Abstract
STAT (signal transducer and activator of transcription) proteins are latent cytoplasmic transcription factors that become activated by tyrosine phosphorylation in response to cytokine stimulation. Tyrosine phosphorylated STATs dimerize and translocate into the nucleus to activate specific genes. Different members of the STAT protein family have distinct functions in cytokine signaling. Biochemical and genetic analysis has demonstrated that Stat1 is essential for gene activation in response to interferon stimulation. Although progress has been made toward understanding STAT activation, little is known about how STAT signals are down-regulated. We report here the isolation of a family of PIAS (protein inhibitor of activated STAT) proteins. PIAS1, but not other PIAS proteins, blocked the DNA binding activity of Stat1 and inhibited Stat1-mediated gene activation in response to interferon. Coimmunoprecipitation analysis showed that PIAS1 was associated with Stat1 but not Stat2 or Stat3 after ligand stimulation. The in vivo PIAS1–Stat1 interaction requires phosphorylation of Stat1 on Tyr-701. These results identify PIAS1 as a specific inhibitor of Stat1-mediated gene activation and suggest that there may exist a specific PIAS inhibitor in every STAT signaling pathway.
The binding of cytokines to their cell surface receptors activates, by tyrosine phosphorylation, a family of latent cytoplasmic transcription factors termed STATs (signal transducer and activator of transcription). After cytokine receptor activation, STATs dimerize and translocate into the nucleus to activate genes. Seven members of the STAT family, activated by a variety of cytokines, have been cloned (1–5). Genetic knockout studies indicate that STATs have highly specific functions. Stat1, the founding member of the STAT family, is essential for innate response to either viral or bacterial infection (6, 7). Stat1 is phosphorylated on a single residue, Tyr-701, in response to stimulation by a number of ligands including interferons (IFNs), interleukin 6 (IL-6), and epidermal growth factor (8–11). The phosphorylation on the Tyr-701 residue of Stat1 is required for its nuclear translocation, dimerization, DNA binding, and gene activation (8, 12).
Although great progress has been made toward the understanding of STAT activation, little is known about how STAT signals are down-regulated. Several mechanisms to down-regulate STAT signaling have been proposed. (i) Because the activities of STATs depend on tyrosine phosphorylation, the precise recognition and dephosphorylation of STATs by their protein tyrosine phosphatases (PTPases) is expected to be crucial for gene regulation (13–16). However, it is not known what and how PTPases can dephosphorylate STATs. (ii) Inhibitors of proteosome activity are shown to prolong the activation of Stat1, implying the involvement of ubiquitination in the degradation of Stat1 (17). However, because these inhibitors also affect the half-life of IFN receptor, the importance of the degradation of Stat1 through ubiquitination pathway remains unclear (4, 16). (iii) Recently, a family of cytokine-inducible inhibitors of signaling have been isolated (18–20). This family of proteins, named SOCS/JAB/SSI, are relatively small protein molecules that contain mainly SH2 domains. SOCS/JAB/SSI proteins can directly bind to JAKs and can inhibit the tyrosine kinase activity of JAKs.
We have identified recently a protein named PIAS3 (protein inhibitor of activated Stat3) that functions as a specific inhibitor of Stat3 signaling (21). We report here the identification of four additional members of the PIAS family. We found that PIAS1 was associated with Stat1, but not with Stat2 or Stat3 in vivo in cells treated with IFN or IL-6. The PIAS1–Stat1 interaction requires the phosphorylation of Stat1 on Tyr-701. Furthermore, PIAS1 but not other PIAS proteins blocked the DNA binding activity of Stat1 and inhibited Stat1-mediated gene activation. Our results suggest that PIAS1 is a specific inhibitor of Stat1-mediated gene activation. The mode of the PIAS-mediated inhibition on STAT activity is distinct from other known inhibitory mechanisms involved in STAT signaling.
MATERIALS AND METHODS
Cells.
U3A and U3A-derived cell lines were maintained in DMEM containing 10% fetal bovine serum at 10% CO2. Human Daudi lymphoblastoid cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum at 5% CO2.
Isolation of PIAS cDNAs.
