Abstract
Following a shift to medium with acetate as the carbon source, a parental yeast strain exhibited a transient moderate 20% reduction in total cellular [NAD+ + NADH] but showed a ~10-fold increase in the ratio of [NAD+]:[NADH] after 36 h. A mutant strain (idhΔ) lacking the tricarboxylic acid cycle enzyme isocitrate dehydrogenase had 50% higher cellular levels of [NAD+ + NADH] relative to the parental strain but exhibited similar changes in cofactor concentrations following a shift to acetate medium, despite an inability to grow on that carbon source; essentially all of the cofactor was in the oxidized form within 36 h. The salvage pathway for NAD(H) biosynthesis was found to be particularly important for viability during early transition of the parental strain to stationary phase in acetate medium. However, oxygen consumption was not affected, suggesting that the NAD(H) produced during this time may support other cellular functions. The idhΔ mutant exhibited increased flux through the salvage pathway in acetate medium but was dependent on the de novo pathway for viability. Long-term chronological lifespans of the parental and idhΔ strains were similar, but viability of the mutant strain was dependent on both pathways for NAD(H) biosynthesis.
Keywords: Redox status in yeast cells, Redox status in stationary phase, NAD(H) levels, Isocitrate dehydrogenase, Tricarboxylic acid cycle, Salvage and de novo pathways for NAD(H), synthesis, Chronological lifespan
During the diauxic shift, as cultures of Saccharomyces cerevisiae exhaust glucose as a carbon source and begin to utilize C2 metabolites (ethanol and acetate) as carbon/energy sources, expression of many mitochondrial proteins is increased to support more rapid flux through oxidative metabolic pathways [1,2]. In addition to changes in expression, allosteric regulation of key enzymes, e.g. mitochondrial NAD+-specific isocitrate dehydrogenase (IDH),1 ensures accelerated flux through the tricarboxylic acid (TCA) cycle as ATP production by glycolysis slows [3–5].
Yeast IDH is an octameric enzyme composed of four IDH1 and four IDH2 subunits [6–8]. Mutants lacking either IDH1 and/or IDH2 lack cellular enzyme activity and are unable to grow with acetate as a carbon source [9]. The acetate growth phenotype is shared with several other yeast mutants containing disruptions in genes encoding TCA cycle enzymes [10–12]. In recent studies [13], we found that levels of some TCA cycle intermediates, e.g. citrate and isocitrate, are substantially elevated in idhΔ mutants grown under permissive conditions (i.e. with glucose or galactose as the carbon source). In the current study, we have extended these studies to examine the effect of loss of IDH on cellular levels of NAD(H) ([NAD+] + [NADH]) and on the relative levels of oxidized to reduced cofactor ([NAD+]/[NADH]). Given that IDH is postulated to regulate rates of oxidative metabolism, we hypothesized that shifting cells lacking IDH from medium with glucose as the carbon source to medium with acetate would have dramatic effects particularly on the latter ratio. Also, for the parental strain, this medium shift recapitulates the growth transition to stationary phase [14], and there have been few rigorous analyses of changes in redox levels during this time.
Previous measurements of pyridine nucleotide levels and ratios in yeast cells have primarily been conducted in the context of lifespan analyses. Lin et al. [15] demonstrated a requirement for respiration to support the extension of replicative lifespan (a measure of the times a single cell can divide) of yeast cells by caloric restriction. Respiration was reported to result in an increase in the cellular ratio of [NAD+]/[NADH] and subsequent stimulation of SIR2, an NAD+-dependent histone deacetylase implicated in regulation of lifespan [15,16]. However, Lin et al. [17] demonstrated a decrease in [NAD+] in older yeast cells, and Kaeberlein et al. [18] showed that severe caloric restriction extended lifespan by some mechanism that is independent of SIR2. Osorio et al. [19] reported a substantial decrease in cellular [NAD+] during the course of chronological lifespan analyses, which measure cellular survival during post-stationary phase growth. However, despite these examples of increasing interest in the relevance of cellular levels of the cofactor NAD(H) to aging, as well as to a number of health-related issues [20–23], there is a general dearth of knowledge about alterations in cellular redox conditions during changes in growth conditions as stated above and about the role of the TCA cycle in controlling the redox state of cells.
We were also interested in determining the biosynthetic pathway(s) responsible for maintaining cellular NAD(H) under our experimental conditions. NAD(H) is synthesized from tryptophan by the cytosolic de novo pathway, catalyzed in yeast by enzymes designated BNA1–BNA6 [24]. NAD(H) can also be recycled from nicotinamide via the salvage pathway catalyzed by enzymes primarily localized in the nucleus [25,26]. The latter include PNC1, a nicotinamidase that converts nicotinamide to nicotinic acid, and NPT1, a nicotinate phosphoribosyltransferase. Codisruption of the BNA6 gene encoding quinolinate phosphoribosyl transferase and of the NPT1 gene was reported to be lethal [24], suggesting that the two pathways are the major cellular sources of net NAD(H). To examine whether the changes in NAD(H) levels and ratios we observed were due to de novo and/or salvage pathway function, we constructed and analyzed strains containing PNC1 or BNA6 gene disruptions in parental and idhΔ strains. This produced novel evidence for differential function of these pathways during entry into stationary phase. Finally, we examined the relevance of control of cellular redox levels to long-term viability using chronological lifespan analyses of stationary phase cultures.
Materials and methods
Yeast strains and cultivation conditions
The parental haploid yeast strain was MMY011 (MATα leu2-3,112 his3-11, ura3-D1 trp1-1 can1–100; [27]). A derivative of this strain (idhΔ) lacking both subunits of IDH was constructed as previously described [28]. The parental and idhΔ strains were each used for deletion/disruption of the BNA6 locus encoding quinolinate phosphoribosyl transferase or of the PNC1 locus encoding pyrazinamidase/ nicotinamidase. For these disruptions, plasmids containing the kan gene or Kluveromyuces lactis URA3 gene [29] were used as templates for polymerase chain reactions (PCR) using oligonucleotides containing 5′- and 3′-sequences from BNA6 and PNC1 genes, respectively. The PCR products were used for transformation [30], and the gene disruptions were confirmed using PCR analyses of genomic DNA samples from yeast transformants.
Yeast strains were cultivated in rich YP medium (1% yeast extract, 2% Bacto-peptone) with 2% glucose (YP glucose, pH 6.8) or 2% sodium acetate (YP acetate, pH 6.0) as the carbon source. For medium shifts, cells were cultivated in YP glucose medium to OD600nm = 0.6–0.9, harvested by centrifugation, washed with water, and diluted into YP acetate medium to OD600nm = 0.6 (≅106 cells/ml). Viable cell numbers (CFUs) were tabulated by plating dilutions of cells taken from cultures at various times onto YP glucose plates. Colonies were counted after 5–7 days of incubation at 30 °C.
