Abstract
PTPRCAP (CD45-AP) is a positive regulator of protein tyrosine phosphatase PTPRC (CD45), which activates Src family kinases implicated in tumorigenesis. Single-nucleotide polymorphism (SNP) rs869736 located at position -309 of the PTPRCAP promoter was associated with susceptibility to diffuse-type gastric cancer in the current case-control study. The minor-allele homozygote was significantly associated with a 2.5-fold increased susceptibility to diffuse-type gastric cancer (P = .0021, n = 252), but not to intestinal-type (P = .30, n = 178), versus the major-allele homozygote, when comparing unrelated Korean patients with healthy controls (n = 406). Nine other SNPs were in nearly perfect linkage disequilibrium (r2 ≥ 0.97) with this SNP, exhibiting the same association, and spread out for 26 kb on chromosome 11q13.1 covering RPS6KB2, PTPRCAP, CORO1B, and GPR152. Among the four genes, however, only PTPRCAP expression was affected by haplotypes of the 10 SNPs. Endogenous transcript levels of PTPRCAP were linearly correlated with copy numbers (0, 1, and 2) of the risk-haplotype (P = .0060) in 12 lymphoblastoid cells derived from blood samples, but those of the other three genes were not. Furthermore, the cancer-risk, minor-allele T of rs869736 increased both promoter activity and specific nuclear protein-binding affinity than the nonrisk, major-allele G in luciferase reporter and electrophoretic mobility shift assays, respectively. Accordingly, the minor allele of rs869736 in the PTPRCAP promoter is associated with increased susceptibility to diffuse-type gastric cancer by increasing PTPRCAP expression, possibly leading to activation of the oncogenic Src family kinases.
Introduction
Gastric cancer is a complex disease, and many environmental and genetic factors are involved in its development. Helicobacter pylori infection, smoking, and dietary habits are well-established environmental risk factors [1–4]. Human genetic polymorphisms of IL1B, IL1R2, IL10, TLR4, TNF, and PSCA have been associated with susceptibility to gastric cancer [5–8]. All of these, except for PSCA, are related to proinflammatory signals and may accelerate the cancerous changes such as inflammation, hypochlorhydria, and atrophic gastritis, which can explain the sequential events of intestinal-type gastric carcinogenesis. PSCA was identified as a diffuse-type gastric cancer gene in a genome-wide association study and is possibly involved in regulating gastric epithelial cell proliferation [8].
Notably, most genetic associations identified to date are specific for histologic subtypes of gastric cancer, and it reflects their distinct features in histologic, pathologic, and epidemiological aspects [9]. Intestinal-type gastric cancer develops through a series of precancerous changes known as Correa sequence, superficial gastritis, chronic atrophic gastritis, intestinal metaplasia, dysplasia, and adenocarcinoma [10]. In contrast, diffuse-type gastric cancer is not associated with atrophic gastritis or intestinal metaplasia, and lacks well-recognized precursor changes [11–13]. H. pylori gastritis is the only common precursor and a major risk factor in both types of gastric cancer [2,11].
Reversible tyrosine phosphorylation, which is regulated by the balanced yin-yang (complementary opposites within a greater whole) action of protein tyrosine kinases and protein tyrosine phosphatases, plays an important role in cell proliferation, adhesion, and migration [14]. The dynamic interactions between protein tyrosine kinases and phosphatases act as molecular switches for signal transduction pathways [15], and deregulated protein tyrosine phosphorylation is associated with tumorigenesis [14]. The protein tyrosine phosphatase receptor type C (PTPRC), also known as CD45, transmits important cellular signals by dephosphorylating the inhibitory tyrosine residue of Src family kinases (SFKs) [16], which are implicated in tumor progression and metastasis [17,18]. The phosphatase activity of PTPRC is enhanced by interaction with a transmembrane protein, PTPRC-associated protein (PTPRCAP), also known as CD45-AP [19–22]. However, little is known about the relation between PTPRCAP and gastric cancer.
In this study, the first evidence for genetic association of PTPRCAP with susceptibility to diffuse-type gastric cancer is presented. A promoter single-nucleotide polymorphism (SNP) G>T at position -309 of PTPRCAP affects the promoter activity and is associated with in vivo expression levels of PTPRCAP and susceptibility to diffuse-type gastric cancer.
Materials and Methods
Study Subjects
This study included 836 unrelated Korean subjects including 430 patients with gastric cancer and 406 healthy controls. The gastric cancers were classified either diffuse- (n = 252) or intestinal-type (n = 178) according to the Lauren system by two independent pathologists, and the mixed-type or ambiguous cases were not included in the study. The tumor stages at diagnosis were diverse: stage IA (n = 42), IB (n = 66), II (n = 86), IIIA (n = 106), IIIB (n = 58), and IV (n = 72) according to the AJCC Cancer Staging Manual, and 58 of them were early and 372 were advanced gastric carcinoma by tumor depth. All the controls were confirmed free of the disease by health examinations including an endoscopy or an upper gastrointestinal track radiography. Patients were aged 57.3 ± 12.9 years (mean ± SD), ranging from 23 to 86 years, whereas the controls were 52.1 ± 8.4 years old, ranging from 39 to 76 years. The male-to-female ratio was 2.0 in patients and 2.3 in controls. These subjects were recruited with written informed consent at Seoul National University Hospital, Hanyang University Guri Hospital, and Inje University Seoul Paik Hospital in the Seoul metropolitan area and at Chungnam National University Hospital and Eulji University Hospital in Daejeon city, with an approval from the institutional review board of Hanyang University Medical Center, and genotyped with an approval from the institutional review board of KAIST.
