Abstract
Several mutations that cause severe forms of the human disease autosomal dominant retinitis pigmentosa cluster in the C-terminal region of rhodopsin. Recent studies have implicated the C-terminal domain of rhodopsin in its trafficking on specialized post-Golgi membranes to the rod outer segment of the photoreceptor cell. Here we used synthetic peptides as competitive inhibitors of rhodopsin trafficking in the frog retinal cell-free system to delineate the potential regulatory sequence within the C terminus of rhodopsin and model the effects of severe retinitis pigmentosa alleles on rhodopsin sorting. The rhodopsin C-terminal sequence QVS(A)PA is highly conserved among different species. Peptides that correspond to the C terminus of bovine (amino acids 324–348) and frog (amino acids 330–354) rhodopsin inhibited post-Golgi trafficking by 50% and 60%, respectively, and arrested newly synthesized rhodopsin in the trans-Golgi network. Peptides corresponding to the cytoplasmic loops of rhodopsin and other control peptides had no effect. When three naturally occurring mutations: Q344ter (lacking the last five amino acids QVAPA), V345M, and P347S were introduced into the frog C-terminal peptide, the inhibitory activity of the peptides was no longer detectable. These observations suggest that the amino acids QVS(A)PA comprise a signal that is recognized by specific factors in the trans-Golgi network. A lack of recognition of this sequence, because of mutations in the last five amino acids causing autosomal dominant retinitis pigmentosa, most likely results in abnormal post-Golgi membrane formation and in an aberrant subcellular localization of rhodopsin.
Rhodopsin and the associated proteins and lipids are delivered by polarized sorting on post-Golgi membranes to the rod outer segment (ROS), a specialized domain of retinal photoreceptor cells (1–4). Once properly localized in ROS, rhodopsin and other factors participate in a cascade of signaling interactions triggered by light (5). Maintaining polarized organization throughout the continuous ROS renewal, which results in the addition of up to 3 μm2/min of membrane (6), is of paramount importance for the health of photoreceptor cells. In some instances, loss of cell polarity heralds the onset of retinal degeneration and blindness (7).
Retinitis pigmentosa encompasses a heterogeneous group of retinal degenerations that are characterized by similar clinical findings but are caused by mutations in over 50 different genes (8). Mutations found in the rhodopsin gene account for 10% of all cases and represent the most common known cause of retinitis pigmentosa (9). More than 70 mutations cause the autosomal dominant form of the disease (ADRP) (10). Although the intracellular fate of mutant rhodopsin is presently unclear, expression of mutant alleles appears to trigger interactions that lead to pathological changes in the photoreceptor cells. The ultimate response is the initiation of an apoptotic cascade that leads to cell death and retinal degeneration (11, 12).
Histopathological findings in patient specimens (7) and studies with transgenic animals (10, 13) suggest that some forms of ADRP result in altered delivery of rhodopsin to ROS. We have been studying rhodopsin trafficking and membrane delivery to ROS at the subcellular level. Recently we reported the characterization of post-Golgi membranes that carry rhodopsin to ROS (1–3, 14–18). A frog retinal cell-free system has been established to study rhodopsin trafficking through various biosynthetic compartments in the photoreceptor cells (2). This system supports formation of rhodopsin-bearing post-Golgi membranes in an ATP-, GTP-, and cytosol-dependent manner (2). After pulse labeling in vivo, the kinetics of movement of radiolabeled rhodopsin through the subcellular compartments can be monitored after equilibrium centrifugation on sucrose density gradients. Because of their unique low buoyant density, rhodopsin-bearing post-Golgi membranes can be enriched >85% and separated away from the Golgi and the trans-Golgi network (TGN) (1). The rhodopsin-bearing post-Golgi membrane-carriers formed in vitro have been found to be indistinguishable from those formed in vivo by several criteria including a distinct profile of membrane-associated small GTPases of the rab family (2).
Here we applied this system to assess the effect of rhodopsin mutations found in ADRP on its sorting and trafficking. Particularly severe forms of ADRP are caused by mutations that cluster within the five C-terminal amino acids of rhodopsin (19, 20). Transgenic animals carrying these mutations show defects in rhodopsin delivery to ROS (10, 13). The C-terminal sequence of rhodopsin QVS(A)PA is highly conserved among different species, and frog and human rhodopsin display an 85% identity and a 95% overall similarity at the amino acid level (21). Furthermore, the amino acids that are mutated in ADRP are conserved in the frog sequence. These amino acids have not been implicated in any light-dependent function (13, 22), and it has been hypothesized that they may participate in rhodopsin sorting (2, 10, 13). We previously have reported that a mAb, whose antigenic site is within nine C-terminal amino acids of rhodopsin, inhibits its post-Golgi trafficking in our cell-free system (2). In this work we investigated the potential targeting function of the C-terminal amino acids of rhodopsin. We mapped the domain that directs intracellular trafficking by using synthetic peptides corresponding to the C termini of the frog and mammalian rhodopsins as competitive inhibitors of rhodopsin trafficking in the cell-free system. We report that the five C-terminal residues QVS(A)PA play a critical role in regulating rhodopsin sorting into specific post-Golgi membranes at the level of TGN. These observations indicate that the defective recognition of mutant rhodopsin in patients with ADRP may result specifically in abnormal post-Golgi membrane formation, or mistargeting of mutant rhodopsin.