Measurements of cellular [NAD+] and [NADH]
Yeast strains were subjected to medium shifts as described above. At times before and following these shifts, culture samples were used to measure OD600nm values and for tabulation of viable cell numbers. Two other samples, each equivalent to OD600nm = 0.5, were harvested and used to prepare acidic and alkaline cellular extracts as previously described [31]. Concentrations of NAD+ in the acidic extract and of NADH in the alkaline extract were measured using enzymatic cycling reactions and standard curves for fluorimetric determination as described by Passonneau and Lowry [32]. Various amounts of each cellular extract were used in cycling reactions to produce at least four values within the linear portions of standard curves. Final experimental values represent averages of such values from at least three independent medium shift experiments. Cofactor levels were expressed relative to viable cell numbers, and a value of 70 fL/cell [33,34] used for conversion to cellular concentrations.
Measurements of respiration
Rates of oxygen consumption were determined using a Clark-type oxygen electrode with 3 ml samples of cell cultures. Cell samples were stirred continuously and maintained at 30 °C. Measurements were conducted in triplicate, and respiration was expressed as pmol oxygen consumed/h/106 viable cells.
Pyrazinamidase activity assays
Yeast NC1 can be assayed using either nicotinamidase or pyrazinamidase activities [35]. Here, procedures described for assays of pyrazinamidase activity in bacterial cells [36] were modified for assays of yeast cell extracts. To prepare cellular protein extracts, ten OD600nm equivalents of harvested cells were lysed using glass beads in 200 µl buffer A (20 mM Tris (pH 7.4), 30 mM NaCl) containing yeast protease inhibitor cocktail (Sigma). The lysates were cleared by centrifugation and used immediately for activity assays. Lysate samples (2.5–20 µl) were incubated with 16 mM pyrazinamide (Sigma) in buffer A (final volume of 200 µl) for 60–90 min at 37 °C. Blank reactions lacking pyrazinamide were similarly prepared for each lysate sample and incubated. Following incubations, 0.8 ml ferrous ammonium sulfate was added to a final concentration of 50 mM. Assays were mixed and incubated at 24 °C for 15 min. Levels of pyrazinoic acid were determined by measuring absorbance at 480 nm and comparison with a standard curve. Protein concentrations were determined using the Bradford assay [37]. Activity values (nmol pyrazinoic acid produced/min/mg protein) represent averages from at least three independent assays.
Chronological lifespan assays
Chronological lifespans were analyzed as described by Fabrizio and Longo [38]. Cells were precultured for 1 day in minimal medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, pH 6.5) containing 2% glucose and supplements (20 µg/ml) to satisfy auxotrophic requirements (including niacin for bna6Δ strains [24]), diluted to OD600nm = 0.1 in the same medium, and cultivated for 3 days to post-stationary growth stages. Cells were harvested and maintained in water at 30 °C. Every 3 days, cells were harvested, washed, resuspended in fresh water. At the same time aliquots were diluted and plated onto YP glucose plates to tabulate viable cell numbers. All experiments were conducted in triplicate.
Results
Growth and changes in redox status with a shift to acetate medium
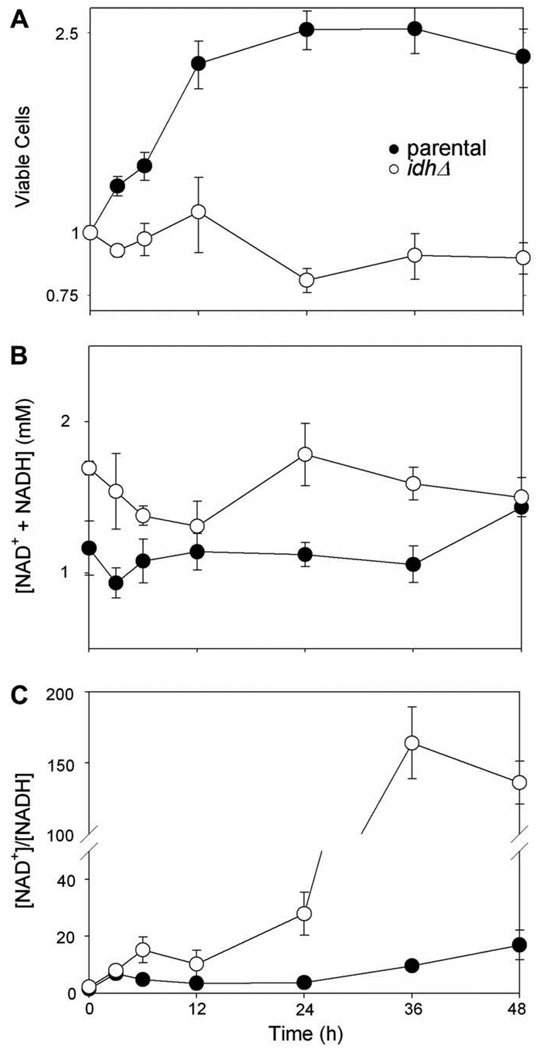
As previously reported [9–12], yeast strains lacking IDH or other TCA cycle enzymes grow at parental strain rates in YP medium with glucose as the carbon source. However, acetate is non-permissive for growth of idhΔ and several other TCA cycle mutant strains. As illustrated in Fig. 1A, viable cell numbers for the parental strain (●) more than double within 12 h after shifting cells growing in logarithmic phase from YP glucose to YP acetate medium. Viable cell numbers then level off from 12 to 48 h as the parental strain enters stationary phase. The idhΔ strain (○), which contains deletion/insertion mutations in genes (IDH1 and IDH2) encoding both subunits of the enzyme, fails to grow following a similar shift to acetate medium but shows little decrease in viable cell numbers over the same period of time.
Fig. 1.
Growth and NAD(H) levels in parental and idhΔ strains following transfer to medium with acetate as the carbon source. Parental (●) and idhΔ (○) yeast strains growing logarithmically in YP glucose medium were harvested and resuspended in YP acetate medium. Samples of the cultures taken before (time 0) or at times shown after the transfer were diluted and plated to determine viable cell numbers (A); these are expressed relative to initial values for each strain set at 1.0. Culture samples were also harvested to prepare extracts for measurements of NAD+ and NADH conducted as described under Materials and methods. Concentrations are expressed as total cofactor concentrations ([NAD+ + NADH]/viable cell/cell volume) (B) and as the ratio of oxidized to reduced cofactor (C).
To monitor changes in cellular redox status, we measured levels of NAD+ and NADH in cellular extracts before and following shifts of cells from YP glucose to YP acetate medium. The total [NAD(H)] in the idhΔ strain logarithmically growing in YP glucose medium was ~50% higher than levels in the parental strain (Fig. 1B, 0 h). Following the shift to acetate medium, the total [NAD(H)] decreased transiently in both strains, by ~20% after 3 h for the parental strain and by ~30% after 12 h for the idhΔ strain. After these initial decreases, both strains regained preshift levels of NAD(H) and the parental strain exhibited a ~20% increase at 48 h relative to the preshift level.
Both the parental and idhΔ strains exhibited substantial increases in the amount of oxidized cofactor (NAD+) relative to reduced cofactor (NADH) following the shift to acetate medium (Fig. 1C). For the parental strain, the [NAD+]/[NADH] ratio increased from ~1.5 in glucose-grown cells (0 h) to 4.7 in cells shifted to acetate medium for 3 h; this ratio further increased to ~17 at 48 h after the shift. This is consistent with an increase in oxidative metabolism following the shift to acetate medium and rapid delivery of NADH to the respiratory chain. For the idhΔ strain, the [NAD+]/[NADH] ratio increased from ~2.5 in glucose-grown cells to 15 at 6 h after the shift to acetate medium. Within 36 h and 48 h, most of the cofactor in the mutant cells was present as the oxidized form. This is consistent with a block in the TCA cycle at the level of the first enzyme (IDH) that normally reduces NAD+ to form NADH.
