Abstract
Pluripotent embryonic stem (ES) cells, a potential source of somatic precursors for cell therapies, cause tumors after transplantation. Studies of mammalian carcinogenesis using nuclear magnetic resonance (NMR) spectroscopy have revealed changes in the choline region, particularly increased phosphocholine (PCho) content. High PCho levels in murine ES (mES) cells have recently been attributed to cell pluripotency. The phosphoinositide 3-kinase (PI3K)/Akt pathway has been implicated in tumor-like properties of mES cells. This study aimed to examine a potential link between the metabolic profile associated with choline metabolism of pluripotent mES cells and PI3K/Akt signaling. We used mES (ES-D3) and murine embryonal carcinoma cells (EC-F9) and compared the metabolic profiles of 1) pluripotent mES (ESD0), 2) differentiated mES (ESD14), and 3) pluripotent F9 cells. Involvement of the PI3K/Akt pathway was assessed using LY294002, a selective PI3K inhibitor. Metabolic profiles were characterized in the extracted polar fraction by 1H NMR spectroscopy. Similarities were found between the levels of choline phospholipid metabolites (PCho/total choline and PCho/glycerophosphocholine [GPCho]) in ESD0 and F9 cell spectra and a greater-than five-fold decrease of the PCho/GPCho ratio associated with mES cell differentiation. LY294002 caused no significant change in relative PCho levels but led to a greater-than two-fold increase in PCho/GPCho ratios. These results suggest that the PCho/GPCho ratio is a metabolic trait shared by pluripotent and malignant cells and that PI3K does not underlie its development. It is likely that the signature identified here in a mouse model may be relevant for safe therapeutic applications of human ES cells.
Introduction
The capacity of embryonic stem (ES) cells for unlimited self-renewal and multilineage differentiation has stimulated unprecedented public and clinical interest as an attractive source of somatic cell precursors for cell-based therapies. However, despite recent advances in the development of ES cell technologies for regenerative medicine, a number of fundamental questions need to be addressed before ES cells can be used therapeutically. The main concern relates to their propensity to produce tumors after transplantation in the absence of any gross chromosomal abnormalities [1].
The similarities between stem and cancer cells have long been recognized, and accumulating evidence suggests that stemness and tumorigenicity might share some phenotypic traits dictated by the same genetic program [2,3]. Despite a wealth of data on the genotypic and phenotypic characteristics of pluripotent self-renewing ES cells, the mechanisms underlying their tumorigenic potential are poorly understood. Searching for genes specifically expressed in murine ES (mES) cells, Takahashi et al. [4] demonstrated that the oncogene ERas is expressed in undifferentiated mES cells and, through its downstream target, the PI3K/Akt pathway, promotes cell proliferation and tumorigenicity. Disruption of ERas does not affect the pluripotency of mES cells but leads to the impairment of cell growth and the ability to produce tumors in vivo. Proliferative and tumorigenic characteristics of the cells can be rescued by forced expression of active PI3K/Akt, implicating the PI3K pathway as a likely mechanism underlying the tumor-like properties of pluripotent ES cells [4].
Nuclear magnetic resonance (NMR) spectroscopy provides unique insight into cell metabolism and has recently been recognized as a powerful tool in phenotypic profiling of human cancers and mammalian cell lines, including stem cells and their derivatives [5–10]. In tumors and associated in vitro models, changes in the choline region of the NMR spectrum arising from an increase of the phosphocholine (PCho) content have been shown to correlate with the stage of tumor development and its invasiveness [8,11–13]. Elevated PCho levels are partially attributed to an increased activity of choline kinase (ChoK) catalyzing the first step in the Kennedy pathway (cytidine diphosphate-choline pathway), an enzyme recently recognized as both a prognostic marker and a therapeutic target in various types of human cancers [14–16]. ChoK expression and activity are regulated by a signaling complex with the PI3K pathway as one of its key components [17,18].
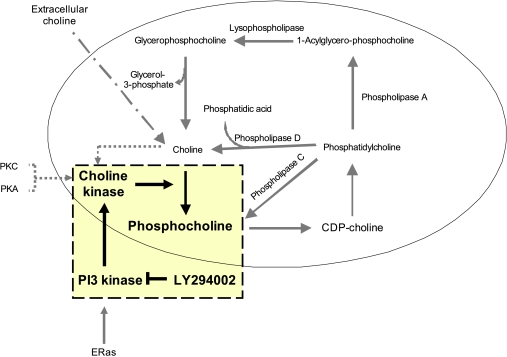
Little is known about the metabolism of ES cells and its changes as differentiation progresses. The only study available to date showed an association between mES cell differentiation status and NMR signals and demonstrated that the NMR profile of pluripotent mES cells is characterized by a very high PCho content [19]. It is therefore likely that there is a link among the metabolic profile, PI3K/Akt signaling, and tumorigenicity of undifferentiated mES cells. As a first step in examining this potential relationship, we have used 1H NMR spectroscopy to assess an involvement of the PI3K/Akt pathway in the mechanism underlying development of a putative metabolic fingerprint of pluripotency/tumorigenicity related to choline metabolism (Figure 1). We used a model of mES cells (ES-D3) and their malignant counterparts, murine embryonal carcinoma (mEC) cells (EC-F9), a well-characterized teratocarcinoma cell line [20]. Pluripotent F9 cells show little spontaneous in vivo or in vitro differentiation and, when injected into adult syngeneic mice, grow rapidly forming malignant tumors. Because lineage commitment is associated with loss of ES cell tumorigenic potential, we compared the genotypic, phenotypic, and metabolic profiles of 1) undifferentiated (ESD0) and 2) differentiated (ESD14) mES cells with those of pluripotent mEC (F9) cells.
