Abstract
ABCG2 is an efflux transporter conferring multidrug resistance (MDR) on cancer cells. However, the initial molecular events leading to its up-regulation in MDR tumor cells are poorly understood. Herein, we explored the impact of drug treatment on the methylation status of the ABCG2 promoter and consequent reactivation of ABCG2 gene expression in parental tumor cell lines and their MDR sublines. We demonstrate that ABCG2 promoter methylation is common in T-cell acute lymphoblastic leukemia (T-ALL) lines, also present in primary T-ALL lymphoblast specimens. Furthermore, drug selection with sulfasalazine and topotecan induced a complete demethylation of the ABCG2 promoter in the T-ALL and ovarian carcinoma model cell lines CCRF-CEM and IGROV1, respectively. This resulted in a dramatic induction of ABCG2 messenger RNA levels (235- and 743-fold, respectively) and consequent acquisition of an ABCG2-dependent MDR phenotype. Quantitative genomic polymerase chain reaction and ABCG2 promoter-luciferase reporter assay did not reveal ABCG2 gene amplification or differential transcriptional trans-activation, which could account for ABCG2 up-regulation in these MDR cells. Remarkably, mimicking cytotoxic bolus drug treatment through 12- to 24-hour pulse exposure of ABCG2-silenced leukemia cells, to clinically relevant concentrations of the chemotherapeutic agents daunorubicin and mitoxantrone, resulted in a marked transcriptional up-regulation of ABCG2. Our findings establish that antitumor drug-induced epigenetic reactivation of ABCG2 gene expression in cancer cells is an early molecular event leading to MDR. These findings have important implications for the emergence, clonal selection, and expansion of malignant cells with the MDR phenotype during chemotherapy.
Introduction
Despite the profound impact that novel treatment strategies may have on some malignancies, chemotherapy continues to be the standard of care in most human cancers. In this respect, multidrug resistance (MDR) remains a major impediment toward curative cancer chemotherapy [1]. The ATP-binding cassette (ABC) superfamily of human membrane transporters consists of 49 members mediating a broad spectrum of physiological functions [2,3]. Moreover, the overexpression of several ABC transporters, including ABCB1 (MDR1/Pgp), members of the ABCC group (MRPs) and ABCG2 (BCRP/MXR), results in ATP-driven efflux of antitumor drugs from cancer cells, thereby leading to decreased intracellular drug accumulation and consequent MDR [4–8]. These drug efflux pumps display a high-transport capacity toward a partially overlapping array of structurally dissimilar cytotoxic agents, thereby creating an active cellular defense network against a multitude of chemotherapeutic drugs [9–11].
DNA methylation within CpG islands is a well-established mechanism mediating epigenetic silencing of gene expression [12]. This process is a prerequisite in vertebrate development and in tissue-specific gene expression [13,14]. CpG island methylation can cause repression of gene expression either directly through steric hindrance of methyl-CpGs with transcription factors such as E2F or CREB [15,16] or indirectly through recruitment of methyl-binding proteins, which actively repress transcription [17]. Further association of methyl-binding proteins with chromatin-modifying proteins produces a complementary repressive heterochromatin environment [18,19]. Simultaneous genomewide hypomethylation and gene-specific hypermethylation events underlie cancer etiology and may account for cancer hallmarks such as global genomic instability and tumor-suppressor gene silencing, respectively [20–22]. CpG island hypermethylation and consequent gene silencing in cancer was found to be mediated by the deregulation of DNA methyltransferases (DNMTs), particularly DNMT1 and DNMT3a/b, which are responsible for maintenance and de novo methylation, respectively [23]. Thus, reactivation of aberrantly silenced tumor-suppressor genes through DNMT inhibition has recently become a relevant molecular target for therapy, currently used for treatment of hematological malignancies [24–26].
Several mechanisms of drug-induced up-regulation of gene expression have been proposed for the extensively studied MDR transporter ABCB1 (P-glycoprotein, MDR1), ranging from karyotypic abnormalities to altered transactivation [27–29]. El-Osta et al. [30] have described epigenetic alterations including promoter demethylation, leading to upregulation of ABCB1 in adriamycin-selected CCRF-CEM cells (CEM-A7R). Similar results were consistently obtained with vincristine-selected KB3-1 cells [31]. In a following study, Baker et al. [32] demonstrated the direct epigenetic up-regulation of the mdr1 gene within a single cell cycle, in antiapoptotic, bcl2-transfected CCRF-CEM-bcl2 cells, occurring in response to chemotherapeutic drug exposure. Recently, To et al. [33] have shown an active CpG island within the proximal ABCG2 promoter region, thereby facilitating its transcriptional silencing in aberrantly methylated renal cell carcinoma lines. A follow-up report by Turner et al. [34] demonstrated that ABCG2 expression in multiple myeloma patients is regulated in part by promoter methylation. Subsequent studies have begun to elucidate the major role that epigenetics play in the transcriptional regulation of the abcg2 gene. Specifically, two recent reports documented the impact of single-step and long-term anticancer drug selection on the association of chromatin-remodeling proteins to the abcg2 locus and concomitant ABCG2 expression, in several carcinoma cell lines [35,36]. However, the role and dynamics of ABCG2 promoter methylation under anticancer drug exposure have not been addressed, hence leaving a crucial part of the early epigenetic machinery governing ABCG2 transcription poorly understood. Toward this end, we here studied the methylation status of the ABCG2 promoter in different malignant cell lines and their ABCG2-overexpressing MDR counterparts. We provide the first evidence that anticancer drug selection of cell lines harboring a transcriptionally silenced ABCG2 results in the complete loss of abcg2 promoter methylation, ultimately leading to a marked transcriptional up-regulation and consequent MDR. Moreover, this transcriptional up-regulation of epigenetically silenced genes is observed as early as 12 hours after drug exposure, hence indicating that drug-induced epigenetic up-regulation of ABCG2 is an upstream event leading ABCG2-mediated MDR.
Materials and Methods
Drugs and Chemicals
Daunorubicin (DNR), doxorubicin (Dox), mitoxantrone (MX), sulfasalazine (SSZ), Taxol, 5-Aza-2′-deoxycytidine (5-Aza-dC), actinomycin D (Act D), trichostatin A (TSA), and zebularine (ZEB) were obtained from Sigma Chemical Co (St Louis, MO). Fumitremorgin C (FTC), topotecan (Topo), and SN-38 were kindly provided by Dr. S. E. Bates, National Cancer Institute, Bethesda, MD.
Tissue Culture
Human malignant cell lines, namely ovarian carcinoma IGROV1, MCF7 breast cancer cells, A549 non-small cell lung cancer cells, T-cell leukemia lines—Jurkat, CCRF-CEM, and MOLT4—and erythroleukemia K562 cells, were grown either under monolayer conditions (attached cell lines) or up to a maximal density of 106 cells/ml (for leukemic cell lines), in RPMI-1640 medium (Invitrogen Gibco, Carlsbad, CA) supplemented with 10% fetal calf serum, 2 mM glutamine, 100 µg/ml penicillin, and 100 µg/ml streptomycin (Biological Industries, Beth-Haemek, Israel) in a humid atmosphere of 5% carbon dioxide. Matches drug-resistant IGROV1/T8, -MX3, MCF7/MR, -FLV1000, A549/K1.5, and CCRF-CEM/SSZ1.5, -2.5 cells, harboring ABCG2 overexpression, were maintained in the previously mentioned conditions, under pulse or continuous drug selection, as described previously [37–40]. IGROV1 and its drug-resistant sublines were generously provided by Dr. Jan H. M. Schellens, Department of Medical Oncology, The Netherlands Cancer Institute, Amsterdam, the Netherlands, and CCRF-CEM and its SSZ-resistant sublines were a gift from Dr. Gerrit Jansen, Department of Rheumatology, VU University Medical Center, Amsterdam, the Netherlands. For cytotoxicity experiments, cells were grown in drug-free medium for at least 1 week before the experiments.