As described above, both the parental and idhΔ strains exhibit an increase in total [NAD(H)] after initial transient decreases following the shift to acetate medium. To investigate the relative contributions of de novo and salvage pathways for NAD(H) synthesis, and also to determine if there are any effects on these pathways due to changes in the cellular [NAD+]/[NADH] ratios, we used the parental and idhΔ strains for disruption of genes (BNA6 or PNC1, respectively) encoding key enzymes in each pathway.
Roles of NAD biosynthetic pathways in glucose-grown cells
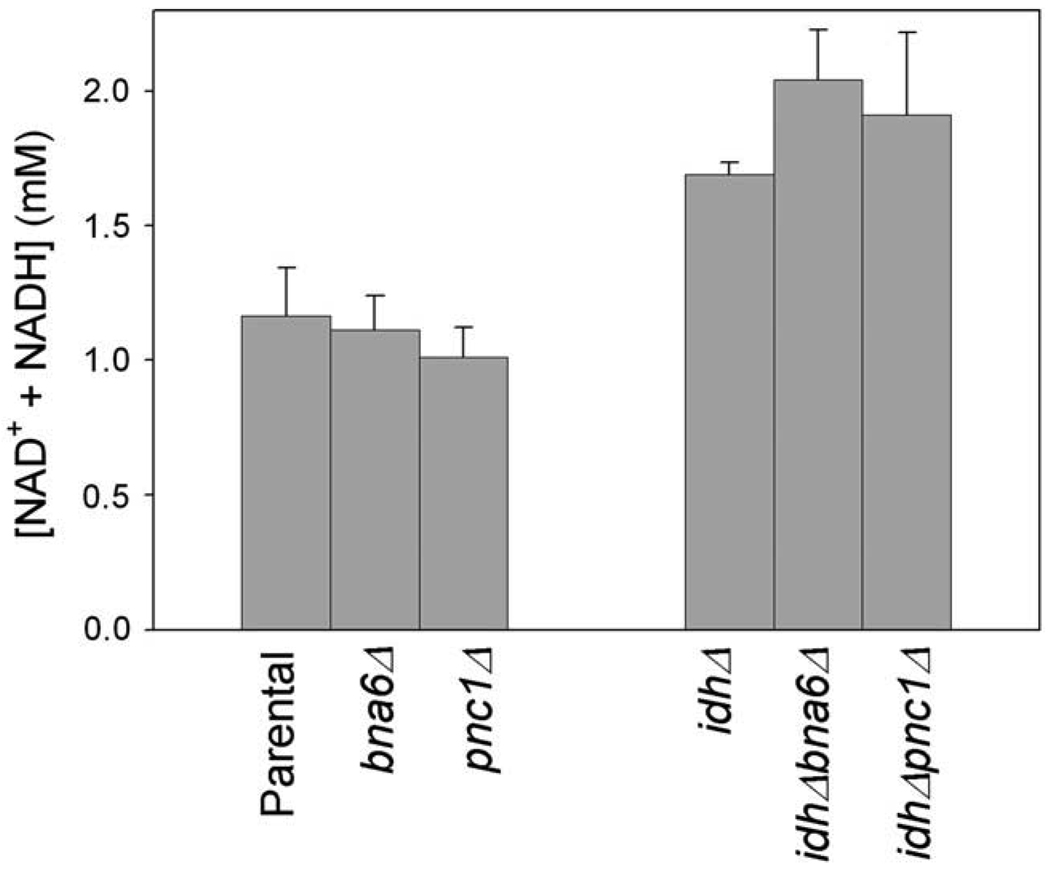
We first examined nucleotide cofactor levels in cells growing logarithmically in YP glucose medium. As shown in Fig. 2, loss of either the de novo pathway (bna6Δ strain) or the salvage pathway (pnc1Δ strain) had little effect on total [NAD(H)]. Thus, as previously reported [24,35,39], either pathway suffices for steady-state growth with glucose. For the idh1Δ strain, loss of either the de novo pathway (idhΔbna6Δ strain) or the salvage pathway (idhΔpnc1Δ strain) also had no significant effect on the total [NAD(H)] in glucose-grown cells, suggesting that either pathway can provide the higher concentrations of cofactor found in the idh1Δ strain relative to the parental level.
Fig. 2.
Concentrations of NAD(H) in glucose-grown cells. Concentrations of NAD+ and NADH were determined using extracts from the indicated yeast strains during logarithmic growth in YP glucose medium.
Roles of NAD biosynthetic pathways in the parental strain following a shift to acetate medium
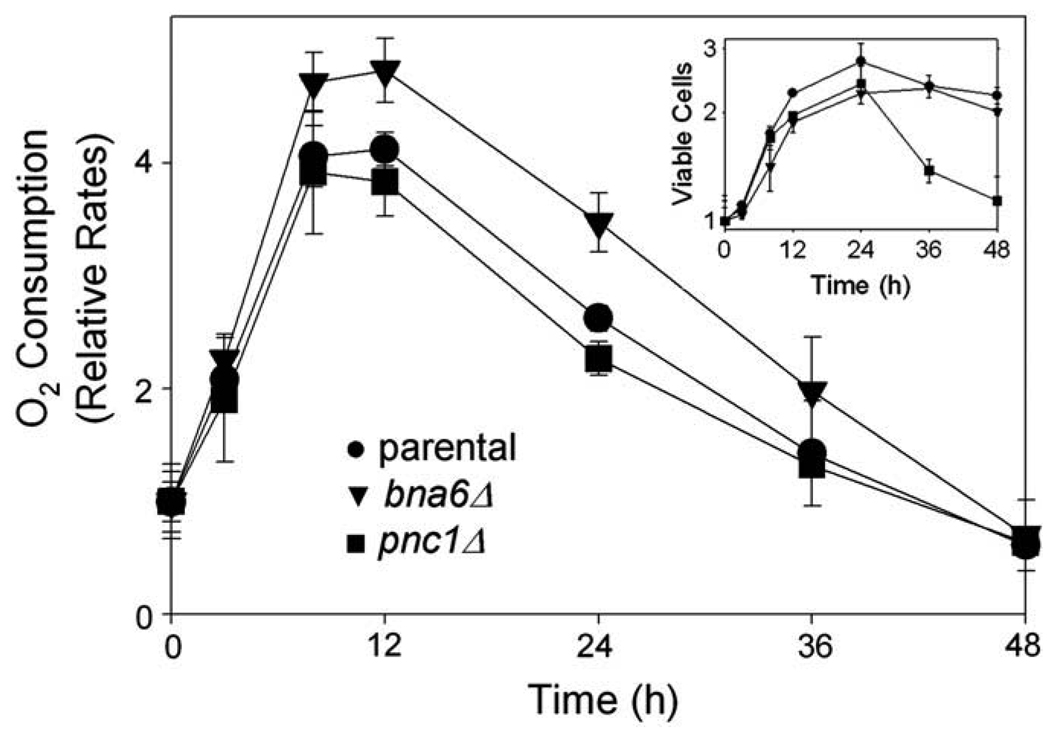
To examine roles of de novo and salvage pathways in redox changes in the parental strain following a shift to acetate medium, we assessed viable cell numbers and cofactor levels in bna6Δ and pnc1Δ strains. As shown in Fig. 3A, for 24 h following a shift to medium with acetate as the carbon source, both bna6Δ (▼) and pnc1Δ (■) mutant strains exhibited increases in viable cell numbers approximately equivalent to numbers for the parental strain (●). After this time, the parental and bna6Δ strains exhibited little further change in cell number, whereas the pnc1Δ strain exhibited a substantial loss in viable cell numbers at 24 h and 48 h following the shift. We determined that these strains differences are transient. With extended times (6–8 days) in expired medium (Supplementary Fig. 1), the parental, bna6Δ and pnc1Δ strains exhibited similar losses in viable cell numbers. Thus, the loss in viability of the pnc1Δ strain as cells enter stationary phase is an acceleration of the loss observed at later times for the other strains.