Figure 1.
Phosphatidylcholine metabolism. The tinted square highlights the pathway of the choline cycle investigated in the study. CDP-choline indicates cytidine diphosphate choline; PKC, protein kinase C; PKA, protein kinase A.
Materials and Methods
Cell Lines
Recent characterization of various stem cell lines demonstrated that major differences in gene expression profiles between pluripotent cells originate from the strain of the source animal [21]. To minimize the impact of this variable on our analysis, we chose closely related cell lines, mES (D3) and mEC (F9), both originating from the 129 strain mouse. Cell lines were purchased from LGC Promochem (Teddington, UK, http://www.lgcstandards-atcc.org). Cell pluripotency was maintained under standardized conditions recommended by the manufacturer. The differentiation protocol for the mES cells did not involve any chemical inducer.
Cell Culture
D3 cells were routinely cultured on 0.1% (vol/vol) gelatin-coated 100-mm tissue culture dishes at a seeding density of 2 x 106 cells per dish, in complete Dulbecco's modified Eagle medium (high-glucose DMEM [Invitrogen, Paisley, UK, http://www.invitrogen.com], 2 mM l-glutamine [Sigma-Aldrich, http://www.sigmaaldrich.com], Primocin [100 µg/ml; AutogenBioclear, Calne, UK, http://www.autogenbioclear.com], 0.1 mM 2-mercaptoethanol [Sigma-Aldrich]) supplemented with 15% (vol/vol) KnockOut Serum Replacement (Invitrogen) and leukemia inhibitory factor (LIF, ESGRO, 1000 U/ml; Millipore, Livingston, UK, http://www.millipore.com). Undifferentiated mES cells (ESD0) at passage 10 to 14 were used. Differentiation of the cells was induced through embryoid body (EB) formation by partial trypsin digestion (0.05% [vol/vol] trypsin/0.53 mM EDTA in 0.1 M PBS without Ca2+ or Mg2+; 2% [vol/vol] chicken serum [Sigma-Aldrich]). EBs were cultured in nonadherent bacterial-grade Petri dishes in complete DMEM supplemented with 10% (vol/vol) batch-tested FBS (Sigma-Aldrich) for 7 days, then transferred to 0.1% (vol/vol) gelatin-coated dishes and grown as adherent cultures for a further 7 days (ESD14) in complete DMEM. The day 14 time point (ESD14), as a differentiation stage of mES cells, was chosen as arbitrary long growth interval based on previous reports demonstrating loss of pluripotency and an almost complete decline in Oct 4 expression in mES (D3) cells cultured for 14 days in the absence of LIF [22].
F9 cells were cultured on 0.1% (vol/vol) gelatin-coated tissue culture dishes (2 x 106/100-mm plate) in complete DMEM supplemented with 10% (vol/vol) batch-tested FBS (Sigma-Aldrich). When 80% confluent, the cells were collected for analyses. All cultures were maintained at 37°C in 10% (vol/vol) carbon dioxide in a humidified tissue culture incubator, and medium was replenished every 24 hours.
In inhibition experiments, 10 µM LY294002 (Merck Chemicals Ltd, Nottingham, UK, http://www.merckbiosciences.co.uk) or vehicle (Me2SO) alone was added to the medium, and cells were incubated for 24 hours before harvesting.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from the cells using Qiagen RNA Miniprep Kit (Qiagen, Crawley, UK, http://www1.qiagen.com), as recommended by the manufacturer. Reverse transcription (RT) and polymerase chain reaction (PCR) with template RNA (0.1 µg per reaction) were carried out using One-Step RT-PCR Kit (Qiagen). The primer sequences were as follows:
alfa-fetoprotein (AFP):
forward 5′-caatctggaagaagaattgc-3′;aps
reverse 5′-acagatccttgtggaagatg-3′ (35 cycles, 428 bp);
T-Brachury (TBra):
forward 5′-atgccaaagaaagaaacgac-3′;
reverse 5′-agaggctgtagaacatgatt-3′ (35 cycles, 835 bp);
neurofilaments-68k (NF-68):
forward 5′-tagagcgcaaagattacctgaag-3′;
reverse 5′-ttgacgttaaggagatcctggta-3′ (35 cycles, 406 bp);
ERas:
forward 5′-aaaaaagcaggctggggaatggctttgccta-3′;
reverse 5′-agaaagctgggtcaaagatcttcaggctacag-3′ (35 cycles, 750 bp) and
β-actin:
forward 5′-aggtcatcactattggcaac-3′;aps
reverse 5′-caaagaaagggtgtaaaacg-3′ (27 cycles, 431 bp).aps
PCR amplification was performed in an Eppendorf Mastercycler (Eppendorf, Cambridge, UK, http://www.eppendorf.co.uk). PCR products were subjected to electrophoresis in 1.5% (wt/vol) Agarose gel in Tris-acetate EDTA buffer and stained with ethidium bromide. Gel images were captured using an ultraviolet ChemiGenius Bioimaging System (Gene-Flow Systems, Fradley, UK, http://www.geneflow.co.uk).
