PPAR-responsive elements after heterodimerization with a partner retinoic X receptor (Bocher et al. 2002; Fruchart and Duriez 2006; Qu et al. 2007). It is considered to be very safe in extensive clinical applications for dyslipidaemia, but the adverse effects of fenofibrate ranged from 2% to 15% and its myotoxicity incidence rate was estimated as 1% (Blane 1987; Ritter and Nabulsi 2001; Ghosh et al. 2004; Holoshitz et al. 2008). Additionally, PPARα agonists induced hepatocarcinogenesis in rodents, but human and non-human primates remain refractory to fibrates-induced peroxisome proliferation and hepatocarcinogenesis (Reddy et al. 1980; Rao and Reddy 1987; Hoivik et al. 2004). The mechanism study of this species-related difference of hepatotoxicity is still ongoing, where the metabolites may play a role (Ward et al. 1998; Peters et al. 2000; Klaunig et al. 2003; Gonzalez and Shah 2008).
The metabolism of fenofibrate in human, rat, guinea pig, dog and also rat hepatocytes has been investigated (Weil et al. 1988, 1990; Cornu-Chagnon et al. 1995). Its metabolites, fenofibric acid (FA) and reduced fenofibric acid (RFA), were reported in rat, pig, dog, human and rat hepatocytes (Weil et al. 1988; Cornu-Chagnon et al. 1995). Its another two metabolites, fenofibric acid ester glucuronide (FAEG) and reduced fenofibric acid glucuronide (RFAEG), were found in rat, pig and human, but not in dog (Cornu-Chagnon et al. 1995). Compound X was only reported as a metabolite in rat hepatocytes. Compounds B and AR were found to induce peroxisomal palmitoyl-CoA oxidation activity significantly, but they were not reported as in vivo metabolites of fenofibrate (Cornu-Chagnon et al. 1995). These three compounds were also structurally part of fenofibrate. Accordingly, they were included as suspected metabolites in this study. The metabolism of clofibrate, which was an analogue of fenofibrate, was well understood and the taurine conjugate of clofibric acid was found in dog, cat, and ferret, but not in human volunteer, rodent, and rabbit (Cayen et al. 1977; Emudianughe et al. 1983). However, no taurine conjugation has ever been reported for metabolism of fenofibrate either in rat or in other species.
Metabolomics (or metabonomics) defines a global quantitative profile of low-molecular-weight metabolites which are context dependent in a cell, tissue or organism varying in response to experimental intervention (Nicholson et al. 1999; Lindon et al. 2004; Weckwerth and Morgenthal 2005). This newly emerged ‘omics’ tool makes full use of nuclear magnetic resonance (NMR) spectroscopy or mass spectometry in conjunction with multivariate data analysis (MDA) such as principal components analysis (PCA) (Chen et al. 2007; Lenz et al. 2007). With the developing technology in metabolomics applications, hybrid time-of-flight mass spectrometry (TOF-MS) instruments such as QTOF-MS instruments are increasingly used in metabolomics due to their high sensitivity, mass resolving power and mass accuracy. Ultra-performance liquid chromatography (UPLC) improves the speed of analysis and, more importantly, provides an excellent chromatographic resolution. The hyphenation of UPLC to QTOF-MS can be advantageous for a better assignment of metabolites from chromatographic mass signals. When combined with MDA, UPLC-QTOF-MS-based metabolomics can yield and recognize a large number of accurate ion-retention time pairs in a single run of sample that describe the differences between different groups (Chen et al. 2007; Zhen et al. 2007). As to pattern recognition, supervised projection to latent structures discriminant analysis (PLS-DA) coupled with Pareto-scaling instead of PCA and unit variance scaling has been increasingly used for the identification of new metabolites and biomarkers (Ward et al. 1998; Peters et al. 2000).
In this study, urine and plasma samples were collected before and after oral dosages of fenofibrate in five Sprague–Dawley rats. The urine samples were subjected to ultra-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOF-MS) analysis and MDA was used for the identification of metabolites. New metabolites were also determined in plasma samples and the metabolism pathway was studied in rat hepatocytes by liquid chromatograph coupled with tandem mass spectrometry (LC-MS/MS). Synthesized and purchased authentic compounds were used for metabolite identification by LC-MS/MS. Five ever reported metabolites were identified in this study (FA, RFA, FAEG, RFAEG and compound X). Another four metabolites, fenofibric acid taurine (FAT), reduced fenofibric acid taurine (RFAT), compound B and compound AR, were also identified as metabolites in rats. FAT and RFAT indicate a new phase II-conjugation metabolism pathway of fenofibrate.
Materials and methods
Chemicals and reagent
Fenofibrate, fenofibric acid (FA) and 4-chloro-4´-hydroxy-benzophenone (compound A) were purchased from Shangqiu Chemry Chemicals Co. Ltd (Shandong, China). Taurine was purchased from Shanghai Jiachen Co. Ltd (Shanghai, China). HPLC-grade solvents (acetonitrile, methanol) were purchased from Sigma Aldrich (MO, USA). Purified water was obtained from a Millipore Elix (Millipore, MA, USA) system. All other chemicals used for experiments were analytical reagent or HPLC grade of commercial resource.
Synthesis of RFA, AR, FAT, RFAT, B, and X
FA was used as a starting material for the synthesis of reduced fenofibric acid (RFA), compound X, fenofibric acid taurine (FAT), reduced fenofibric acid taurine (RFAT). Compound A was used for compound A reduced (AR) and 4-chloro-4´-isopropox benzophenone (compound B). Taurine was used as an intermediate for the synthesis of FAT and RFAT. Synthesized compounds were subjected to 1H-NMR (Bruker AV-400, Faellanden, Switzerland) for structure and purity confirmation (detailed procedures and conditions for RFA and compound X are not shown).