Fig. 3.
Effects of loss of de novo or salvage pathway on growth and NAD(H) levels in a parental yeast strain. Parental (●), bna6Δ (▼), and pnc1Δ (■) yeast strains growing logarithmically in YP glucose medium were harvested and resuspended in YP acetate medium. Samples of the cultures taken before (time 0) or at times shown after the transfer were diluted and plated to determine viable cell numbers (A); these are expressed relative to initial values set at 1.0. Culture samples were also harvested to prepare extracts for measurements of NAD+ and NADH conducted as described under Materials and methods. Concentrations are expressed as total cofactor concentrations ([NAD+ + NADH]/viable cell/cell volume) (B) and as the ratio of oxidized to reduced cofactor (C).
During the phase of rapid growth following the shift to acetate medium, the parental and bna6Δ and pnc1Δ mutant strains exhibited similar reductions in total [NAD(H)] 3 h after a shift to acetate medium and similar increases in these levels at 6 h and 12 h time points (Fig. 3B). Thereafter, as cells entered stationary phase, levels were approximately constant for the parental and bna6Δ strains, whereas total [NAD(H)] in the pnc1Δ strain decreased dramatically to a level at 48 h 10-fold below the preshift level.2 The [NAD+]/ [NADH] ratio increased, as described above for the parental strain, and similarly in the bna6Δ strain (Fig. 3C; note differences in scales for Fig. 1C and Fig. 3C). For the pnc1Δ strain, following a transient increase at 3 h following the shift, this ratio dropped at 12 h to preshift levels, and the low total level of [NAD(H)] in the pnc1Δ strain precluded accurate measurement of the ratio after 24 h.
These results suggest that either the de novo or salvage pathway is sufficient to support logarithmic growth in acetate medium (0– 12 h following the shift to acetate medium), but that entry into stationary phase (12–48 h following the shift to acetate medium) involves synthesis of NAD(H) primarily via the salvage pathway. Entry into stationary phase also involves an increase in the [NAD+]/[NADH] ratio, presumably due to depletion of various respiratory substrates from the medium.
To examine possible reasons for the loss of viability of the pnc1Δ strain during entry into stationary phase, we examined the effects of changes in [NAD(H)] or [NAD+]/[NADH] ratios on respiration. Rates of oxygen consumption were measured using samples of cultures taken during similar medium shifts. As shown in Fig. 4, relative to rates for cultures growing logarithmically with glucose (0 h), oxygen consumption increased ~4-fold within 9–12 h for the parental and pnc1Δ strains, and ~5-fold for the bna6Δ strain following the shift to acetate medium. Rates of oxygen consumption then declined steadily for all three strains as they entered stationary phase (inset) to levels after 48 h comparable to those observed prior to the shift to acetate medium. Thus, the overall decline in cellular [NAD(H)] observed for the pnc1Δ strain during this time (Fig. 3B) does not appear to impact the relative rate of oxygen consumption measured for viable cells. In fact, as shown in Supplementary Fig. 2C, the pnc1Δ mutant actually has higher cellular levels of NADH, relative to parental and bna6 strains, that likely support normal respiratory rates.
Fig. 4.
Respiration of parental and mutant yeast strains following transfer to YP acetate medium. Parental (●), bna6Δ (▼), and pnc1Δ (■) yeast strains growing logarithmically in YP glucose medium were harvested and resuspended in YP acetate medium. Culture samples were used at indicated times for measurement of oxygen consumption and for determination of viable cell numbers (inset). Respiration was determined as pmol oxygen consumed/h/106 viable cells, and values are expressed relative to initial values set at 1.0.
We also examined if loss of viability of the pnc1Δ strain during entry into stationary phase could be due to an intracellular accumulation of the nicotinamide substrate of PNC1. Since addition of this metabolite to cultures of yeast has previously been reported to inhibit growth [40,41], we reasoned that the pnc1Δ strain might be more sensitive than the other strains to addition of nicotinamide. As shown in Supplementary Fig. 3, relative to viability with no addition to the medium, the addition of 7.5 mM nicotinamide to the medium during a glucose to acetate shift produced similar reductions in viable cell numbers for parental and bna6Δ strains, and delayed growth of the pnc1Δ strain in acetate medium. Addition of 15 mM nicotinamide further reduced growth of parental and bnc6Δ strains and essentially eliminated growth of the pnc1Δ strain in acetate medium. These data suggest that the pnc1Δ strain is more sensitive than the other strains to exogenous addition of nicotinamide as might be expected since intracellular levels of this metabolite accumulate in strains lacking the nicotinamidase [42].
