Abstract
Although inflammatory cytokines and obesity-associated serum proteins have been reported as biomarkers of colorectal adenoma risk in humans, little is known of biomarkers of response to interventions that attenuate tumorigenesis. Dietary navy beans and their fractions attenuate colon carcinogenesis in carcinogen-induced genetically obese mice. We hypothesized that this attenuation would be associated with changes in inflammatory cytokines and obesity-related serum proteins that may serve as measures of efficacy. ob/ob mice (n = 160) were injected with the carcinogen azoxymethane (AOM) to induce colon cancer and randomly placed on one of four diets (control, whole navy bean, bean residue fraction, or bean extract fraction) for 26 to 28 wk. Serum was analyzed for 14 inflammation-or obesity-related proteins, and colon RNA was analyzed for expression of 84 inflammation-associated genes. Six of 14 serum proteins were increased [i.e., interleukin (IL)-4, IL-5, IL-6, IL-10, IFNγ, granulocyte macrophage colony-stimulating factor] in hyperplastic/dysplastic stages of colon carcinogenesis. Bean-fed mice had significantly higher monocyte chemoattractant protein-1 and lower IL-6 levels in serum. In colon mucosa, 55 of 84 inflammation-associated genes differed between AOM-induced and noninduced mice. Of the 55 AOM-induced genes, 5 were counteracted by bean diets, including IL-6 whose increase in expression levels was attenuated by bean diets in AOM-induced mice. In summary, IL-6 emerged as a serum protein that was increased in hyperplastic/dysplastic stages of colon carcinogenesis, but attenuated with bean-based diet in serum and colon mucosa. Changes in a subset of inflammation-associated serum proteins and colon gene expression may serve as response indicators of dietary attenuation of colon carcinogenesis.
Colorectal cancer is the third most common cancer found in men and women (1, 2). The American Cancer Society estimates that there will be 148,810 new cases of colorectal cancer in the United States in 2008. Furthermore, colorectal cancer is estimated to result in ~49,960 deaths in 2008 (2). Prevention strategies are needed because although the 5-year survival rate for those with early-stage disease is 90% (2), more than half (i.e., 57%) of new colorectal cancer cases have regional or distant metastasis at the time of diagnosis (3). Prognosis becomes significantly worse the more advanced the disease, with those with distant metastases having a 5-year survival rate of 10%. Thus, there exists a need for interventions that show efficacy for preventing early stages of carcinogenesis and for methods to monitor the efficacy. To detect this efficacy calls for the identification of early biomarkers that indicate attenuation of carcinogenesis.
Dietary interventions to attenuate colon carcinogenesis may be promising. Previous studies in humans and animals have shown the preventive effects of beans on recurrence of colon cancer and on colon carcinogenesis (4–7). An epidemiologic study by Lanza et al. (7) has shown that consuming a diet high in dry beans reduced the risk of developing advanced colorectal adenoma recurrence among participants in the Polyp Prevention Trial. In addition, we and other groups have found with animal studies that consuming a bean-based diet reduces colon cancer in azoxymethane (AOM)-induced rats and mice (4–6). Specifically, our previous study (4) showed that navy beans and their fractions attenuated colon carcinogenesis in AOM-induced genetically obese mice. Attenuation of colon carcinogenesis was observed in all bean groups (whole bean, residue, and extract), with the extract showing the greatest attenuation. The bean equivalency was similar in the three bean groups. The relative number of bean equivalents for whole bean, residue, and extract fractions was 1.00, 1.14, and 1.47, respectively. That is, the residue fraction group consumed 14% more and the extract fraction group consumed 47% more whole bean equivalents than the whole bean group (4).
The rapid development of technologies such as proteomics and tissue arrays presents an opportunity to identify potential biomarkers for early detection of colorectal cancer (8). In addition, these tools can be used to identify biomarkers for efficacious response to dietary interventions and may facilitate ascertaining the effectiveness of agents in a shorter and less expensive study (9). Among the biomarkers that may indicate attenuation of colon carcinogenesis are inflammation- and obesity-related serum proteins and gene transcripts. Inflammation has been proposed as an important mechanism by which cancer develops (10–13). Obesity is a risk factor for the development of colorectal cancer (14). Two processes may link body size (e.g., obesity) and dietary status to the development of colorectal cancer. These are insulin resistance and chronic inflammation. Markers of interest include the adipokines [e.g., leptin, resistin, adiponectin, adipsin, visfatin, and monocyte chemoattractant protein-1 (MCP-1); ref. 14] and proinflammatory cytokines [e.g., interleukin (IL)-4, IL-5, IL-6, IL-10, IL-13, and IFN-γ; ref. 12]. Dietary attenuation of the expression of these markers may associate with reduced risk of developing colorectal cancer.
Studies are needed to identify noninvasive biomarkers that are indicative of early-stage colorectal carcinogenesis and dietary attenuation thereof. For the current study, the objective was to analyze serum and colon mucosa from the initial study (4) to identify indicators of response to the bean diet intervention under conditions of efficacy.
We aimed to identify indicators of bean diet attenuation of colon carcinogenesis in AOM-induced ob/ob mice. ob/ob mice have a single mutation within the Ob gene, which results in development of obesity, hyperinsulinemia, hyperphagia, and hyperglycemia (15–17). We tested the hypothesis that the bean-based diets would produce changes in inflammation- and obesity-associated serum proteins and colonic mucosa gene expression and that some of these changes would counteract carcinogen-induced changes. Mice on bean-based diets showed significant changes in a subset of inflammation-associated proteins and transcripts that may serve as measures of efficacy.
Materials and Methods
Animals and housing
The main study was carried out at Michigan State University and involved exclusively AOM-induced mice, which were sacrificed at 29 to 37 wk of age (or 27–29 wk following the last AOM injection; ref. 4). Briefly, male ob/ob mice (4–5 wk of age) were received from The Jackson Laboratory and were housed four per cage. Mice were housed in temperature- and humidity-controlled rooms that were on a 12-h light/12-h dark cycle. The AOM-induced carcinogenesis study was conducted with approval from the Michigan State University Animal Care and Use Committee. To evaluate the effect of AOM induction, a group of male ob/ob mice (housed at The Jackson Laboratory) did not receive AOM (i.e., No AOM). Briefly, n = 10 male ob/ob mice were housed until they were ~20 wk of age. All mice had access to food and water ad libitum. Although the No AOM comparison group (20 wk of age) was not age matched to the AOM ob/ob group (29–37 wk of age), every effort was made to have a comparison group as close to the AOM ob/ob group as possible.
Diet preparations
Diets for the AOM-induced mice were prepared and given as previously reported (4). Briefly, 682 kg of raw navy beans grown in Michigan were used to prepare the diets. Beans were soaked overnight and then cooked for 1 h in steam kettles. To prepare whole beans, ~250 kg of the 682 kg were dried overnight in a large oven at 75 ± 5°C. The beans were then ground and passed through a screen with 1.6-mm holes. The remaining 432 kg of the 682 kg were used to prepare the bean residue and extract fractions.