The yeast two-hybrid method (22) was used to identify proteins that can interact with Stat1. Stat1β, an alternatively spliced form of Stat1 lacking the COOH-terminal transcriptional activation domain (23), was used as a bait to screen a human JY112 B cell library for Stat1 interacting proteins (24). Fifty positive clones were identified from 3 × 106 primary transformants. Of these clones, 40 were partial-length cDNAs from the same gene. The corresponding full-length cDNA encoding a human protein of 650 aa, which we named as PIAS1, was generated by fusing a longer cDNA clone obtained by screening a human K562 cDNA library with the expressed sequence tag (EST) clone 301840. By EST database searching and library screening, the full-length murine PIAS1 (mPIAS1) as well as four other related clones that encode putative new members of the PIAS family were identified. The database search was done with the Baylor College of Medicine Search Launcher. The murine full-length PIAS1 cDNA was obtained by sequencing EST clone 930725. EST clone 785675 was used to screen a human testis library to obtain PIASxα and PIASxβ. EST clone 59244 was sequenced and identified as PIASy. The nucleotide sequences of each of these genes has been deposited in the GenBank database [accession nos. AF077951 (hPIAS1), AF077950 (mPIAS1), AF077953 (hPIASxα), AF077954 (hPIASxβ), AF077952 (hPIASy)].
Plasmid Constructions.
FLAG–PIAS1 was constructed by insertion of the murine PIAS1 cDNA into the BglII and SalI sites of pCMV–FLAG vector. GST–PIASxα and GST–PIAS1 constructs were prepared by insertion of human PIASxα cDNA or murine PIAS1 cDNA into the EcoRI and NotI sites of p4T-1 (Pharmacia).
Luciferase Assays.
Calcium phosphate was used for transfection (8). Twenty-four hours after transfection, cultures were either left untreated or treated with IFN-γ for 6 h, and cell extracts were prepared and measured for luciferase activity (Promega). The relative luciferase units were corrected for relative expression of β-galactosidase.
Electrophoretic Mobility-Shift Assays (EMSA).
EMSA analysis was performed as described (25). Briefly, protein extracts were mixed with or without glutathione S-transferase (GST) fusion proteins and incubated for 5 min at room temperature in gel shift buffer. The samples were then mixed with the 32P-labeled probe derived from a high-affinity Stat1–DNA binding site (10) for 18 min and were then analyzed on nondenaturing 4% polyacrylamide gels.
Coimmunoprecipitation Analysis.
Immunoprecipitation and protein immunoblotting were done as described (21). Whole-cell extracts were prepared by using lysis buffer containing 1% Brij, 50 mM Tris (pH 8), 150 mM NaCl, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 3 μg/ml aprotinin, 1 μg/ml pepstatin, and 0.1 mM sodium vanadate. The mixture was rotated at 4°C for 30 min and centrifuged at 13,000 × g for 5 min. The supernatant was used for immunoprecipitation with anti-PIAS1n (1:100 dilution). Anti-PIAS1n was raised against a GST fusion protein containing 119 aa residues from the NH2-terminal region of PIAS1 (aa 50–168).
RESULTS
Isolation of PIAS1 and Other PIAS Family Members.