Genotyping and Sequencing
Genomic DNA was extracted from blood samples using the Puregene DNA purification kit (Gentra, Minneapolis, MN) or from frozen nontumorous gastric tissues using DNeasy tissue kit (Qiagen, Hilden, Germany), and quantified using the double-stranded DNA-specific fluorescent dye, PicoGreen (Molecular Probes, Eugene, OR). The final concentration of each DNA sample was adjusted to 2.5 to 10 ng/µl for genotyping assays. SNPs were genotyped using the MassARRAY system (Sequenom, San Diego, CA) according to the manufacturer's instructions. The overall genotyping success rate was 99.6% for the 23 SNPs, and genotype frequencies of the control subjects in all SNPs were under Hardy-Weinberg equilibrium.
Polymerase chain reaction (PCR) was performed using Pfu DNA polymerase (Solgent, Daejeon, Korea), and the products were purified from agarose gels using AccuPrep gel purification kit (Bioneer, Daejeon, Korea). Genomic DNA from two subjects, a younger (43 years old) patient with an advanced (stage IV) diffuse-type gastric cancer and an older (69 years old) healthy control were sequenced by the Solgent Corp. All primers for genotyping and sequencing analysis were synthesized by the Bioneer Corp.
Statistical Analyses
Allelic association of individual SNP with susceptibility to diffuse or intestinal-type gastric cancer and Hardy-Weinberg equilibrium in the control subjects were assessed by χ2 tests. Haplotypes were constructed using PHASE 2.1 and linkage disequilibrium (LD) maps using Haploview 4.1 [23–25]. In logistic regression analyses of genotype and diplotype associations, odds ratio (OR) and 95% confidence interval (CI) were adjusted for age and sex using SPSS 11.5 because the age distribution and sex ratio were different between the cases and controls.
Real-time Reverse Transcription-PCR
Genomic DNA was isolated from 12 lymphoblastoid cell lines and genotyped for rs1808279. Total RNA was extracted using NucleoSpin (Macherey-Nagel, Düren, Germany), and the first-strand complementary DNA was synthesized using an oligo(dT) primer and Improm-II reverse transcription (Promega, Madison, WI). Real-time PCR was performed using the Bio-Rad iQ SYBR Green Supermix and iCycler iQ5 machine. The first-strand complementary DNA and RNA (as a negative control) samples were amplified for four genes (PTPRCAP, RPS6KB2, CORO1B, and GPR152), and the amounts were normalized against that of GAPDH.
Luciferase Reporter Assays
DNA segments containing one of the two alleles in an SNP was amplified by PCR using Pfu DNA polymerase (Solgent) and cloned into the KpnI and XhoI sites of pGL3-Basic vector (Promega) or to the XbaI site of pGL3-Promoter vector. Especially, a PTPRCAP promoter region was amplified using previously reported primer sequences [26]. All the recombinant constructions were confirmed by sequencing analysis.
MKN28 cells (2 x 105), maintained in RPMI1640 (Gibco, Rockville, MD) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), were transfected with 1 µg of plasmid and 50 ng of pRL-CMV control vector in 12-well tissue culture plates using Lipofectamine LTX reagent (Invitrogen, Carlsbad, CA). Cells were harvested at 36 hours after transfection, and luciferase activities were measured using the dual-luciferase reporter assay system (Promega). All experiments were performed in triplicate.
Electrophoretic Mobility Shift Assays
Double-stranded oligonucleotides for electrophoretic mobility shift assay (EMSA) were generated using complementary oligonucleotides 5′-AACCTGGAGACAGAGGATGTCACAGGAGT-3′ and 5′-ACTCCTGTGACATCCTCTGTCTCCAGGTT-3′ for the -309G carrier, and 5′-AACCTGGAGACAGATGATGTCACAGGAGT-3′ and 5′-ACTCCTGTGACATCATCTGTCTCCAGGTT-3′ for the -309T carrier (the SNP-corresponding position is underlined). Double-stranded probes were radioactively labeled with [γ-32P]dATP (Perkin Elmer, Wellesley, MA) at the 5′ ends using the T4 polynucleotide kinase (Takara Bio, Shiga, Japan), and unincorporated [γ-32P]dATP was removed using the PROBER probe DNA purifying system (iNtRON Biotechnology, Seongnam, Korea).
A nuclear extract (8 µg) from MKN28 cells and 0.6 µg of poly(dIdC) were preincubated with or without 200-fold molar excess of unlabeled competitor in 1x EMSA reaction buffer (10 mM HEPESKOH, pH 7.9, 60 mM KCl, 1 mM EDTA, 10% glycerol, 10 mM MgCl2, and 200 µM dithiothreitol) for 20 minutes at room temperature. Incubation was continued with addition of a radioactively labeled probe (80 fmol) for another 20 minutes at room temperature. The whole 20 µl of the reaction product was subjected to gel electrophoresis (6% polyacrylamide gel in 0.25x Tris/borate/EDTA running buffer at 180 V for 2.5 hours). Finally, the gel was scanned by BAS- 3000 (Fujifilm, Tokyo, Japan).