MATERIALS AND METHODS
Southern leopard frogs, Rana berlandieri, (100–250 g) purchased from Rana (Brownsville, TX), were maintained in a 12-h light/dark cycle. [35S]-Express protein labeling mixture (1,000 Ci/mmol), was from DuPont/NEN. ATP, creatine phosphate, creatine phosphokinase (800 units/mg), and hexokinase (450 units/mg) were from Boehringer Mannheim.
Peptide Synthesis.
Peptides corresponding to the frog (21) and bovine rhodopsin sequence (23, 24) were synthesized by solid phase technique using fluorenylmethoxycarbonyl derivatives of amino acids with an automatic synthesizer (model 431A, Applied Biosystems). After deprotection, peptides were purified by preparative reverse-phase liquid chromatography on a 2.5 × 25 cm Partisil-10 ODS-3 column (Whatman). The following peptides were tested: bovine amino acids 3–14, 63–75, 141–153, 234–245, 231–252, 310–321, 324–348, 330–348, and 331–342, and frog amino acids 330–354, 330–349, 330–354 V351M, and 330–354 P353S.
Pulse Labeling of Frog Retinas and Preparation of Photoreceptor-Enriched Postnuclear Supernatant (PNS).
All experiments were conducted under dim red light. Frogs were dark-adapted for 2 h before the experiment for two reasons: (i) isolation of the retina is facilitated by the retraction of pigment epithelium in the dark, and (ii) subcellular organelle distribution has been well established for the dark-adapted rhodopsin-containing membranes (1). Isolated frog retinas were incubated in oxygenated medium with [35S]-Express protein-labeling mixture (25 μCi/retina) at 22°C for 1 h. Retinal fractionation and preparation of PNS enriched in photoreceptor biosynthetic membranes was performed as described (2).
In Vitro Incubation of Photoreceptor-Enriched PNS.
Radiolabeled PNS was preincubated with synthetic peptides (dissolved in 50 μl of 10 mM Hepes-KOH, pH 7.0) for 30 min on ice. The standard assay for cell-free post-Golgi membrane formation was as follows: to 1 ml of PNS in 0.25 M sucrose (obtained from seven radiolabeled retinas) 100 μl of 10× concentrated buffer stock solution was added to give a final concentration of 25 mM Hepes-KOH, pH 7.0, 25 mM KCl, and 2.5 mM MgAc2. The assay was initiated by the addition of 50 μl of an ATP regenerating system and by transfer to 22°C (2). The assay was terminated by the addition of 0.2 M EDTA to a final concentration of 3 mM.
Upon completion of cell-free post-Golgi membrane formation, assay mixtures were overlaid on 10-ml linear 20–39% (wt/wt) sucrose gradients. After centrifugation at 28,000 rpm (100,000 gav) in an SW 40 rotor (Beckman) for 15 h at 4°C, fourteen 0.9-ml fractions were collected from the top of the gradient. Subcellular fractions were pooled according to the kinetics of their acquisition of radiolabeled-rhodopsin as described (2) as follows: pool 1 = fractions 1–3, 2 = 4–6, 3 = 7–8, 4 = 9–10, 5 = 11–12, and 6 = 13–14, diluted with 10 mM Tris acetate, pH 7.4 and centrifuged at 70,000 rpm (336,000 gav) for 30 min in a 70.1 Ti rotor (Beckman). Pellets were resuspended in 10 mM Tris acetate, pH 7.4 and aliquoted for analysis by SDS/PAGE.
Gel Electrophoresis and Quantitative Analysis of Radiolabeled Rhodopsin in Retinal Subcellular Fractions.
SDS/PAGE was performed as described (2). Dried SDS gels were subjected to quantitative analysis of 35S-labeled rhodopsin in retinal subcellular fractions in a PhosphorImager (Molecular Dynamics), while the images of the gels were generated by autoradiography at −85°C using Kodak BioMax MR film.
RESULTS
Retinal Cell-Free System for Rhodopsin Trafficking.
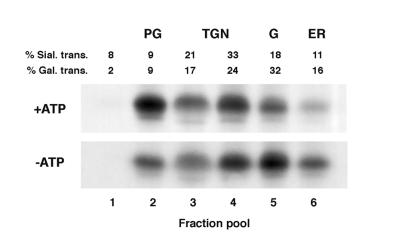
Rhodopsin-bearing post-Golgi membranes are formed after pulse labeling in vivo followed by in vitro chase in a frog retinal cell-free system derived from photoreceptor-enriched PNS in an ATP-, GTP- and cytosol-dependent manner (2). Fig. 1 illustrates the ATP-dependent transfer of radiolabeled rhodopsin to the post-Golgi fraction during incubation in the cell-free system. In this assay, isolated frog retinas first are pulse-labeled for 1 h, so that the bulk of the newly synthesized rhodopsin is localized in the Golgi. Therefore, the majority of the detectable radiolabeled rhodopsin corresponds to the Golgi form with trimmed oligosacharides. Golgi complex is identified by the galactosyltransferase activity of the trans-Golgi cisternae, as previously described (1). In the presence of ATP and an ATP-regenerating system, intracellular trafficking continues in vitro. As shown in Fig. 1 (+ATP), after 2-h incubation with ATP the Golgi and endoplasmic reticulum (ER) contents of radiolabeled rhodopsin were significantly reduced, TGN membranes became enriched in newly synthesized rhodopsin and post-Golgi membranes contained high levels of radiolabeled rhodopsin, consistent with the previously reported findings (2). In the control sample containing an ATP-depleting system, Fig. 1 (−ATP), radiolabeled rhodopsin did not redistribute to TGN and post-Golgi membranes, indicating a complete arrest of trafficking in the absence of ATP, consistent with the previously reported characterization of the system (2).