To synthesize FAT, FA (318.5 mg, 1 mmol) and triethylamine (0.14 ml, 1.2 mmol) was dissolved in 5 ml acetone at −5°C (ice bath). Methylchloroformate (0.1 ml, 5 mM) in 1 ml dichloromethane was added dropwise. The mixture was stirred at −5°C to approximately 0°C until the disappearance of the starting material was noted by TLC (5–10 min). Taurine (125 mg, 1 mmol) in 2 ml aqueous sodium hydroxide (2 M) was then added over a period of 10 min. After the reaction was complete (noted by TLC, 3–4 h), the mixture was evaporated at ambient temperature using a rotary evaporator. Purified FAT was produced by preparative TLC on a silica gel plate using a mobile phase composed of dichloromethane, methanol and acetic acid (15:1:0.1). For RFAT, purified FAT (160 mg, 0.5 mmol) was dissolved in 2 ml 3:1 tetrahydrofuran–methanol solution in an ice bath. NaBH4 (95 mg, 2.5 mmol) was then added portion-wise over 30 min. The mixture was stirred at 0–25°C until the disappearance of the starting material was noted by TLC (2–3 h). The reaction mixture was concentrated on a rotary evaporator. The residue was dissolved in 4 ml water and the solution acidified to pH 6 with the slow addition of 6 M citric acid and then extracted with ethyl acetate three times. The ethyl acetate layers were dried and concentrated by rotary evaporation to yield the product.
For compound AR, NaBH4 (4 mmol) was added portion-wise to solution of compound A (1 mmol) in methanol (6 ml) at 0°C over 30 min. After stirring at room temperature overnight, the reaction mixture was acidified to pH 5 with 5% H2SO4 and extracted with CH2Cl2. The organic phase was dried with MgSO4, and the solvent was evaporated in vacuo to yield compound AR. To synthesize compound B, compound A (0.9 mmol), 2-bromopropane (1 mmol) and anhydrous potassium carbonate (0.9 mmol) were dissolved in DMF (0.9 ml). After stirring at 90°C for 3 h, the reaction mixture was cooled to room temperature, diluted with water, poured into 1 N sodium hydroxide (0.5 ml), and extracted with ether (three times using 8 ml). The combined extracts were dried (Na2SO4), filtered, and evaporated in vacuo. The residue was chromatographed on silica gel using hexane/EtOAc as eluent to yield the product of compound B.
Animals treatments and collection of samples
Five male Sprague–Dawley rats, 8 weeks old and weighing 200 ± 10 g, were originally provided by the Experimental Animal Center, Southern Medical University, and maintained under specific pathogen-free conditions in the Animal Center of Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (GIBH). Rats were housed in plastic cages (one for each) and maintained under a standard 12-h light/12-h dark cycle with free access to purified water and commercial diet. Environmental controls for the animal rooms were set to maintain 18–29°C and a relative humidity of 30–70%. Before the experiment, rats were allowed to acclimatize to the animal facility environment for at least 7 days. Animal handling adhered to the Principles of Laboratory Animal Care and all experimental procedures were approved by the Laboratory Animal Committee (LAC) of GIBH.
The rats were dosed with fenofibrate twice daily for 3 days by oral gavage and the dosage was 2500 mg kg−1 day−1. Four days before the start of fenofibrate dosing, vehicle of 0.5% (w/v) aqueous sodium carboxymethylcellulose was given by individualized volume. Urine and plasma samples were collected on the day before fenofibrate dosing and were used as blank control. Twelve hours after the last dosage, urine and plasma samples were collected. All the samples collected were immediately placed in dry ice and stored at −80°C until analysis.
UPLC-ESI-QTOF-MS analyses and MDA for metabolite variables
Urine samples were diluted (1:3) with acetonitrile/water (50:50) and centrifuged at 16 000g 4°C for 30 min to remove particulates and proteins. The aliquots of 5 µl of each samples were subjected to chromatography on a 50 mm*2. 1 mm ACQUITY 1.7 µm BEH C18 column (Waters Corporation, Milford, MA, USA) with a gradient mobile phase containing 0.1% formic acid. The flow rate was set to 0.5 ml min−1 and the time for a single run was 10 min. The eluent was introduced directly into the mass spectrometer by electrospray. Mass spectrometry was performed on a Waters Q-TOF Micro coupled with an ACQUITY UPLC system (Waters Corporation) operating in both positive- and negative-ion modes. The desolvation gas flow was set to 650 l h−1 at a temperature of 350°C with the cone gas set to 50 l h−1 and the source temperature at 120°C. The capillary voltage and the cone voltage were set to 3000 V and 30 V, respectively. Bezafibrate was used as the lock mass (FW: 361.1081) for accurate mass calibration and introduced by a Lockspray interface at 50 µl min−1 with a concentration of 0.5 ng ml−1 in 50% aqueous acetonitrile. In mass spectrometry scanning, data were recorded in centroid mode from 100 to 900 m/z.
After UPLC-ESI-QTOF-MS analyses, centroided and integrated mass chromatographic data were deconvoluted and processed by MarkerLynx mass spectrometry software (Waters Corporation) to generate multivariate data matrices. The positive and negative data matrices were respectively exported into SIMCA-P+11 (Umetrics AB, Umea, Sweden) for MDA. Pareto-scaling and PLS-DA were selected for data mining and pattern recognition after optimization.
Identification of fenofibrate metabolites by LC-MS/MS
From urine negative data matrix, suspected metabolites were selected by analysing the score plots and the corresponding loadings plots, which were FA, RFA, FAT and FAEG. For a positive data matrix, no other new metabolite variables could be found. These suspected metabolites were identified by LC-MS/MS analysis of urine samples. For compound B, X, RFAT, RFAEG, which were suspected as metabolites, plasma samples were used for the identification by LC-MS/MS determination in addition to urine samples. For compound AR, it was identified by LC-MS/MS analysis after compound B was metabolized by hepatocytes.
When the metabolites were synthesized and characterized by NMR, or the authentic compounds were purchased, MS/MS spectra of those compounds were determined using an ABI3000 mass spectrometer (Applied Biosystems, CA, USA) and ion-pairs were selected for multiple reactions monitoring (MRM) analysis of those compounds. For FAEG and RFAEG, two abundant fragments were determined by MS/MS spectra of plasma samples, which apparently come from FA/RFA and its ligand glucuronide. MRM with transitions mass-to-charge ratio (m/z) of 317.0/230.9 for FA, 319.2/233.1 for RFA, 233.1/215.0 for compound AR, 426.3/192.2 for RFAT, and 424.2/192.2 for FAT were used for identification. Double ion-pairs 493.5/317.1 and 493.5/175.4 for FAEG, and 495.5/319.1 and 495.4/175.4 for RFAEG were used for the identification because no pure authentic compounds were synthesized. All the above detections were performed in negative MRM mode. Transitions m/z 275.3/233.1 for compound B and 333.3/233.1 for compound X were used in positive detection mode.