Western Blot Analysis
Cells grown in 100-mm culture plates were washed with ice-cold PBS, lysed in 0.5 ml of cold lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA. 1 mM EGTA, 1% [vol/vol] Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin 1 mM PMSF [Cell Signaling, Hitchin, UK, http://www.cellsignal.com]) for 5 minutes on ice, scraped, and sonicated. The lysates were then cleared by centrifugation at 14,000g for 10 minutes at 4°C. Equal amounts of cell lysate proteins (30 µg) were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane, blocked, and probed with the following antibodies: rabbit anti-Oct 4 (1:1000; Santa Cruz, Heidelberg, Germany, http://www.scbt.com), rabbit anti-Akt (1:1000; Cell Signaling), rabbit anti-phospho (S473) Akt (1:1000; Cell Signaling), and mouse anti-β-actin (1:4000; Abcam, Cambridge, UK, http://www.abcam.com). Membranes were then washed and incubated with secondary antibodies conjugated with donkey antirabbit or antimouse horseradish peroxidase (1:5000; GE Healthcare, Chalfont St Giles, UK, http://www.gehealthcare.com) and visualized by enhanced chemiluminescence applied following the manufacturer's instructions (GE Healthcare). Blots were developed using an automated Kodak Film Developer System (Kodak, Hemel Hempstead, UK, http://www.kodak.com).
NMR Sample Preparation
Cells grown to 70% to 80% confluence were trypsinized, washed twice in ice-cold PBS, and harvested by centrifugation. Three replicate samples of approximately 15 x 106 cells each were prepared per time point and cell line. The pellets were immediately frozen in liquid nitrogen and stored at -80°C. The extraction of intracellular metabolites from cell pellets was performed using a modified Bligh-Dyer procedure [23]. Samples were dried overnight in a centrifugal vacuum concentrator. The dried polar extracts were redissolved in 90% H2O/10% D2O (GOSS Scientific Instruments Ltd, Essex, UK) prepared as 100 mM phosphate buffer (pH 7.0), containing 0.5 mM sodium 3-(trimethylsilyl)-propionate-2,2,3,3-d4 (TMSP; Cambridge Isotope Laboratories, Inc, Andover, MA) as an internal chemical shift reference.
NMR Data Acquisition
A 500-MHz DRX Bruker spectrometer equipped with a cryogenically cooled probe was used for one-dimensional 1H and two-dimensional 1H J-resolved (J-RES) spectra. In both cases, the water resonance was suppressed using excitation sculpting [24]. One-dimensional spectra were acquired using a 30° pulse, with a 5-kHz spectral width, a relaxation delay of 15 seconds, and 256 transients. Two-dimensional J-RES spectra were collected using a double spin-echo sequence with 32 transients per increment and 64 increments. Strong coupling artifacts were suppressed by phase cycling [25].
NMR Data Processing and Postprocessing
NMR data were processed using NMRLaB [26] in the MATLAB (The MathWorks, Natick, MA, http://www.mathworks.com) programming environment. Before Fourier transformation, two-dimensional J-RES spectra were multiplied by a combined sine bell/exponential window function in the direct dimension and by a sine bell function in the incremented dimension [27]. After apodization, two-dimensional J-RES spectra were tilted in the frequency space by 45°, and skyline projections were calculated using NMRLaB to simplify one-dimensional spectra and reduce peak congestion. One-dimensional and projected J-RES spectra were aligned, signals arising from residual solvents, and TMSP were excluded, and the total spectral area was normalized to unity. NMRs of metabolites were assigned and quantified using the Chenomx NMR Suite (version 5.1; Chenomx Inc, Edmonton, Canada). The final concentration of selected metabolites was adjusted according to the coefficients of total spectral area normalization obtained from TMSP peak area-normalized spectra.
Statistical Analysis
The concentrations of metabolites are reported as mean values ± SD. Statistical comparison between data obtained from undifferentiated and differentiated ES and F9 cells was performed using an unpaired Student's t-test (statistical significance: *P < .05, **P < .01, and ***P < .001).
Results
Expression of Markers of Pluripotency and Differentiation
To assess the status of pluripotency and/or differentiation of ESD0, ESD14, and F9 cells, we used a panel of commonly accepted markers.