Figure 1.
Rhodopsin trafficking to the post-Golgi membranes in the retinal cell-free system. Isolated frog retinas were pulse-labeled for 60 min, and radiolabeled PNS was incubated for 2 h in the presence or absence of ATP. After sucrose gradient separation subcellular fractions were pooled according to the kinetics of their acquisition of newly synthesized rhodopsin as previously described (2). Aliquots corresponding to radiolabeled membrane proteins from two retinas were separated by SDS/PAGE and autoradiographed. In the absence of ATP (−ATP), rhodopsin remains in the Golgi (G) during in vitro chase, indicating that the assay is ATP dependent (2). Upon addition of ATP (+ATP), radiolabeled rhodopsin exits the Golgi and appears in the TGN and in low density post-Golgi fractions (PG). The distribution of Golgi and TGN membranes has been determined by their galactosyltransferase or sialyltransferase activities, respectively, (1, 2, 14). Activities of pooled membrane fractions are indicated. The distribution of transferase activities is independent of rhodopsin content and is not affected by post-Golgi membrane budding (2).
A Synthetic Peptide Corresponding to the C Terminal of Rhodopsin Inhibits Its Post-Golgi Trafficking in the Cell-Free System.
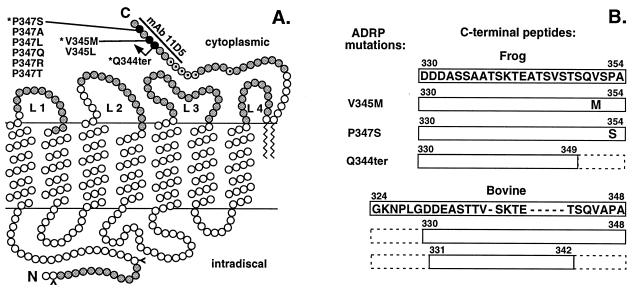
Post-Golgi trafficking in the frog retinal cell-free system is inhibited by anti-rhodopsin mAb 11D5, or its Fab fragments, and rhodopsin is arrested in the compartment identified by its sialyltransferase activity as the TGN (2). The antigenic epitope of inhibitory mAb 11D5 has been mapped within the nine amino acids comprising the C terminal of rhodopsin (1). Fig. 2 schematically represents the transmembrane model of rhodopsin showing the mAb 11D5 antigenic site and the locations of mutations in the C-terminal domain identified in patients with ADRP. Because the inhibitory anti-C-terminal antibody also may interfere with the neighboring regions, we wanted to test whether the C-terminal domain of rhodopsin contains the signal that regulates its trafficking. To study the role of the C-terminal domain we synthesized several peptides corresponding to the cytoplasmic or intradiscal domains of rhodopsin (as shown in Fig. 2) and tested their effect on rhodopsin trafficking in vitro.
Figure 2.
Model of rhodopsin showing locations of ADRP mutations in the C-terminal domain, the mAb 11D5 antigenic site, and locations and the sequences of the peptides used in this study. (A) Peptides synthesized for this study are highlighted in gray. L1-L4 are cytoplasmic loops. C-terminal amino acids mutated in retinitis pigmentosa are shown in black. Mutations modeled in this study are indicated with ∗. (B) Sequences of the C-terminal peptides from frog and bovine rhodopsin used in this study. Because frog rhodopsin contains two (one and five amino acids) insertions in this region, these are represented by dashes in the bovine peptide sequence and with the dotted open circles in the model.