Drug Treatment with Zebularine, 5-Aza-dC, or TSA
IGROV1, CCRF-CEM cells, and their MDR sublines were grown in medium containing 2 µM 5-Aza-dC (Sigma) for 72 hours, and the 5-Aza-dC-containing medium was replaced every 24 hours. TSA or Act D (Sigma) were added to the appropriate cultures at a concentration of 75 ng/ml or 0.5 µg/ml, respectively, and incubated for the final 20 hours. Alternatively, sequential treatment of CCRF-CEM cells with 50 µM zebularine was carried out for 30 days as previously described [41]. Cell viability was assessed using trypan blue staining, and drug concentrations were optimized to achieve maximal transcriptional change while minimally affecting cell viability. After treatment, genomic DNA or RNA were isolated, and bisulfite conversion or reverse transcription-polymerase chain reaction was performed, respectively, as described later.
Anticancer Drug Treatment
CCRF-CEM, Jurkat, and K562 cells were grown in medium containing 0.01 to 15 µM MX, DNR, or Dox; 0.001 to 2 mM SSZ; 0.01 to 10 µM topotecan; or 0.001 to 1 µM SN-38. Drug concentration range was calibrated to span the reported drug concentration required to inhibit cell growth by 50% of the different drugs. Cells were harvested at variable time points up to 48 hours, and RNA was isolated as described later. Concurrent addition of Act D (0.5 µg/ml) or FTC (5 µM) to the cell culture was carried out at the optimal (ABCG2-inducing) drug concentration for each drug at 20-hour incubations. Cell death was assessed using trypan blue.
Cytotoxicity Assay
Parental CCRF-CEM cells, their SSZ-resistant subline, and zebularine-treated cells were seeded in 96-well plates (3 x 104 cells per well) in growth medium (100 µl per well) containing various concentrations of MX or SSZ for 72 hours at 37°C in the presence or absence of the specific ABCG2 transport inhibitor FTC. Viable cell numbers were determined using a hemocytometer after trypan blue staining. The 50% inhibition of cell growth was calculated relative to untreated controls. Results presented are means of at least three independent experiments.
RNA Extraction and Quantification of Gene Expression by Real-time PCR
RNA extraction and complementary DNA synthesis were carried out as previously described [37]. The levels of ABCG2, ABCB1, ABCC1, and PCFT gene expression were determined using a quantitative realtime PCR method as previously described [37]. Expression levels were normalized using the β2-MICROGLOBULIN (B2M) gene as an internal control. QPCR primers (Table 1) were obtained from Harvard Primer Bank (http://pga.mgh.harvard.edu/primerbank/index.html). Results represent means ± SD of three independent experiments.
Table 1.
Summarizing Table of Primers Used in This Study.
| Genomic PCR Primers for Bisulfite-Treated DNA | ||||
| Fragment | Sense Primer | Antisense Primer | Length (bp) | Tm (°C) |
| ABCG2 (A) | 5′-TTTGTGATTGGGTAATTTGTG-3′ | 5′-TCCCTCAAAACTAAAATCACCCTA-3′ | 221 | 53 |
| ABCG2 (B)* | 5′-GGAGTGTTTGGTTTGTTTTTG-3′ | 5′-CAATAACCCCTCCCCAA-3′ | 223 | 52 |
| PCFT† | 5′-TTTTTGTTATTTGTGGTGTGTT-3′ | 5′-CAAAATTAACCAAAAAAACCA-3′ | 414 | 56 |
| ABCB1‡ | 5′-GGAAGTTAGAATATTTTTTTTGGAAAT-3′ | 5′-ACCTCTACTTCTTTAAACTTAAAAAAACC-3′ | 223 | 48 |
| Primers for Quantitative Real-time PCR | ||||
| Name | Sense Primer | Antisense Primer | Length (bp) | Tm (°C) |
| ABCG2§ | 5′-CCCGCGACAGCTTCCAATGA-3′ | 5′-GGCGTTGAGACCAGGTTTCA-3′ | 171 | 60 |
| ABCB1¶ | 5′-GTGGTGGGAACTTTGGCTG-3′ | 5′-TACCTGGTCATGTCTTCCTCC-3′ | 188 | 60 |
| ABCC1¶ | 5′-GAGGAACCATATTACAGGTCCGT-3′ | 5′-AGGGGATCATCGAAGAAGTAAAT-3′ | 188 | 60 |
| PCFT† | 5′-CACTCTACCCAGCCACTCTGAAC-3′ | 5′-GATCAGCCTTTTCCAGCATCC-3′ | 109 | 60 |
| B2M† | 5′-GGCTATCCAGCGTACTCCAAA-3′ | 5′-CGGCAGGCATACTCATCTTTTT-3′ | 246 | 60 |
To et al. [33].
Gonen et al. [14].
Lee et al. [42].
Bram et al. [37].
Harvard Primer Bank (http://pga.mgh.harvard.edu/primerbank/index.html).
Isolation of Genomic DNA and Bisulfite Conversion of DNA
Genomic DNA was isolated using The DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA) followed by bisulfite conversion of 2 µg of genomic DNA using the EpiTect Bisulfite Kit (QIAGEN) according to the instructions of the manufacturer.
DNA Methylation Analysis
The methylation status of abcg2, mdr1, and pcft promoter was examined using combined bisulfite restriction analysis (COBRA) as well as bisulfite DNA sequencing as described previously [14]. Sodium bisulfite-treated DNA was amplified in two subsequent rounds using primers specifically designed for this purpose (Table 1); some primers were designed using the Methyl Primer Express software v1.0 (Applied Biosystems, Foster City, CA) and others were obtained from previous reports [14,33,42]. COBRA restriction used BstUI and Taq1α restriction enzymes for a 200-fold overdigestion period on abcg2 and pcft, and mdr1 bisulfite converted PCR fragments, respectfully, according to the manufacturer's instructions (New England Biolabs, Ipswich, MA).
In Vitro Methylation
In vitro methylation was performed using CpG methyltransferases (M.SssI) according to the instructions of the manufacturer (New England Biolabs, Ipswich, MA), whereas a mock assay was performed at the same conditions without the enzyme. After the assay, plasmids were purified using the Wizard SV Gel and PCRClean-up kit according to the instructions of the manufacturer (Promega, Madison, WI), and the methylation percentage was evaluated using the methylation-sensitive restriction enzymes SalI and NotI present on the pGl3 plasmid.
Transient Transfections with pGL3-abcg2 Promoter Expression Vectors and Luciferase Reporter Gene Assay
For the luciferase reporter assay, IGROV1 and IGROV1/T8 cells were grown in 24-mm dishes (3 x 104 cells per well) for 24 hours, after which cells were transiently cotransfected with either the pGL3-abcg2 constructs (generously provided by Dr. Douglas D. Ross) [43], pGL3-Basic empty vector, or the in vitro-methylated vectors described, along with the extensively used pROL control plasmid (Renilla luciferase), using the jetPEI transfection reagent (Polyplus-transfection; Genesee Scientific, San Diego, CA) according to the instructions of the manufacturer. Twenty-four hours after transient transfection, cells were harvested and tested for luciferase and Renilla activities using the Dual-Luciferase Reporter Assay System (Promega) as described in the manufacturer's protocol. Results presented were obtained from at least three independent experiments performed in duplicate cultures.