Oct 4
Oct 4 is a mammalian POU (pit, oct, unc are the subgroup domain transcription factors of homeodomain proteins) transcription factor expressed in the early embryo whose transcriptional activity is tightly controlled throughout embryogenesis. Oct 4 plays a critical role in maintaining pluripotency of ES cells [28]. As its down-regulation is required for differentiation into somatic lineages [29], Oct 4 is widely regarded as a marker of pluripotent stem cells.
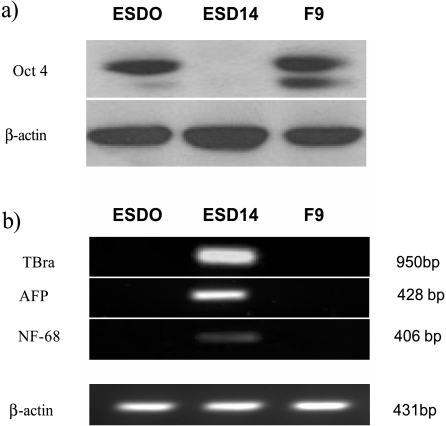
Immunoblot analysis showed strong expression of the protein in both ESD0 and F9 and its absence from ESD14 cells (Figure 2a).
Figure 2.
Pluripotency and differentiation. (a) Western blot analysis for Oct 4 shows strong expression in ESD0 and F9 cells and its absence from ESD14 cells. Expression of β-actin confirms equal loading. (b) Semiquantitative RT-PCR analysis shows expression of transcripts of TBra, AFP, and NF-68, markers of mesoderm, endoderm, and ectoderm, respectively, in differentiated ESD14 cells. In contrast, these transcripts are not detected in undifferentiated ESD0 and F9 cells. β-actin was used as loading control.
Lineage markers
Withdrawal of LIF from the culture medium and cell aggregation into EBs leads to ES cell differentiation and lineage commitment [30]. This is characterized by an up-regulation of transcript markers of early germ layers. Expression of T-Brachury (TBra), alfa-fetoprotein (AFP), and neurofilaments-68k (NF-68), markers of mesoderm, endoderm, and ectoderm, respectively, was studied. In undifferentiated ESD0 and F9 cells, transcripts of TBra, AFP, and NF-68 were not detected. Differentiated ESD14 cells expressed all three lineage markers (Figure 2b).
The inverse relationship between Oct 4 and lineage markers' expression in the three cell lines confirmed the pluripotency of ESD0 and F9 cells and its loss associated with mES cell differentiation (ESD14).
PI3K/Akt Signaling
Murine ES cells maintain a pluripotent phenotype when cultured in the presence of LIF, which acts through the gp130 receptor component and activates the PI3K/Akt cascade [31,32]. In F9 cells, the PI3K/Akt pathway is activated constitutively [33]. Akt is a mediator of many events downstream of PI3K, and its phosphorylation at Ser473 and/or Thr308 is associated with PI3K activity. We compared the involvement of PI3K/Akt to cell pluripotency between normal and malignant mES cells using antibodies to phospho-Akt (Ser473) (pAkt).
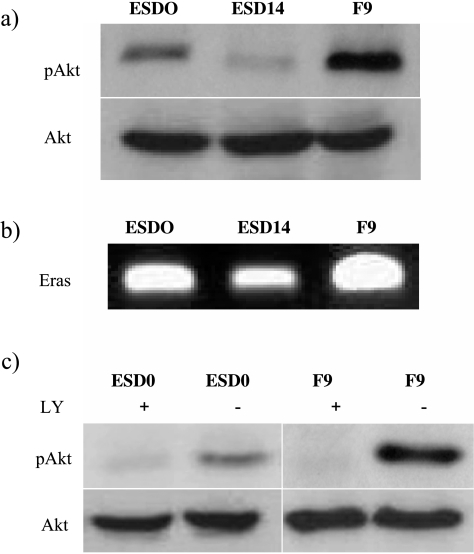
The steady-state levels of PI3K/Akt activity differed between cell lines, with basal levels of pAkt being much higher in F9 cells. In mES cells, a marked decrease of the level of Akt phosphorylation followed cell differentiation (Figure 3a).
Figure 3.
PI3K/Akt signaling. (a)Western blot analysis for pAkt shows its strongest expression in F9 cells. For mES cells, Akt phosphorylation is high in ESD0 cells and decreases with cell differentiation (ESD14). (b) Semiquantitative RT-PCR for Eras transcript shows its strongest expression in F9 cells and weakest in differentiated ESD14 cells. (c) Treatment of cells with 10 µM LY294002 for 24 hours caused an almost complete attenuation of Akt phosphorylation.
Akt is a downstream effector of ERas/PIK3 pathway implicated in tumor-like properties of mES cells [4]. Using RT-PCR, we examined the level of ERas transcripts. Eras showed a gradient of expression from the highest in F9 cells to the lowest in ESD14 cells (Figure 3b), reflecting the pattern observed for pAkt (Figure 3a). These observations are consistent with the suggested role of ERas/PI3K in the functional phenotype of the cells.