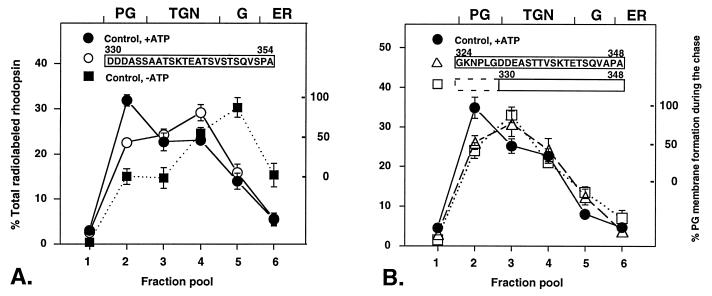
The orderly progression of radiolabeled rhodopsin through the biosynthetic compartment during the cell-free chase was significantly affected by the addition of the 50 μM frog C-terminal peptide (amino acids 330–354) (Fig. 3A). Newly synthesized rhodopsin was predominantly associated with the membranes identified by their sialyltransferase activity as the TGN, and its appearance in the post-Golgi membranes was reduced by 60%. This inhibition was concentration dependent as 25 μM peptide inhibited post-Golgi transport by 47% in the same series of experiments (data not shown). The distribution of radiolabeled rhodopsin closely resembled its previously described distribution in the presence of mAb 11D5 (2), suggesting a common mechanism for inhibition at the level of the TGN. This finding is significantly different from the previously reported rhodopsin distribution in the presence of rab GDI (2). The inhibition of trafficking by rab GDI is nearly identical to that in the absence of ATP and reflects an early and complete block of trafficking in all compartments during in vitro chase. In contrast, in the presence of rhodopsin C-terminal peptide some rhodopsin trafficking initially occurred, and approximately 25% of the radiolabeled rhodopsin redistributed from the ER and the Golgi at the end of the chase period. The rate of clearance from these compartments was comparable to the control with ATP. This finding suggests that rhodopsin trafficking at the level of the ER and the Golgi is unaffected by the C-terminal peptide and that its inhibitory effect is exhibited once rhodopsin reaches the TGN.
Figure 3.
Inhibition of post-Golgi rhodopsin trafficking with C-terminal synthetic peptides in the frog retinal cell-free system. (A) Radiolabeled frog retinal PNS was preincubated with the frog rhodopsin C-terminal peptide 330–354 before cell-free chase in the presence of ATP. The distribution of radiolabeled rhodopsin was measured in a PhosphorImager in nine (control, +ATP and peptide) or three (control, −ATP) separate experiments. The data are presented as the means ± SE In the control sample without peptide added (control, +ATP), in addition to the post-Golgi membranes already formed in the isolated retinas during the pulse (≈15%) (2), an additional ≈16% of total radiolabeled rhodopsin accumulates in the post-Golgi membrane fraction (fraction pool 2) during the cell-free chase. ATP depletion (−ATP) completely inhibits additional post-Golgi membrane formation during the chase (2). In the absence of ATP further trafficking does not occur, therefore the residual levels of radiolabeled rhodopsin in the post-Golgi fraction are caused by the trafficking that occurred in the living retinas, before the cell-free chase. ATP-dependent appearance of rhodopsin in post-Golgi membranes during the chase is inhibited by 60% in the presence of the 50 μM C-terminal peptide and rhodopsin is arrested in the TGN. Under these conditions C-terminal peptide is estimated to be in a 100-fold excess over rhodopsin. Inhibition of rhodopsin trafficking is assessed over the −ATP background; the ordinate on the right refers only to the ATP-dependent appearance of rhodopsin in the post-Golgi membrane fraction, when this background is subtracted. (B) The inhibition of post-Golgi trafficking (≈50%) and the distribution of radiolabeled rhodopsin was nearly identical for the two bovine peptides tested (a 25-mer and a 19-mer). The peptide corresponding to the C-terminal 19 amino acids of bovine rhodopsin fully retains the inhibitory effect on rhodopsin trafficking. The data are presented as the means ± SE of three separate experiments.
Post-Golgi Trafficking of Frog Rhodopsin Also Is Inhibited by Bovine C-Terminal Peptides.
Because the C-terminal domain is highly conserved among rhodopsin species (25) we tested the effect of the bovine peptide (amino acids 324–348) on rhodopsin trafficking in the frog retinal system. A comparison of the frog and bovine C-terminal sequence is shown in Fig. 2. At the same concentration (50 μM) the bovine peptide inhibited post-Golgi trafficking by ≈50% and radiolabeled-rhodopsin accumulated predominantly in the light TGN fraction (fraction pool 3) (Fig. 3B). This compartment is rich in small GTPase rab6 (it contains nearly 30% of the total membrane-bound rab6) but has a lower sialyltransferase activity than the TGN membranes that sediment in fraction pool 4 (see Fig. 1) (14).
A shorter bovine C-terminal peptide (amino acids 330–348) inhibited post-Golgi trafficking to a similar extent as the longer one, which suggests that amino acids 324–330 do not participate in this recognition (six of seven of these amino acids are identical in frog and bovine sequence with one conservative substitution). These amino acids are also absent in the frog C-terminal peptide that inhibits rhodopsin trafficking (see Fig. 2). The primary sequences of frog and bovine rhodopsin in the C-terminal domain are highly homologous, but frog rhodopsin contains two (one and five amino acids) insertions (21) (see Fig. 2). The three-dimensional structure of the bovine C-terminal domain has been determined by using NMR (26, 27). The comparison of frog and bovine sequence suggests that if the frog peptide assumes a similar conformation the conserved amino acids cluster on one face of this domain.
Peptides Corresponding to the Cytoplasmic Loops of Rhodopsin Have No Effect on Its Trafficking.