Healthy Donor T-cell and T-Acute Lymphoblastic Leukemia Lymphoblast Specimens
Analysis of ABCG2 promoter methylation status in T-cell lymphoblasts was performed on stored genomic DNA samples previously obtained from adult T-cell acute lymphoblastic leukemia (T-ALL) patients who were treated according to the UKALL12/ECOG 2993 protocol [44] at the Department of Hematology, Rambam Medical Center. The samples were previously derived as part of the routine clinical management and were used in the current study after receiving approval from the local institutional review board (study no. 2902) at the Rambam Medical Center and informed consent in accordance with the Declaration of Helsinki. Percent purity of peripheral T-ALL lymphoblasts for all patient samples tested was greater than 90%. For genomic DNA extraction, lymphoblasts were isolated from patients' peripheral blood by a standard Ficoll-Hypaque (Sigma) gradient density centrifugation. In addition, healthy volunteers' primary T cells were isolated using the Rosette Human T-Cell Enrichment kit (Stem Cell Technologies, Vancouver, British Columbia, Canada), according to the manufacturer's instructions. Total genomic DNA was purified using The DNeasy Blood&Tissue Kit (QIAGEN) according to the instructions of the manufacturer.
Statistical Analyses
We used a 1-tailed nonpaired Student's t-test to examine the significance of the difference between two populations for a certain variable. A difference was considered significant if the P value obtained was less than .05.
Results
ABCG2 Is Transcriptionally Upregulated in MDR Tumor Cell Lines
Real-time PCR analysis revealed a substantial variability in ABCG2 gene expression in tumor cell lines of epithelial and hematological origins; furthermore, a marked transcriptional up-regulation of ABCG2 was observed in their drug-selected counterparts (Figure 1, black bars). This up-regulation resulted in a consistent plasma membrane overexpression of functional ABCG2 and a consequent MDR phenotype [37–40]. Remarkably, when comparing the extent of ABCG2 overexpression (expressed as fold difference) in these ABCG2-overexpressing MDR cells, a differential pattern of up-regulation was apparent; the MDR cell lines A549/K1.5, MCF7/MR, and MCF7/FLV1000, in which ABCG2 overexpression was associated with gene amplification ([37]; see Supplementary Data), displayed the highest absolute levels of ABCG2 messenger RNA (mRNA; Figure 1, black bars). Conversely, the maximal fold increase in ABCG2 mRNA levels relative to parental cells was found in the MDR sublines CCRF-CEM/SSZ2.5 and IGROV1/T8 (Figure 1, gray bars) and may be attributed to the extremely low basal ABCG2 mRNA levels observed in parental IGROV1 and CCRF-CEM cells (Figure 1, black bars). Interestingly, ABCG2 overexpression in these MDR cell lines was not due to genomic amplification or differential transcriptional transactivation (see Supplementary Data).
Figure 1.
ABCG2 mRNA levels in paired parental and MDR tumor cell lines assessed by QPCR. ABCG2 mRNA levels are expressed as fold of expression relative to the basal transcript levels of CCRF-CEM cells (black bars) or as fold of expression of the MDR sublines over their matched parental cells (gray bars).
The ABCG2 Promoter Undergoes Demethylation in MDR Cell Lines Lacking ABCG2 Gene Amplification
Recently it was shown that the abcg2 promoter undergoes CpG methylation in the renal carcinoma cell lines UOK121 and UOK143, thereby resulting in its transcriptional silencing [33]. We therefore examined the possibility that the extremely low levels of ABCG2 transcripts in human T-cell leukemia CCRF-CEM and ovarian cancer IGROV1 cells are due to transcriptional silencing of the abcg2 gene through promoter methylation. Furthermore, the lack of abcg2 gene amplification or transactivation in their moderately ABCG2-expressing MDR counterparts, suggested the possibility that the dramatic upregulation of ABCG2 in these drug-resistant cell lines may be due to the loss of transcriptional silencing of this gene. This hypothesis was tested using COBRA methodology on a 223-bp fragment of the reported CpG island of the ABCG2 promoter (termed fragment B), digested with BstUI. Indeed, parental ABCG2 mRNA levels inversely correlated with the methylation status of the ABCG2 promoter; accordingly, A549 and MCF7 cells, which exhibited relatively high basal levels of ABCG2 mRNA, were devoid of ABCG2 promoter methylation (Figure 2A). In contrast, both CCRF-CEM and IGROV1 cells displayed high levels of promoter methylation, with greater than 87% and greater than 50% of the ABCG2 genes being methylated, respectively (Figure 2A). Remarkably, inspection of the ABCG2 CpG island in the corresponding MDR sublines CCRF-CEM/SSZ, IGROV1/T8, and IGROV1/MX3 revealed a prominent promoter demethylation (Figure 2B). These results were further corroborated using direct bisulfite sequencing of a 386-bp fragment (ranging from -380 to +6 relative to the transcription initiation site) [43] previously reported to be a functional CpG island that undergoes a dense methylation in the ABCG2 promoter (Figure 2C) [33,43].
Figure 2.
Quantitative determination of promotermethylation status in parental and MDR cell lines assessed by COBRA and direct bisulfite sequencing. Methylation levels were assessed using a PCR-amplified 223-bp fragment of bisulfite-converted genomic DNA from within the CpG island of the abcg2 promoter of parental A549, MCF7, CCRF-CEM, and IGROV1 cells (A) or their corresponding A549/K1.5, MCF7/MR/FLV1000, CCRF-CEM/SSZ1.5/2.5, and IGROV1/T8/MX3 MDR cells (B). In addition, two consecutive fragments (termed A and B) from the abcg2 promoter's CpG island were subjected to direct bisulfite sequencing (C). Schematic representation of the sequenced genomic fragments including all CpG sites (line representation) is displayed (top). Eight representative clones from parental CCRF-CEM and IGROV1 cells and their MDR CCRF-CEM/SSZ1.5/2.5 and IGROV1/T8 sublines are presented in dot display (black dot represents a methylated CpG). Also, the promoter methylation status of the pcft and mdr1 genes in parental CCRF-CEM cells and their MDR CCRF-CEM/SSZ2.5 counterparts were assessed using a 414- and 223-bp PCR-amplified fragment of bisulfite-converted genomic DNA from within their CpG island, respectively (D). PCR fragments were subjected to 200-fold BstUI (abcg2 and pcft) or TaqIα (mdr1) overdigestion and then compared to various dilutions of the uncut DNA.
ABCG2 Demethylation in Nongenomically Amplified MDR Cell Lines Is Not Due to Global Demethylation
Drug-induced DNA demethylation is known to be either global or site-specific [25,30,45]. Promoter methylation status of ABCB1 (MDR1/P-glycoprotein) and proton-coupled folate transporter (PCFT/SLC46A1), the promoters of which were reported to be methylated in CCRF-CEM cells [14,46], were evaluated in conjunction with their MDR subline CCRF-CEM/SSZ2.5. As evident from the COBRA results depicted in Figure 2D, the hypermethylation pattern of both the mdr1 and pcft promoters observed in parental CCRF-CEM cells remained intact in CCRF-CEM/SSZ2.5 cells, suggesting that the marked demethylation of the abcg2 locus under SSZ drug exposure was presumably a result of a site-specific demethylation event and not part of a global demethylation process (Figure 2D).