Both the urine samples and plasma samples were diluted 1:3 with methanol for LC-MS/MS identification. For LC-MS/MS comparison, the gradient was applied as follows: 50% B (methanol containing 0.1% formic acid) for 0.3 min, then a linear gradient to 100% B for 2.2 min and held for 3.0 min, lastly dropped to 50% within 0.1 min, and then equilibrated for 0.2 min. LC-MS/MS analysis of those newly found metabolites was performed on an ABI3000 mass spectrometer coupled with a Shimadzu 10A LC pump (Shimadzu, Japan) and MPS3C autosampler (Gerstel, Germany), which were controlled by Analyst 1.4.2 workstation software. The compounds were chromatographed on a Capcell Pak C18 column (5 µm, 2. 0 mm ID* 50 mm) at room temperature with a flow rate of 0.2 ml min−1. The mass spectrometer was operated in MRM mode using a turbo ion spray where 350°C and 4500 V were applied to the spray needle. Nitrogen was used as nebulizer gas, curtain gas and collision gas. All the raw data were processed using the Analyst software 1.4.2.
Metabolites and metabolism pathways in rat hepatocytes
Male Sprague–Dawley rats (200–300 g) were used for the preparation of isolated hepatocytes. For the primary culture of rat hepatocyte, cells were isolated by a two-step in-situ perfusion technique as previously reported with minor modifications. Aseptic procedures were followed throughout the isolation process. The viability of isolated hepatocytes preparations was ≥ 85% determined by trypan blue dye exclusion.
A total of 2 × 106 viable hepatocytes were seeded in 24-well culture plates precoated with collagen. DMEM containing 10% heat-inactivated FBS, 580 mg l−1 glutamine, 10−6 M insulin, 10−5 M dexamethasone, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin were used as the culture medium. The incubation was carried out under an atmosphere of 95%O2/5% CO2 at 37°C in a humidified incubator. Medium replacement was performed 24 h after seeding when good adherence of hepatocytes was reached.
Compounds fenofibrate, A, AR, B, X, FA, RFA, FAT, and RFAT were dissolved in DMSO in 1 mg ml−1 and 10 µl of each were added to the monolayer with the final concentration of about 10 µg ml−1 24 h after cell seeding and medium replacement. After metabolism for 24 h, the medium was collected and mixed with 2 vols of iced acetonitrile. For control wells, 10 µl of each compound were added after the medium was mixed with 2 vols of iced acetonitrile. All these samples were centrifuged at 16 000g 4°C for 30 min to remove particulates and proteins. A total of 10 µl of the supernatant were subjected to LC-MS/MS analysis for the detection of all these compounds with the same method as metabolite identification. Result data from the hepatocytes of three rats (including male and female) were combined for metabolism pathway conclusion.
Results
Synthesis of fenofibrate metabolites
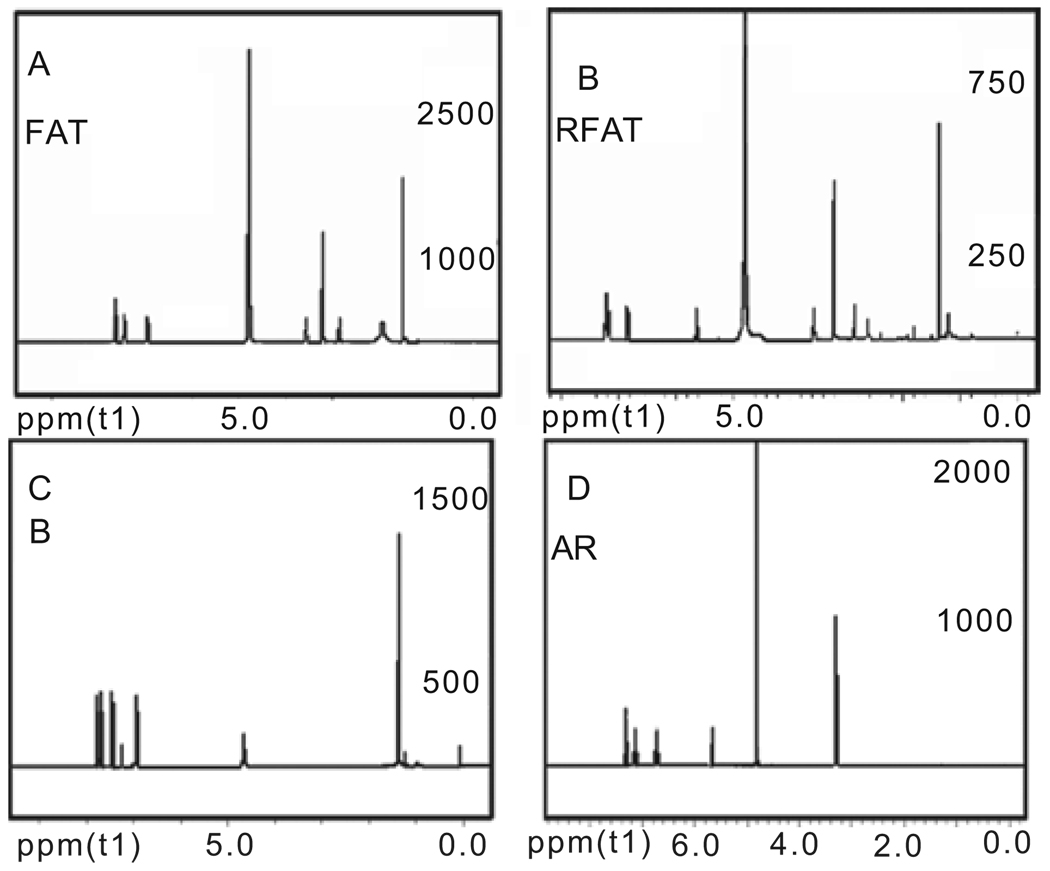
After synthesis as given in the Materials and Methods section, the NMR spectra for compounds reduced fenofibric acid (RFA), compound A reduced (AR), fenofibric acid taurine (FAT), reduced fenofibric acid taurine (RFAT), 4-chloro-4´-isopropox benzophenone (compound B), and 2-[4-(4-chloro-benzoyl)-phenoxy]-2-methyl-propionic acid methyl ester (compound X) were acquired. The purity of these chemicals by MS validation was ≥ 95% according to the proton NMR analyses (NMR spectra of FAT, RFAT, compound B and AR shown in Figure 1, and that of RFA and compound X not shown).
Figure 1.