Because the C terminal comprises only a part of the cytoplasmic surface of rhodopsin we wanted to test whether neighboring cytoplasmic loops contribute to the sorting signal. Addition of the peptide corresponding to the third cytoplasmic loop (amino acids 234–245) or a longer peptide (amino acids 231–252), known to contain sites of interaction with transducin (28), arrestin (29), and rhodopsin kinase (30), had no effect on post-Golgi trafficking (30.6 ± 0.9% of total radiolabeled rhodopsin in post-Golgi membrane fraction in control vs. 30.6 ± 2.1% in the presence of the peptide 234–245, four separate experiments). Several other peptides, including the second cytoplasmic loop (amino acids 141–153) and a control N-terminal (intradiscal) domain (amino acids 3–14) also had no effect (data not shown). Peptides corresponding to the first (amino acids 63–75) and the fourth loop (amino acids 310–321) showed minimal inhibition (≈20%). Because these loops are in close proximity to the C-terminal domain in the three-dimensional structure (31) it is possible that they make some contribution to the recognition site. However, the last 24 C-terminal amino acids of frog rhodopsin (or the last 19 amino acids of bovine rhodopsin) form the major part of the domain responsible for its sorting.
Deletion of QVSPA from the Inhibitory C-Terminal Peptide Eliminates Its Effect on Rhodopsin Trafficking.
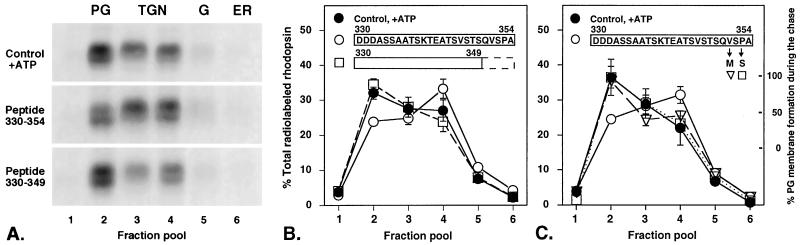
The five amino acids QVS(A)PA are the most conserved in the C-terminal domain. To test the importance of these amino acids we modeled a naturally occurring stop codon mutation that removes the last five amino acids of rhodopsin Q344ter (32, 33). We synthesized a truncated frog C-terminal peptide (amino acids 330–349) and followed its effect on post-Golgi trafficking of rhodopsin. As shown in Fig. 4 A and B, deletion of QVSPA from the inhibitory C-terminal peptide eliminated its effect on rhodopsin trafficking (23.8 ± 1.3% of radiolabeled rhodopsin in the post-Golgi membrane fraction in the presence of the C-terminal peptide vs. 34.4 ± 1.6% in the presence of the truncated peptide, P = 1.8 × 10−3, control = 32.0 ± 1.7%, three separate experiments). The subcellular distribution of radiolabeled rhodopsin completely paralleled its distribution in the control. A similar truncated bovine C-terminal peptide (amino acids 331–342) had no significant effect on rhodopsin transport (data not shown). This finding suggests that C-terminal peptides lacking amino acids QVS(A)PA cannot successfully compete with rhodopsin for the binding to the specific recognition factor(s) and therefore cannot affect its intracellular trafficking.
Figure 4.
ADRP mutations Q344ter (lacking amino acids QVAPA), V345M, and P347S introduced into the frog C-terminal peptide eliminate its inhibitory effect on rhodopsin trafficking. (A) Autoradiograms of the SDS/PAGE showing subcellular distribution of radiolabeled rhodopsin after the chase in the control with ATP (control, +ATP), in the presence of 50 μM frog C-terminal peptide (peptide 330–354), and in the presence of 50 μM truncated frog C-terminal peptide (peptide 330–349). Radiolabeled rhodopsin appears as a doublet in this gel. However, both forms migrate faster than the untrimmed rhodopsin present in the ER (data not shown and ref. 1). It is possible that the two detected forms are resolved based on differential glycosylation in the Golgi and/or the TGN, but the origin of these differences is unknown at present. Importantly, both of these forms are found in the post-Golgi fraction (fraction pool 2) in the control and are delivered to the ROS in vivo (not shown). (B) The distribution of radiolabeled rhodopsin measured in a PhosphorImager in three separate experiments and presented as the means ± SE. Truncation of the last five amino acids reduces the inhibitory effect of the C-terminal peptide. (C) The distribution of radiolabeled rhodopsin was measured in four separate experiments. Substitution of the amino acid V351M (−3 from the C terminal) or P353S (−1) eliminates the inhibitory effect of the C-terminal peptide. Other details are as in Figs. 1 and 3.
Substitutions that Mimic ADRP Mutations V345M and P347S Completely Eliminate the Inhibitory Activity of the C-Terminal Peptide.
To further define amino acids critical for the recognition of the C-terminal sequence of rhodopsin, we synthesized two peptides corresponding to amino acids 330–354 of frog rhodopsin with single amino acid substitutions of V351M and P353S. These substitutions correspond to mutations V345M and P347S found in patients with ADRP (34–36). Addition of these peptides to the cell-free system had no effect on rhodopsin trafficking while in the same series of experiments the parent peptide 330–354 reduced its exit from the TGN by 60% (Fig. 4C). The peptide-dependent inhibition was not complete in our experiments, but the presence of the conserved last five amino acids of rhodopsin was critical for inhibitory activity. Collectively our data suggest that the last five amino acids of rhodopsin contain a signal that is specifically recognized in the TGN. Amino acids valine (−3 from the C terminal) and proline (−1) are absolutely required for signal recognition, while serine (−2) can be substituted by alanine as evidenced by a successful competition of the bovine peptide with frog rhodopsin.