Treatment with Demethylating Agents Results in ABCG2 Promoter Demethylation and Consequent Restoration of Gene Expression
The high correlation between abcg2 promoter methylation and lack of gene expression prompted us to further explore the role of epigenetics in ABCG2 gene expression using these ABCG2-methylated MDR tumor cell lines. Previous reports demonstrated that treatment with the demethylating agent 5-Aza-dC and the histone deacetylase inhibitor (HDACi) depsipeptide, which induces histone acetylation, resulted in restoration of ABCG2 gene expression in promoter-silenced tumor cell lines [33,36]. We thus explored the responsiveness of IGROV1 and CCRF-CEM cells harboring a methylated abcg2 promoter, to 5-Aza-dC and TSA; as evident from Figure 3, treatment with 5-Aza-dC resulted in an approximately 9- to 11-fold increase in ABCG2 transcript levels, relative to untreated cells (P = .006 and P = .004, respectively; Figure 3, A and B). A further increase in ABCG2 gene reactivation in CCRF-CEM and IGROV1 cells was obtained when combining both treatments with 5-Aza-dC and the HDACi, TSA (P = .03 and P = .02, respectively; Figure 3, A and B). However, neither 5-Aza-dC alone nor its combination with TSA induced a significant alteration in ABCG2 gene expression in the corresponding MDR cell lines CCRF-CEM/SSZ2.5 and IGROV1/T8 (P > .21; Figure 3, C and D), which initially displayed substantial ABCG2 overexpression. Furthermore, the use of TSA alone did not increase ABCG2 gene expression in any of the parental or ABCG2-overexpressing tumor cell lines (P > .24; Figure 3, A–D). To verify that restoration of ABCG2 gene expression is due to transcriptional up-regulation rather than alterations in ABCG2 mRNA stability, we used the transcription inhibitor Act D. Concurrent addition of Act D to 5-Aza-dC- and TSA-treated tumor cells resulted in the loss of ABCG2 gene expression in both parental and MDR cells (Figure 3, A–D).
Figure 3.
The effect of epigenetic modifying agents on the status of ABCG2 promoter methylation and consequent transcription in parental cells and the correspondingMDR cells. QPCR analysis of ABCG2 transcript in CCRF-CEM(A), IGROV1 (B) cells, and their MDR sublines CCRF-CEM/SSZ2.5 (C) and IGROV1/T8 (D) that were treated for 72 hours with 2 µM of the demethylating agent 5-Aza-dC, in the presence or absence of 75 ng/ml of theHDACi-TSA. To some samples, 0.5 µg/ml of the transcription inhibitor ActDwas added in combination with the latter. abcg2 promoter methylation status was assessed using COBRA in parental CCRF-CEM and MDR CCRF-CEM/SSZ2.5 cells as well as CCRF-CEM cells treated with 5-Aza-dC+zebularine (E; see Materials and Methods for details). The corresponding ABCG2mRNA expression levels were assessed using QPCR (F); all QPCR results are expressed as fold of expression relative to the corresponding untreated or parental cells.
We further evaluated the ability of demethylating agents to confer drug resistance on abcg2 promoter-methylated parental cell lines, thereby mimicking the epigenetic events underlying the observed MDR phenotype of the corresponding sublines, using a 72-hour cytotoxicity assay with known ABCG2 substrates. However, the demethylating agent 5-Aza-dC is unstable in cell culture conditions and is known to cause a marked cellular toxicity under long-term exposure [26]. Hence, to eliminate the component of cell toxicity while achieving steady-state demethylation, we used a combined demethylation protocol (see Materials and Methods) [41] with the stable and mild nucleoside analog zebularine for a period of 30 days. Zebularine is a novel DNA methylation inhibitor that forms a covalent complex with DNMTs. Application of this protocol to parental CCRF-CEM cells, which originally contained ABCG2 promotermethylation, resulted in a greater than 20% demethylation of ABCG2 gene copies in the population (Figure 3E) and in a consequent 10-fold increase in ABCG2 transcript levels, relative to untreated parental CCRF-CEM cells (Figure 3F). Accordingly, a 72-hour cytotoxicity assay revealed a significant (P = .0006) three-fold increase in cellular drug resistance of the zebularine-treated cells to MX, a well-established ABCG2 transport substrate, relative to untreated cells (Table 2). This resistance was completely reversed using the ABCG2-specific transport inhibitor FTC. However, despite the observed FTC-dependent five-fold resistance of CCRF-CEM/SSZ2.5 cells to the ABCG2 transport substrate SSZ, zebularine-treated cells retained parental cell sensitivity to this drug, possibly because SSZ is a relatively low-affinity ABCG2 transport substrate (Table 2) [40].
Table 2.
Summary of Growth Inhibition Studies on Parental CCRF-CEM, MDR CCRF-CEM/SSZ2.5 Subline, and 5-Aza-dC (AZA) + Zebularine (ZEB)-Treated CCRF-CEM Cells, on 72 Hours of Exposure to Established ABCG2 Substrate Drugs in the Presence or Absence of the ABCG2 Inhibitor FTC.
| Drug | Cell Line | |||||
| IC50 | ||||||
| CCRF-CEM | CCRF-CEM/SSZ2.5 | CCRF-CEM/AZA + ZEB | ||||
| -FTC | +FTC | -FTC | +FTC | -FTC | +FTC | |
| MX (nM) | 18 ± 3 (1.0) | 15 ± 5 (0.8) | 172 ± 16 (9.5) | 32 ± 7 (1.8) | 55 ± 7 (3.0) | 20 ± 4 (1.1) |
| SSZ (µM) | 627 ± 47 (1.0) | 733 ± 91 (1.2) | 2860 ± 212 (4.6) | 995 ± 83 (1.6) | 687 ± 41 (1.1) | 702 ± 88 (1.1) |
IC50 was evaluated using a hemocytometer after trypan blue staining.
In Vitro Promoter Methylation Accounts for the Complete Silencing of ABCG2 Promoter Activity
Despite the cumulative effect of 5-Aza-dC and zebularine on the restoration of ABCG2 gene expression in parental cells harboring a methylated ABCG2 promoter, the degree of ABCG2 overexpression in the corresponding MDR sublines remained substantially higher, with an additional 183- and 14-fold of ABCG2 overexpression in IGROV1/T8 and CCRF-CEM/SSZ2.5, respectively. To preclude the existence of additional mechanisms underlying ABCG2 overexpression in these MDR cell lines, we explored the ability of in vitro methylation to account for the attenuation of ABCG2 transcription using a luciferase reporter assay with the 1285/+362 ABCG2 promoter construct in IGROV1 and IGROV1/T8 cells. This assay demonstrated that in vitro methylation of the previously mentioned ABCG2 promoter construct resulted in the complete loss of luciferase activity in both IGROV1 and IGROV1/T8 cells (P < .004; Figure 4). These findings indicate that ABCG2 promoter methylation mediates a robust gene silencing capacity, regardless of the underlying transcriptional potential of the cells.
Figure 4.
Effect of in vitro methylation on ABCG2 promoter activity. Methylated and nonmethylated -1285/+362 ABCG2 promoter luciferase reporter constructs were transiently transfected into parental IGROV1 and MDR IGROV1/T8 cells and the relative luciferase activity was determined. Results are expressed as the average (±SD) luciferase-to-Renilla ratio of three experiments.
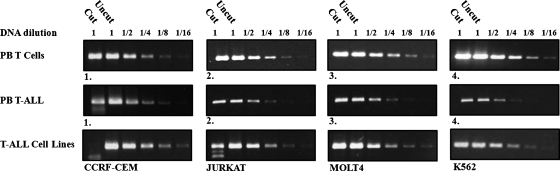
ABCG2 Promoter Methylation Is Observed in T-ALL Cell Lines and Some T-ALL Samples
Our current findings suggest that abcg2 promoter methylation is an inherent characteristic of tumor cell lines from different tissue origins. We specifically find here that abcg2 promoter methylation appears to be present in several T-ALL cell lines including CCRF-CEM and Jurkat cells (Figure 5). To expand the validity of our findings, the status of ABCG2 promotermethylation was assessed using the COBRA technique on genomic DNA from primary T cells and T-ALL lymphoblasts obtained from peripheral blood of four healthy individuals and four adult T-ALL patients at diagnosis, respectively. abcg2 promoter methylation was not observed in any of the healthy blood samples; however, one of the T-ALL patients displayed approximately 50% methylation of the CpG island in the abcg2 promoter (Figure 5).