Nuclear magnetic resonance (NMR) spectra of synthesized FAT, RFAT, compound B, and AR. FAT: 1H-NMR (400 MHz, MeOD) δ1.530 (s, 6H), δ2.825–2.858 (t, 2H), δ3.530–3.575 (m, 2H), δ6.939–6.960 (d, 2H), δ7.432–7.453 (d, 2H), δ7.629–7.667 (m, 4H). RFAT: 1H-NMR (400 MHz, MeOD) δ1.362 (s, 6H), δ2.843–2.876 (t, 2H), δ3.542–3.575 (t, 2H), δ5.623 (s, 1H), δ6.814–6.835 (d, 2H), δ7.148–7.253 (m, 6H). Compound B: 1H-NMR (400 MHz, CDCl3) δ1.371–1.386 (d, 6H), δ4.632–4.692 (m, 1H), δ6.918–6.940 (d, 2H), δ7.432–7.453 (d, 2H), δ7.694–7.784 (m, 4H). Compound A: 1H-NMR (400 MHz, MeOD) δ5.666 (s, 1H), δ6.710–6.732 (d, 2H), δ7.119–7.140 (d, 2H), δ7.269–7.307 (m, 4H).
MDA of data matrices
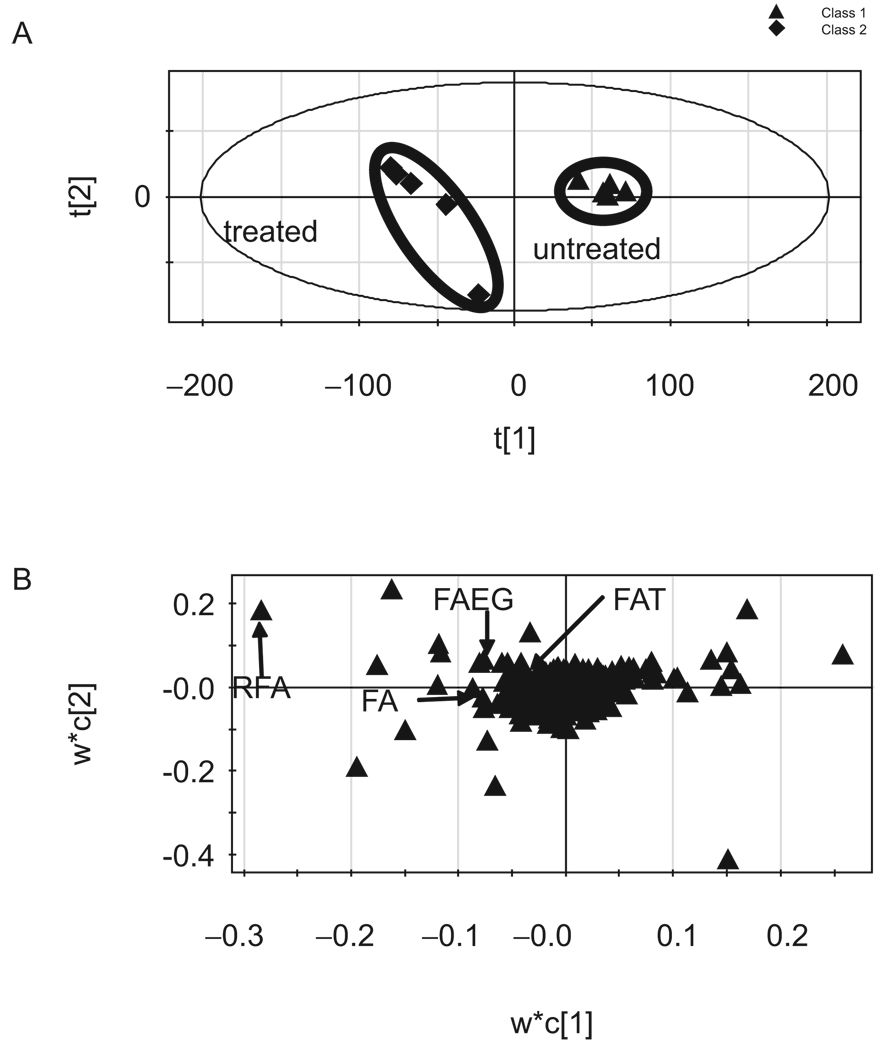
After UPLC-ESI-QTOF-MS analysis of the urine samples and pretreatment of the raw information by MarkerLynx, PLS-DA coupled with Pareto-scaling was used to profile the metabolome changes in negative data matrix (R2X(cum) = 0.668, R2 (cum) = 0.984, and Q2(cum) = 0.832 for the first two components). Data before and after fenofibrate treatment were respectively clustered together by score plot (Figure 2A). The loading plots revealed those variables that contributed most to group separation (Figure 2B). Similarly, the positive mode data were used to produce the score plot and loading plot (R2X(cum) = 0.623, R2Y(cum) = 0.993 and Q2(cum) = 0.916 for the first two components; data not shown). It can be seen that the two clusters were discriminated along the first component for both negative- and positive-score plots and the treated samples located on the left. Thus, the ions that deviated from the cloud of ions on the left of the first component in the loading plot represented those changed urine metabolome, which included elevated biomarkers and fenofibrate-related metabolites (Figure 2).
Figure 2.
Score plot and loading plot of urine samples after MDA by SIMCA-P+11. Pareto-scaling and supervised PLS-DA were used for data pretreatment and pattern recognition (R2X(cum) = 0.668, R2Y(cum) = 0.984, and Q2(cum) = 0.832). The score plot indicated the two clusters of the treated and untreated groups, and the first component separated the two clusters. The loading plot indicated the variables along the first component which contributed for grouping. Here, R2 was the fraction of sum of squares of all the X’s or Y’s explained by the current component, and Q2 was the fraction of the total variation of X’s that can be predicted by a component determined by cross-validation.
Selection of suspected metabolite signals
When best-score plots and loading plots were reached, the variation lists were created by ascending the p-value of the first component and lists of the first 50 variations were exported for signal selection. Metabolite-related variations should be zero or almost zero in the blank observations and significantly positive in the fenofibrate-treated observations. By examining the structure similarities, signals had almost the same retention time (RT), but related mass data would be produced because of in-source fragmentation. Isotope-related signals have the same RT and the mass data accord with those calculated molecular weights. Under these assumptions, many signals were excluded and only four variables were selected as metabolite-related information which might come from FA, RFA, FAT and FAEG (Table 1). By the same approach, the positive data matrix was processed, but no new metabolites variables other than those above the four variable-related signals were indicated.