DISCUSSION
Rhodopsin interacts through its cytoplasmic domain with a number of cytosolic proteins. Many of these interactions lead to activation and deactivation of the enzymatic cascade triggered by light (5). Interestingly, one of the most conserved regions in the cytoplasmic C-terminal of rhodopsin, QVAPA (QVSPA in frog, fish, and lamprey) appears not to be engaged in such light-dependent functions (13, 22). Nevertheless, this highly conserved domain is very important as mutations in these amino acids cause severe forms of ADRP (19, 20, 37). We previously have shown that a mAb with an antigenic site within nine amino acids at the C terminal of rhodopsin inhibits its trafficking in the frog retinal cell-free system (2). This initial observation suggested to us that a part of the signal responsible for polarized sorting of rhodopsin in photoreceptor cells may be within this domain. Independent observations by several groups studying mutations known to cause ADRP in transgenic animals support this possibility. Mice expressing the Q344ter transgene (missing amino acids QVAPA) show nearly normal light response but accumulate truncated rhodopsin in the plasma membrane of photoreceptor inner segments (13). Abnormal accumulation of extracellular membranes containing rhodopsin has been observed in mice carrying the proline 347 to serine (P347S) mutation (10). Studies in transgenic animals suggest that rhodopsin transport may be defective but have not defined the specific step affected by the mutations.
In the present study we defined the C terminal of rhodopsin as the domain that regulates its TGN to post-Golgi trafficking. We used an in vitro assay that reconstitutes intra-Golgi trafficking as well as post-Golgi membrane formation from the TGN (2). The distribution of biosynthetic membranes and subcellular organelles in this system has been previously defined by a combination of morphological and biochemical approaches (1, 2, 14, 16, 17). By using this system, we chose synthetic peptides to explore further the role of the cytoplasmic domain of rhodopsin. The majority of the peptides that we have tested in this assay have been successfully used in other studies delineating functional domains of rhodopsin (28, 38). These are highly polar water-soluble peptides that can assume proper conformation in solution and act as competitive inhibitors of physiological processes involving rhodopsin (28, 38). Moreover, NMR analysis of the bovine C-terminal peptide has revealed a compact globular structure potentially comprising a separate domain of rhodopsin (26, 27). Both frog and bovine C-terminal peptides arrested rhodopsin in the TGN. They had no significant effect on rhodopsin transit through other compartments distinguishable in this system, which suggests that the structure and the function of these compartments was unaffected by the peptides. This finding is important because certain peptides are capable of assuming a conformation that increases their affinity for membranes and inhibiting intracellular trafficking in a nonspecific fashion (39). This does not appear to be the case with the rhodopsin C-terminal peptides.
The inhibitory C-terminal peptides did not interfere with the exit of newly synthesized rhodopsin from the ER or its progression through the Golgi complex in the frog retinal cell-free system. In the presence of the peptide both the Golgi and the ER-enriched fractions had a content of newly synthesized rhodopsin nearly identical to that of the control with ATP. The untrimmed form of rhodopsin with slower electrophoretic mobility previously has been found associated with the ER in untreated cells, or located in a fused Golgi-ER compartment upon short treatment with brefeldin (1). This rhodopsin precursor, specific for early biosynthetic compartments, was not detected in our inhibition experiments with peptides. Instead, processed rhodopsin was found in the fractions enriched in TGN membranes (see Fig. 4A). This suggests that the recognition of the C-terminal sequence occurs in the TGN, past the ER and Golgi compartments, and before post-Golgi membrane formation. The addition of the C-terminal peptides interferes with these processes most likely competing for the sorting factors and affecting the rate of rhodopsin exit from the TGN.
The most significant finding reported here is that the C-terminal sequence QVS(A)PA appears to be critical for the regulatory function of this domain. Peptides lacking this sequence, or containing single amino acid substitutions, do not inhibit post-Golgi trafficking of rhodopsin. This indicates that putative recognition factor(s) may bind directly to the C-terminal sequence when rhodopsin appears in the TGN, although other domains of rhodopsin also may contribute to the overall binding affinity. The most frequent ADRP mutations in the C-terminal domain involve substitutions of proline 347 (equivalent to 353 in frog) where six different missense mutations at this position have been identified (37). This penultimate proline may be critical for positioning the last amino acid, which is invariably alanine, for binding within the hydrophobic pocket of the putative interacting protein involved in sorting events. Hydrophobic pockets that selectively recognize the four-residue C-terminal consensus sequence (X-T/S-X-V-COO−) of membrane proteins recently have been described in a family of PDZ domain-containing proteins that cluster membrane receptors and signaling molecules at specific subcellular sites (40, 41) and also may regulate their subcellular targeting (42). A Drosophila PDZ domain protein, InaD, organizes rhodopsin RH1 with multiple components of the phototransduction cascade into a signaling complex, and mutation in one of its PDZ domains causes retinal degeneration (43, 44). A protein with PDZ-like or other recognition domains may regulate rhodopsin sorting in vertebrate photoreceptor cells or may be important to recruit other components of visual transduction cascade such as transducin and cGMP phosphodiesterase to the rhodopsin-bearing post-Golgi membranes, as they have been localized to these membranes in frog photoreceptors (1).