The whole bean and bean residue diets contained similar protein and fiber concentrations. The bean extract diet contained similar oligosaccharides and phenolic components similar to those found in the whole bean. The bean extract diet included bean extract, cellulose, casein, and cornstarch in proportions of 33.7:5.1:6.3:13.0 (w/w/w/w), respectively (4). The phenolic content of the whole bean, residue, and bean extract fractions was previously measured and found to be 8.36, 3.55, and 11.56 mg of (+)-catechin equivalents per kilogram of diet (4). All diets were formulated to be isocaloric and isonitrogenous.
Administration of diets and AOM carcinogen
For AOM-induced mice, diets and AOM were administered as previously stated (4). Briefly, ob/ob mice were placed on control diet (modified AIN-93G: 16.7% fat, 14% fiber) on receipt from The Jackson Laboratory. Mice received s.c. injections of AOM (7 mg/kg body weight) after a 3-wk acclimation period. Mice received two separate AOM injections, 1 wk apart. One week after last injection, mice were randomly assigned to one of four experimental groups (control, whole bean, bean residue, or bean extract) for 26 to 28 wk (4). Body weights of mice were measured throughout the course of the study and have been previously reported (4). For noninduced ob/ob mice that were sacrificed at 20 wk of age, the diet was a standard AIN-93G diet (7% fat, 5% fiber; ref. 18). Although the fat and energy content differed between the two control diets, no difference in weight gain was observed (not shown), indicating that weight gain occurred as a function of the ob/ob genotype. Although the similar weight gains at The Jackson Laboratory (No AOM) and Michigan State University (+AOM) suggest no measurable difference due to the modified AIN-93G diet at Michigan State University, we cannot exclude the possibility of diet contribution to any observed gene expression changes unrelated to weight gain.
Necropsy and pathology
Mice were euthanized using CO2 inhalation and exsanguination. Before necropsy, serum samples were collected from all animals. For necropsy, the colons were cut, rinsed, and grossly examined for lesions. Suspected lesions and representative normal colon tissue (mid-colon) were fixed in neutral buffered formalin, processed, embedded in paraffin, and H&E stained. Histopathologic examination of colon tissue was done by a pathologist. From a subset of mice, colon mucosa was scraped from the remaining tissue and snap frozen.
RNA isolation
RNA was isolated using the RNeasy Mini Kit from Qiagen. Briefly, colon mucosa samples were disrupted and homogenized in a 2-mL O-ring tube containing 1-mL Buffer RLT and 1.0-mm glass beads filled up to 1-mL mark. The Mini-Beadbeater instrument from Biospec was used to disrupt and homogenize colon mucosa samples. Next, samples were centrifuged for 3 min and the supernatant was transferred to a new 2-mL O-ring tube for further processing. Sample lysates were washed (in 70% ethanol) by running them through an RNeasy spin column and were further purified through subsequent washes with Buffer RW1 and Buffer RPE. Resultant RNA was eluted from column in 50 µL of RNase-free water. All samples were DNase treated during the on-column RNA isolation steps. RNA concentration and integrity were determined by measurement at 260/280 and 260/230 absorbance wavelengths by using the Ultrospec 2000 spectrophotometer (Pharmacia Biotech). The RNA Cleanup kit from Qiagen was used to purify RNA samples that did not pass the A260/230 test (samples whose ratios were <1.7, which would indicate high carbohydrate and protein content in RNA sample). All samples were run on a 1% Tris-borate EDTA agarose gel to check for sample integrity.
Quantitative real-time PCR array
Two micrograms of total RNA from each colon mucosa sample were used to synthesize cDNA using the Superarray RT2 First Strand Kit (C-02). Briefly, 1.0 µL of a primer and external control mixture was added to each RNA sample and total volume was brought up to 10 µL using RNase-free water. The contents of the tube were then placed in a thermal cycler for 3 min at 70°C. Samples were then chilled on ice and 10 µL of the reverse transcriptase (RT) cocktail were added to each sample.
RT cocktail for each sample included 5× RT buffer (4 µL), RNase-free water (4 µL), RNase inhibitor (1 µL), and RT Enzyme Mix II (1 µL). Samples were mixed and then incubated at 37°C for 60 min for cDNA synthesis. This was followed by heat inactivation at 95°C for 5 min to inactivate reverse transcriptase and degrade any remaining RNA. To dilute samples for subsequent Superarray analysis, 91 µL of double-distilled water were added to the newly synthesized cDNA. Samples were stored at −20°C until ready for further Superarray analysis. All Superarray analysis was done using the Mouse Th1 Th2 Th3 RT2 Profiler PCR Array profiles for expression of 84 inflammation-associated genes. For all Superarray analysis, a cocktail was prepared containing the following: 2× Superarray SYBR Green PCR Master Mix, diluted synthesized cDNA, and double-distilled water. Cocktail was mixed in a 50-mL tube and then transferred to a multi-channel reservoir. Twenty-five microliters of the cocktail mixture were loaded per well, after which each plate was loaded into the Bio-Rad iQ5 cycler to perform real-time detection. Real-time detection was done using the following two-step cycling program: cycle 1 for 10 min at 95°C, 40 cycles (15 s at 95°C followed by 1 min at 60°C).
Serum analysis of inflammation-associated cytokines
Serum analyses were done on the Meso Scale Sector 6000 instrument (Meso Scale Discovery). Briefly, the Mouse Th1 Th2 panel (9-plex) contained the following proteins: IFNγ, IL-1β, IL-2, IL-4, IL-5, KC, IL-10, IL-12, and tumor necrosis factor (TNF)-α. The remaining proteins were contained in the Mouse Custom Metabolic panel. These proteins included IL-6, granulocyte macrophage colony-stimulating factor (GM-CSF), insulin, MCP-1, resistin, and TNFα. All samples were run in duplicates across two plates (i.e., single run per plate). Data were similar in most cases, with coefficient of variation −20% for most proteins of interest.
Calibration curves were generated for standards (run in duplicate) in the range of 10,000 to 2.4 pg/mL (for Mouse Th1 Th2 panel and MCP-1), 37,500 to 2.4 pg/mL (for resistin), and 300,000 to 2.4 pg/mL (for insulin). All standards were diluted in MSD Mouse Serum Cytokine Assay Diluent. Twenty-five microliters of MSD Mouse Serum Cytokine Assay Diluent were added to each well. Plates were then sealed and incubated for 30 min with vigorous shaking (300–1,000 rpm) at room temperature. Next, 10 µL of each calibrator or sample were added to a separate well. The plate was then sealed and incubated for 2 h with vigorous shaking (300–1,000 rpm) at room temperature. Then plates were washed and detection antibody added. This was followed by a washing step and subsequent reading using the Sector 6000 instrument. The majority of samples were run using original concentrations. However, if sample volume was too low, samples were diluted 1:2.