Table 1.
Variables of identified metabolites and their responses extracted from negative data matrix from ultra-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOF-MS).
| Retention time (min) | m/z detected | Rank | Blank | Dosage | Proposed metabolites |
|---|---|---|---|---|---|
| 7.4749 | 319.075 | 1 | 0 ± 0 | 713.4 ± 418.7 | Reduced fenofibric acid (RFA) |
| 8.3111 | 317.061 | 11 | 0 ± 0 | 63.9 ± 57.3 | Fenofibric acid (FA) |
| 7.0779 | 493.12 | 12 | 0 ± 0 | 58.6 ± 46.8 | Fenofibric acid ester glucuronide (FAEG) |
| 6.4846 | 424.077 | 46 | 0 ± 0 | 14.3 ± 8.9 | Fenofibric acid taurine (FAT) |
Responses are expressed as mean ± standard deviation (SD) (n = 5).
Compound X was reported as a metabolite in rat hepatocytes. The authors suspected it might be a metabolite in vivo. For compounds AR, B, RFAT and RFAEG, they were not revealed by this metabolomics approach. Since compounds B and AR were found to induce peroxisomal palmitoyl-CoA oxidation activity significantly, it will be interesting to see if they are in vivo metabolites. Considering that FA, FAT, FAEG and RFA were identified in urine and that RFAEG was not found in urine by UPLC-ESI-QTOF-MS, it is reasonable to suspect RFAT as a metabolite. Hence, for these compounds that were not indicated by UPLC-ESI-QTOF-MS, LC-MS/MS analysis of urine samples, plasma samples and hepatocyte incubation medium were performed for their identification (Table 2).
Table 2.
Metabolites identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of urine, plasma and hepatocytes.
| Metabolites | Mode | Urine (UPLC-ESI-QTOF-MS) |
Urine (LC-MS/MS) |
Plasma (LC-MS/MS) |
Hepatocyte (LC-MS/MS) |
|---|---|---|---|---|---|
| Reduced fenofibric acid taurine (RFAT) | [M − H]− | n.d. | Low | Yes | Yes |
| Reduced fenofibric acid glucuronide (RFAEG) | [M − H]− | n.d. | Low | Yes | Yes |
| Compound X | [M + H]+ | n.d. | Low | Yes | Yes |
| Compound B | [M + H]+ | n.d. | n.d. | Yes | Yes |
| Compound AR | [M − H]− | n.d. | n.d. | n.d. | Yes |
n.d., Not detected because of low abundance or interference. For low abundance, RFAT, RFAEG, and compound X in urine were not detected or listed as significant variables contributing to grouping. However, they could be identified by LC-MS/MS in both urine and plasma. Compound B was not detected in urine even by LC-MS/MS but was found in plasma and hepatocyte. Compound AR could only be detected in hepatocyte by an in vitro approach. UPLC-ESI-QTOF-MS, ultra-performance liquid chromatography-electrospray ionization quadrupole time-of-flight mass spectrometry.
Identification of suspected metabolites
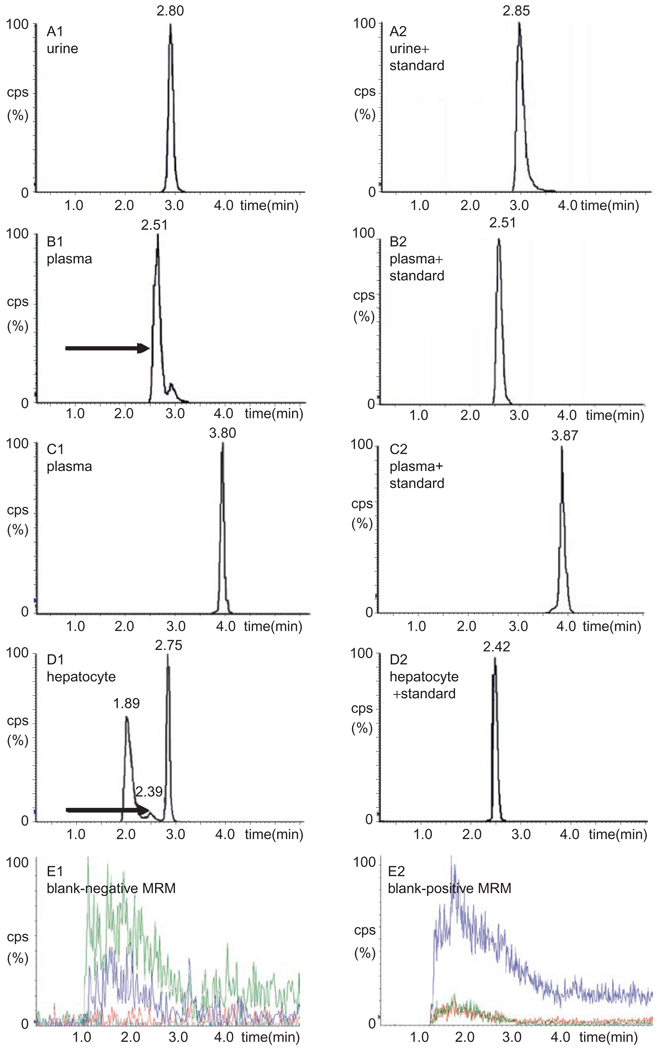
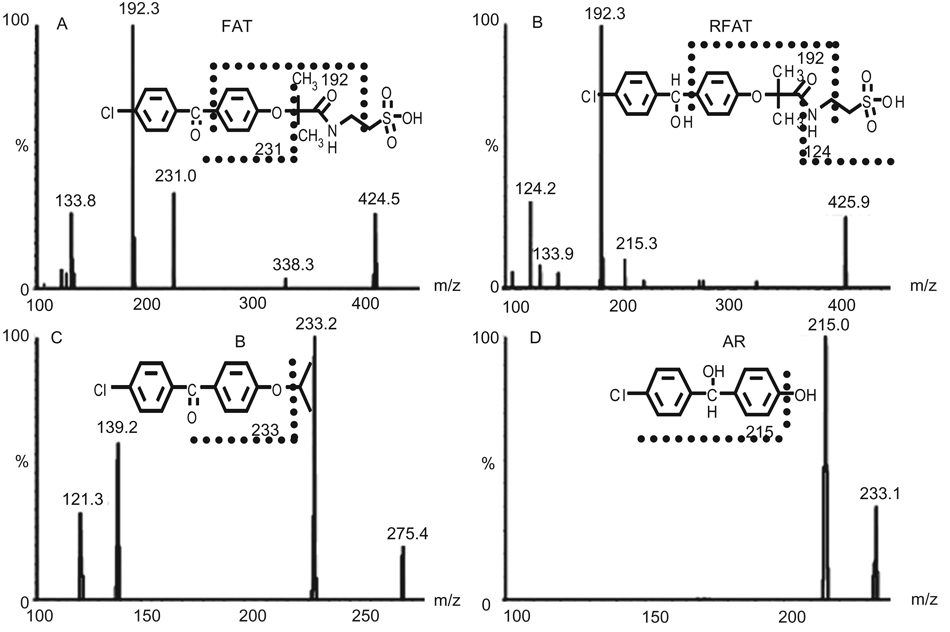
LC-MS/MS analysis of urine samples identified FA, RFA, FAT and FAEG as metabolites of fenofibrate. Their RT were identical with authentic compounds, and for FAEG, double ion-pairs were used for comparison. In addition, their abundances were very high in urine (data for FAT in urine samples are shown in Figure 3A and its fragmentation in Figure 4; data for other ever reported metabolites in rodent are not shown).