The post-Golgi membranes carrying rhodopsin do not contain clathrin and do not have a morphologically distinguishable coat (1, 4). However, a nonclathrin coat could be involved in some stages in their formation. In general, membrane trafficking from the ER and within the Golgi complex is regulated by nonclathrin-associated coat protein complexes COPI and COPII that interact with specific sorting signals in the cargo molecules (45). Because nearly nothing is known at present about nonclathrin-associated coat proteins that regulate post-Golgi trafficking, COP proteins, a lace-like coat that has been microscopically observed bound to the TGN of NRK cells (46) and a newly described TGN-associated adaptor-related protein complex AP3 with neuron-specific isoforms (47, 48), are potential candidates that could play a role in protein sorting in a compartment relevant for rhodopsin trafficking.
Although the identification of the protein(s) that recognize the QVS(A)PA sequence of rhodopsin remains a future challenge compounded by the current dearth of information on the role of nonclathrin coat proteins in post-Golgi trafficking, we propose that the polarized sorting of rhodopsin at the exit from the TGN is mediated by the interaction of the C-terminal sequence QVS(A)PA with a coat protein that may contain a PDZ-like or other recognition domain. Such a protein coat could allow entrance of rhodopsin molecules into the specific post-Golgi carriers. Although the precise interactions underlying these phenomena remain to be delineated, the data presented in this work distinguish between several models, including potential events upstream and downstream of the specific sorting processes described here, which have been proposed to explain the effects of mutations in the C terminus of rhodopsin. Based on the presented results, the mutations investigated in this work and elsewhere (10, 13) most likely interfere with the proper sorting of rhodopsin and result in abnormal post-Golgi membrane formation.
Acknowledgments
We are grateful to Dr. David Papermaster for his continuous support, thoughtful discussions, and critical reading of the manuscript. We thank Belen Puleo-Scheppke and Claudia Trippe for their invaluable help at the initial stages of this project. This work was supported by National Institutes of Health Grant EY-6891 and the Core Grant for Vision Research EY-07003 (D.D.) and EY-6226 and an unrestricted departmental award from Research to Prevent Blindness (P.A.H.).
ABBREVIATIONS
- ROS
rod outer segment(s)
- TGN
trans-Golgi network
- PNS
postnuclear supernatant
- ADRP
autosomal dominant retinitis pigmentosa
- ER
endoplasmic reticulum
References
- 1.Deretic D, Papermaster D S. J Cell Biol. 1991;113:1281–1293. doi: 10.1083/jcb.113.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic D, Puleo Scheppke B, Trippe C. J Biol Chem. 1996;271:2279–2286. doi: 10.1074/jbc.271.4.2279. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez de Turco E B, Deretic D, Bazan N G, Papermaster D S. J Biol Chem. 1997;272:10491–10497. doi: 10.1074/jbc.272.16.10491. [DOI] [PubMed] [Google Scholar]
- 4.Papermaster D S, Schneider B G, Defoe D, Besharse J C. J Histochem Cytochem. 1986;34:5–16. doi: 10.1177/34.1.2934469. [DOI] [PubMed] [Google Scholar]
- 5.Hargrave P A, McDowell J H. FASEB J. 1992;6:2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- 6.Besharse J C. In: The Retina: A Model for Cell Biological Studies. Adler R, Farber D, editors. New York: Academic; 1986. pp. 297–352. [Google Scholar]
- 7.Li Z Y, Kljavin I J, Milam A H. J Neurosci. 1995;15:5429–5438. doi: 10.1523/JNEUROSCI.15-08-05429.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan L S, Daiger S P. Mol Med Today. 1996;2:380–386. doi: 10.1016/s1357-4310(96)10037-x. [DOI] [PubMed] [Google Scholar]
- 9.Berson E L. Proc Natl Acad Sci USA. 1996;93:4526–4528. doi: 10.1073/pnas.93.10.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Snyder W K, Olsson J E, Dryja T P. Proc Natl Acad Sci USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papermaster D S, Windle J. Invest Ophthalmol Visual Sci. 1995;36:977–983. [PubMed] [Google Scholar]
- 12.Adler R. Arch Ophthalmol. 1996;114:79–83. doi: 10.1001/archopht.1996.01100130075012. [DOI] [PubMed] [Google Scholar]
- 13.Sung C H, Makino C, Baylor D, Nathans J. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic D, Papermaster D S. J Cell Sci. 1993;106:803–813. doi: 10.1242/jcs.106.3.803. [DOI] [PubMed] [Google Scholar]