Statistical analysis
Data were analyzed using SAS Software version 8.2. All serum data were transformed to the log (X + 0.5) scale because these data were approximately normally distributed on this scale. In most cases, we used the actual values of measurements below the level of detection in the analysis. Geometric means and corresponding 95% confidence intervals were estimated to interpret the results on the original scale (Fig. 1 and Fig. 2). In this case, a Wilcoxon rank sum test was used to test for significant differences between groups.
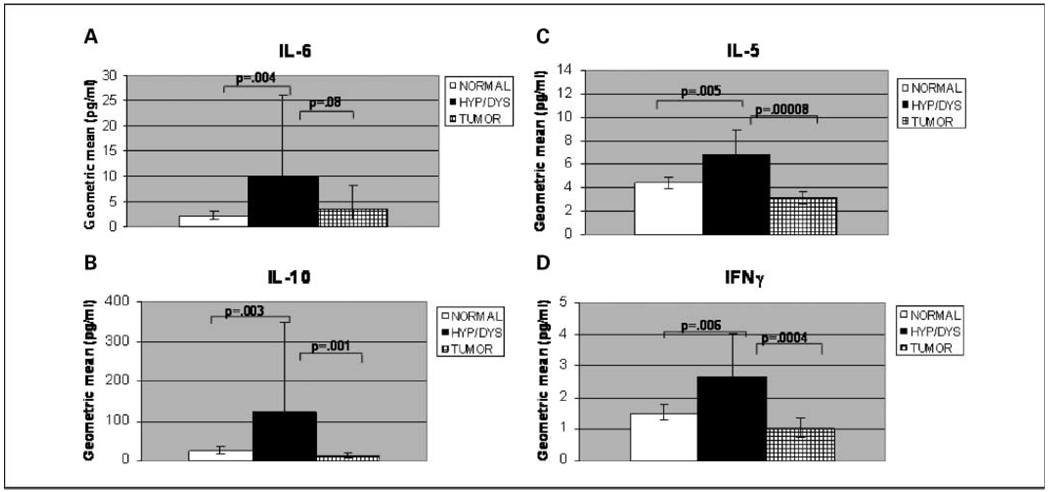
Fig. 1.
Serum levels of inflammation-associated proteins were increased during preneoplastic stages of colon carcinogenesis in AOM-induced ob/ob mice. Serum levels of IL-6 (A), IL-10 (B), IL-5 (C), and IFNγ (D) were increased in hyperplastic and dysplastic stages of colon carcinogenesis. Columns, geometric mean (pg/mL); bars, 95% confidence interval. Total mice per group: normal, n = 111; hyperplasia/dysplasia (hyp/dys), n = 12; tumor-bearing (tumor), n = 14. Normal is classified as any ob/ob mouse that was free of tumors or preneoplastic lesions. Tumor-bearing includes mice with adenomas or adenocarcinomas. Comparisons are shown for hyperplasia/dysplasia versus normal and hyperplasia/dysplasia versus tumor. Similar changes were observed in stages of colon carcinogenesis whether or not bean-fed mice were excluded from the comparison.
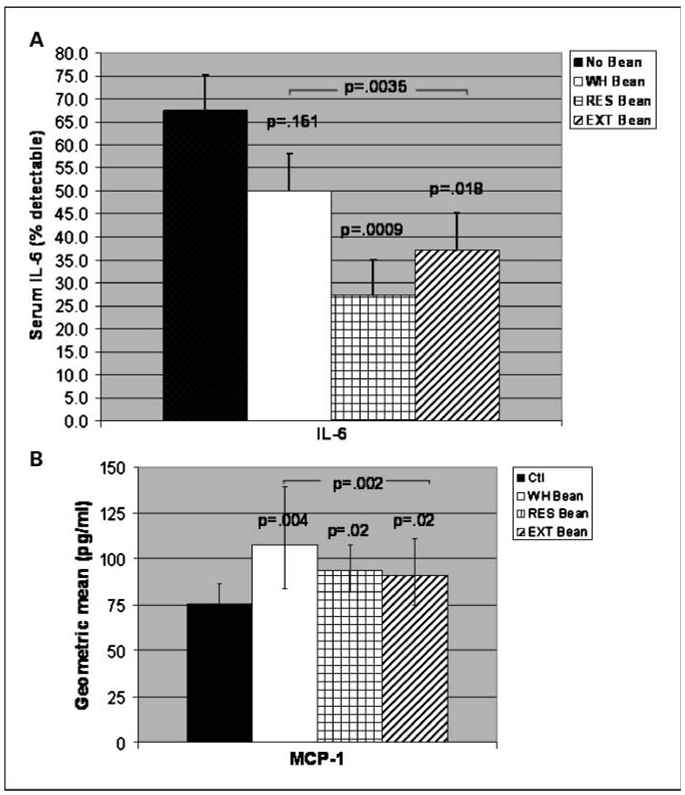
Fig. 2.
Bean diets produced changes in serum levels of inflammation-associated proteins in AOM-induced ob/ob mice. A, mice on bean-based diets overall (P < 0.0035) showed significant decrease in serum levels of IL-6. Significant reductions in IL-6 were seen in mice on residue (P = 0.009) and bean extract (P = 0.018) diets. Columns, percent detectable IL-6; bars, SEM. B, mice on bean-based diets overall (P = 0.002) and on whole bean (P = 0.004) showed significant increase in serum levels of MCP-1. A significant trend in MCP-1 increase was seen in mice on residue (P = 0.02) and bean extract (P = 0.015) diets. Columns, geometric mean (pg/mL); bars, 95% confidence interval. For all figures, P values spanning bar graphs represent overall bean diets (i.e., whole bean, residue, and extract groups combined). All comparisons are to mice on control (no bean) diet. Total mice per group: control, n = 37; whole bean, n = 32; residue, n = 33; and extract, n = 35. The above decreases and increases were observed with bean diets whether or not tumor-bearing mice were excluded from the comparison.
For serum measurements that had a large proportion of values below the level of detection, we estimated the proportion that were above detectable limits in each group (% detectable data ± SE) and compared these proportions across groups using Fisher's exact test (two-sided).
For the Superarray quantitative PCR data, Ct (cycle threshold) values were measured for the genes of interest as well as the house-keeping genes. Data were normalized against three housekeeping genes [hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1), heat shock protein 90kDa α (cytosolic) class B member 1 (Hsp90ab1), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh)] by subtracting the average Ct value for three housekeeping genes from the average Ct value of gene of interest (i.e., calculating ΔΔCt). We compared ΔΔCt across groups using t tests.
Although the expression of housekeeping genes is expected not to vary across samples, previous studies have shown that sometimes they do vary, especially across stages in cancer or in response to carcinogens. Thus, to have one or more genes usable for normalization, it becomes necessary to use more than one housekeeping gene. In this study, five housekeeping genes were run with every sample. The criterion for selection of the housekeeping genes was the Ct value, which should not differ by more than 1 cycle across the groups of interest. In this case, the average Ct value for three housekeeping genes met this criterion, varying by ≤1 cycle. This generated a normalization factor with minimal variability.
Due to the large number of comparisons made in our analyses, differences in serum measurements or gene expression between groups were considered statistically significant when P < 0.01. The false discovery rate was estimated as (level of significance) × (number of genes evaluated) / (number of significant genes).