Figure 3.
LC-MS/MS chromatographs of metabolites of fenofibrate newly identified in Sprague–Dawley rats. A1: FAT in urine by UPLC-ESI-QTOF-MS coupled with LC-MS/MS. B1: RFAT in plasma by LC-MS/MS. C1: compound B in plasma by LC-MS/MS. D1: compound AR in hepatocytes by LC-MS/MS. A2, B2, C2 and D2 were blank matrix spiked with standard compounds, respectively. E1 and E2 were blank plasma detected in negative and positive MRM.
Figure 4.
MS/MS spectra and proposed fragmentation patterns of the four newly found metabolites. A: FAT; B: RFAT; C: compound B; and D: compound AR.
For compounds X, RFAT and RFAEG, they were not listed as discriminate variables by UPLC-ESI-QTOF-MS analysis in either a negative or a positive data matrix. Even they could not be found by examining the raw data because of their low abundance in urine samples and limited power of trace detection of QTOF-MS analysis. However, LC-MS/MS analysis could identify them as metabolites in both urine and plasma samples and their abundance were really lower in urine. For compound B, both UPLC-ESI-QTOF-MS and LC-MS/MS analysis of urine samples could not find the information. By LC-MS/MS analysis of plasma samples, it was finally identified as a metabolite, even though its abundance was very low (data for RFAT and compound B in plasma samples are shown in Figure 3B and 3C, their fragmentation is shown in Figure 4; data for other ever reported metabolites in rodent are not shown).
For compound AR, fragmentation from structure-related metabolites was high, which might mask its low abundance. Thus, both urine and plasma samples by UPLC-ESI-QTOF-MS and LC-MS/MS provided no information. It was finally identified by LC-MS/MS analysis of the hepatocyte metabolism system. Metabolism of compound B by hepatocytes produced metabolite AR, but no other pathways could be confirmed because of interference from severe in-source fragmentation (data for compound AR by hepatocyte metabolism are shown in Figure 3D and its fragmentation is shown in Figure 4).
Metabolism pathway by hepatocyte
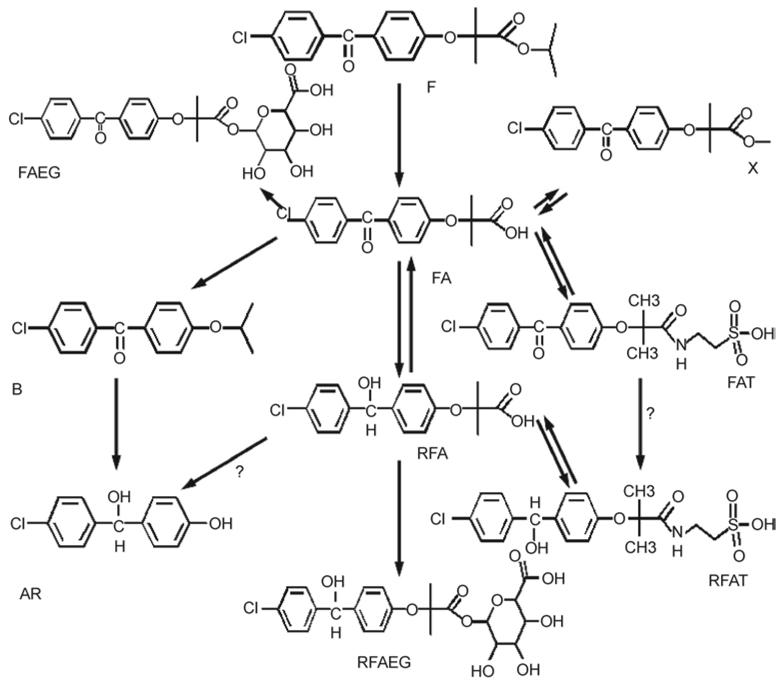
Metabolism of fenofibrate, compounds AR, B, X, FA, RFA, FAT, RFAT in rat hepatocytes, and LC-MS/MS detection of those above metabolites revealed the metabolism relationship of these compounds (Table 3). Combining data by the metabolomics analysis of urine samples, LC-MS/MS analysis of plasma samples and the metabolism relationship by rat hepatocytes, the metabolism map of fenofibrate is proposed as shown in Figure 5.
Table 3.
Metabolism results from in vitro rat hepatocytes.
| Multiple reactions monitoring (MRM) positive | MRM negative | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrates | FA | RFA | Compound AR | FAT | RFAT | FAEG | RFAEG | Compound B | Compound X |
| Fenofibrate | + | + | ? | + | + | − | − | − | − |
| FA | S | + | ? | + | + | + | + | + | − |
| RFA | + | S | ? | + | + | − | + | − | − |
| Compound AR | − | − | S | − | − | − | − | − | − |
| FAT | + | + | ? | S | + | − | − | − | − |
| RFAT | + | + | − | − | S | − | − | − | − |
| Compound B | − | − | + | − | − | − | − | S | − |
| Compound X | + | + | − | + | + | + | − | + | S |
After each substrate was added to cultured hepatocytes and incubated for the indicated time, supernatant was subjected to MRM detection in positive and negative modes, respectively, by LC-MS/MS. +, Detected positive by LC-MS/MS; −, detected negative by LC-MS/MS. A question mark means detection was masked or interfered.