Roles of NAD biosynthetic pathways in the idhΔ strain following a shift to acetate medium
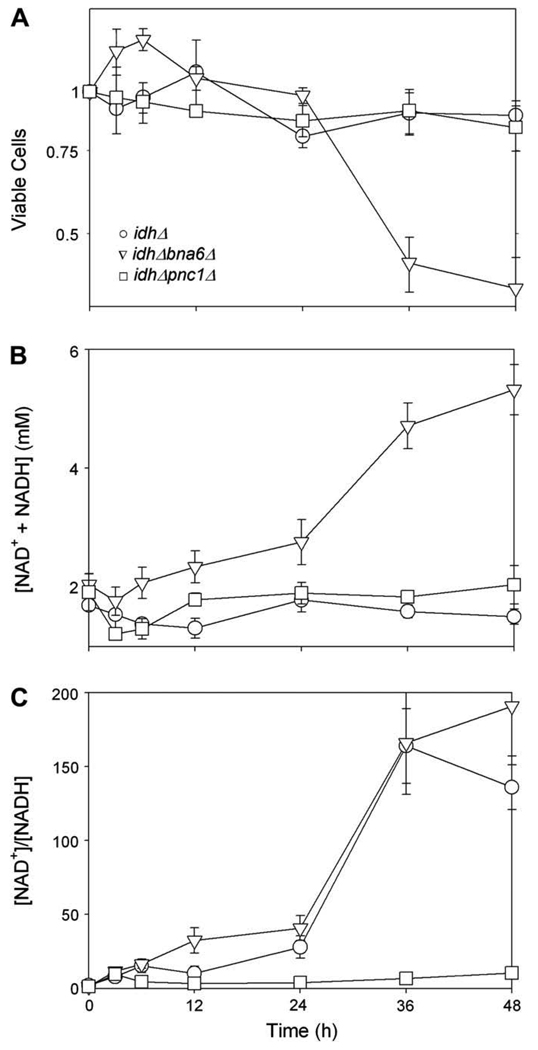
Although the idhΔ mutant strain does not grow with medium containing acetate as the carbon source (Fig. 1A), it does show a transient drop (to 12 h) then a restoration of total [NAD(H)] (Fig. 1B) following a shift to acetate medium. We therefore examined viability and cofactor levels in idhΔ bna6Δ and idhΔ pnc1Δ strains following such a shift to determine the contributions of residual salvage and de novo pathways, respectively. As shown in Fig. 5A, the idhΔ (○) and idhΔ pnc1Δ (□) strains exhibited no growth but little loss in viability 48 h following the medium shift, whereas the idhΔ bna6Δ strain (▽) lost viability after 24 h. This loss in viability was accompanied by a significant 2.6-fold increase in the total [NAD(H)] at 48 h (Fig. 5B) in remaining viable cells, suggesting significant flux through the residual salvage pathway. For both the idhD and idhΔ bna6Δ strains, most of the cofactor was in the oxidized form (Fig. 5C), whereas the increase in the [NAD+]/[NADH] ratio for the idhΔ pnc1Δ strain was quite similar to that observed for the parental strain (Fig. 3C; note differences in scales for Fig. 3C and Fig. 5C). [In further comparisons, we found that all strains lacking IDH failed to consume oxygen when shifted to medium with acetate as the carbon source. Also, experiments to test sensitivity to exogenous nicotinamide were not conducted with these strains due to their inability to grow or respire in acetate medium.]
Fig. 5.
Effects of loss of de novo or salvage pathway on growth and NAD(H) levels in an idhΔ yeast strain. The idhΔ (○), idhΔ bna6Δ (▽), and idhΔ pnc1Δ (□) yeast strains growing logarithmically in YP glucose medium were harvested and resuspended in YP acetate medium. Samples of the cultures taken before (time 0) or at times shown after the transfer were diluted and plated to determine viable cell numbers (A); these are expressed relative to initial values set at 1.0. Culture samples were also harvested to prepare extracts for measurements of NAD+ and NADH as described under Materials and methods. Concentrations are expressed as total cofactor concentrations ([NAD+ + NADH]/viable cell/cell volume) (B) and as the ratio of oxidized to reduced cofactor (C).
Thus, the relative contributions of salvage and de novo pathways following shifts of the parental and idhΔ mutant strains to acetate medium are quite different. While the parental strain appears to depend more on salvage pathway function for entry into stationary phase, the idhΔ strain appears to depend on the de novo pathway for viability when shifted to acetate medium. Flux through the residual salvage pathway in the idhΔ bna6Δ strain as it loses viability after the shift to acetate medium, however, is clearly substantial. One possibility is that the redox imbalance in the idhΔ strain following such a shift, i.e. the relatively high level of NAD+ and low level of NADH (Fig. 5C), acts as a signal for increased flux through the salvage pathway. The absence of the salvage pathway in the idhΔ pnc1Δ strain does not reduce viability of the strain and even appears to prevent the large increase in the [NAD+]/[NADH] ratio observed for the idhΔ strain, potentially due to antagonistic effects of loss of IDH and of PNC1 (see Discussion).
PNC1 activity
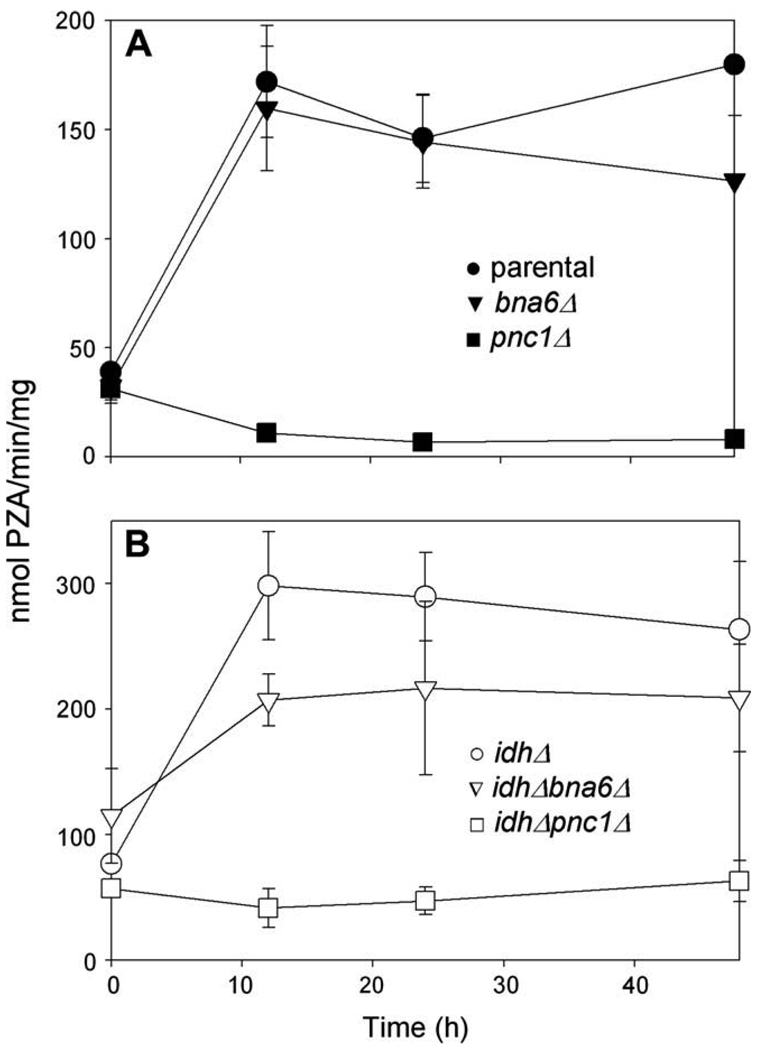
Since the parental strain appears to depend on salvage pathway function for entry into stationary phase, we measured PNC1 activity in various strains before and following a shift to medium with acetate as the carbon source. As shown in Fig. 6A, for cells growing logarithmically with glucose as the carbon source (0 h), the levels of PNC1 activity in parental (●) and bna6Δ (▼) strains were low and comparable to background values measured for the pnc1Δ strain (■). PNC1 activity increased more than 3-fold in parental and bna6Δ strains within 12 h following a shift to acetate medium. This is consistent with salvage pathway function during logarithmic growth in acetate medium and during early entry into stationary phase, and is also consistent with previous reports of increased expression of PNC1 with growth of yeast in media with non-fermentable carbon sources [25,35].
Fig. 6.