A P value between 0.01 and 0.05 was considered to be a statistical trend. In addition, a fold difference of ≥1.7 was required in screening for differences between groups in gene expression.
Results
Serum levels of IL-6, IL-10, IL-5, IL-4, IFNγ, and GM-CSF were increased during preneoplastic stages of colon carcinogenesis
We first wanted to determine the stagewise changes in serum because, ultimately, we are interested in intervening to attenuate early stages of carcinogenesis. To identify possible indicators of colon carcinogenesis risk, a multiplex-based electrochemiluminescence assay to measure serum levels of 14 proteins was used. Serum samples from AOM-induced ob/ob mice were analyzed in mice that were tumor-bearing (i.e., adenomas, adenocarcinomas), bore only preneoplastic lesions (i.e., hyperplasia, dysplasia), or bore no lesions at the end of the study. Because the number of lesion-bearing mice on bean diets was not sufficient to compare stages within diet groups, we initially compared stagewise changes in the colon carcinogenesis process (i.e., normal versus hyperplastic/dysplastic versus tumor) without regard to intervention. Serum levels of IL-6 (P = 0.004), IL-10 (P = 0.003), IL-5 (P = 0.005), and IFNγ (P = 0.006) were statistically significantly increased or had a statistical trend (i.e., IL-4: P = 0.026, GM-CSF: P = 0.021) during preneoplastic stages of colon carcinogenesis compared with serum from AOM-induced ob/ob mice with normal colons (Fig. 1A–D; data not shown for IL-4 and GM-CSF). Serum levels of IL-10 (P = 0.001), IL-5 (P = 0.00008), IFNγ (P = 0.0004), and GM-CSF (P = 0.009) were significantly reduced or had a significant trend in reduction (i.e., IL-4: P = 0.011) in tumor-bearing, AOM-induced ob/ob mice compared with serum from mice with hyperplastic or dysplastic colons (Fig. 1B–D; data not shown for IL-4 and GM-CSF), to levels similar to or less than those seen in mice with normal colons. Similar changes were observed when comparing exclusively stagewise changes in mice on control diet (data not shown).
Serum levels of IL-6 were not significantly reduced in tumor-bearing mice compared with mice with hyperplastic or dysplastic colons (Fig. 1A). IL-5 serum levels had a statistical trend toward lower levels in tumor-bearing mice than in mice with normal colons (P = 0.037; Fig. 1C). No significant differences in serum levels of IL-1β, IL-2, KC, IL-12, insulin, resistin, or MCP-1 were seen among mice in normal versus hyperplastic/dysplastic versus tumor groups (data not shown). TNFα levels were not sufficiently detectable to permit analysis. Thus, serum levels of IL-10, IL-5, IL-4, IFNγ, and GM-CSF were increased during preneoplastic stages of colon carcinogenesis relative to tumor-bearing stages.
Bean-based diets produce changes in serum levels of IL-6 and MCP-1 in AOM-induced ob/ob mice
We next wanted to know which serum protein levels would be attenuated by the bean diet. Although the number of mice per group was sufficient to compare serum levels by stage, the number of lesion-bearing mice was not sufficient for subdividing the stages into those that did or did not receive the bean diets.
Thus, we ascertained changes in inflammation-associated serum proteins that occurred in response to bean diet interventions (Fig. 2). Serum levels of the inflammation-associated protein IL-6 were significantly decreased in AOM-induced ob/ob mice on bean-based diets overall (P = 0.0035) and in mice on residue (P = 0.009) and had a statistical trend in decreasing IL-6 in mice on extract fraction (P = 0.018) bean diets compared with controls (Fig. 2A). Serum levels of IL-6 were less detectable in AOM-induced ob/ob mice on bean-based diets (27% and 37% for residue and extract fraction, respectively) compared with mice on control diet (68%; Fig. 2A). Serum levels of the chemokine MCP-1 were significantly increased in AOM-induced ob/ob mice on bean-based diets overall (P = 0.002) and in mice on whole bean diet (P = 0.004) and had a statistical trend in increase in mice on residue and extract diets (P = 0.02 and P = 0.02, respectively; Fig. 2B). Thus, bean-based diets seem to target serum IL-6 and MCP-1.
Bean diets change gene expression of a subset of inflammation-related cytokines in colon mucosa of AOM-induced ob/ob mice
To identify possible indicators of response to bean diets in colon mucosa, the expression of 84 inflammation-associated genes was queried using real-time quantitative PCR array analysis. Cd4 expression changes were observed in both whole bean (4.97-fold) and bean residue (5.25-fold) fractions, suggesting that this residue-associated gene may contribute to the cancer-preventive effect of the whole beans (Fig. 3). Sftpd transcript changes were observed with both whole bean (−6.37-fold) and bean extract (−5.04-fold), suggesting that extract components provided the source of the whole bean effect on this gene. We also observed Tnfrsf8 transcript changes in both residue (−1.81-fold) and extract (−2.16-fold) bean fractions. Fold differences that were statistically significant at the P < 0.01 level included Tnfrsf8 (−2.16-fold for bean extract; P = 0.005) and Stat4 (−5.56-fold for bean extract; P = 0.007). The remaining fold differences had a significant trend where P values ranged from P = 0.012 (Tnfrsf8; bean residue) to P = 0.047 (Icos).
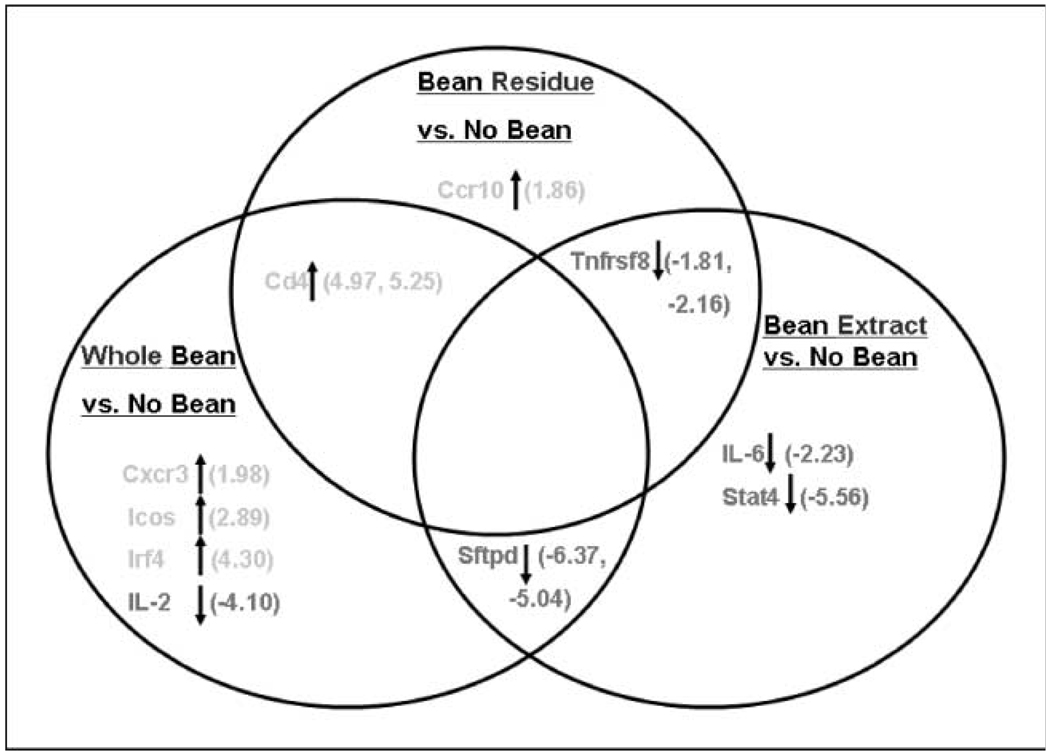
Fig. 3.