Figure 5.
Quantitative determination of abcg2 promoter methylation status in hematolymphoid cells. Genomic DNA was extracted from peripheral blood T cells of healthy volunteers (top lane 1–4), peripheral blood lymphoblasts fromadult T-ALL patients (middle lane 1–4), T-ALL cell lines CCRF-CEM, JURKAT, and MOLT4 (bottom lane 1–3), and the erythroleukemia cell line K562. A 223-bp fragment (fragment B) of bisulfite-converted genomic DNA from within the abcg2 promoter CpG island was PCR-amplified and subjected to COBRA (as described above).
Anticancer Drug Exposure Induces a Dose-Dependent Restoration of ABCG2 Gene Expression in Transcriptionally Silenced Tumor Cell Lines within a Single Cell Cycle
The observed chemoresistance in IGROV1/T8, IGROV1/MX3, and CCRF-CEM/SSZ2.5 cells may have emerged owing to the selection of either a preexisting variant or a phenotype induced by the mutagenic effect of the cytotoxic agent itself. Thus, to pinpoint the upstream molecular events that occur after drug exposure, dose- and time-dependent exposures of the abcg2-methylated cell line CCRF-CEM to various chemotherapeutics were assessed, and the consequent relative change in ABCG2 transcript level was assessed by QPCR. We found that several antitumor agents administered at clinically relevant doses induced a marked increase in ABCG2 transcript levels, whereas DNR induced the highest increase (55-fold) in ABCG2 transcript levels, other anthracyclines tested including Dox and MX induced a 14- and 17-fold elevation of ABCG2 transcript levels, respectively (P < .002). This increase in ABCG2 mRNA levels was observed within a defined concentration range, in a dose-dependent manner (Figure 6A); cytotoxicity was evident at the relatively higher drug concentrations, thereby severely affecting cellular viability and subsequently impairing ABCG2 overexpression (Figure 6A). This drug-induced increase in ABCG2 mRNA was observed as early as 12 hours after treatment, peaking at 24 hours and diminishing gradually thereafter owing to a marked drug-induced toxicity and subsequent cell death (Figure 6B). Drug-induced increase in ABCG2 transcript levels, albeit at lower levels, was also observed when using SSZ (P = .0048; Figure 6C). However, several antitumor agents including the camptothecins topotecan and SN-38 as well as the antimitotic agent paclitaxel (Taxol) failed to induce a significant change in ABCG2 mRNA levels, when compared with untreated controls (P > .06; Figure 6C). The observed drug-induced increase in ABCG2 mRNA was maintained on the addition of the ABCG2-specific inhibitor FTC but completely abolished by the addition of the transcriptional inhibitor Act D (Figure 6C). Comparable results, albeit at lower levels, were achieved when treating the other abcg2-methylated Jurkat cell line (P < .01; Figure 6D); however, drug treatment of the nonmethylated K562 cell line failed to induce alterations in ABCG2 transcript levels (P > .1; Figure 6D). Similar results were obtained when treating non-methylated MCF7 and A549 cells with DNR (data not shown). Interestingly, QPCR analysis of gene expression of several other genes in CCRF-CEM cells revealed a differential pattern of drug-induced upregulation, which correlated with their promoter CpG methylation status. Both pcft and mdr1 promoters, shown to be methylated in CCRF-CEM cells, displayed a marked transcriptional up-regulation under comparable drug exposure (P < .008; Figure 6E). However, the MDR transporter ABCC1 (MRP1), which is highly expressed in these cells [47], did not undergo transcriptional up-regulation under drug exposure (Figure 6E). Similar results were observed for the ubiquitously expressed genes—histone 3.3, β2-microglobulin, hypoxanthine-guanine phosphoribosyltransferase (HGPRT), and glucose-6-phosphate dehydrogenase (G6PD) transcripts in DNR-treated CCRF-CEM cells (data not shown). Thus, drug-induced overexpression of ABCG2 and other methylated genes occurs only in transcriptionally silenced tumor cell lines through active transcriptional up-regulation. This process seems to be independent of the transport function of ABCG2 (Figure 6C).
Figure 6.
The effect of short-term drug exposure on ABCG2 gene expression. CCRF-CEM cells were subjected to MX, Dox, and DNR in a dose- (A) and time-dependent (B) manner. Cells were harvested, and relative abcg2 mRNA levels were assessed using QPCR. Furthermore, the effect of functional inhibition of ABCG2 using the transport inhibitor FTC and transcriptional inhibition using Act D on drug-induced ABCG2 gene expression in CCRF-CEM cells was assessed (C). Drug-induced ABCG2 expression was evaluated in abcg2 methylated and nonmethylated cell lines Jurkat and K562, respectively (D). Drug-induced relative up-regulation of ABCG2 gene expression was expanded to other known silenced and nonsilenced genes in CCRF-CEM cells (E). Drug concentration and incubation times were set at an optimal induction capacity. QPCR expression levels are presented as fold expression relative to untreated cells.
Discussion
Anticancer drug resistance mediated by the overexpression of ABCG2 is an important modality of MDR in multiple tumor cell lines selected with various chemotherapeutic agents [7]. Several mechanisms have been previously described that facilitate transcriptional up-regulation of ABCG2 in different MDR cell line models: 1) ABCG2 gene amplification, 2) chromosomal rearrangements, and 3) alternative 3′-untranslated-mediated mRNA stability [48]. It has been recently shown that the ABCG2 promoter undergoes CpG island methylation in renal cell carcinoma lines, thereby resulting in epigenetic silencing [33]. Thus, drug-induced epigenetic modulation of ABCG2 gene expression has gained increasing focus in a number of recent publications [35,36]. Here, we show for the first time that ABCG2 promoter demethylation is a key mechanism underlying the reactivation of ABCG2 transcription and subsequent ABCG2-dependent MDR phenotype in the T-ALL, drug-selected cell line CCRF-CEM/SSZ2.5 and the ovarian carcinoma MDR cell lines IGROV1/T8 and IGROV1/MX3. Importantly, our present results suggest that this mode of restoration of ABCG2 gene expression in epigenetically silenced tumor cell lines may be directly induced by chemotherapeutic drug exposure as early as 12 hours after exposure, hence occurring within a single cell cycle.