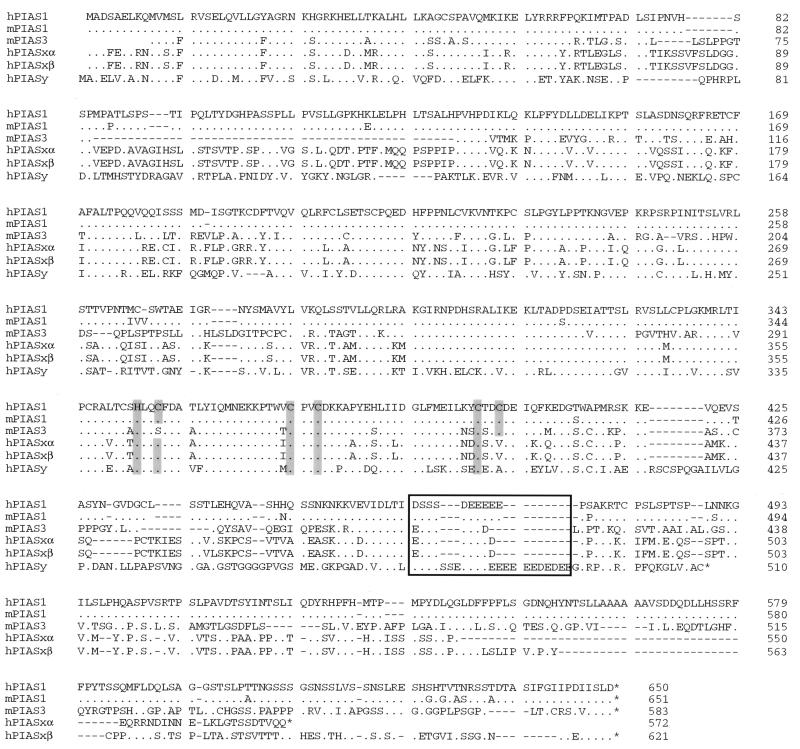
We used the yeast two-hybrid method (22) to identify proteins that can interact with Stat1. Stat1β, an alternatively spliced form of Stat1 lacking the COOH-terminal transcriptional activation domain (23), was used as a bait to screen a human B cell library for Stat1 interacting proteins. A human cDNA encoding a protein of 650 aa, which we named as PIAS1, was obtained. By EST database searching and library screening, the murine PIAS1 as well as four related clones that encode putative new members of the PIAS family were identified. One member of this family, PIAS3 is a specific inhibitor of Stat3 signaling (21). The PIAS family of proteins show significant homology (>50%). PIAS proteins have several highly conserved domains, including a putative zinc binding motif and a highly acidic region (Fig. 1). The COOH-terminal regions of PIAS proteins are the least conserved. PIASxα and PIASxβ are identical, except their COOH-terminal regions, and are probably the products of differentially spliced messages from the same gene. Sequence analysis indicates that human PIAS1 is almost identical to a previously reported human protein named GBP (Gu/RH-II binding protein) (26). GBP lacks 9 aa residues at the NH2 terminus and displays a few amino acid differences when compared with PIAS1.
Figure 1.
Sequence comparison of the PIAS family of proteins. The predicted amino acid sequences of PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy are shown. h, Human; m: mouse. Cysteine and histidine residues that are predicted to form a zinc finger are shaded. The conserved acidic region is boxed. Dots indicate amino acid identity.
PIAS1 Inhibits Stat1-Mediated Gene Activation.
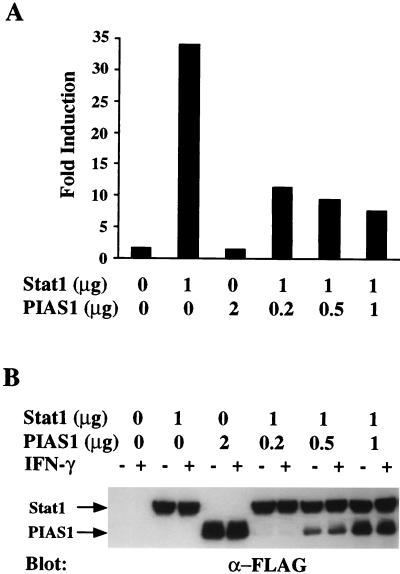
To test whether PIAS1 is indeed involved in regulating Stat1 activity, we examined the effect of PIAS1 on Stat1-mediated gene activation by luciferase assays. A luciferase reporter construct [(3x)Ly6] containing three copies of the Stat1 binding sequence from the murine Ly-6A/E gene was used (25, 27). Human 293 cells were transfected with expression vectors encoding FLAG-tagged Stat1 and PIAS1 together with [(3x)Ly6] in various combinations as indicated (Fig. 2A). The transfected cells were then treated with or without IFN-γ. Cells cotransfected with Stat1 and [(3x)Ly6] reporter construct showed a 35-fold increase of luciferase expression in response to IFN-γ. In the presence of an increasing amount of PIAS1, the Stat1-activated luciferase expression in response to IFN-γ stimulation was dramatically inhibited (Fig. 2A). The proper expression of Stat1 and PIAS1 in these transfections was confirmed by Western blot analysis of the same extracts with anti-FLAG antibody (Fig. 2B). In contrast, neither PIAS3 or PIASx was able to inhibit the Stat1-mediated gene activation (ref. 21, unpublished data) These results demonstrated that PIAS1 can inhibit Stat1-mediated gene activation.