FA, fenofibric acid; FAEG, fenofibric acid ester glucuronide; FAT, fenofibric acid taurine; RFA, reduced fenofibric acid; RFAEG, reduced fenofibric acid glucuronide; RFAT, reduced fenofibric acid taurine.
Figure 5.
Proposed metabolism map of fenofibrate in Sprague–Dawley rats by combining the metabolites ever reported and newly identified in this study. These metabolism pathways were validated by in vitro metabolism using rat hepatocytes from three individuals. A question mark indicates that the pathway was not confirmed.
Discussion
Fenofibrate has been reported to produce hepatocarcinogenesis and peroxisome proliferation in some rodents, but not in human and non-human primates (Hoivik et al. 2004; Yang et al. 2007). PPARa was found to mediate hepatocarcinogenesis of xenobiotics (Gonzalez and Shah 2008), and activation and proliferation of hepatic non-parenchymal cells were involved in peroxisome proliferators-induced hepatocarcinogenesis (Peters et al. 2000). In rat skeletal muscle cultures, PPARa agonism was found to mediate in part the myotoxicity response to PPARa agonists (Johnson et al. 2005). Fenofibrate also induces mitochondrial dysfunction by inhibition of complex I of the respiratory chain in homogenates of rat skeletal muscle (Brunmair et al. 2004). Although clinically used for more than 30 years, metabolism of fenofibrate has not been fully researched. Exploration of new metabolites of fenofibrate in rodent and primate may help us understand those safety issues.
FA, RFA, FAEG, and RFAEG were identified as metabolites of fenofibrate in human volunteers, dog, rat, and guinea pig (Weil et al. 1988, 1990). In rat hepatocytes, compound X was found to be another metabolite (Cornu-Chagnon et al. 1995). In this study, by UPLC-ESI-QTOF-MS-based metabolomics coupled with LC-MS/MS analysis, all those metabolites ever reported were identified in rats. More importantly, because taurine conjugate of fenofibric acid was found in this metabolomics approach according to the molecular weight, a tauring conjugation pathway of fenofibrate was suspected. By synthesis of authentic compounds, FAT and RFAT were finally identified by LC-MS/MS. By in vitro metabolism of structure-related compounds in rat hepatocytes, the metabolism pathway of fenofibrate was better understood and several reversible processes were revealed.
The metabolism of clofibrate, another PPARα agonist, indicated significant species differences as indicated. It was found that taurine conjugation of clofibrate occurred in dog, cat, and ferret, but not in human volunteers, rodents, and rabbits (Cayen et al. 1977; Emudianughe et al. 1983). In this study, taurine conjugates of FA and RFA were identified as metabolites of fenofibrate by comparison of RT and fragmentation profiles with synthesized authentic compounds. It seemed different from the metabolism of clofibrate where taurine conjugated metabolites were not in rodent. However, in this study, RFAT and RFAEG, which were phase II conjugates of RFA, were much lower (in QTOF-MS response) than their unreduced forms in rats. They were detected in rat urine by LC-MS/MS analysis of urine and plasma, but not by the UPLC-ESI-QTOF-MS approach. For compound B, its abundance was too low to be detected in urine even by LC-MS/MS analysis, but it was identified in plasma. These results may be due to species-related difference of metabolism in terms of quantity coupled with a difference of excretion. The contribution of this kind of metabolism difference to the safety problems, especially those indicating species differences, remains for investigation.
Recent advances in mass spectrometric techniques coupled with UPLC have resulted in the substantial development of robust methods for low molecular mass organic molecules in complex biological matrices (Chen et al. 2007). Time-of-flight mass spectrometry (TOF-MS) can deduce the empirical formulae of metabolite candidates with an excellent mass accuracy. UPLC-ESI-QTOF-MS-based metabolomics has been used for the investigation of new metabolites and unknown biomarkers (Lenz et al. 2007; Zhen et al. 2007). Because of different applications and their original design, LC-MS/MS are more sensitive and selective for a known or proposed compound than QTOF-MS. However, in this study the power of this platform was limited when detecting metabolites of low abundance. Synthesis of authentic compounds coupled with LC-MS/MS determination is a sensitive supplementary approach for the identification of metabolites of trace amount. Due to the excretion property of an individual compound, the concentration in plasma may be higher than that in urine, which makes it possible for metabolite identification. The concentrations of substrates in in vitro metabolism by hepatocytes can reach higher levels than those of the in vivo study via absorption and the detective background is usually low. This is helpful for metabolite identification and metabolism pathway research. The identification of metabolite AR was an example that could only be found in hepatocyte metabolism because of low abundance in plasma and urine.
Conclusively, in this study the metabolism of fenofibrate was better understood by in vivo and in vitro metabolism approaches. Nine metabolites of fenofibrate were identified by UPLC-ESI-QTOF-MS-based metabonomics coupled with LC-MS/MS, of which four were found to be new in Sprague–Dawley rats. Among these new metabolites, FAT and RFAT indicate a new phase II conjugation pathway of fenofibrate, which may shed light on the investigation of species-related safety issues.
Acknowledgement
This paper includes a word that is, or is asserted to be, a proprietary term or trade mark. Its inclusion does not imply it has acquired for legal purposes a non-proprietary or general significance, nor is any other judgement implied concerning its legal status.
Footnotes
Declaration of interest: The authors report no conflicts of interest.