To date, the most frequently occurring mechanism of ABCG2 overexpression in MDR tumor cell lines is abcg2 gene amplification [49–51]. Interestingly, MDR cells exhibiting this modality of up-regulation were all derived from solid tumors of epithelial origin, thereby harboring distinct basal ABCG2 transcription. Accordingly, ABCG2 transcript levels in the carcinoma cell lines studied here were several orders of magnitude higher than those observed in human leukemia CCRF-CEM and ovarian carcinoma IGROV1 cells, harboring abcg2 promoter methylation. This dramatic difference in basal ABCG2 gene expression leads us to explore an alternative paradigm for the epigenetic mode of up-regulation of ABCG2, emerging in drug-selected malignant cells displaying poor basal levels of ABCG2 transcripts. We hypothesized that the lack of basal transcription in these epigenetically silenced tumor cell lines may impede their ability to undergo abcg2 gene amplification under drug-selective pressure, using two complementary mechanisms: 1) Heterochromatin-associated, increased genomic stability at the genomic ABCG2 locus. 2) Under drug-selective pressure, the complete lack of ABCG2 function in these silenced cells deprives them of a crucial MDR driving and protective force. Thus, these silenced tumor cells may lack the crucial molecular tools facilitating drug-induced genomic amplification; as such, these cells must rely on a different mode for ABCG2 up-regulation presumably occurring at the early stages of drug exposure. Reactivation of transcription in epigenetically silenced genes due to loss of promoter methylation may meet this exact need. This is in accord with our current findings: 1) chemotherapeutic drug-induced transcriptional up-regulation of ABCG2 and other methylated genes occurs as early as 12 hours after initial drug exposure, hence resulting in as much as 55-fold increased ABCG2 transcript levels within a single cell cycle. This early upregulation of ABCG2 under drug exposure may provide the necessary initial driving force for acquisition of the MDR phenotype under drug selection, endogenously lacking in these ABCG2-silenced cells. 2) Previous analysis of the abcg2 promoter revealed multiple binding sites for strong and constitutive transcriptional factors [43], which, on loss of repression through demethylation, may readily restore promoter binding, thereby resulting in the dramatic increase in ABCG2 overexpression, ultimately leading to the MDR phenotype. The striking (>700-fold) up-regulation of ABCG2 transcript due to the elimination of CpG island promoter methylation in our drug-selected cells may portray exactly this scenario. This is also corroborated in our ABCG2 promoter activity assay displaying equal and significant transcriptional activity in both parental (silenced) cells and their paired MDR sublines. 3) The robust silencing produced by our in vitro methylation assay on the previously reported ABCG2's proximal promoter [43], in both IGROV1 and IGROV1/T8 cell lines, demonstrates its ABCG2-silencing capacity, which, on demethylation, unleashes the observed high transcriptional potential of this MDR gene. 4) The marked difference in basal ABCG2 expression between promoter methylated and nonmethylated tumor cell lines revealed differential characteristics of ABCG2 up-regulation in their corresponding MDR sublines. Accordingly, genomically amplified MDR carcinoma sublines displayed the absolute highest ABCG2 overexpression levels, consistent with their degree of abcg2 gene amplification. In contrast, the demethylated MDR sublines displayed only moderate ABCG2 expression while producing the most prominent relative increase in ABCG2 transcript levels when compared with their parental cells. Thus, as opposed to the linear transcriptional effect of genomic amplification, alterations in the status of promotermethylation may dramatically modulate ABCG2 gene expression in an “all-or-none” fashion, thereby conferring sufficient levels of MDR under drug-selective pressure.
It is noteworthy that neither treatment with bona fide epigenetics modifiers including 5-Aza-dC and TSA nor treatment with various antitumor agents, all of which produced a marked induction of methylated ABCG2 transcription, resulted in a noticeable increase in ABCG2 protein levels or drug efflux function, as assessed by Western blot analysis and flow cytometry-based chromophore drug accumulation assay (data not shown). This may be explained by the extremely low basal levels of ABCG2 mRNA in methylated parental cells, hence limiting the ability to detect quantitative changes in protein levels. Although the relative drug-induced up-regulation is remarkable (up to 55-fold), the absolute levels of ABCG2 transcript in these drug-treated cells are more than an order of magnitude lower than ABCG2 expression in various parental cell lines such as MCF7 and A549 (compare relative ABCG2 mRNA levels in Figures 1 to 3, A and F). The latter parental cells display only low ABCG2 protein levels and, accordingly, barely detectable efflux function [37]. Nevertheless, although the absolute drug-induced up-regulation of ABCG2 may seem extremely low, our results reveal a significant three-fold ABCG2-mediated resistance to the anticancer agent MX (Table 2). Thus, these results demonstrate the ABCG2-dependent chemoprotective function achieved from levels as low as 10-fold up-regulation of ABCG2 mRNA produced by treatment with demethylating agents (Figure 3F). ABCG2 expression and function in the drug-selected sublines CCRF-CEM/SSZ2.5 and IGROV1/T8 are indeed markedly high (i.e., corresponding to >200- and 700-fold mRNA overexpression, respectively). However, this is a result of the prolonged multistep drug selection protocol used to achieve this MDR phenotype. Thus, although our results display only low levels of ABCG2-mediated resistance, they may bear important implications for the early molecular events and dynamics of induction of MDR gene expression, in particular the dynamics of restoration of silent MDR gene expression under cytotoxic stress with chemotherapeutic agents. These early quantitative alterations in ABCG2 gene expression are relatively small and are therefore detectable only using very high sensitivity analyses such as realtime PCR. However, these early changes can be the very beginning of the multistage process that presumably leads to the ultimate ABCG2 overexpression-dependent MDR phenotype.
The aberrant methylation of CpG islands is a hallmark of cancer, leading to the epigenetic silencing of genes that normally function in all different aspects of suppression of malignant transformation, tumor cell growth, and metastasis [21]. This process is considered to occur owing to the deregulation of DNMT activity [23], thereby leading to the stochastic methylation of dense CpG islands throughout the cancer cell genome. Moreover, CpG hypermethylation is considerably augmented during the process of tumor cell line establishment by yet unknown mechanisms [52]. Thus, the random silencing of ABCG2 gene expression through CpG island promoter methylation may be a common event in cancer cell lines and possibly human malignancies. Our results focused on several different cell lines harboring ABCG2 promoter methylation. Interestingly, our ABCG2 methylation analysis revealed that promoter methylation seems to be more common within T-ALL-derived cell lines, when compared with the array of solid tumor cell lines tested. This observation is in accord with the physiological tissue distribution and expression levels of ABCG2. Active transcription has been reported to repress de novo CpG methylation through transcription factor protection [53]. Conversely, histone modifications such as deacetylation promote DNA condensation and contribute to DNA methylation [18]. Therefore, relatively low levels of ABCG2 transcription observed in primary T-cell lineage [54] may reflect a general state of tightly packed DNA at this genomic locus, thereby contributing to the susceptibility of the ABCG2 promoter to undergo hypermethylation during malignant transformation. Indeed, ABCG2 promoter methylation was observed in T-ALL clinical samples. This preliminary observation warrants a large cohort study to reliably determine the prevalence and possible therapeutic implications of ABCG2 promoter methylation in T-ALL and other leukemia. A model summarizing the putative dynamics of ABCG2 promoter methylation in tumor progression and under anticancer drug exposure is depicted in Figure 7.
Figure 7.
Schematic representation of the putative dynamics of abcg2 promoter methylation in tumor progression and under anticancer drug treatment.
It has been previously suggested that the therapeutic use of demethylating agents for the treatment of hematological malignancies may serve as a double-edged sword which, while inducing the reexpression of the necessary silenced tumor-suppressor genes, might also provoke the reexpression of proto-oncogenes [55]. In light of our current findings, ABCG2 may also be reexpressed under combination chemotherapeutic regimens, which may include demethylating agents and ABCG2 transport substrates; this could ultimately lead to the generation of highly MDR clones in the clinical setting. Also, the use of methylated oligonucleotides has been reported to specifically target and induce transcriptional silencing of methylation-sensitive genes in both in vitro and in vivo models, through complementary association to the target gene followed by recruitment of DNMT machinery [56]. It is likely that the strong susceptibility of the ABCG2 gene to silencing through promoter methylation may render it an excellent candidate for in vivo methylated oligonucleotide treatment in MDR tumor models in conjunction with chemotherapy and possibly subsequent to treatment with demethylating agents.