Figure 2.
PIAS1 inhibits Stat1-mediated gene activation. (A) Luciferase reporter assays. Human 293 cells were transiently transfected with [(3x)Ly6] luciferase reporter construct together with empty expression vector, FLAG–Stat1 or various amounts of FLAG–PIAS1 vectors, alone or in combination as indicated. Cells were either treated or untreated with IFN-γ for 6 h. Shown is the fold increase after IFN-γ treatment. (B) Western blot analysis. Equal amounts of protein extracts from A were analyzed by immunoblot with anti-FLAG (Sigma).
PIAS1, but Not Other PIAS Proteins, Can Inhibit the DNA Binding Activity of Stat1.
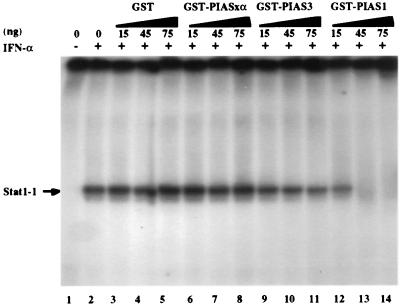
We next tested whether PIAS1 could affect the DNA binding activity of Stat1 by EMSA analysis. We prepared and purified a recombinant fusion protein of GST with PIAS1 (GST–PIAS1) as well as GST–PIAS3 and GST–PIASxα. The effect of these fusion proteins on the DNA binding activity of Stat1 was tested. Nuclear extracts from human Daudi B cells treated with IFN-α were incubated with GST or GST–PIASxα or GST–PIAS3 or GST–PIAS1 (15–75 ng). The mixtures were then analyzed by gel retardation assays by using the Stat1 DNA binding site as the probe (Fig. 3). GST–PIAS1 (45 ng) completely blocked the DNA binding activity of Stat1. In contrast, GST, GST–PIASxα, and GST–PIAS3 had little effect on Stat1 binding. These results suggest that PIAS1, but not other PIAS proteins, can inhibit the DNA binding activity of Stat1.
Figure 3.
PIAS1 inhibits the DNA binding activity of Stat1. EMSA analysis was performed with nuclear extracts prepared from Daudi cells with (+) or without (−) IFN-α treatment in the absence or presence of GST, GST–PIASxα, GST–PIAS3, or GST–PIAS1 proteins (15–75 ng) as indicated. The probe used is a high-affinity Stat1–DNA binding site (10). The concentrations of GST fusion proteins were estimated on 7% SDS/PAGE with various dilutions of BSA as the standard.
PIAS1 Is Associated with Stat1, but Not Stat2 or Stat3 in Vivo.