NMR Spectra
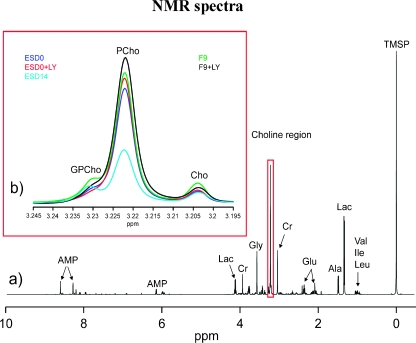
A NMR-based approach was used to search for traits common to pluripotency and tumorigenicity that would encompass the complexity of cell metabolism. We monitored and recorded the changes associated with the loss of pluripotency of mES cells in comparison to the metabolic profile of their pluripotent malignant counterparts. Figure 4a shows a representative one-dimensional NMR spectrum of a ESD0 cell extract. The spectrum is dominated by the resonances of amino acids (e.g., valine, leucine and isoleucine, alanine, glycine, and glutamate), intermediate metabolites of the glycolytic pathway (e.g., lactate), and other compounds related to energy metabolism (e.g., AMP, Figure 4a).
Figure 4.
NMR spectra. (a) Representative 1H NMR spectrum of a mES cell extract. Predominant metabolite resonances are labeled on the spectrum. Val indicates valine; Ile, isoleucine; Leu, leucine; Lac, lactate; Ala, alanine; Gluc, glucose; Cr, creatine; AMP, adenosine monophosphate. (b) Section of 1H NMR spectra (expanded between 3.195 and 3.245 ppm) of mES cells at day 0 with (ESD0 + LY, red line) and without (ESD0, blue line) LY treatment, mES cells after 14 days of growth (cyan line) and teratocarcinoma cell line F9 with (F9 + LY, black line) and without (F9, green line) LY294002 treatment.
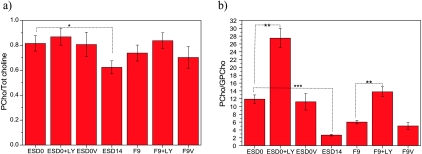
An expanded section of the 1H NMR spectrum relates to choline metabolism, that is, choline, PCho, and GPCho resonances, as shown in Figure 4b (3.195–3.245 ppm). The results demonstrate that the concentration of PCho in pluripotent mES cells (ESD0) was noticeably higher than that in their differentiated progenies (ESD14) and comparable to the PCho content of F9 cells. The relative contribution of PCho to the total choline pool and the PCho/GPCho ratios are depicted in Figure 5, a and b, respectively. Data show that the choline phospholipid profile of mES cells changed considerably with cell differentiation. In ESD14 cells, an overall decrease of choline metabolites was observed (Figure 4b). In addition, the PCho/GPCho ratio showed a dramatic (approximately five-fold) decrease as compared with that of ESD0 cells (Figure 5b).
Figure 5.
Choline phospholipid metabolites. (a) PCho and total choline (Tot choline) ratios. PCho / (GPCho + PCho + Cho) for mES cells at day 0 with (ESD0 + LY) and without (ESD0, ESD0V) LY294002 treatment, mES cells after 14 days of differentiation (ESD14) and teratocarcinoma cells with (F9 + LY) and without (F9, F9V) LY treatment. Differentiation is associated with a significant reduction of PCho/Tot (total) choline (ESD0 vs ESD14, P < .05). In both cell lines, treatment with LY294002 caused observable but not significant increases in the PCho/Tot choline ratio. Vehicle alone (ESD0V; F9V) did not have any impact on PCho/Tot choline ratios. ESD0V and F9V—cells treated with vehicle alone. (b) PCho and GPCho ratios. PCho/GPCho for mES cells at day 0 with (ESD0 + LY) and without (ESD0, ESD0V) LY294002 treatment, mES cells after 14 days of differentiation (ESD14) and teratocarcinoma cells with (F9 + LY) and without (F9, F9V) LY294002 treatment. In differentiated mES cells, there is a significant decrease in PCho/GPCho ratios compared with undifferentiated cells (ESD0 vs ESD14, P < .0001). In both pluripotent cell lines (ESD0 and F9), treatment with LY294002 caused a significant (P < .001) increase in PCho/GPCho ratios. Vehicle alone (ESD0V and F9V) did not have any impact on PCho/GPCho concentrations.
To explore a potential contribution of PI3K/Akt signaling to the metabolic signature of pluripotent normal and malignant cells, we measured the concentrations of choline-related metabolites in cells cultured in the presence of LY294002, a specific PI3K inhibitor (Figure 3c). Figure 5a shows that in cells incubated with LY294002, there was an observable but not significant increase in the relative concentration of PCho. However, in both pluripotent cell lines, inhibition of PI3K activity caused a greater than two-fold increase in the PCho/GPCho ratios, suggesting that PI3K/Akt contributes to the choline cycle and PCho synthesis through mechanisms other than the Kennedy pathway. Addition of vehicle alone to the cell culture medium did not have any significant effect on NMR spectra (Figure 5, a and b).