Supplementary Material
Supplementary Data
Material and Methods
Determination of abcg2 gene amplification using quantitative genomic PCR
Quantification of abcg2 gene copy number in MDR sublines relative to parental counterparts was performed. Serial dilutions of template DNA in a PCR were compared using the previously described settings, primers, and internal controls [37].
Transient transfections with pGL3-abcg2 promoter expression vectors and luciferase reporter gene assay
For the luciferase reporter assay, IGROV1 and IGROV1/T8 cells were grown in 24-mm dishes (3 x 104 cells per well) for 24 hours, after which cells were transiently cotransfected with pGL3-abcg2 constructs (generously provided by Dr. Douglas D. Ross) [43], including the pGL3-Basic empty vector, along with the extensively used pROL control plasmid (Renilla luciferase) using the jetPEI transfection reagent (Polyplus-transfection) according to the instructions of the manufacturer. Twenty-four hours after transient transfection, cells were harvested and tested for luciferase and Renilla activities using the Dual-Luciferase Reporter Assay System (Promega) as described in the manufacturer's protocol. Results presented were obtained from at least three independent experiments performed in duplicate cultures.
Result
Lack of abcg2 gene amplification in IGROV1- and CCRF-CEM- derived MDR cell lines
Previous studies have shown that the marked overexpression of ABCG2 in drug selected tumor cell lines of different tissue origin is frequently due to abcg2 gene amplification [49–51]. Consistently, approximately 5-, 7-, and 16-fold abcg2 gene amplification was found in MCF7/FLV1000, MCF7/MRi and A549/K1.5 cells, respectively (Figure W1) [37]. In contrast, semiquantitative genomic PCR analysis revealed that the abcg2 gene was not amplified in the ABCG2-overexpressing IGROV1/T8, IGROV1/MX3, and CCRF-CEM/SSZ2.5 cells (Figure W1). Thus, the molecular mechanism underlying ABCG2 overexpression in these MDR cell lines is not genomic amplification of the abcg2 gene.
Transacting factors do not contribute to up-regulation of ABCG2 in IGROV1-derived MDR cell lines
To date, no molecular mechanism of transcriptional transactivation has yet been associated with up-regulation of ABCG2 in drug-selected cell lines. However, this is not the case with the extensively studied MDR transporters ABCB1 (Pgp, MDR1) and ABCC1 (MRP1), which exhibit various mechanisms of transactivation in drug selected tumor cell lines [28,57,58]. We hence investigated the possible involvement of transacting factors in this up-regulation of ABCG2 in the nongenomically amplified MDR cell line IGROV1/T8. To this end, a luciferase reporter assay was used on transient transfection in both parental IGROV1 and MDR IGROV1/T8 cells, using the previously described [43] -1285/+362, -312/+362, and -115/+362 consecutive ABCG2 promoter deletion constructs (Figure W2). In contrast to the transcriptional upregulation of ABCG2 in IGROV1/T8 cells, all ABCG2 promoter constructs demonstrated slightly lower activity in IGROV1/T8 cells than their activity in parental IGROV1 cells (Figure W2). Yet, the positive control construct harboring activating transcription factor maintained parental ABCG2 expression levels. Thus, changes in transacting factors do not seem to contribute to ABCG2 up-regulation in IGROV1/T8 cells.
Acknowledgments
The authors thank Irit Avivi from the Department of Hematology, Rambam Medical Center, Haifa, Israel, for generously providing the primary T-ALL specimens. The authors also thank Hila Novak for her help with the isolation of primary T-cells.
Abbreviations
- ABC
ATP-binding cassette
- Act D
actinomycin D
- ALL
acute lymphoblastic leukemia
- COBRA
combined bisulfite restriction analysis
- DNMT
DNA methyltransferase
- DNR
daunorubicin
- Dox
doxorubicin
- FTC
fumitremorgin C
- HDACi
histone deacetylase inhibitor
- MDR
multidrug resistance
- MX
mitoxantrone
- SSZ
sulfasalazine
- TSA
trichostatin A
- QPCR
quantitative polymerase chain reaction
- 5-Aza-dC
5-Aza-2′-deoxycytidine
Footnotes
This work was supported by a grant from The Fred Wyszkowski Cancer Research Fund (to Y.G.A.).
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 2.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 3.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 4.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92:1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 5.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 6.Litman T, Druley TE, Stein WD, Bates SE. From MDR to MXR: new understanding of multidrug resistance systems, their properties and clinical significance. Cell Mol Life Sci. 2001;58:931–959. doi: 10.1007/PL00000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Sarkadi B, Ozvegy-Laczka C, Nemet K, Varadi A. ABCG2 — a transporter for all seasons. FEBS Lett. 2004;567:116–120. doi: 10.1016/j.febslet.2004.03.123. [DOI] [PubMed] [Google Scholar]
- 9.Colabufo NA, Berardi F, Contino M, Niso M, Perrone R. ABC pumps and their role in active drug transport. Curr Top Med Chem. 2009;9:119–129. doi: 10.2174/156802609787521553. [DOI] [PubMed] [Google Scholar]
- 10.Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9:105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 11.Sarkadi B, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Gonen N, Bram EE, Assaraf YG. PCFT/SLC46A1 promoter methylation and restoration of gene expression in human leukemia cells. Biochem Biophys Res Commun. 2008;376:787–792. doi: 10.1016/j.bbrc.2008.09.074. [DOI] [PubMed] [Google Scholar]
- 15.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 17.Kudo S. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118(5):549–565. doi: 10.1007/s00412-009-0221-9. [Epub 2009 June 9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 21.Plass C, Smiraglia DJ. Genome-wide analysis of DNA methylation changes in human malignancies. Curr Top Microbiol Immunol. 2006;310:179–198. doi: 10.1007/3-540-31181-5_9. [DOI] [PubMed] [Google Scholar]
- 22.Socha MJ, Said N, Dai Y, Kwong J, Ramalingam P, Trieu V, Desai N, Mok SC, Motamed K. Aberrant promoter methylation of SPARC in ovarian cancer. Neoplasia. 2009;11:126–135. doi: 10.1593/neo.81146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 24.Claus R, Almstedt M, Lubbert M. Epigenetic treatment of hematopoietic malignancies: in vivo targets of demethylating agents. Semin Oncol. 2005;32:511–520. doi: 10.1053/j.seminoncol.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 26.Jung Y, Park J, Kim TY, Park JH, Jong HS, Im SA, Robertson KD, Bang YJ, Kim TY. Potential advantages of DNA methyltransferase 1 (DNMT1)-targeted inhibition for cancer therapy. J Mol Med. 2007;85:1137–1148. doi: 10.1007/s00109-007-0216-z. [DOI] [PubMed] [Google Scholar]
- 27.Assaraf YG, Molina A, Schimke RT. Sequential amplification of dihydrofolate reductase and multidrug resistance genes in Chinese hamster ovary cells selected for stepwise resistance to the lipid-soluble antifolate trimetrexate. J Biol Chem. 1989;264:18326–18334. [PubMed] [Google Scholar]
- 28.Borellini F, Aquino A, Josephs SF, Glazer RI. Increased expression and DNA-binding activity of transcription factor Sp1 in doxorubicin-resistant HL-60 leukemia cells. Mol Cell Biol. 1990;10:5541–5547. doi: 10.1128/mcb.10.10.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgnia MJ, Eytan GD, Assaraf YG. Competition of hydrophobic peptides, cytotoxic drugs, and chemosensitizers on a common P-glycoprotein pharmacophore as revealed by its ATPase activity. J Biol Chem. 1996;271:3163–3171. doi: 10.1074/jbc.271.6.3163. [DOI] [PubMed] [Google Scholar]