Bean diets change gene expression of a subset of inflammation-related cytokines in colon mucosa of AOM-induced ob/ob mice. RNA isolated as described in Materials and Methods. The expression of 84 Th1 Th2 Th3 inflammation-associated genes was analyzed using Superarray real-time quantitative PCR arrays. Shown are Venn diagrams depicting bean-induced gene expression changes that were overlapping or not overlapping between diet groups. Arrows, direction of fold change depicted in parentheses. Criteria for gene selection were P < 0.05 for the difference between bean fed and control, and fold change −1.7. Fold change range for all genes was −2.0 to +6.0. n = 6 mice per group. Analysis was done on non–tumor-bearing mice. Comparison groups included whole bean versus no bean, bean residue versus no bean, and bean extract versus no bean, respectively. Statistical significance: Tnfrsf8 (Bean Extract; P = 0.005) and Stat4 (Bean Extract; P = 0.007). All other genes in the figure had a significant trend where P values ranged from P = 0.012 (Tnfrsf8; Bean Residue) to P = 0.047 (Icos). Genes include CD4 antigen (Cd4), surfactant-associated protein D (Sftpd), TNF receptor superfamily member 8 (Tnfrsf8), chemokine C-X-C motif receptor 3 (Cxcr3), inducible T-cell costimulator (Icos), IFN regulatory factor 4 (Irf4), IL-2, chemokine C-C motif receptor 10 (Ccr10), IL-6, and signal transducer and activator of transcription 4 (Stat4).
The expression of 10 genes was changed overall in AOM-induced ob/ob mice consuming bean-based diets (Fig. 3). Five of these genes were up-regulated (Cxcr3, Icos, Irf4, Cd4, and Ccr10) and five were down-regulated (IL-2, Sftpd, Tnfrsf8, IL-6, and Stat4). Fold differences for the up-regulated genes ranged from 1.86 to 5.25, whereas fold difference for down-regulated genes ranged from −1.81 to −6.37. A trend emerged in the direction of gene change. Four of five up-regulations occurred in mice consuming whole bean diet. These up-regulated genes are largely proinflammatory. Two of three up-regulated genes were seen in mice consuming bean residue diet with one of the two gene changes (Cd4) in common with whole bean. The third gene expression change (i.e., down-regulation of Tnfrsf8) occurred in common with mice consuming bean extract diet. Four of five gene down-regulations occurred in mice consuming bean extract diet, the most efficacious of the three bean diets (4). Because these down-regulated genes are involved principally in the proinflammatory response, this down-regulation may contribute to the greater efficacy of the bean extract compared with the other bean diets.
There were n = 6 mice per comparison group unless otherwise noted. This sample size was less than the total number of samples used for the serum analysis because colon tissues were not collected from every mouse. Colon samples were selected from mice that had conclusive pathology and serum analysis data. Every effort was made to ensure that the colon samples collected were representative of the animals in each group.
In addition to evaluating gene expression changes in each respective bean group, we wanted to determine if there were gene expression differences in tumor-bearing versus non–tumor-bearing mice. To carry this out, all three bean groups (i.e., All Bean) were combined and compared with the No Bean group. The results indicate that there was a distinct set of genes whose expressions changed based on tumor status, with only one gene in common between the two groups (Maf; Fig. 4). The up-regulation of Maf in both groups is consistent with the possibility that Maf contributes to but is not sufficient to account for the efficacy of the beans. The set of gene changes occurring in non–tumor-bearing mice may or may not be sufficient to protect against colon tumorigenesis. In contrast, the set of gene changes occurring in tumor-bearing mice is not a sufficient set to protect against colon tumorigenesis.
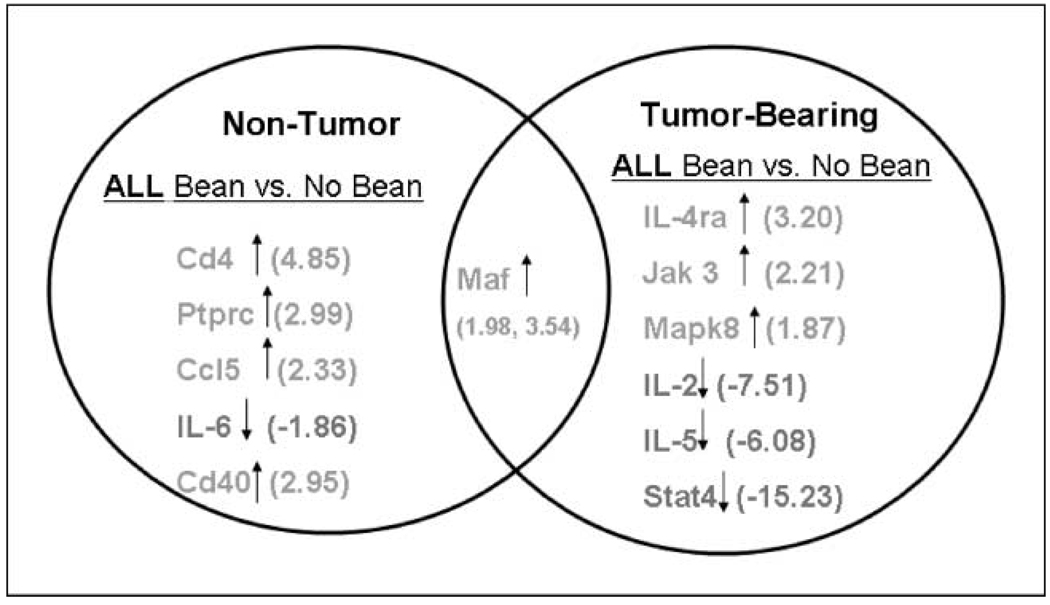
Fig. 4.