Prediction for Transcription Factor Binding
Transcription factors that can bind to the PTPRCAP SNP rs869736 at position -309 in an allele-specific manner were predicted for the two 30-bp sequences, 5′-AACCTGGAGACAGA[G/T]GATGTCACAGGAGTC-3′ using seven different programs: AliBaba 2.1 (BIOBASE, Wolfenbuettel, Germany), Match 1.0 (BIOBASE), MatInspector (Genomatix, Munich, Germany), Patch 1.0 (BIOBASE), TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess), TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html), and Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl). Their default settings were used, but target organisms were restricted to vertebrates or mammalians.
Results
A 26-kb Locus on 11q13.1 Associated with Diffuse-type Gastric Cancer
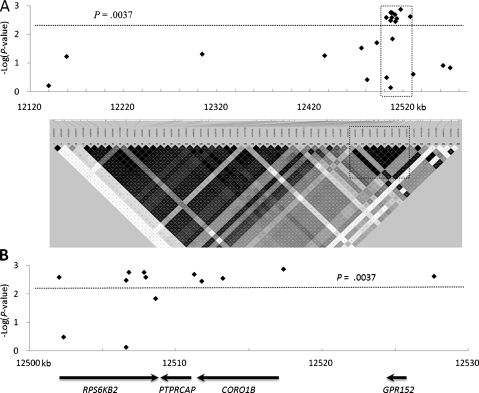
Many SNPs around the PTPRCAP gene on human chromosome 11q13.1 were in high LD with one another according to the genotype data from the International HapMap Project (Figure 1A). From a 430-kb region surrounding PTPRCAP, 18 SNPs were selected and genotyped for 252 unrelated Korean patients with diffuse-type gastric cancer and 406 healthy controls. Among them, seven SNPs (rs1476792 and rs1790753 in RPS6KB2; rs869736 in PTPTCAP, rs872375, rs2302264, and rs1808279 in CORO1B; and rs1790761 in GPR152) showed significant allelic associations with susceptibility to diffuse-type gastric cancer (Table 1).
Figure 1.
Association profile and LD map of all 23 tested SNPs. (A) P values in negative logarithm (y axis) for allelic associations with diffuse-type gastric cancer susceptibility are shown against chromosome positions (x axis) of 23 SNPs in the upper half. An LD map of 430-kb region generated by the genotype data of 56 SNPs with minor allele frequencies of 0.1 or higher in the International HapMap CHB + JPT population is shown in the lower half. (B) A detailed view of the 30-kb region boxed in A. The region contains four genes and the 10 SNPs significantly associated with the susceptibility to diffuse-type gastric cancer (.0013 ≤ P ≤ .0036).
Table 1.
Allelic Association Tests of 23 SNPs with Diffuse-type Gastric Cancer.
| SNP | Gene | Function | Allele* | MAF† | OR (95% CI)‡ | P‡ |
| rs2282502 | RHOD | Asp88 | C>T | 0.42 | 0.95 (0.75–1.18) | .62 |
| rs10791899 | Intergenic | A>C | 0.22 | 1.28 (0.99–1.66) | .060 | |
| rs3927807 | FBXL11 | Intron | G>A | 0.23 | 1.29 (1–1.67) | .050 |
| rs1790740 | Intergenic | T>C | 0.23 | 1.28 (0.99–1.65) | .056 | |
| rs7480390 | Intergenic | C>G | 0.23 | 1.33 (1.03–1.71) | .030 | |
| rs10896172 | TBC1D10C | Intron | T>C | 0.18 | 1.13 (0.85–1.5) | .39 |
| rs1790733 | KIAA1394 | Intron | T>C | 0.25 | 1.34 (1.05–1.72) | .020 |
| rs1476792 | RPS6KB2 | Intron | T>C | 0.29 | 1.44 (1.13–1.82) | .0026 |
| rs917570 | RPS6KB2 | Intron | C>G | 0.18 | 1.16 (0.87–1.54) | .32 |
| rs55987642 | RPS6KB2 | Pro267Leu | C>T | 0.07 | 1.08 (0.7–1.67) | .74 |
| rs4930427 | RPS6KB2 | Phe269 | C>T | 0.29 | 1.43 (1.12–1.81) | .0033 |
| rs1790753 | RPS6KB2 | Intron | G>A | 0.29 | 1.46 (1.15–1.85) | .0017 |
| rs1790752 | RPS6KB2 | Intron | C>T | 0.29 | 1.46 (1.15–1.85) | .0017 |
| rs13859 | RPS6KB2 | Ala420Val | C>T | 0.29 | 1.44 (1.13–1.82) | .0026 |
| rs10274 | RPS6KB2 | 3′ UTR | G>A | 0.25 | 1.36 (1.06–1.75) | .014 |
| rs869736 | PTPRCAP | Promoter | G>T | 0.29 | 1.45 (1.14–1.83) | .0020 |
| rs872375 | CORO1B | Intron | G>A | 0.29 | 1.42 (1.12–1.8) | .0036 |
| rs2302264 | CORO1B | Intron | G>A | 0.29 | 1.43 (1.13–1.81) | .0028 |
| rs1808279 | CORO1B | Promoter | A>G | 0.29 | 1.47 (1.16–1.86) | .0013 |
| rs1790761 | GPR152 | Promoter | A>G | 0.29 | 1.44 (1.14–1.82) | .0024 |
| rs17501521 | CABP4 | Intron | C>T | 0.11 | 1.22 (0.87–1.72) | .25 |
| rs4084113 | AIP | Intron | G>A | 0.18 | 1.24 (0.94–1.64) | .12 |
| rs2276120 | PITPNM1 | Intron | C>T | 0.18 | 1.23 (0.93–1.62) | .15 |
Significant values after correction for multiple testing (P ≤ .0037) are underlined.