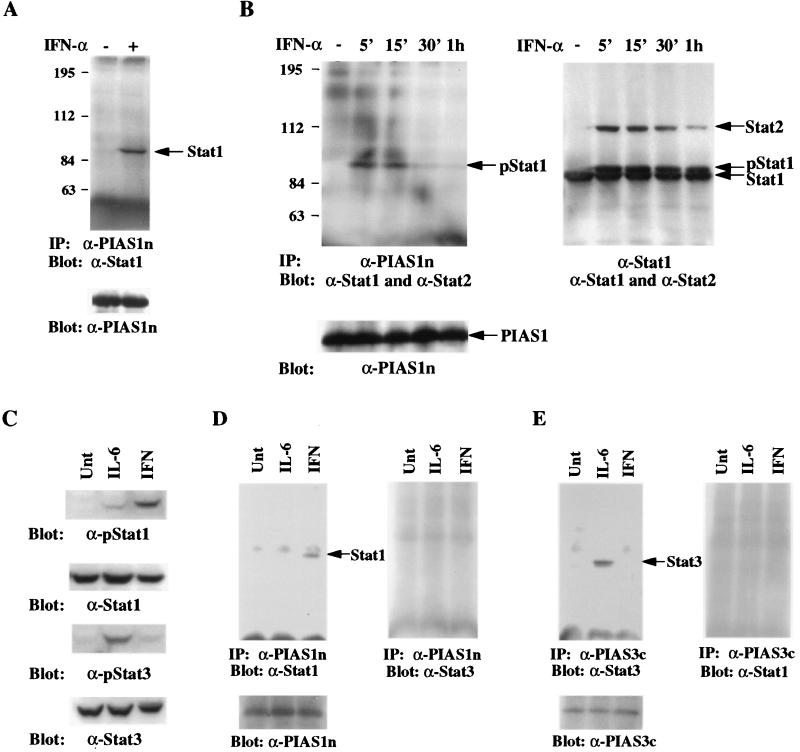
We next wished to examine the specificity of PIAS–STAT interaction. Because it has been well documented that the specificity of protein–protein interaction may be lost when examined in vitro or when assayed under overexpression situation, we wished to analyze the PIAS–STAT interaction in vivo. We prepared a specific antiserum (anti-PIAS1n) against a GST fusion protein containing 119 aa residues from the NH2-terminal region of PIAS1 (amino acids 50–168). This antibody specifically recognized a protein with the molecular weight of 78 kDa, a predicted size of PIAS1, in a number of human and murine cell lines tested (unpublished observation). We first wanted to determine whether PIAS1 is associated with Stat1 in vivo. Protein extracts prepared from human Daudi B cells untreated or treated with IFN-α for 15 min were used for immunoprecipitation with anti-PIAS1n. The immunoprecipitates were then washed and analyzed by Western blot with anti-Stat1 antibody. Stat1 was present in the PIAS1 immunoprecipitate from cells treated with IFN-α but not from untreated cells (Fig. 4A). These results suggest that PIAS1 is associated with Stat1 in vivo and that the PIAS1–Stat1 interaction depends on ligand stimulation.
Figure 4.
Specificity of PIAS–STAT interaction. (A) PIAS1 interacts with Stat1 in vivo. Protein extracts from Daudi cells untreated (−) or treated (+) with IFN-α for 15 min were prepared and used for immunoprecipitation with anti-PIAS1n. Immunoprecipitates were then analyzed on SDS/PAGE and the blot was probed anti-Stat1. The filter was washed and reprobed with anti-PIAS1 (Lower). (B) PIAS1 does not interact with Stat2. Daudi cells were treated with IFN-α for the times indicated. Whole-cell extracts were prepared and one-half of these extracts were used for immnunoprecipitation with anti-PIAS1n (Left). Immunoprecipitation was carried out as described in A and the filter was probed with anti-Stat1 and anti-Stat2. The same filter was washed and reprobed with anti-PIAS1n (Lower). The other half of protein extracts were subjected to immunoprecipitation with anti-Stat1 (Right). The filter was probed with a mixture of anti-Stat1 and anti-Stat2 antibodies. p-Stat1, phosphorylated Stat1. (C) Western blot analysis. HepG2 cells were untreated or treated with IL-6 or IFN-γ for 15 min, and protein extracts were prepared and analyzed by immunoblot with anti-Stat1, anti-pStat1 (New England Biolabs), anti-Stat3 (Santa Cruz Biothechnology), or anti-pStat3 (New England Biolabs) as indicated. (D) PIAS1 interacts with Stat1 but not Stat3. Protein extracts from C were immunoprecipitated with anti-PIAS1n. The precipitates were subjected to electrophoresis, and the filter was blotted with anti-Stat1 (Left). The same filter was washed and reprobed with anti-Stat3 (Left) or reprobed with anti-PIAS1n (Lower). (E) Same as D except that anti-PIAS3c was used for immunoprecipitation. The filter was probed with anti-Stat3 (Left) or anti-Stat1 (Right) or PIAS3c (Lower).