PNC1 activity. The pyrazinamidase activity of PNC1 (nmol pyrazinoic acid produced/min/mg cellular protein) was measured using extracts from (A) parental (●), bna6Δ (▼), and pnc1Δ (■) yeast strains and (B) idhΔ (○), idhΔ bna6Δ (▽), and idhΔ pnc1Δ (□) yeast strains growing logarithmically in YP glucose medium (0 h) and at times following a shift to YP acetate medium.
We similarly examined PNC1 activity in idhΔ mutant strains (Fig. 6B). This activity also increased within 12 h following a shift to acetate medium in both the idhΔ (○) and idhΔ bna6Δ (▽) strains to levels, respectively, substantially (~73%) and slightly (~20%) higher than observed for the parental strain at the same time. This is also consistent with salvage pathway function during this time. For the idhΔ bna6Δ strain, PNC1 activity is presumably solely responsible for the substantial increase in total [NAD(H)] observed 24–48 h following the shift (Fig. 5B) as cells lose viability.
Effects on long-term viability
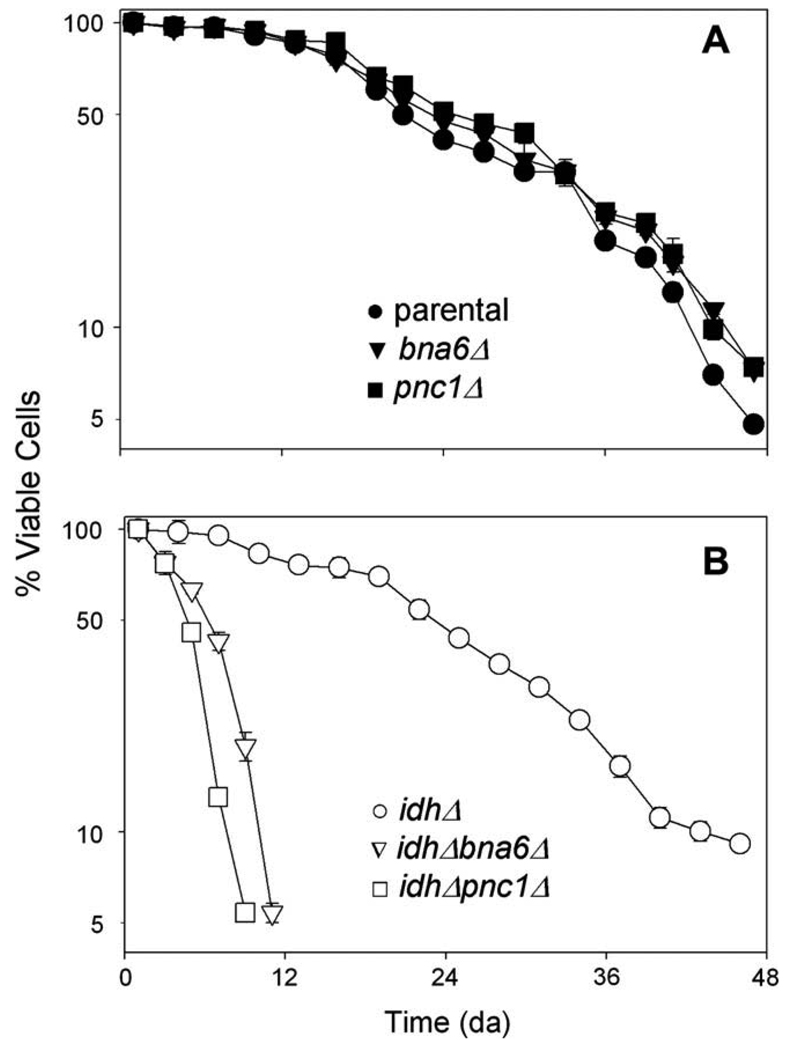
Since we observed transient differences in viability 24–48 h following a shift to acetate medium, with losses observed for the pnc1Δ strain (relative to parental and bna6Δ strains) and for the idhΔ bna6Δ strain (relative to idhΔ and idhΔ pnc1Δ strains), we compared all strains for long-term survival using standard chronological lifespan assays [38]. For this, viability assays were initiated following growth of strains to stationary phase in minimal medium with glucose as the carbon source and subsequent transfer to water. Thus, in contrast to medium shifts conducted as described above, chronological lifespan assays examine post-stationary phase viability. Also, since cells are washed during the lifespan analyses, effects of dying cells including nutrient release and accumulation of toxic compounds [43,44] are minimized.
As shown in Fig. 7A, the parental (●), bna6Δ (▼), and pnc1Δ (■) strains exhibited similar chronological lifespans, with 50% of the cells surviving at days 21, 23, and 24, respectively. Thus, the transient loss of viability of the pnc1Δ strain following entry into stationary phase (Fig. 3A and Supplementary Fig. 1) does not affect the long-term survival of this strain. The idhΔ strain (○, Fig. 7B) showed a similar half life as the parental strain, with 50% of the cells surviving at day 22. In contrast, the idhΔ bna6Δ (▽) and idhΔ pnc1Δ strains (□) died rapidly (Fig. 7B), with respective cell survival levels of 50% on days 5 and 6. Thus, both the salvage and de novo pathways for NAD(H) synthesis are essential for long-term survival of the idhΔ strain.
Fig. 7.
Chronological lifespan analyses. (A) Parental (●), bna6Δ (▼), and pnc1Δ (■) yeast strains, and (B) idhΔ (○), idhΔ bna6Δ (▽), and idhΔ pnc1Δ (□) yeast strains were grown to stationary phase in minimal glucose medium prior to transfer to water for chronological lifespan assays conducted as described under Materials and methods. Viable cell numbers are expressed as a percentage of the initial value for each strain.