Major allele > minor allele.
Minor allele frequency in controls.
Crude OR, 95% CI, and P for allelic association test.
These seven associated SNPs were very tightly linked to each other (r2 ≥ 0.97), together constituting an LD block of 26 kb, and other 11 SNPs located outside this LD block were not associated (Figure 1A). Thus, a gastric cancer susceptibility locus was localized to a 26-kb block containing four genes, RPS6KB2, PTPRCAP, CORO1B, and GPR152 by the seven SNPs very tightly linked together (Figure 1B).
Haplotypes Associated with Diffuse-type Gastric Cancer
To find more polymorphisms that are tightly linked to the seven associated SNPs and potentially affect protein activity or gene expression, all the exons, exon-intron junctions, and promoters (1–1.5 kb from the transcription start site) of the four genes in genomic DNA from two extreme individuals were sequenced. The two individuals were a younger patient with an advanced diffuse-type gastric cancer who was homozygous for the seven risk alleles and an older healthy control homozygous for the nonrisk alleles. Between the two individuals, the alleles were also different in five additional SNPs, all located in RPS6KB2: rs55987642, rs13859, rs4930427, rs10274, and rs1790752. Genotyping of these additional five SNPs disclosed that only three of them, rs4930427, rs1790752, and rs13859, were significantly associated with the diffuse-type gastric cancer (Table 1). From a total of 23 SNPs tested, 10 SNPs were significantly associated with diffuse-type gastric cancer and in nearly perfect LD with each other (r2 ≥ 0.97).
All the 10 SNPs passed a significance level of association adjusted for multiple testing. The adjusted significance level (α = 1 - (1 - 0.05)1/13.86 = 0.0037) was calculated with the effective degree of freedom estimated for the set of SNP genotypes in LD, using the EDF software [27]. Moreover, four SNPs of these passed even the significance level of Bonferroni correction (α = 1 - (1 - 0.05)1/23 = 0.0022) for multiple testing; however, the Bonferroni method could be an overcorrection in cases with very tightly linked SNPs, similar to this case.
When haplotypes were constructed with the 10 significantly associated SNPs (their P values are underlined in Table 1), two haplotypes, TCGCCGGGAA (designated as H1) and CTATTTAAGG (H2), in the order of SNP nucleotide positions listed in Table 1, constituted 98.1% of entire haplotypes, and only a small portion of the subjects had other haplotypes (1.4% in controls and 2.8% in diffuse-type cancer patients). The H2 haplotype with the risk allele at every SNP was significantly associated with susceptibility to diffuse-type cancer (OR = 1.43, P = .00340) versus the H1 with the nonrisk alleles at all SNPs. Logistic regression analysis of diplotype association revealed 2.50- and 1.46-fold higher odds of having diffuse-type cancer in the H2/H2 homozygotes (P = .0021) and H1/H2 heterozygotes (P = .030), respectively, in comparison with the H1/H1 homozygotes (Table 2). Thus, the 10-SNP haplotypes were associated with susceptibility to diffuse-type gastric cancer.
Table 2.
Association of Haplotypes with Diffuse-type Gastric Cancer.
| Category | Control | Diffuse-type | OR (95% CI)* | P* |
| Haplotype† | 2n = 812 | 2n = 504 | ||
| H1 (TCGCCGGGAA) | 571 (70.3%) | 311 (61.7%) | 1 | |
| H2 (CTATTTAAGG) | 230 (28.3%) | 179 (35.5%) | 1.43 (1.12–1.82) | .0034 |
| Others | 11 (1.4%) | 14 (2.8%) | ||
| Diplotype | n = 406 | n = 252 | ||
| H1/H1 | 196 (48.3%) | 92 (36.5%) | 1 | |
| H1/H2 | 176 (43.3%) | 118 (46.8%) | 1.46 (1.04–2.07) | .030 |
| H2/H2 | 27 (6.7%) | 30 (11.9%) | 2.50 (1.39–4.47) | .0021 |
| Others | 7 (1.7%) | 12 (4.8%) |
Crude OR, 95% CI, and P values calculated in χ2 tests for haplotype association, and age/sex-adjusted OR, 95% CI, and P values calculated in logistic regression analysis for diplotype association.
In each haplotype, the alleles are listed in the SNP order of rs1476792, rs4930427, rs1790753, rs1790752, rs13859, rs869736, rs872375, rs2302264, rs1808279, and rs1790761.