- 30.El-Osta A, Kantharidis P, Zalcberg JR, Wolffe AP. Precipitous release of methyl-CpG binding protein 2 and histone deacetylase 1 from the methylated human multidrug resistance gene (MDR1) on activation. Mol Cell Biol. 2002;22:1844–1857. doi: 10.1128/MCB.22.6.1844-1857.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusaba H, Nakayama M, Harada T, Nomoto M, Kohno K, Kuwano M, Wada M. Association of 5′ CpG demethylation and altered chromatin structure in the promoter region with transcriptional activation of the multidrug resistance 1 gene in human cancer cells. Eur J Biochem. 1999;262:924–932. doi: 10.1046/j.1432-1327.1999.00469.x. [DOI] [PubMed] [Google Scholar]
- 32.Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene. 2005;24:8061–8075. doi: 10.1038/sj.onc.1208955. [DOI] [PubMed] [Google Scholar]
- 33.To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26:8572–8585. doi: 10.1128/MCB.00650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner JG, Gump JL, Zhang C, Cook JM, Marchion D, Hazlehurst L, Munster P, Schell MJ, Dalton WS, Sullivan DM. ABCG2 expression, function, and promoter methylation in human multiple myeloma. Blood. 2006;108:3881–3889. doi: 10.1182/blood-2005-10-009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calcagno AM, Fostel JM, To KK, Salcido CD, Martin SE, Chewning KJ, Wu CP, Varticovski L, Bates SE, Caplen NJ, et al. Single-step doxorubicin-selected cancer cells overexpress the ABCG2 drug transporter through epigenetic changes. Br J Cancer. 2008;98:1515–1524. doi: 10.1038/sj.bjc.6604334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.To KK, Polgar O, Huff LM, Morisaki K, Bates SE. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res. 2008;6:151–164. doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bram EE, Ifergan I, Grimberg M, Lemke K, Skladanowski A, Assaraf YG. C421 allele-specific ABCG2 gene amplification confers resistance to the antitumor triazoloacridone C-1305 in human lung cancer cells. Biochem Pharmacol. 2007;74:41–53. doi: 10.1016/j.bcp.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 38.Ifergan I, Scheffer GL, Assaraf YG. Novel extracellular vesicles mediate an ABCG2-dependent anticancer drug sequestration and resistance. Cancer Res. 2005;65:10952–10958. doi: 10.1158/0008-5472.CAN-05-2021. [DOI] [PubMed] [Google Scholar]
- 39.Maliepaard M, van Gastelen MA, de Jong LA, Pluim D, van Waardenburg RC, Ruevekamp-Helmers MC, Floot BG, Schellens JH. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected ovarian tumor cell line. Cancer Res. 1999;59:4559–4563. [PubMed] [Google Scholar]
- 40.van der Heijden J, de Jong MC, Dijkmans BA, Lems WF, Oerlemans R, Kathmann I, Schalkwijk CG, Scheffer GL, Scheper RJ, Jansen G. Development of sulfasalazine resistance in human T cells induces expression of the multidrug resistance transporter ABCG2 (BCRP) and augmented production of TNFa. Ann Rheum Dis. 2004;63:138–143. doi: 10.1136/ard.2002.005249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, Jones PA. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TB, Park JH, Min YD, Kim KJ, Choi CH. Epigenetic mechanisms involved in differential MDR1 mRNA expression between gastric and colon cancer cell lines and rationales for clinical chemotherapy. BMC Gastroenterol. 2008;8:33. doi: 10.1186/1471-230X-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey-Dell KJ, Hassel B, Doyle LA, Ross DD. Promoter characterization and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta. 2001;1520:234–241. doi: 10.1016/s0167-4781(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 44.Goldstone AH, Richards SM, Lazarus HM, Tallman MS, Buck G, Fielding AK, Burnett AK, Chopra R, Wiernik PH, Foroni L, et al. In adults with standard-risk acute lymphoblastic leukemia, the greatest benefit is achieved from a matched sibling allogeneic transplantation in first complete remission, and an autologous transplantation is less effective than conventional consolidation/maintenance chemotherapy in all patients: final results of the International ALL Trial (MRC UKALL XII/ECOG E2993) Blood. 2008;111:1827–1833. doi: 10.1182/blood-2007-10-116582. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26:5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 46.Kantharidis P, El-Osta A, deSilva M, Wall DM, Hu XF, Slater A, Nadalin G, Parkin JD, Zalcberg JR. Altered methylation of the human MDR1 promoter is associated with acquired multidrug resistance. Clin Cancer Res. 1997;3:2025–2032. [PubMed] [Google Scholar]
- 47.Assaraf YG, Rothem L, Hooijberg JH, Stark M, Ifergan I, Kathmann I, Dijkmans BA, Peters GJ, Jansen G. Loss of multidrug resistance protein 1 expression and folate efflux activity results in a highly concentrative folate transport in human leukemia cells. J Biol Chem. 2003;278:6680–6686. doi: 10.1074/jbc.M209186200. [DOI] [PubMed] [Google Scholar]
- 48.To KK, Zhan Z, Litman T, Bates SE. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28:5147–5161. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen JD, Brinkhuis RF, Wijnholds J, Schinkel AH. The mouse Bcrp1/Mxr/Abcp gene: amplification and overexpression in cell lines selected for resistance to topotecan, mitoxantrone, or doxorubicin. Cancer Res. 1999;59:4237–4241. [PubMed] [Google Scholar]
- 50.Knutsen T, Rao VK, Ried T, Mickley L, Schneider E, Miyake K, Ghadimi BM, Padilla-Nash H, Pack S, Greenberger L, et al. Amplification of 4q21-q22 and the MXR gene in independently derived mitoxantrone-resistant cell lines. Genes Chromosomes Cancer. 2000;27:110–116. [PubMed] [Google Scholar]
- 51.Rao VK, Wangsa D, Robey RW, Huff L, Honjo Y, Hung J, Knutsen T, Ried T, Bates SE. Characterization of ABCG2 gene amplification manifesting as extrachromosomal DNA in mitoxantrone-selected SF295 human glioblastoma cells. Cancer Genet Cytogenet. 2005;160:126–133. doi: 10.1016/j.cancergencyto.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 53.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 54.Plasschaert SL, van der Kolk DM, de Bont ES, Kamps WA, Morisaki K, Bates SE, Scheffer GL, Scheper RJ, Vellenga E, de Vries EG. The role of breast cancer resistance protein in acute lymphoblastic leukemia. Clin Cancer Res. 2003;9:5171–5177. [PubMed] [Google Scholar]
- 55.Cui W, Wang L-H. DNA methylation: a two-edged sword as a target for anti-cancer drugs. Biosci Hypotheses. 2008;1:334–335. [Google Scholar]
- 56.Yao X, Hu JF, Daniels M, Shiran H, Zhou X, Yan H, Lu H, Zeng Z, Wang Q, Li T, et al. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J Clin Invest. 2003;111:265–273. doi: 10.1172/JCI15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogura M, Takatori T, Sugimoto Y, Tsuruo T. Identification and characterization of three DNA-binding proteins on the promoter of the human MDR1 gene in drug-sensitive and -resistant cells. Jpn J Cancer Res. 1991;82:1151–1159. doi: 10.1111/j.1349-7006.1991.tb01770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shinoda C, Maruyama M, Fujishita T, Dohkan J, Oda H, Shinoda K, Yamada T, Miyabayashi K, Hayashi R, Kawagishi Y. Doxorubicin induces expression of multidrug resistance-associated protein 1 in human small cell lung cancer cell lines by the c-jun N-terminal kinase pathway. Int J Cancer. 2005;117:21–31. doi: 10.1002/ijc.21094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.