Discussion
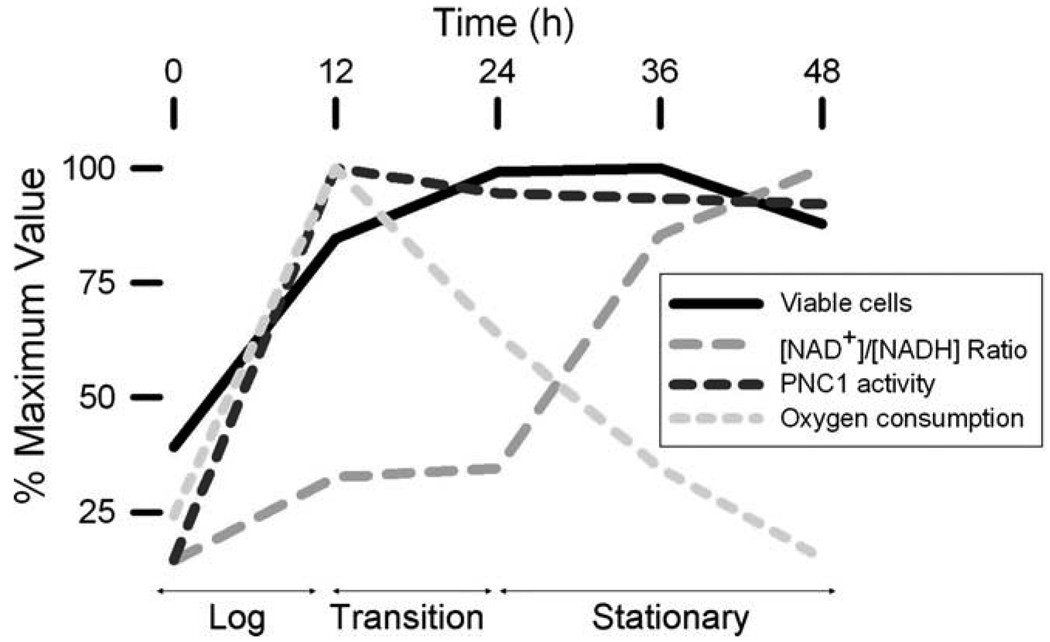
We examined central aspects of redox levels in yeast cells by quantifying levels of reduced and oxidized NAD(H). Although not a direct measurement of flux, sampling cells at times following a change in cultivation conditions allows assessment of changes in cellular pools. We found that alterations in total cellular levels of NAD(H) in a parental yeast strain were relatively minor following a shift from medium with glucose to acetate as the carbon source, with a transient decline followed by an increase to preshift levels as cells entered stationary phase. As summarized in Fig. 8, we also found that as the parental strain entered stationary phase in acetate medium, levels of oxidized:reduced cofactor increased ~10-fold, presumably reflecting depletion of NADH reducing equivalents for oxidative metabolism as well as exhaustion of respiratory substrates from the medium. Rates of oxygen consumption (which had increased during logarithmic growth) decreased proportionally during this time and were independent of functional de novo or salvage pathway synthesis of cofactor. The salvage pathway for NAD(H) production appeared to be particularly important for maintenance of optimum viability and cellular [NAD(H)] during the transition into stationary phase. However, lack of either the de novo or salvage pathway had no significant effect on respiratory rates during these transitions, suggesting that the transient NAD(H) requirement for optimum viability of a pnc1Δ mutant lacking the salvage pathway may be related to factors other than mitochondrial oxygen consumption. Since stationary phase growth of yeast is characterized by a slowing of metabolism, chromatin condensation, and decreased transcription [14], it is possible that the transient reduction in viability and cellular [NAD(H)] noted for the pnc1Δ mutant during entry into stationary phase may correlate with adverse effects on chromatin remodeling likely mediated by Sirtuins, NAD-dependent histone deacetylases.
Fig. 8.
Summary of redox changes in a parental yeast strain during the transition from logarithmic to stationary phase growth. Various parameters measured for the parental strain in experiments described in the text are directly compared. The stages of growth were defined on the basis of viable cell numbers.
Several studies have demonstrated that the activities of Sirtuins are important for cellular lifespan and survival during stress conditions [20,21,45]. In some parental yeast strains, the major yeast Sirtuin, SIR2, is reported to be necessary for extension of replicative lifespan during caloric restriction due, in part, to NAD+-dependent histone deacetylation which regulates silencing and ribosomal DNA recombination [46–48]. Although SIR2 protein levels are not altered in response to growth with non-fermentable carbon sources, enzymatic activity is proposed to be altered by changes in substrate [NAD+] or by induction of PNC1, which removes nicotinamide, an inhibitor of SIR2 [25,26,41,42,49], and which replenishes [NAD+]. Our data are consistent with the latter proposal, i.e. that a 3- to 4-fold increase in PNC1 levels in parental cells following a shift to acetate medium may not dramatically change cellular levels of NAD(H) but may transiently remove inhibition of SIR2 and may facilitate entry into stationary phase. Our other results showing that the pnc1Δ mutant is more sensitive than the parental strain to exogenous addition of nicotinamide during this time are also consistent with this interpretation.
We found that cellular levels of NAD(H) were ~50% higher in an idhΔ mutant than in the parental strain during logarithmic growth with glucose as the carbon source, and that these higher levels persisted in the absence of either the de novo or salvage pathways. While cellular concentrations of NAD(H) changed little following a shift of the idhΔ mutant to non-permissive medium with acetate as the carbon source, the cofactor was almost entirely oxidized within 36 h after the shift. Thus, loss of IDH and, consequently, of a functional TCA cycle largely precludes reduction of NAD+ under these conditions. An idhΔ mutant lacking the de novo pathway for NAD(H) synthesis (idhΔ bna6Δ strain) exhibited a substantial loss in viability 36–48 h after a shift to acetate medium and a concomitant increase in the cofactor (almost all as the oxidized form) in remaining viable cells. Thus, the de novo pathway is important for viability, and flux through the salvage pathway appears to be substantially increased in response to the loss in viability of the idhΔ bna6Δ strain and/or as a mechanism to compensate for low levels of reduced cofactor in this strain. We previously reported a similar compensatory increase in cellular levels of NADP+ when a mutant lacking major cellular enzymatic sources of NADPH was shifted to non-permissive growth conditions [31]. In contrast to the idhΔ bna6Δ strain, an idhΔ mutant lacking the salvage pathway for NAD(H) synthesis (idhΔ pnc1Δ strain) exhibited similar cellular levels of NAD(H) as did the idhΔ mutant, but maintained parental ratios of oxidized to reduced cofactor. One possibility is that the mutations may have antagonistic effects, since loss of IDH alone produces an increase in [NAD+] at the expense of [NADH] (Fig. 1C), whereas loss of PNC1 alone reduces the [NAD+]:[NADH] ratio, at least as measured at 12 h and 24 h time points following a shift to acetate medium (Fig. 3C). Production of NAD+ in different cellular compartments may also have an impact. As illustrated in Supplementary Fig. 2, levels of NADH in the pnc1Δ mutant are ~2-fold higher than in the parental strain 12 h and 24 h following a shift to acetate medium. Similarly, levels of NADH in the idhΔ pnc1Δ mutant are ~3-fold higher than in the idhΔ strain 12 h and 24 h following a shift to acetate medium. This suggests that the NAD+ produced by the residual de novo pathway may be accessible for reduction in the cytosol to produce the excess NADH. This is not the case for strains lacking the de novo pathway, suggesting that the NAD+ produced by the residual salvage pathway, which is presumed to be primarily catalyzed by nuclear enzymes [25,26], is not readily accessible for reduction. With respect to compartmentalization, yeast lack the mitochondrial NAD(H) salvage pathway described for mammalian cells [20]. Thus, the cytosolic de novo biosynthetic pathway may contribute substantially to cell survival during conditions when nutrients are limiting. Future plans include investigation of compartmentalized pools of NAD(H) in yeast.
Mutants lacking either NAD(H) biosynthetic pathway (pnc1Δ or bna6Δ strains) exhibited chronological lifespans, which are measured after stationary phase is attained, equivalent to that of the parental strain. This suggests that rates of NAD(H) synthesis and/or turnover are substantially slowed during the lifespan assays. Osório et al. [19] previously reported a reduction in cellular levels of NAD+ in a parental strain during chronological lifespan assays by 79% at the time (day 28 for their strain) when 50% of the cells remained viable. Consistent with this, we found rates of respiration for both the parental and idhΔ strains, which exhibit similar chronological lifespans, were similar at the outset of lifespan assays and were reduced to comparable, essentially immeasurable levels by day 5 after transfer to water, a time when both strains retain >90% viability (data not shown). In contrast, loss of IDH and either of the NAD(H) biosynthetic pathways (idhΔ pnc1Δ or idhΔ bna6Δ strains) produced very short chronological lifespans, suggesting that both biosynthetic pathways are essential in the absence of IDH. Since rates of respiration are so low during these assays, the need for NAD(H) biosynthesis to maintain parental levels of viability of the idh Δ strain likely reflects support of other cellular processes.