- 15.Deretic D, Papermaster D S. In: Methods for the Study of Photoreceptor Cells. Hargrave P A, editor. Vol. 15. New York: Rockefeller Univ. Press; 1993. pp. 108–120. [Google Scholar]
- 16.Deretic D, Huber L A, Ransom N, Mancini M, Simons K, Papermaster D S. J Cell Sci. 1995;108:215–224. doi: 10.1242/jcs.108.1.215. [DOI] [PubMed] [Google Scholar]
- 17.Deretic D, Papermaster D S. In: Progress in Retinal and Eye Research. Osborne N N, Chader G J, editors. Vol. 14. New York: Pergamon; 1995. pp. 249–265. [Google Scholar]
- 18.Deretic D. Electrophoresis. 1997;18:2537–2541. doi: 10.1002/elps.1150181408. [DOI] [PubMed] [Google Scholar]
- 19.Berson E L, Rosner B, Sandberg M A, Weigel DiFranco C, Dryja T P. Am J Ophthalmol. 1991;111:614–623. doi: 10.1016/s0002-9394(14)73708-0. [DOI] [PubMed] [Google Scholar]
- 20.Sandberg M A, Weigel DiFranco C, Dryja T P, Berson E L. Invest Ophthalmol Visual Sci. 1995;36:1934–1942. [PubMed] [Google Scholar]
- 21.Pittler S J, Fliesler S J, Baehr W. FEBS Lett. 1992;313:103–108. doi: 10.1016/0014-5793(92)81422-i. [DOI] [PubMed] [Google Scholar]
- 22.Weiss E R, Hao Y, Dickerson C D, Osawa S, Shi W, Zhang L, Wong F. Biochem Biophys Res Commun. 1995;216:755–761. doi: 10.1006/bbrc.1995.2686. [DOI] [PubMed] [Google Scholar]
- 23.Hargrave P A, McDowell J H, Curtis D R, Wang J K, Juszczak E, Fong S L, Rao J K, Argos P. Biophys Struct Mech. 1983;9:235–244. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- 24.Nathans J, Hogness D S. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 25.Hargrave P A, McDowell J H. Int Rev Cytol. 1992;137:49–97. doi: 10.1016/s0074-7696(08)62600-5. [DOI] [PubMed] [Google Scholar]
- 26.Yeagle P L, Alderfer J L, Albert A D. Nat Struct Biol. 1995;2:832–834. doi: 10.1038/nsb1095-832. [DOI] [PubMed] [Google Scholar]
- 27.Yeagle P L, Alderfer J L, Albert A D. Mol Vision. 1996;2:12. [PubMed] [Google Scholar]
- 28.Konig B, Arendt A, McDowell J H, Kahlert M, Hargrave P A, Hofmann K P. Proc Natl Acad Sci USA. 1989;86:6878–6882. doi: 10.1073/pnas.86.18.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krupnick J G, Gurevich V V, Schepers T, Hamm H E, Benovic J L. J Biol Chem. 1994;269:3226–3232. [PubMed] [Google Scholar]
- 30.Thurmond R L, Creuzenet C, Reeves P, Khorana H G. Proc Natl Acad Sci USA. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unger V M, Hargrave P A, Baldwin J M, Schertler G F. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 32.Sung C H, Davenport C M, Hennessey J C, Maumenee I H, Jacobson S G, Heckenlively J R, Nowakowski R, Fishman G, Gouras P, Nathans J. Proc Natl Acad Sci USA. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobson S G, Kemp C M, Sung C H, Nathans J. Am J Ophthalmol. 1991;112:256–271. doi: 10.1016/s0002-9394(14)76726-1. [DOI] [PubMed] [Google Scholar]
- 34.Dryja T P, McGee T L, Hahn L B, Cowley G S, Olsson J E, Reichel E, Sandberg M A, Berson E L. N Engl J Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- 35.Dryja T P, Hahn L B, Cowley G S, McGee T L, Berson E L. Proc Natl Acad Sci USA. 1991;88:9370–9374. doi: 10.1073/pnas.88.20.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berson E L, Sandberg M A, Dryja T P. Trans Am Ophthalmol Soc. 1991;89:117–128. and 128–130. [PMC free article] [PubMed] [Google Scholar]
- 37.Macke J P, Hennessey J C, Nathans J. Hum Mol Genet. 1995;4:775–776. doi: 10.1093/hmg/4.4.775. [DOI] [PubMed] [Google Scholar]
- 38.Palczewski K, Arendt A, McDowell J H, Hargrave P A. Biochemistry. 1989;28:8764–8770. doi: 10.1021/bi00448a013. [DOI] [PubMed] [Google Scholar]
- 39.Weidman P J, Winter W M. J Cell Biol. 1994;127:1815–1827. doi: 10.1083/jcb.127.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 41.Gomperts S N. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 42.Staudinger J, Lu J, Olson E N. J Biol Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- 43.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker C S. Nature (London) 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 44.Chevesich J, Kreuz A J, Montell C. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- 45.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 46.Ladinsky M S, Kremer J R, Furcinitti P S, McIntosh J R, Howell K E. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson F, Peden A A, Christopoulou L, Robinson M S. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dell’Angelica E C, Ohno H, Ooi C E, Rabinovich E, Roche K W, Bonifacino J S. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]