Discussion
Pluripotent ES cells have a propensity to produce tumors after transplantation, and identification of markers of their tumorigenic potential is of prime importance for their prospective therapeutic use. Here, we provide evidence that metabolic profiling of pluripotent mES cells and their malignant counterparts by 1H NMR spectroscopy identifies features in the choline region of the spectra observed previously in cancer cells and attributed to malignancy [6,11]. Loss of pluripotency and cell lineage commitment is associated with loss of this distinct “cancer-like” metabolic fingerprint.
PCho is a precursor of phosphatidylcholine (PtdCho), the key phospholipid of biologic membranes. An increase in the intracellular pools of PtdCho is required for membrane synthesis and, thus, is believed to be associated with cell proliferation [34].
Our results show that NMR spectra of ESD0 and F9 cells were dominated by high PCho contents and that the rapid growth characteristics of pluripotent ES cells could account for these observed elevated PCho levels. However, comparative studies of human normal and malignant epithelial cells with similar doubling times demonstrated that accelerated cell growth alone cannot justify the high concentrations of choline phospholipid metabolites, suggesting a close link between the rise in PCho metabolism and malignancy [11,35]. In addition to an increase in phospholipid biosynthesis and breakdown, malignant transformation is also associated with alterations in relative concentrations of choline-containing metabolites. In particular, “a glycerophosphocholine to phosphocholine switch” is proposed as a hallmark of malignant phenotype [8,11–13]. Our results show an overall decline of choline metabolites and, more significantly, a decrease of the relative concentration of PCho in differentiated (ESD14) mES cells and are in agreement with the recently reported NMR profiling of the same mES cell line [19]. Comparing the PCho content of pluripotent mES cells with the database of transformed somatic cell lines, Jansen et al. [19] have implied that mES and cancer cells may share a choline metabolome. Our study was carried out in a model system of closely related cell lines to minimize the possible impact of the cell genetic background on their metabolic profiles and its results provide support for this hypothesis. The data point at striking similarities between the choline regions of pluripotent mES and F9 cell spectra and demonstrate a greater-than five-fold decrease of PCho/GPCho ratio in response to cell differentiation. It is tempting to speculate that this inversion of the GPCho to PCho switch reflects loss of mES cell tumorigenicity and reversal to a “normal” phenotype.
We investigated the potential link between distinct features of choline phospholipid metabolism shared by pluripotent mES and F9 cells and the PI3K/Akt pathway. The PI3K/Akt signaling, long recognized as a survival pathway in mammalian tumorigenesis, is also a known regulator of self-renewal in various stem cell systems [31,36–38]. Recent work by Takahashi et al. [4] has provided evidence that signaling through the PI3K pathway promotes tumorigenicity of pluripotent mES cells. The PI3K pathway has also been associated with choline phosopholipid metabolism and shown to mediate Ras-induced activation of ChoK [18], the key enzyme of the dominant pathway for PCho [34], that is strongly implicated in mammalian carcinogenesis [14]. Although the regulation of PI3K activity in ES cells, as well as the downstream signaling targets, differ from those in somatic cells [31,32,36–39], a link between PI3K activity, tumorigenicity of pluripotent cells, and PCho biosynthesis seemed very plausible. Our data show that loss of pluripotency is associated with decreases in both PCho levels and PI3K/Akt activity. However, inhibition of PI3K in pluripotent mES and F9 cells did not have any significant effect on relative PCho concentration and, in both cell lines, caused a similar, greater-than two-fold increase in the PCho/GPCho ratio. These results show that in our in vitro model, the PI3K/Akt signaling pathway is not involved in the de novo synthesis of PCho and PtdCho through the Kennedy pathway, ruling out the PI3K/PCho as a mechanism underlying the putative metabolic signature of pluripotency/tumorigenicity.
ChoK activation may be stimulated by a number of other mechanisms including availability of choline [40], phospholipase D [18], protein kinase C, or protein kinase A [17]. Interestingly, aberrant activities of these enzymes have been associated with malignant transformation of mammalian somatic cells and human cancer [41–43]. Furthermore, PCho is a product of both PtdCho biosynthesis and catabolism, and besides increased activity of ChoK, its accumulation may also be a consequence of an activation of hydrolytic enzymes of the choline cycle [34]. In particular, up-regulation of PtdCho-specific phospholipase C (PC-PLC) catalyzing the breakdown of PtdCho to PCho has recently been reported in human ovarian cancer and demonstrated to correlate with tumor progression [13,44]. Thus, it seems that alternative or combined activation of various pathways of phospholipid metabolism is responsible for the development of the distinct metabolic trait observed in our study. Whatever the mechanism, our results demonstrate that PI3K/Akt, thus far the only mechanism positively identified as a link between pluripotency and tumorigenicity, does not underlie the metabolic signature believed to reflect tumorigenic potential of pluripotent cells.