Because IFN-α also activates Stat2 (28, 29), we tested whether PIAS1 could interact with Stat2 upon IFN-α stimulation. Human Daudi B cells were untreated or treated with IFN-α for various time periods. Protein extracts were prepared and subjected to immunoprecipitation with anti-PIAS1n followed by immunoblot with both anti-Stat1 and Stat2 antibodies. Although the association of Stat1 and PIAS1 was observed in cells treated with IFN-α for 5 or 15 min and was significantly decreased after 30 min, Stat2 was absent in PIAS1 immunoprecipitates with or without IFN-α treatment (Fig. 4B). In contrast, Stat2 was found to be present in Stat1 immunoprecipitates (Fig. 4B). This result is consistent with the known fact that upon IFN-α stimulation, a fraction of Stat1 and Stat2 proteins can form heterodimers (30). These results suggest that PIAS1 interacts with Stat1 but not Stat2.
We have recently reported that PIAS3 is a specific inhibitor of Stat3 signaling (21). To examine the specificity of PIAS–STAT interactions, we carried out in vivo coimmunoprecipitation analysis with protein extracts prepared from human HepG2 cells untreated or treated with IFN-γ or IL-6. IFN-γ treatment activates Stat1, but not Stat3, in HepG2 cells—as shown by immunoblot analysis with antisera that can specifically recognize tyrosine phosphorylated Stat1 or Stat3 (Fig. 4C). IL-6 treatment strongly induces the tyrosine phosphorylation of Stat3 but only weakly stimulates phosphorylation of Stat1 in HepG2 cells. Samples of the same protein extracts were subjected to immunoprecipitation analysis with anti-PIAS1n or anti-PIAS3c [an antiserum against the COOH-terminal 79 aa residues of PIAS3 (21)]. The immunoprecipitates were then analyzed by protein blot with anti-Stat1 or anti-Stat3. PIAS1 was found to be associated with Stat1 but not Stat3 (Fig. 4D). [Because the activation of Stat1 by IL-6 was weak, the association of PIAS1 with Stat1 in IL-6 treated HepG2 cells was observed only when the blot was overexposed (unpublished observation).] In contrast, PIAS3 was found to be associated with Stat3 but not Stat1 (Fig. 4E). These experiments suggest that individual PIAS proteins display specificity for STATs in vivo.
The in Vivo PIAS1–Stat1 Association Requires the Phosphorylation of Stat1 on Tyr-701.
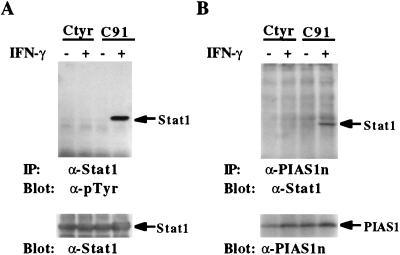
Upon IFN stimulation, Stat1 becomes tyrosine phosphorylated on a single tyrosine residue Tyr-701. This phosphorylation is required for the dimerization, nuclear translocation, and DNA binding activity of Stat1 (8, 12). Because PIAS1 is associated with Stat1 only in ligand-stimulated cells, we wished to determine whether IFN-induced tyrosine phosphorylation of Stat1 is required for the in vivo PIAS1-Stat1 interaction. Two stable cell lines, Ctyr and C91, derived from U3A cells that do not express Stat1 protein (31) were used for coimmunoprecipitation analysis. C91 and Ctyr cell lines were established by complementing U3A cells with the wild type Stat1 and a mutant Stat1 (Tyr-701 → Phe), respectively (8). Phosphotyrosine blot analysis confirmed that the T701F Stat1 mutant protein was not tyrosine phosphorylated in response to IFN-γ stimulation (Fig. 5A). Stat1 was clearly coimmunoprecipitated by anti-PIAS1 from C91 cells treated with IFN-γ (Fig. 5B). In contrast, anti-PIAS1 antibody failed to coimmunoprecipitate the T701F Stat1 mutant protein from Ctyr cells. These results suggest that IFN-induced phosphorylation on Tyr-701 of Stat1 is required for PIAS1-Stat1 interaction.
Figure 5.