Bean diets as a group change the expression of a subset of inflammation-related cytokine genes in colon mucosa of AOM-induced ob/ob mice. The fold change range for all genes was −15.00 to +5.0. Cd4 (P = 0.003) and IL-4ra (P = 0.005) were statistically significant by t test analysis. All other genes in the figure had a statistical trend with P values ranging from P = 0.010 (IL-5) to P = 0.046 (Cd40). Total mice per group: All Bean, n = 18; No Bean, n = 6. Comparison groups include All Bean versus No Bean among non–tumor-bearing mice and All Bean versus No Bean among tumor-bearing mice. All Bean group is combination of mice in whole, residue, and extract bean fed groups. Genes include CD4antigen (Cd4), protein tyrosine phosphatase receptor type C (Ptprc), chemokine C-C motif ligand 5 (Ccl5), IL-6, CD40 antigen (Cd40), IL-4 receptor α (IL-4ra), Janus kinase 3 (Jak3), mitogen-activated protein kinase 8 (Mapk8), IL-2, IL-5, signal transducer and activator of transcription 4 (Stat4), and avian musculoaponeurotic fibrosarcoma v-maf AS42 oncogene homologue (Maf).
Among non–tumor-bearing mice, the overall bean diets (n = 24 mice: whole bean, residue, and extract groups combined) significantly changed expression of Cd4 (P = 0.003) and had a statistical trend in the remaining genes (Ptprc, Ccl5, IL-6, Cd40, and Maf) compared with mice on control diet (n = 6) showing −1.86- to 4.85-fold differences (Fig. 4). Statistical trend ranged from P = 0.014 (Ccl5) to P = 0.046 (Cd40). Among tumor-bearing mice, the bean diets overall (n = 3 mice) significantly changed gene expression of IL-4ra (P = 0.005) and had a statistical trend in the remaining genes (Jak3, Mapk8, IL-2, IL-5, Stat4, and Maf) compared with tumor-bearing mice on control diet (n = 4 mice) showing −15.23-to 3.54-fold differences (Fig. 4). Statistical trend ranged from P = 0.010 (IL-5) to P = 0.045 (Mapk8).
The false discovery rate was estimated for the gene expression changes observed. For genes that were statistically significant at the 0.01 significance level (P < 0.01), false discovery rate = 0.01(84)/2 = 42%. Therefore, 58% of our significant findings are expected to be correct. For genes that showed a statistical trend (P < 0.05), false discovery rate = 0.05(84)/8 = 53%. Therefore, 47% of the significant findings are expected to be correct.
Taken together, the results of gene expression analysis suggest several bean diet-induced changes in inflammation-associated genes. Those expression changes shared in mice on bean extract and whole beans (i.e., Sftpd) and those shared in mice on bean residue and whole beans (i.e., Cd4) implicate extract or residue, respectively, as the source of the changes seen with whole beans.
Bean diets attenuate the effects of AOM on a subset of inflammation-related genes in AOM-induced ob/ob mice
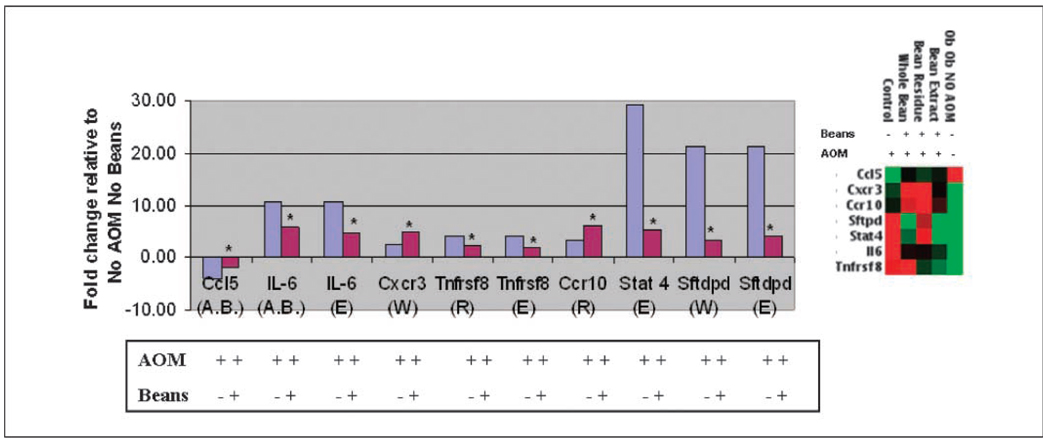
To ascertain whether bean diets counteract gene expression changes induced by the carcinogen, colon mucosa RNAs from no-carcinogen controls (i.e., No AOM No Bean) were compared with those treated with AOM. No AOM group was used as the reference group to which all other samples were compared. That is, both AOM-induced and bean attenuated values are compared with a constant denominator to determine a ratio. This calculation was carried out to determine which of the AOM-induced gene expression events were counteracted by bean diet (Fig. 5). The AOM-induced change was then compared with the AOM plus bean diet change. Figure 5 shows genes whose expression was altered by AOM in the absence of bean diet and genes whose expression was altered by bean diets in AOM-induced mice. All fold changes are relative to expression levels in the group of mice that did not receive AOM or bean diets [i.e., AOM (−), bean (−)]. The total number of genes found to change significantly or show a significant trend in response to AOM was 55. The expression of only five of the AOM-altered genes was counteracted by bean diets. IL-6,Tnfrsf8, Stat4, and Sftpd showed expression levels that were induced by AOM, but attenuated (i.e., less induced) in ob/ob mice on overall bean, whole bean, residue, or extract diet. Expression of Ccl5 was repressed by AOM, but attenuated by bean diets. The changes in Stat4 and Tnfrsf8 were statistically significant at the P < 0.001 level, whereas changes in IL-6, Ccl5, and Sftpd showed a significant trend (Fig. 5). Furthermore, the false discovery rate of the genes that were significant at the 0.01 significance level (P < 0.01) in the assessment of AOM-treated mice versus the reference No AOM group was estimated as 0.01(84)/39 = 2%. Therefore, 98% of the significant findings are expected to be correct. Thus, several inflammation-associated gene expression changes induced during AOM-induced carcinogenesis seem to be counteracted by bean diets.
Fig. 5.