Development of molecular fingerprints of stemness/tumorigenicity may have implications reaching far beyond regenerative medicine. According to the cancer stem (CS) cell hypothesis, recently revived with a new impetus, these cancer-initiating cells share many features with normal stem cells, and their biology is governed by self-renewal pathways [45]. A comprehensive analysis by Wong et al. [46] supports this hypothesis and demonstrates that a transcriptional ES cell-like program is indeed reactivated in multiple epithelial cancers and may be important for the CS cell function. Because CS cells are believed to be responsible not only for sustained cancer progression but also for the failure of eradicating therapies [47], considerable effort is being made in the search for a fingerprint of stemness common to both normal and malignant stem cells. Exhaustive data generated by gene and protein expression analyses are inconsistent and, thus far, do not provide the basis for a molecular definition of stemness [48,49]. It seems that integration of various approaches, including metabolomics reflecting the outcome of multiple molecular signaling pathways [50], is required for establishing the criteria for identification of true stem cells. Our study contributes to this worldwide search and suggests that PCho and PCho/GPC levels might provide surrogate metabolic markers of stemness/tumorigenicity common to pluripotent stem and cancer cells. Understanding of the mechanisms underlying the development of this signature, demonstration of its potential existence in human ES cells, as well as in vivo validation require further studies.
Footnotes
The authors thank the EU for supporting S.T. through the Marie Curie Transfer of Knowledge award MOTET (MTKD-CT-2004-014434). The authors also thank the Wellcome Trust and the EU in the context of the EU-NMR grant (RII3-026145) for supporting the HWB-NMR facility in Birmingham.
References
- 1.Takahashi K, Ichisaka T, Yamanaka S. Identification of genes involved in tumor-like properties of embryonic stem cells. Methods Mol Biol. 2006;329:449–458. doi: 10.1385/1-59745-037-5:449. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Armstrong SA. Cancer: inappropriate expression of stem cell programs? Cell Stem Cell. 2008;2:297–299. doi: 10.1016/j.stem.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 5.Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, Reid LM. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26:1547–1555. doi: 10.1634/stemcells.2007-0863. [DOI] [PubMed] [Google Scholar]
- 6.Ackerstaff E, Glunde K, Bhujwalla ZM. Choline phospholipid metabolism: a target in cancer cells? J Cell Biochem. 2003;90:525–533. doi: 10.1002/jcb.10659. [DOI] [PubMed] [Google Scholar]
- 7.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 8.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001;61:3599–3603. [PubMed] [Google Scholar]
- 9.Tiziani S, Lodi A, Khanim FL, Viant MR, Bunce CM, Günther UL. Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS ONE. 2009;4:e4251. doi: 10.1371/journal.pone.0004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiziani S, Lopes V, Günther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 12.Ronen SM, Jackson LE, Beloueche M, Leach MO. Magnetic resonance detects changes in phosphocholine associated with Ras activation and inhibition in NIH 3T3 cells. Br J Cancer. 2001;84:691–696. doi: 10.1054/bjoc.2000.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D'Ascenzo S, Millimaggi D, Pavan A, Dolo V, Canevari S, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–9376. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 14.Glunde K, Bhujwalla ZM. Choline kinase α in cancer prognosis and treatment. Lancet Oncol. 2007;8:855–857. doi: 10.1016/S1470-2045(07)70289-9. [DOI] [PubMed] [Google Scholar]
- 15.Ramirez de Molina A, Sarmentero-Estrada J, Belda-Iniesta C, Taron M, Ramirez dMV, Cejas P, Skrzypski M, Gallego-Ortega D, de Castro J, Casado E, et al. Expression of choline kinase a to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 16.Nimmagadda S, Glunde K, Pomper MG, Bhujwalla ZM. Pharmacodynamic markers for choline kinase down-regulation in breast cancer cells. Neoplasia. 2009;11:477–484. doi: 10.1593/neo.81430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi MG, Kurnov V, Kersting MC, Sreenivas A, Carman GM. Phosphorylation of the yeast choline kinase by protein kinase C. Identification of Ser25 and Ser30 as major sites of phosphorylation. J Biol Chem. 2005;280:26105–26112. doi: 10.1074/jbc.M503551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002;21:937–946. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- 19.Jansen JF, Shamblott MJ, van Zijl PC, Lehtimaki KK, Bulte JW, Gearhart JD, Hakumaki JM. Stem cell profiling by nuclear magnetic resonance spectroscopy. Magn Reson Med. 2006;56:666–670. doi: 10.1002/mrm.20968. [DOI] [PubMed] [Google Scholar]
- 20.Solter D, Shevinsky L, Knowles BB, Strickland S. The induction of antigenic changes in a teratocarcinoma stem cell line (F9) by retinoic acid. Dev Biol. 1979;70:515–521. doi: 10.1016/0012-1606(79)90043-5. [DOI] [PubMed] [Google Scholar]