Association of the Risk Haplotype with Increased In Vivo Transcript Levels of PTPRCAP
As explained in the previous section, a diffuse-type gastric cancer susceptibility locus was mapped to a 26-kb region, which contains four genes, RPS6KB2, PTPRCAP, CORO1B, and GPR152 (Figure 1B). Because the strong LD of this locus did not allow for a further fine mapping by association analysis, functional analyses were then tried to identify the susceptibility gene(s). Abnormal protein activity or altered gene expression may cause an increased susceptibility to cancer. In this susceptibility locus, two coding SNPs in RPS6KB2 showed significant association, but there was no amino acid change (Phe269) in rs4930427 or the change was too small (Ala420Val) in rs13859 to alter protein activity dramatically.
The next step involved investigating whether expression of any of the four genes was altered by the risk haplotype in vivo. Twelve lymphoblastoid cell lines derived from healthy individuals were genotyped for rs1808279, which was almost perfectly linked to all the other haplotype-constituent SNPs (r2 ≥ 0.97) and was most significantly associated with the diffuse-type cancer (P = .0013). They were categorized into three groups according to their diplotypes that were estimated by their rs1808279 genotypes: five samples were homozygous for the nonrisk haplotype (H1/H1), five samples were heterozygous (H1/H2), and two samples were homozygous for the risk haplotype (H2/H2). Total RNA was isolated from each cell line and expression levels of the four endogenous genes were measured by real-time reverse transcription-PCR, except for those of GPR152, which were too low to make reliable measurements.
As shown in Figure 2, the risk-haplotype copy number was significantly correlated with the endogenous transcript levels of PTPRCAP (P = .0060) in linear regression analysis but not with those of RPS6KB2 (P = .29) or CORO1B (P = .53). The linear correlation was positive, supporting for association of PTPRCAP expression with the risk haplotype in an additive genetic model (Figure 2). PTPRCAP transcript levels of the H2/H2 homozygotes and H1/H2 heterozygotes were 35% and 18% higher than those of the H1/H1 homozygotes, respectively. Thus, the diffuse-type gastric cancer risk haplotype was associated with increased endogenous transcript levels of PTPRCAP but not with those of RPS6KB2, CORO1B, or GPR152.
Figure 2.
Relationship between the haplotypes and gene expression levels. Relative endogenous transcript levels of RPS6KB2 (A), PTPRCAP (B), and CORO1B (C) were measured in five lymphoblastoid cell lines homozygous for the nonrisk haplotype (H1/H1), five heterozygotes (H1/H2), and two homozygotes with the risk haplotype (H2/H2) using real-time reverse transcription-PCR and displayed in box plots. The linear regression coefficient (R2) and P values are shown. The haplotypes were estimated by the genotypes of rs1808279.
A Promoter SNP at Position -309 in PTPRCAP Affecting Promoter Activity
The alteration of PTPRCAP transcript levels depending on the risk haplotype copy number could be attributed to one SNP, rs869736 G>T, which was cancer-associated and located at position -309 of the PTPRCAP promoter. Contribution of this SNP to the altered gene expression was evaluated using luciferase reporter assays. A 610-bp DNA segment (from position -580 to +30) with either risk or nonrisk allele at the SNP was cloned upstream of the luciferase gene in the pGL3-Basic plasmid, and luciferase activities were measured in MKN28 gastric cancer cells (Figure 3). The luciferase levels relative to the empty vector were 55% higher in the construct of the risk allele T-carrying promoter than in the nonrisk G carrier (P = .00039 in one-way analysis of variance). Thus, the risk allele T at position -309 of PTPRCAP increased the promoter activity and gene expression.
Figure 3.
Luciferase reporter assays for evaluating allelic difference in regulatory activity of the associated SNPs. A promoter fragment with the risk or nonrisk allele in rs869736 (PTPRCAP) or rs1808279 (CORO1B) was cloned upstream of the luciferase gene in pGL3-Basic vector, and a 3′-UTR fragment with either allele in rs10274 (RPS6KB2) downstream of the gene in pGL3-Promoter vector. Relative luciferase activities were measured in the MKN28 gastric cancer cell line in triplicate. P values were calculated by one-way analysis of variance tests.
Three other cancer-associated SNPs, namely, rs10274, rs1808279, and rs1790761, were located in the 3′ untranslated region (UTR) of RPS6KB2 and in the promoter regions of CORO1B and GPR152, respectively, and were also tested for possibility of altering gene expression. The DNA segments containing a promoter SNP were cloned into the pGL3-Basic upstream of the luciferase gene, and those containing a 3′-UTR SNP were cloned into the pGL3-Promoter plasmid downstream of the luciferase gene. No allelic difference was detected in luciferase activity with respect to the UTR SNP in RPS6KB2 or the promoter SNP in CORO1B (Figure 3), and no luciferase activity was observed with the constructs of the promoter SNP in GPR152. Thus, alternations of gene expression by the three SNPs were not observed.