References
- Blane GF. Comparative toxicity and safety profile of fenofibrate and other fibric acid derivatives. Am J Med. 1987;83(5B):26–36. doi: 10.1016/0002-9343(87)90868-0. [DOI] [PubMed] [Google Scholar]
- Bocher V, Chinetti G, Fruchart JC, Staels B. Role of the peroxisome proliferator-activated receptors (PPARS) in the regulation of lipids and inflammation control. J Soc Biol. 2002;196(1):47–52. [PubMed] [Google Scholar]
- Brunmair B, Lest A, Staniek K, Gras F, Scharf N, Roden M, Nohl H, Waldhausl W, Furnsinn C. Fenofibrate impairs rat mitochondrial function by inhibition of respiratory complex I. J Pharmacol Exp Ther. 2004;311(1):109–114. doi: 10.1124/jpet.104.068312. [DOI] [PubMed] [Google Scholar]
- Cayen MN, Ferdinandi ES, Greselin E, Robinson WT, Dvornik D. Clofibrate and clofibric acid: comparison of the metabolic disposition in rats and dogs. J Pharmacol Exp Ther. 1977;200(1):33–43. [PubMed] [Google Scholar]
- Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39(2–3):581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu-Chagnon MC, Dupont H, Edgar A. Fenofibrate: metabolism and species differences for peroxisome proliferation in cultured hepatocytes. Fundam Appl Toxicol. 1995;26(1):63–74. doi: 10.1006/faat.1995.1075. [DOI] [PubMed] [Google Scholar]
- Emudianughe TS, Caldwell J, Sinclair KA, Smith RL. Species differences in the metabolic conjugation of clofibric acid and clofibrate in laboratory animals and man. Drug Metab Disposit Biol Fate Chem. 1983;11(2):97–102. [PubMed] [Google Scholar]
- Fruchart JC, Duriez P. Mode of action of fibrates in the regulation of triglyceride and HDL-cholesterol metabolism. Drugs Today (Barc) 2006;42(1):39–64. doi: 10.1358/dot.2006.42.1.963528. [DOI] [PubMed] [Google Scholar]
- Ghosh B, Sengupta S, Bhattacharjee B, Majumder A, Sarkar SB. Fenofibrate-induced myopathy. Neurol India. 2004;52(2):268–269. [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YA. PPAR alpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246(1):2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Hoivik DJ, Qualls CW, Jr, Mirabile RC, Cariello NF, Kimbrough CL, Colton HM, Anderson SP, Santostefano MJ, Morgan RJ, Dahl RR, et al. Fibrates induce hepatic peroxisome and mitochondrial proliferation without overt evidence of cellular proliferation and oxidative stress in cynomolgus monkeys. Carcinogenesis. 2004;25(9):1757–1769. doi: 10.1093/carcin/bgh182. [DOI] [PubMed] [Google Scholar]
- Holoshitz N, Alsheikh-Ali AA, Karas RH. Relative safety of gemfibrozil and fenofibrate in the absence of concomitant cerivastatin use. Am J Cardiol. 2008;101(1):95–97. doi: 10.1016/j.amjcard.2007.07.057. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Zhang X, Shi S, Umbenhauer DR. Statins and PPARalpha agonists induce myotoxicity in differentiated rat skeletal muscle cultures but do not exhibit synergy with co-treatment. Toxicol Appl Pharmacol. 2005;208(3):210–221. doi: 10.1016/j.taap.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Babich MA, Baetcke KP, Cook JC, Corton JC, David RM, DeLuca JG, Lai DY, McKee RH, Peters JM, et al. PPARalpha agonist-induced rodent tumors: modes of action and human relevance. Crit Rev Toxicol. 2003;33(6):655–780. doi: 10.1080/713608372. [DOI] [PubMed] [Google Scholar]
- Lenz EM, Williams RE, Sidaway J, Smith BW, Plumb RS, Johnson KA, Rainville P, Shockcor J, Stumpf CL, Granger JH, et al. The application of microbore UPLC/oa-TOF-MS and 1H-NMR spectroscopy to the metabonomic analysis of rat urine following the intravenous administration of pravastatin. J Pharmaceut Biomed Anal. 2007;44(4):845–852. doi: 10.1016/j.jpba.2007.04.035. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Holmes E, Nicholson JK. Metabonomics: systems biology in pharmaceutical research and development. Curr Opin Mol Ther. 2004;6(3):265–272. [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Peters JM, Rusyn I, Rose ML, Gonzalez FJ, Thurman RG. Peroxisome proliferator-activated receptor alpha is restricted to hepatic parenchymal cells, not Kupffer cells: implications for the mechanism of action of peroxisome proliferators in hepatocarcinogenesis. Carcinogenesis. 2000;21(4):823–826. doi: 10.1093/carcin/21.4.823. [DOI] [PubMed] [Google Scholar]
- Qu S, Su D, Altomonte J, Kamagate A, He J, Perdomo G, Tse T, Jiang Y, Dong HH. PPAR{alpha} mediates the hypolipidemic action of fibrates by antagonizing FoxO1. Am J Physiol. 2007;292(2):E421–E434. doi: 10.1152/ajpendo.00157.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Reddy JK. Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis. 1987;8(5):631–636. doi: 10.1093/carcin/8.5.631. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Azarnoff DL, Hignite CE. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature. 1980;283(5745):397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- Ritter JL, Nabulsi S. Fenofibrate-induced elevation in serum creatinine. Pharmacotherapy. 2001;21(9):1145–1149. doi: 10.1592/phco.21.13.1145.34623. [DOI] [PubMed] [Google Scholar]
- Ward JM, Peters JM, Perella CM, Gonzalez FJ. Receptor and nonreceptor-mediated organ-specific toxicity of di(2-ethylhexyl) phthalate (DEHP) in peroxisome proliferator-activated receptor alpha-null mice. Toxicol Pathol. 1998;26(2):240–246. doi: 10.1177/019262339802600208. [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Morgenthal K. Metabolomics: from pattern recognition to biological interpretation. Drug Disc Today. 2005;10(22):1551–1558. doi: 10.1016/S1359-6446(05)03609-3. [DOI] [PubMed] [Google Scholar]
- Weil A, Caldwell J, Strolin-Benedetti M. The metabolism and disposition of fenofibrate in rat, guinea pig, and dog. Drug Metab Disp Biol Fate Chem. 1988;16(2):302–309. [PubMed] [Google Scholar]
- Weil A, Caldwell J, Strolin-Benedetti M. The metabolism and disposition of 14C-fenofibrate in human volunteers. Drug Metab Disp Biol Fate Chem. 1990;18(1):115–120. [PubMed] [Google Scholar]
- Yang Q, Ito S, Gonzalez FJ. Hepatocyte-restricted constitutive activation of PPAR alpha induces hepatoproliferation but not hepatocarcinogenesis. Carcinogenesis. 2007;28(6):1171–1177. doi: 10.1093/carcin/bgm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Krausz KW, Chen C, Idle JR, Gonzalez FJ. Metabolomic and genetic analysis of biomarkers for peroxisome proliferator-activated receptor alpha expression and activation. Mol Endocrinol (Balt) 2007;21(9):2136–2151. doi: 10.1210/me.2007-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]