Bean diet attenuates some of the gene expression changes induced by AOM in ob/ob mice. Graph represents fold changes relative to non–AOM induced (i.e., −AOM) ob/ob mice, which was the overall comparison group for this analysis. Right, clustergram of fold changes depicted in graph (red, high-expressing genes; green, low-expressing genes). All mice from The Jackson Laboratory (No AOM) were free of colon lesions and tumors, showing normal colon on histopathologic examination. As expected in ob/ob mice, some fat infiltration was observed. In contrast, the AOM-treated mice included some with preneoplastic lesions or tumors as indicated in Fig. 1 (4). Blue columns, ratio of +AOM to −AOM in the absence of beans. Red columns, ratio of +beans to −beans following AOM exposure. The determined (blue columns) and calculated (red columns) fold change values have been ascertained as follows: To identify genes whose expression is induced by AOM (A) and attenuated/counteracted by the bean diets (or repressed by AOM and attenuated by the bean diets) (B), all expression levels were normalized to the level found without AOM or bean diet. The following calculations were done using Superarray results. Expression values were measured on Superarray Th1 Th2 Th3 array plates with one sample run per plate. Calculations were determined using the following x, y, and z groups: x = AOM (+) Beans (−), y = AOM (+) Beans (+), and z = AOM(−) Beans (−). Two ratios were determined: (a) y/x or AOM (+) Beans (+)/AOM (+) Beans (−) and (b) x/z or AOM (+) Beans (−)/AOM (−) Beans (−). To calculate the ratio of expression with AOM with beans to expression without AOM without beans (y/z), the following equation was used: y/z = y/x × x/z. Letter(s) in parentheses following the gene indicates the bean group: A.B., all bean; W, whole bean; R, bean residue; E, bean extract. Total mice per group: No AOM No Bean, n = 5; ob/ob mice, +AOM No Bean, n = 6; +AOM All Bean, n = 18; +AOM whole bean, n = 6; +AOM residue, n = 6; +AOM extract, n = 6. Values for AOM + beans (red columns) were significantly different or had a statistical trend from those for AOM alone (blue columns). Stat4 and Tnfrsf8 genes were significant at the P < 0.001 level. *, genes statistically significant or having a statistical trend. Genes include chemokine C-C motif ligand 5 (Ccl5), IL-6, chemokine C-X-C motif receptor 3 (Cxcr3), TNF receptor superfamily member 8 (Tnfrsf8), chemokine C-C motif receptor 10 (Ccr10), signal transducer and activator of transcription 4 (Stat4), and surfactant associated protein D (Sftpd).
Discussion
Bean-based diets produce changes in inflammation-associated serum proteins and colon gene expression under conditions in which these bean diets attenuate colon carcinogenesis in AOM-induced ob/ob mice. This is among the first identification of AOM-induced serum protein and gene expression changes in the colon of ob/ob mice. Moreover, the current findings identify bean diet-induced changes that associate with efficacy for cancer prevention in this genetically obese mouse model of colon carcinogenesis. Among the serum protein changes seen in mice on bean-based diets were decreases in IL-6 and an increase in MCP-1 levels. Noteworthy, colon gene expression changes were seen for Cd4, Tnfrsf8, and Sftpd with these changes distinguishing extract, residue, and whole bean diets, all of which showed efficacy. A subset of AOM-induced gene expression changes was counteracted by bean diets. Bean diets decreased the levels of IL-6 protein in serum and decreased the level of its gene expression in the colon, counteracting the AOM induction of IL-6.
Serum protein changes during multistage carcinogenesis: possible risk indicators
Recent studies in humans have identified serum proteins that increase with stage and colon tumor size (19, 20). Those studies, which identified galanin and defensin α6, sought early detection markers of colon carcinogenesis. Other studies evaluating circulating levels of inflammatory cytokines as biomarkers of colorectal adenoma risk in humans (21) identified IL-6 as did the current study. Our observations that IL-6 was AOM-induced in the colon and up-regulated in serum from mice with hyperplastic/dysplastic colons suggest IL-6 as a risk indicator in AOM-induced ob/ob mice. A study using a similar model, dextran sodium sulfate–induced acute and chronic colitis, also identified IL-6 as a risk indicator (22). This study reported elevated IL-6 expression in colon culture supernatants from ob/ob and wild-type mice following in vivo dextran sodium sulfate exposure. The set of serum protein changes seen during the preneoplastic stages may reflect changes in colon or microenvironment expression that contribute to hyperplastic/dysplastic stages, or they may be consequential. If causal, they may be needed for tumor induction but not for tumor maintenance. Additional studies are needed to determine the functional significance of these serum changes.
Our findings suggest that the bean-based diets may be altering levels of proteins such as IL-6 involved in the preneoplastic stages of the colon carcinogenesis process in these ob/ob mice. If so, these proteins may serve not only as risk indicators but also as response indicators of efficacy to this dietary intervention.
Identification of response indicators for bean-based dietary interventions to prevent colon cancer
Although protective effects of bean-based diets have been seen in rat and mouse colon carcinogenesis models (5, 6, 23, 24) and in a human study (7), few studies have sought to identify markers of response to those dietary interventions. Pinto bean–induced changes in the blood of humans were reported for an intervention that did not reduce colon cancer risk (25). In this case, the serum changes, which included changes in serum lipids (i.e., total, high-density lipoprotein, and low-density lipoprotein cholesterol), would either be indicators (but insufficiently changed) or nonindicators of efficacy. The Polyp Prevention Trial (7) and the current mouse study, which showed efficacy (4), present opportunities for ascertaining response indicators of consuming a bean-based diet that attenuates carcinogenesis.
To date, a number of animal and human studies have attempted to follow both biomarkers of dietary intake and biomarkers of risk. Lin et al. (26) assessed the response to a low-fat low-glycemic load diet. This study of men with prostate cancer identified a subset of genes that were differentially expressed pre-diet versus post-diet (i.e., indicators of dietary intake). Powers et al. (27) measured the response to folate and riboflavin supplementation among healthy and colorectal polyp patients. Although intake of folate and riboflavin was confirmed, the interventions did not change the biomarkers of risk measured (i.e., promoter methylation of tumor suppressor genes such as p16). In the case of this dietary supplementation, risk indicators did not constitute response indicators but efficacy was also not determined. Whereas biomarkers of risk may also constitute markers of response, as seems to be the case for IL-6 in ob/ob mice, studies to ascertain this need to be undertaken.
Serum IL-6 and MCP-1 as response indicators
Mice on bean-based diets had lower serum levels of IL-6 and higher levels of MCP-1 compared with controls. Others have observed reductions in IL-6 (28) for dietary interventions. The study by Roy et al. (28) detected these changes in an animal inflammatory bowel disease model. In another study, luteolin consumption reduced plasma levels of IL-6 in mice treated with lipopolysaccharide (29). Luteolin reduced IL-6 production in microglia by inhibiting c-jun NH2-terminal kinase phosphorylation and activation of activator protein 1.
We cannot exclude the possibility that beans decrease adiposity as has been reported for phytoestrogens (30). However, the fact that the mice on the bean diet had higher body weights than the mice in the control groups would argue against the beans reducing adiposity. Sources of IL-6 might be adipocytes (31) or macrophages (32, 33), cell types that were not measured in this study. IL-6 is known to be an important contributor to carcinogenesis and has been shown to be regulated by transcription factors that play a role in regulating cell proliferation, differentiation, and apoptosis. For example, a study by Yu et al. (34) showed that IL-6 induces transformation of JB6 P+ mouse skin epithelial cells. In that model, IL-6 functions as a tumor promoter in a Stat3-dependent manner. Other studies have shown IL-6 to stimulate the silencing of p53 by maintaining the methylated state of p53 in a multiple myeloma cell line (35). Possible mechanisms of action of IL-6 include activation of Janus kinases, resulting in subsequent activation of the mitogen-activated protein kinase, phosphatidylinositol 3-kinase, and signal transducer and activator of transcription pathways (36, 37).