Specific Nuclear Protein Binding to the -309 SNP in PTPRCAP
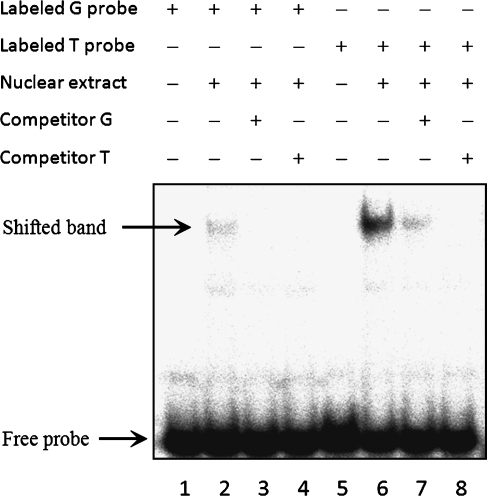
Because the SNP at position -309 of PTPRCAP affected the promoter activity, a possibility of its binding to a nuclear transcription factor was tested using EMSA. Nuclear extracts from MKN28 cells and two radioactively labeled oligonucleotides of 29 bp carrying G or T in the SNP-corresponding position were used for the assays (Figure 4). A shifted band was observed with both variants, but the band intensity was much higher with the T-carrying variant (lane 6) than the G variant (lane 2). With 200-fold excess amounts of unlabeled probes of the two variants used in separate assays, the band intensities were markedly reduced for both variants (lanes 3, 4, 7, and 8), but the T variant showed a stronger competition than G variant (lanes 7 vs 8); this was consistent with the observation made earlier that the T variant had a stronger promoter activity than the G variant. Thus, the risk allele Tat position -309 of PTPRCAP increased the specific affinity to a nuclear protein present in MKN28 cells versus the nonrisk allele G.
Figure 4.
Electrophoretic mobility shift assays using DNA probes with the -309 SNP in PTPRCAP. A nuclear extract (8 µg) from MKN28 cells was incubated with a 29-bp [γ-32P]dATP-labeled DNA probe with either major (G) or minor (T) allele at the SNP-corresponding position (lanes 2 and 6). For competition assays, unlabeled DNA (200-fold excess) were pre-incubated with the nuclear extract before addition of the labeled probes (lanes 3, 4, 7, and 8).
No Association with Intestinal-type Gastric Cancer
To test association of rs869736 at position -309 of PTPRCAP with susceptibility to intestinal-type gastric cancer, 178 unrelated patients with intestinal-type gastric cancer were additionally genotyped and compared with the 406 healthy controls. Either alleles or genotypes were not significantly associated with susceptibility to intestinal-type gastric cancer (Table 3). Although the minor allele T was more frequent in both diffuse- (37.1%) and intestinal-type (33.7%) cancers than in the controls (28.9%), the difference was significant only in diffuse-type cancer (diffuse-type, OR = 1.45, P = .0020, intestinal-type, OR = 1.25, P = .10). In logistic regression analysis, TT and GT genotypes showed 2.47- and 1.45-fold higher odds of having diffuse-type cancer than the GG genotype, respectively (P = .0021 and P = .031, respectively); however, the genotypes were not significantly associated with intestinal-type cancer risk (P = .30 and P = .87, respectively; Table 3). Thus, the association of rs869736 was specific for diffuse-type gastric cancer.
Table 3.
Type Specificity in Genotypic Association of rs869736 with Gastric Cancer Susceptibility.
| Genotype | Control (n = 406) | Diffuse-type Gastric Cancer Patient | Intestinal-type Gastric Cancer Patient | ||||
| (n = 252) | OR (95% CI)* | P | (n = 178) | OR (95% CI)* | P | ||
| GG | 198 (48.8%) | 96 (38.1%) | 1 | 76 (42.7%) | 1 | ||
| GT | 181 (44.6%) | 125 (49.6%) | 1.45 (1.04–2.03) | .031 | 84 (47.2%) | 1.03 (0.68–1.56) | .87 |
| TT | 27 (6.7%) | 31 (12.3%) | 2.47 (1.39–4.40) | .0021 | 18 (10.1%) | 1.52 (0.72–3.20) | .30 |
Age- and sex-adjusted OR and 95% CI in multivariate logistic regression.
Discussion
This study is the first to find that PTPRCAP is a gastric cancer susceptibility gene specific to diffuse-type. The minor allele in SNP rs869736 (G>T) located at position -309 of the PTPRCAP promoter was significantly associated with a 1.45-fold increased susceptibility to diffuse-type gastric cancer versus the major allele (P = .0020). The minor-allele homozygote had a 2.47-fold higher odds than the major-allele homozygote (P = .0021). In contrast, either the alleles or genotypes were not significantly associated with susceptibility to intestinal-type gastric cancer. The results are consistent with a previous study that stated that the two types of gastric cancer develop through distinct pathways [11], although this type-specific association needs to be confirmed in other populations.
When compared with the nonrisk major allele G, the cancer-risk minor allele T at this SNP was observed to increase both promoter activity and the affinity to a nuclear protein, and as a consequence the gene expression in vitro and in vivo. These results confirm that PTPRCAP expression is higher in human diffuse-type gastric cancer tissues compared with corresponding normal tissues [28]. The 1.55-fold difference in promoter activity between the two alleles observed in the luciferase assays conducted for this study using MKN28 gastric cancer cell lines is qualitatively consistent with the 2- and 1.3-fold differences in previous assays performed by other researchers using HEK297T and TE671 cell lines, respectively [26]. Several transcription activators such as AP-1, c-Jun, c-Fos, and CREB are predicted to prefer the Tallele to G allele at this SNP based on the known sequence specificities of these transcription factors. Thus, the risk allele at position -309 of the PTPRCAP promoter could increase cancer susceptibility by enhancing PTPRCAP expression.