Phosphorylation on Tyr-701 of Stat1 is required for PIAS1–Stat1 interaction. U3A cells complemented with Stat1 or Stat1(Tyr-701 → Phe) mutant protein were untreated or treated with IFN-γ for 15 min, and protein extracts were prepared. (A) Immunoprecipitation was performed with anti-Stat1 followed by blotting with a specific phosphotyrosine antiserum [anti-pTyr (Transduction Laboratories, Lexington, KY)]. The same filter was washed and reprobed with anti-Stat1 (Lower). (B) Immunoprecipitation was performed with anti-PIAS1n antiserum and blotted with anti-Stat1. The filter was washed and reprobed with anti-PIAS1 (Lower).
DISCUSSION
PIAS1 was identified by yeast two-hybrid screening for its ability to interact with Stat1. Sequence analysis indicates that human PIAS1 is almost identical to a previously reported human protein named GBP (26). GBP lacks 9 aa residues at the NH2 terminus and displays a few amino acid differences when compared with PIAS1. GBP was identified, by the yeast two-hybrid screening, as a putative interaction protein of Gu/RNA helicase II (26). The interaction of GBP with Gu/RNA helicase II has not been demonstrated in systems other than yeast. Based on results from in vitro assays using a crude yeast supernatant containing GBP, it was proposed that GBP may contain proteolytic activity toward Gu/RNA helicase II (26). However, purified GBP protein cannot cleave Gu/RNA helicase II (B. Valdez, personal communication). Thus, the previously observed proteolytic cleavage of Gu/RNA helicase is likely caused by a contaminating protease activity present in yeast supernatant.
Results from three independent assays presented in this paper suggest that PIAS1 is a specific inhibitor of Stat1 signaling. (i) Coimmunoprecipitation analysis suggests that PIAS1 is associated with Stat1 but not Stat2 or Stat3 in vivo upon cytokine stimulation. (ii) PIAS1 but not other PIAS proteins blocked the DNA binding activity of Stat1 in EMSA analysis. (iii) PIAS1 inhibited Stat1-mediated gene activation in luciferase reporter assays. PIAS1 represents a protein that can directly inhibit Stat1-mediated gene activation. We have recently reported the identification of PIAS3, which functions as a specific inhibitor of Stat3 signaling (21). PIAS3 interacts with Stat3 but not Stat1 and inhibits Stat3 but not Stat1-mediated gene activation. The identification of a family of PIAS proteins and the striking specific in vivo association between individual PIAS and STAT proteins shown in this paper suggest the possible involvement of a specific PIAS inhibitor in every STAT signaling pathway.
Our conclusion that PIAS1 has specificity toward Stat1 is largely based on the in vivo coimmunoprecipitation analysis. Interestingly, we noticed that PIAS1 displayed only partial specificity toward STAT proteins when analyzed in vitro or under overexpression conditions. Although PIAS1 but not other PIAS proteins can inhibit Stat1 DNA binding activity and Stat1-mediated gene activation, we found that PIAS1 can block the DNA binding activity of Stat3 in vitro and can inhibit Stat3-mediated gene activation when overexpressed in 293 cells (unpublished observation). The significance of the inhibitory activity of PIAS1 toward Stat3 observed in vitro and under overexpression conditions is not clear at the present. It is possible that PIAS1 can inhibit Stat3 function in cells where PIAS1 and Stat3 are highly expressed.
An interesting feature of PIAS proteins is that their association with STATs is cytokine-dependent. PIAS proteins seem to have higher binding affinity toward tyrosine phosphorylated than unphosphorylated STATs. Because PIAS proteins do not contain phosphotyrosine binding domains such as SH2 or PTB (32, 33), it seems likely that tyrosine phosphorylation of STATs may induce a protein conformational change, resulting the exposure of the PIAS interacting domain. This conformational change model may also explain our observation that partial PIAS1 but not full-length PIAS1 is able to interact with Stat1 in yeast two-hybrid assays (unpublished observation).
Our data suggest that PIAS proteins block STAT-mediated gene activation through the inhibition of STAT–DNA binding activity. The inhibitory effect of a PIAS protein on STAT–DNA binding activity can be achieved by two possible mechanisms: (i) the binding of a PIAS protein to a STAT dimer may mask the DNA binding domain of STAT, or (ii) PIAS proteins may bind to STATs and prevent the dimerization of STATs.