Although we saw no dramatic effects of idhΔ on chronological lifespan, others have reported that loss of IDH increases yeast replicative lifespan [50,51], a measure of the number of times a specific cell can divide. In contrast, we have found that loss of mitochondrial malate dehydrogenase (MDH1) substantially reduces chronological lifespan (data not shown), while Easlon et al. [52] found no extension of replicative lifespan in an mdh1Δ strain. Thus, TCA cycle defects have different effects on lifespan. This is likely due to the accumulation and/or deficiency of different key metabolites caused by different blocks in the cycle ([13] and unpublished results). For idhΔ, data presented here suggest that the absence of detrimental effects on chronological lifespan and the extension of replicative lifespan is unlikely to be related to any effects on redox state. Instead, the accumulation of isocitrate in an idhΔ mutant [13] might be advantageous for production by NADP+-specific isocitrate dehydrogenases of NADPH for various biosynthetic and antioxidant functions [31]. Because of the differences observed for idhΔ and mdh1Δ mutants, it will clearly be of future interest to compare cellular redox balances and lifespans for these and other TCA cycle mutants.
Supplementary Material
Acknowledgments
This work was supported by NIH RO1 AG017477 and NIH RO1 GM051265. We thank Sondra L. Anderson for assistance with chronological lifespan assays.
Footnotes
Abbreviations used: IDH, isocitrate dehydrogenase; TCA, tricarboxylic acid; PCR, polymerase chain reactions.
We were concerned that loss of viability for the pnc1Δ strain might compromise our measurements of total [NAD+ + NADH]. However, as shown in subsequent experiments (Fig. 5), we observed a similar loss of viability for another strain but a concomitant increase in total [NAD+ + NADH]. Thus, measures of cofactor levels do not appear to be influenced by cell viability in these experiments.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.abb.2008.12.014.
References
- 1.Johnston M, Carlson M. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones EW, Pringle JR, Broach JR, editors. vol. 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 2.DeRisi JL, Iyer VR, Brown PO. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 3.Hathaway JA, Atkinson DE. J. Biol. Chem. 1963;238:2875–2881. [PubMed] [Google Scholar]
- 4.Chen RF, Plaut GWE. Biochemistry. 1963;2:1023–1032. doi: 10.1021/bi00905a020. [DOI] [PubMed] [Google Scholar]
- 5.Gancedo C, Serrano R. In: The Yeasts. second ed. Rose AH, Harrison JS, editors. vol. 3. San Diego: Academic Press; 1989. pp. 205–259. [Google Scholar]
- 6.Keys DA, McAlister-Henn L. J. Bacteriol. 1990;172:4280–4287. doi: 10.1128/jb.172.8.4280-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cupp JR, McAlister-Henn L. J. Biol. Chem. 1991;266:22199–22205. [PubMed] [Google Scholar]
- 8.Cupp JR, McAlister-Henn L. J. Biol. Chem. 1992;267:16417–16423. [PubMed] [Google Scholar]
- 9.Cupp JR, McAlister-Henn L. Biochemistry. 1993;32:9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- 10.Thompson LM, Sutherland P, Steffan JS, McAlister-Henn L. Biochemistry. 1988;27:8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- 11.Kim KS, Rosenkrantz MS, Guarente L. Mol. Cell Biol. 1986;6:1936–1942. doi: 10.1128/mcb.6.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCammon MT. Genetics. 1996;144:57–69. doi: 10.1093/genetics/144.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin AP, Hakala KW, Weintraub ST, McAlister-Henn L. Arch. Biochem. Biophys. 2008;474:205–212. doi: 10.1016/j.abb.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. Microbiol. Mol. Biol. Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 16.Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 17.Lin SS, Manchester JK, Gordon JI. J. Biol. Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osorio H, Silles E, Maia R, Peleteiro B, Moradas-Ferreira P, Gunther Sillero MA, Sillero A. FEMS Yeast Res. 2005;5:387–398. doi: 10.1016/j.femsyr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Veer E, Ho C, O’Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. J. Biol. Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 22.Lin SJ, Guarente L. Curr. Opin. Cell Biol. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 23.Belenky P, Bogan KL, Brenner C. Trends Biochem. Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Panozzo C, Nawara M, Suski C, Kucharczyka R, Skoneczny M, Becam AM, Rytka J, Herbert CJ. FEBS Lett. 2002;517:97–102. doi: 10.1016/s0014-5793(02)02585-1. [DOI] [PubMed] [Google Scholar]
- 25.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandmeier JJ, Celic I, Boeke JD, Smith JS. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCammon MT, Veenhuis M, Trapp SB, Goodman JM. J. Bacteriol. 1990;172:5816–5827. doi: 10.1128/jb.172.10.5816-5827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao WN, McAlister-Henn L. J. Biol. Chem. 1997;272:21811–21817. doi: 10.1074/jbc.272.35.21811. [DOI] [PubMed] [Google Scholar]
- 29.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gietz D, St Jean A, Woods RA, Schiestl RH. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minard KI, McAlister-Henn L. J. Biol. Chem. 2005;280:39890–39896. doi: 10.1074/jbc.M509461200. [DOI] [PubMed] [Google Scholar]
- 32.Passonneau JV, Lowry OH. Enzymatic Analysis: A Practical Guide. Totowa, NJ: Humana Press; 1993. pp. 1–110. [Google Scholar]
- 33.Sherman F. In: Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology. Guthrie C, Fink GR, editors. vol. 194. San Diego: Academic Press; 1991. pp. 1–21. [PubMed] [Google Scholar]
- 34.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghislain M, Talla E, Francois JM. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- 36.Frothingham R, Meeker-O’Connell WA, Talbot EA, George JW, Kreuzer KN. Antimicrob. Agents Chemother. 1996;40:1426–1431. doi: 10.1128/aac.40.6.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Fabrizio P, Longo VD. Methods Mol. Biol. 2007;371:89–95. doi: 10.1007/978-1-59745-361-5_8. [DOI] [PubMed] [Google Scholar]
- 39.Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA. Mol. Cell. Biol. 2003;23:7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 41.Gallo CM, Smith DL, Jr., Smith JS. Mol. Cell. Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauve AA, Moir RD, Schramm VL, Willis IM. Mol. Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 43.Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, Diaspro A, Dossen JW, Gralla EB, Longo VD. J. Cell Biol. 2004;166:1055–1067. doi: 10.1083/jcb.200404002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zambrano MM, Kolter R. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Lavu S, Sinclair DA. Exp. Gerontol. 2006;41:718–726. doi: 10.1016/j.exger.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guarente L. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 47.Rusche LN, Kirchmaier AL, Rine J. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 48.Blander G, Guarente L. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 49.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 50.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 51.Kaeberlein M, Burtner CR, Kennedy BK. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.