PTPRCAP (or CD45-AP) are functionally linked to the protooncogenic SFKs, which, in an activated state, transmit various cellular signals toward aberrant proliferation, increased motility and invasiveness, resistance to apoptosis, and increased angiogenesis [17]. SFK can be inhibited by phosphorylation of a carboxyl-terminal tyrosine residue, but this inhibitory phosphate group can be removed by the protein tyrosine phosphatase PTPRC (or CD45), leading to activation of SFK [29–31]. The phosphatase activity of PTPRC is enhanced [32], and its interaction with SFK is stabilized when PTPRC dimer formation is inhibited by binding to PTPRCAP [22,33]. Thus, PTPRCAP binding to PTPRC can result in activation of SFK.
Overproduction or hyperactivation of SFK has been observed in a variety of human epithelial cancers [17] and is known to disrupt cell-cell adhesion [34,35], by inducing impairment in the membrane localization of E-cadherin (CDH1) [36], and to promote gain of invasive properties of epithelial cells [37]. Notably, down-regulation of CDH1 is a recognized feature of diffuse-type gastric cancer [38], and mutations in CDH1 have been found in hereditary diffuse-type gastric cancer [39]. Accordingly, a possible hypothesis is that enhanced levels of PTPRCAP facilitate activation of SFK and, consequently, impair E-cadherin function to increase susceptibility to diffuse-type gastric tumorigenesis.
In H. pylori infection, SFK can phosphorylate CagA of H. pylori to activate a cellular oncoprotein PTPN11 (or SHP2) [40], which is known to induce the sequential changes of intestinal-type gastric carcinogenesis [41]. Alternatively, PTPRCAP has been shown to enhance the sensitivity of T-cell receptor signaling through activation of SFK in hematopoietic cells [20,21,32,42], which may exacerbate inflammation and offer an attractive microenvironment for tumor growth, facilitate genomic instability, and promote angiogenesis [43,44]. However, these possibilities are yet unknown for diffuse-type gastric tumorigenesis.
Ten SNPs of the cancer-associated haplotype discovered in this study are dispersed in four genes, RPS6KB2, PTPRCAP, CORO1B, and GPR152 (Figure 1B). Among them, PTPRCAP was identified to be functionally associated with the gastric cancer because 1) the endogenous transcript levels of PTPRCAP alone were linearly correlated with the risk haplotype copy number and 2) the promoter activity was increased by the risk allele at a promoter SNP of PTPRCAP compared with the nonrisk allele. Associations of the other nine SNPs, especially those showing no functional variation in this study, were probably because of their very high linkage disequilibria with the PTPRCAP promoter SNP. However, a possibility of their functional relevance in the cancer susceptibility should not be ruled out because not all of these SNPs were examined for every possible function.
In conclusion, a promoter SNP rs869736 at position -309 of PTPRCAP is associated with susceptibility to diffuse-type gastric cancer rather than to intestinal-type gastric cancer. Its minor allele T increases both promoter activity and promoter affinity to a nuclear protein (presumably a transcription factor) and, as a consequence, the gene expression. The resulting increase in cellular levels of PTPRCAP can increase the susceptibility to gastric cancer, as it might result in activation of the oncogenic SFKs and disruption of cell-cell contacts mediated by E-cadherin, among other possibilities.
Acknowledgments
The authors thank Sang-Cheol Bae for lymphoblastoid cell lines derived from healthy individuals, Jongkyeong Chung and Dae-Sik Lim for their critical reading of the manuscript, Kyu-Sang Song for assistance in sample collection, and Yikyeong Kim for technical and administrative assistance.
Abbreviations
- CI
confidence interval
- EMSA
electrophoretic mobility shift assay
- LD
linkage disequilibrium
- OR
odds rati
- PCR
polymerase chain reaction
- SFK
Src family kinase
- SNP
single-nucleotide polymorphism
- UTR
untranslated region
Footnotes
This work was supported by a grant of the Korea Healthcare Technology R&D Project (A084417). This funding sponsor had no role or involvement in the study design, the collection, analysis, and interpretation of data, or in writing and submission of the manuscript.