PIAS proteins contain a conserved putative zinc binding motif that is present in many proteins including transcription factors. Interestingly, a N-terminal truncated mutant PIASxβ protein (from amino acids 134 to 621) was recently shown to interact with a homeobox DNA binding protein Msx2 (34). In addition, this mutant protein named as Miz1 was shown to have sequence specific DNA binding activity (34). Thus, a PIAS protein may play a dual functional role by inhibiting the expression of genes containing STAT binding sites while activating another distinct set of genes.
Acknowledgments
We thank G. Stark for U3A cells, O. Witte for the cDNA library, and H. Herschman for reading the manuscript. This work was supported by a University of California, Los Angeles, Jonsson Cancer Center Postdoctoral Fellowship (to B.L.); National Institutes of Health National Institute of General Medical Sciences Training Grant GM08042, the Medical Scientist Training Program, and the Aesculapians Fund of the University of California, Los Angeles, School of Medicine (to S.A.K.); and by grants from the National Institutes of Health (AI39612), the Margaret E. Early Medical Research Trust, and the Council for Tobacco Research, U.S.A. (to K.S.).
ABBREVIATIONS
- IFN
interferon
- STAT
signal transducer and activator of transcription
- PIAS
protein inhibitor of activated STAT
- IL-6
interleukin 6
- GST
glutathione S-transferase
- GBP
Gu/RH-II binding protein
- EMSA
electrophoretic mobility shift assay
- EST
expressed sequence tag
Footnotes
References
- 1.Darnell J E, Jr, Kerr I M, Stark G M. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi T. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 4.Darnell J E., Jr Science. 1997;277:1630–1636. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea J J. Immunity. 1997;7:1–11. doi: 10.1016/s1074-7613(00)80505-1. [DOI] [PubMed] [Google Scholar]
- 6.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 7.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 8.Shuai K, Stark G R, Kerr I M, Darnell J E., Jr Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 9.Shuai K, Ziemiecki A, Wilks A F, Harpur A G, Sadowski H B, Gilman M Z, Darnell J E., Jr Nature (London) 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- 10.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 11.Fu X-Y, Zhang J-J. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 12.Shuai K, Horvath C M, Tsai-Huang L H, Qureshi S, Cowburn D, Darnell J E., Jr Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 13.David M, Grimley P M, Finbloom D S, Larner A C. Mol Cell Biol. 1993;13:7515–7521. doi: 10.1128/mcb.13.12.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque S J, Flati V, Deb A, Williams B R G. J Biol Chem. 1995;270:25709–25714. doi: 10.1074/jbc.270.43.25709. [DOI] [PubMed] [Google Scholar]
- 15.Shuai K, Liao J, Song M M. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haspel R L, Salditt-Georgieff M, Darnell J E., Jr EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T K, Maniatis T. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 18.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 19.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 20.Starr R, Willson T A, Viney E M, Murray L J L, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 21.Chung C D, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 22.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 23.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang D D, Wong C, Smith H, Liu J. J Cell Biol. 1997;138:1149–1157. doi: 10.1083/jcb.138.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahn K D, Shuai K, Lindwall G, Maher S E, Darnell J E, Jr, Bothwell A L M. Proc Natl Acad Sci USA. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdez B C, Henning D, Perlaky L, Busch R K, Busch H. Biochem Biophys Res Commun. 1997;234:335–340. doi: 10.1006/bbrc.1997.6642. [DOI] [PubMed] [Google Scholar]
- 27.Wen Z, Darnell J E., Jr Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindler C, Shuai K, Prezioso V R, Darnell J E., Jr Science. 1992;257:809–815. [PubMed] [Google Scholar]
- 29.Fu X-Y. Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 30.Improta T, Schindler C, Horvath C M, Kerr I M, Stark G R, Darnell J E., Jr Proc Natl Acad Sci USA. 1994;91:4776–4780. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen G B, Ren R, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 33.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 34.Wu L, Wu H, Sangiorgi F, Wu N, Bell J R, Lyons G E, Maxson R. Mech Dev. 1997;65:3–17. doi: 10.1016/s0925-4773(97)00032-4. [DOI] [PubMed] [Google Scholar]