- 21.Sharova LV, Sharov AA, Piao Y, Shaik N, Sullivan T, Stewart CL, Hogan BL, Ko MS. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toumadje A, Kusumoto K-I, Parton A, Mericko P, Dowell L, Ma G, Chen L, Barnes DW, Sato JD. Pluripotent differentiation in vitro of murine ES-D3 embryonic stem cells. In Vitro Cell Dev Biol Anim. 2003;39:449–453. doi: 10.1290/1543-706X(2003)039<0449:PDIVOM>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Southam AD, Hines A, Viant MR. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal Biochem. 2008;372:204–212. doi: 10.1016/j.ab.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Hwang TL, Shaka AJ. Water suppression that works—excitation sculpting using arbitrary waveforms and pulsed field gradients. J Magn Reson A. 1995;112:275–279. [Google Scholar]
- 25.Thrippleton MJ, Edden RA, Keeler J. Suppression of strong coupling artefacts in J-spectra. J Magn Reson. 2005;174:97–109. doi: 10.1016/j.jmr.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Günther UL, Ludwig C, Rüterjans H. NMRLaB-advanced NMR data processing in MATLAB. J Magn Reson. 2000;145:201–208. doi: 10.1006/jmre.2000.2071. [DOI] [PubMed] [Google Scholar]
- 27.Tiziani S, Lodi A, Ludwig C, Parsons HM, Viant MR. Effects of the application of different window functions and projection methods on processing of 1H J-resolved nuclear magnetic resonance spectra for metabolomics. Anal Chim Acta. 2008;610:80–88. doi: 10.1016/j.aca.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 28.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:37–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 29.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 30.Wobus AM, Holzhausen H, Jakel P, Schoneich J. Characterization of a pluripotent stem cell line derived from a mouse embryo. Exp Cell Res. 1984;152:212–219. doi: 10.1016/0014-4827(84)90246-5. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe S, Umehara H, Murayama K, Okabe M, Kimura T, Nakano T. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 32.Jirmanova L, Afanassieff M, Gobert-Gosse S, Markossian S, Savatier P. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene. 2002;21:5515–5528. doi: 10.1038/sj.onc.1205728. [DOI] [PubMed] [Google Scholar]
- 33.Lianguzova MS, Chuikin IA, Pospelov VA. PI 3-kinase activity is necessary for F9 mouse embryonic carcinoma cell proliferation. Tsitologiia. 2004;46:26–34. [PubMed] [Google Scholar]
- 34.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–439. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 35.Ting YL, Sherr D, Degani H. Variations in energy and phospholipid metabolism in normal and cancer human mammary epithelial cells. Anticancer Res. 1996;16:1381–1388. [PubMed] [Google Scholar]
- 36.Takahashi K, Murakami M, Yamanaka S. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem Soc Trans. 2005;33:1522–1525. doi: 10.1042/BST0331522. [DOI] [PubMed] [Google Scholar]
- 37.Paling NR, Wheadon H, Bone HK, Welham MJ. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J Biol Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 38.Kim SJ, Cheon SH, Yoo SJ, Kwon J, Park JH, Kim CG, Rhee K, You S, Lee JY, Roh SI, et al. Contribution of the PI3K/Akt/PKB signal pathway to maintenance of self-renewal in human embryonic stem cells. FEBS Lett. 2005;579:534–540. doi: 10.1016/j.febslet.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 39.Savatier P, Lapillonne H, Jirmanova L, Vitelli L, Samarut J. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol. 2002;185:27–33. doi: 10.1385/1-59259-241-4:27. [DOI] [PubMed] [Google Scholar]
- 40.Lacal JC. Choline kinase: a novel target for antitumor drugs. IDrugs. 2001;4:419–426. [PubMed] [Google Scholar]
- 41.Reich R, Blumenthal M, Liscovitch M. Role of phospholipase D in laminin induced production of gelatinase A (MMP-2) in metastatic cells. Clin Exp Metastasis. 1995;2:134–140. doi: 10.1007/BF00133618. [DOI] [PubMed] [Google Scholar]
- 42.Tortora G, Ciardiello F. Antisense targeting protein kinase A type I as a drug for integrated strategies of cancer therapy. Ann N Y Acad Sci. 2003;1002:236–243. doi: 10.1196/annals.1281.026. [DOI] [PubMed] [Google Scholar]
- 43.Mackay HJ, Twelves CJ. Targeting the protein kinase C family: are we there yet? Nat Rev Cancer. 2007;7:554–562. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 44.Spadaro F, Ramoni C, Mezzanzanica D, Miotti S, Alberti P, Cecchetti S, Iorio E, Dolo V, Canevari S, Podo F. Phosphatidylcholine-specific phospholipase C activation in epithelial ovarian cancer cells. Cancer Res. 2008;68:6541–6549. doi: 10.1158/0008-5472.CAN-07-6763. [DOI] [PubMed] [Google Scholar]
- 45.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 46.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 48.Baharvand H, Fathi A, Gourabi H, Mollamohammadi S, Salekdeh GH. Identification of mouse embryonic stem cell-associated proteins. J Proteome Res. 2008;7:412–423. doi: 10.1021/pr700560t. [DOI] [PubMed] [Google Scholar]
- 49.Fortunel NO, Otu HH, Ng HH, Chen J, Mu X, Chevassut T, Li X, Joseph M, Bailey C, Hatzfeld JA, et al. Comment on “‘Stemness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- 50.Kell DB. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discov Today. 2006;11:1085–1092. doi: 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]