References
- 1.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 2.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 3.Tsugane S, Sasazuki S, Kobayashi M, Sasaki S. Salt and salted food intake and subsequent risk of gastric cancer among middle-aged Japanese men and women. Br J Cancer. 2004;90:128–134. doi: 10.1038/sj.bjc.6601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palli D. Epidemiology of gastric cancer: an evaluation of available evidence. J Gastroenterol. 2000;35(Suppl. 12):84–89. [PubMed] [Google Scholar]
- 5.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 6.El-Omar EM. The importance of interleukin 1β in Helicobacter pylori associated disease. Gut. 2001;48:743–747. doi: 10.1136/gut.48.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, Risch HA, Vaughan TL, McColl KE, Lissowska J, Zatonski W, et al. A functional polymorphism of toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology. 2007;132:905–912. doi: 10.1053/j.gastro.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, Kato S, et al. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 9.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 10.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554–3560. [PubMed] [Google Scholar]
- 11.Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Pract Res Clin Gastroenterol. 2006;20:651–674. doi: 10.1016/j.bpg.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]
- 13.Correa P, Shiao YH. Phenotypic and genotypic events in gastric carcinogenesis. Cancer Res. 1994;54:1941s–1943s. [PubMed] [Google Scholar]
- 14.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T, Cooper JA. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 16.Penninger JM, Irie-Sasaki J, Sasaki T, Oliveira-dos-Santos AJ. CD45: new jobs for an old acquaintance. Nat Immunol. 2001;2:389–396. doi: 10.1038/87687. [DOI] [PubMed] [Google Scholar]
- 17.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 18.Barraclough J, Hodgkinson C, Hogg A, Dive C, Welman A. Increases in c-Yes expression level and activity promote motility but not proliferation of human colorectal carcinoma cells. Neoplasia. 2007;9:745–754. doi: 10.1593/neo.07442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura K, Maiti A, Ng DH, Johnson P, Maizel AL, Takeda A. Characterization of the interaction between CD45 and CD45-AP. J Biol Chem. 1995;270:21151–21157. doi: 10.1074/jbc.270.36.21151. [DOI] [PubMed] [Google Scholar]
- 20.Motoya S, Kitamura K, Matsuda A, Maizel AL, Yamamoto H, Takeda A. Interaction between CD45-AP and protein-tyrosine kinases involved in T cell receptor signaling. J Biol Chem. 1999;274:1407–1414. doi: 10.1074/jbc.274.3.1407. [DOI] [PubMed] [Google Scholar]
- 21.Veillette A, Soussou D, Latour S, Davidson D, Gervais FG. Interactions of CD45-associated protein with the antigen receptor signaling machinery in T-lymphocytes. J Biol Chem. 1999;274:14392–14399. doi: 10.1074/jbc.274.20.14392. [DOI] [PubMed] [Google Scholar]
- 22.Takeda A, Matsuda A, Paul RM, Yaseen NR. CD45-associated protein inhibits CD45 dimerization and up-regulates its protein tyrosine phosphatase activity. Blood. 2004;103:3440–3447. doi: 10.1182/blood-2003-06-2083. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O'Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- 27.Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu CW, Kao HL, Li AF, Chi CW, Lin WC. Protein tyrosine-phosphatase expression profiling in gastric cancer tissues. Cancer Lett. 2006;242:95–103. doi: 10.1016/j.canlet.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 29.Mustelin T, Coggeshall KM, Altman A. Rapid activation of the T-cell tyrosine protein kinase pp56lck by the CD45 phosphotyrosine phosphatase. Proc Natl Acad Sci USA. 1989;86:6302–6306. doi: 10.1073/pnas.86.16.6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustelin T, Pessa-Morikawa T, Autero M, Gassmann M, Andersson LC, Gahmberg CG, Burn P. Regulation of the p59fyn protein tyrosine kinase by the CD45 phosphotyrosine phosphatase. Eur J Immunol. 1992;22:1173–1178. doi: 10.1002/eji.1830220510. [DOI] [PubMed] [Google Scholar]
- 31.Trowbridge IS, Thomas ML. CD45: an emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu Rev Immunol. 1994;12:85–116. doi: 10.1146/annurev.iy.12.040194.000505. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda A, Motoya S, Kimura S, McInnis R, Maizel AL, Takeda A. Disruption of lymphocyte function and signaling in CD45-associated proteinnull mice. J Exp Med. 1998;187:1863–1870. doi: 10.1084/jem.187.11.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeda A, Maizel AL, Kitamura K, Ohta T, Kimura S. Molecular cloning of the CD45-associated 30-kDa protein. J Biol Chem. 1994;269:2357–2360. [PubMed] [Google Scholar]
- 34.Owens DW, McLean GW, Wyke AW, Paraskeva C, Parkinson EK, Frame MC, Brunton VG. The catalytic activity of the Src family kinases is required to disrupt cadherin-dependent cell-cell contacts. Mol Biol Cell. 2000;11:51–64. doi: 10.1091/mbc.11.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irby RB, Yeatman TJ. Increased Src activity disrupts cadherin/catenin- mediated homotypic adhesion in human colon cancer and transformed rodent cells. Cancer Res. 2002;62:2669–2674. [PubMed] [Google Scholar]
- 36.Avizienyte E, Wyke AW, Jones RJ, McLean GW, Westhoff MA, Brunton VG, Frame MC. Src-induced de-regulation of E-cadherin in colon cancer cells requires integrin signalling. Nat Cell Biol. 2002;4:632–638. doi: 10.1038/ncb829. [DOI] [PubMed] [Google Scholar]
- 37.Behrens J, Vakaet L, Friis R, Winterhager E, Van Roy F, Mareel MM, Birchmeier W. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/β-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J Cell Biol. 1993;120:757–766. doi: 10.1083/jcb.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boussioutas A, Li H, Liu J, Waring P, Lade S, Holloway AJ, Taupin D, Gorringe K, Haviv I, Desmond PV, et al. Distinctive patterns of gene expression in premalignant gastric mucosa and gastric cancer. Cancer Res. 2003;63:2569–2577. [PubMed] [Google Scholar]
- 39.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 40.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 41.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 42.Leitenberg D, Falahati R, Lu DD, Takeda A. CD45-associated protein promotes the response of primary CD4 T cells to low-potency T-cell receptor (TCR) stimulation and facilitates CD45 association with CD3/TCR and lck. Immunology. 2007;121:545–554. doi: 10.1111/j.1365-2567.2007.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mumm JB, Oft M. Cytokine-based transformation of immune surveillance into tumor-promoting inflammation. Oncogene. 2008;27:5913–5919. doi: 10.1038/onc.2008.275. [DOI] [PubMed] [Google Scholar]
- 44.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]