MCP-1 levels in plasma and adipose tissue are known to be elevated in ob/ob mice (38). The bean intervention did not decrease but increased serum levels of MCP-1. It is important to note that MCP-1 expression changes are associated with changes in obesity and obesity-related genes (38–41). Thus, the increased MCP-1 levels may not be surprising because mice on bean-based diets had increased body weights (4) compared with mice consuming the control diet. Whether the increased expression of Cd4, Cxcr3, Ccr10, Icos, and Irf4 in the colon mucosae of mice on bean-based diets is associated with increased body weight is unknown. Obesity is thought to be a state of chronic inflammation whereby these genes (Cd4, Cxcr3, Ccr10, Icos, and Irf4) play a role (42). For example, Cxcr3 infiltrates T-lymphocytes in colorectal carcinomas in humans (43).
Ten of 84 genes showed expression changes in response to efficacious bean interventions
In addition to serum changes, the bean-based diets changed the expression of a subset of inflammation-associated cytokines in colon mucosa of AOM-induced ob/ob mice. Cd4 transcript changes were common in whole bean and residue fractions, implicating Cd4 as a residue-responsive gene. Similarly, Sftpd gene changes were common in whole bean and extract fractions, implicating this gene as an extract-responsive gene. Cd4 and Sftpd may be genes that contribute to the cancer-preventive effect of the whole beans. More studies are needed to assess this phenomenon.
Five of 55 genes whose expression was altered by AOM showed attenuation by bean diets
This observation supports the hypothesis that bean diets target the expression of a subset of inflammation-associated genes. This expression analysis showed that the following genes may be indicators of AOM-induced carcinogenesis whose expression changes are counteracted by the bean-based intervention: Ccl5, IL-6, Tnfrsf8, Stat4, and Sftpd. These changes may also reflect diet differences (i.e., standard AIN-93G versus modified AIN-93G).
It should be noted that due to the large number of statistical comparisons between the 84 genes, it is possible that some of the reported significant results may be false positives. If the observed AOM-induced, bean-counteracted changes in gene expression emerge as functionally significant in contributing to colon carcinogenesis, their attenuation by bean diets may suggest a mechanism by which these diets protect against carcinogenesis.
Potential for application to other interventions and other models
Obese mice have increased levels of proinflammatory proteins such as MCP-1, IL-1β, TNFα, and IL-6 (44–47). Some of these findings have been translated to humans (48–50). One of these (i.e., IL-6) seems to be target of our bean diets.
Whether these observed changes in serum proteins and gene expression apply only to bean interventions or will generalize to other efficacious interventions will be of interest to ascertain. If diet-specific indicators emerge, using these indicators may facilitate the development of combination interventions. The question of generalizability to other inflammation-associated models and cancer sites will also be of interest.
Future studies should determine how early during multistage carcinogenesis these inflammation-associated serum protein and gene expression changes may be occurring, with the goal of identifying early biomarkers of efficacy that might translate to human dietary intervention studies.
Acknowledgments
We thank the Michigan Bean Commission for the donation of navy beans and Helen Rager for her help in running the Meso Scale Discovery serum assays.
Grant support: NIH Intramural Research Program, National Cancer Institute; the United States Agency for International Development through Bean/Cowpea Collaborative Research Support Program; and Michigan Agricultural Experiment Station.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2006;15:389–394. doi: 10.1158/1055-9965.EPI-05-0678. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Ries LA, Wingo PA, Miller DS, et al. The annual report to the nation on the status of cancer, 1973–1997, with a special section on colorectal cancer. Cancer. 2000;88:2398–2424. doi: 10.1002/(sici)1097-0142(20000515)88:10<2398::aid-cncr26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Bobe G, Barrett KG, Mentor-Marcel RA, et al. Dietary cooked navy beans and their fractions attenuate colon carcinogenesis in azoxymethane-induced ob/ob mice. Nutr Cancer. 2008;60:373–381. doi: 10.1080/01635580701775142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hangen L, Bennink MR. Consumption of black beans and navy beans (Phaseolus vulgaris) reduced azoxymethane-induced colon cancer in rats. Nutr Cancer. 2002;44:60–65. doi: 10.1207/S15327914NC441_8. [DOI] [PubMed] [Google Scholar]
- 6.Hughes JS, Ganthavorn C, Wilson-Sanders S. Dry beans inhibit azoxymethane-induced colon carcinogenesis in F344 rats. J Nutr. 1997;127:2328–2333. doi: 10.1093/jn/127.12.2328. [DOI] [PubMed] [Google Scholar]
- 7.Lanza E, Hartman TJ, Albert PS, et al. High dry bean intake and reduced risk of advanced colorectal adenoma recurrence among participants in the polyp prevention trial. J Nutr. 2006;136:1896–1903. doi: 10.1093/jn/136.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward DG, Suggett N, Cheng Y, et al. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94:1898–1905. doi: 10.1038/sj.bjc.6603188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breikers G, van Breda SG, Bouwman FG, et al. Potential protein markers for nutritional health effects on colorectal cancer in the mouse as revealed by proteomics analysis. Proteomics. 2006;6:2844–2852. doi: 10.1002/pmic.200500067. [DOI] [PubMed] [Google Scholar]
- 10.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allavena P, Garlanda C, Borrello MG, Sica A, Mantovani A. Pathways connecting inflammation and cancer. Curr Opin Genet Dev. 2008;18:1–8. doi: 10.1016/j.gde.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Moss SF, Blaser MJ. Mechanisms of disease: inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005;2:90–97. doi: 10.1038/ncponc0081. [DOI] [PubMed] [Google Scholar]
- 13.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 14.Gunter MJ, Leitzmann MF. Obesity and colorectal cancer: epidemiology, mechanisms and candidate genes. J Nutr Biochem. 2006;17:145–156. doi: 10.1016/j.jnutbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Garthwaite TL, Martinson DR, Tseng LF, Hagen TC, Menahan LA. A longitudinal hormonal profile of the genetically obese mouse. Endocrinology. 1980;107:671–676. doi: 10.1210/endo-107-3-671. [DOI] [PubMed] [Google Scholar]
- 16.Bray GA, York DA. Hypothalamic and genetic obesity in experimental animals: an autonomic and endocrine hypothesis. Physiol Rev. 1979;59:719–809. doi: 10.1152/physrev.1979.59.3.719. [DOI] [PubMed] [Google Scholar]
- 17.Chen NG, Swick AG, Romsos DR. Leptin constrains acetylcholine-induced insulin secretion from pancreatic islets of ob/ob mice. J Clin Invest. 1997;100:1174–1179. doi: 10.1172/JCI119629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 19.Kim KY, Kee MK, Chong SA, Nam MJ. Galanin is up-regulated in colon adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:2373–2378. doi: 10.1158/1055-9965.EPI-06-0740. [DOI] [PubMed] [Google Scholar]
- 20.Nam MJ, Kee MK, Kuick R, Hanash SM. Identification of defensin α6 as a potential biomarker in colon adenocarcinoma. J Biol Chem. 2005;280:8260–8265. doi: 10.1074/jbc.M410054200. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Keku TO, Martin C, et al. Circulating levels of inflammatory cytokines and risk of colorectal adenomas. Cancer Res. 2008;68:323–328. doi: 10.1158/0008-5472.CAN-07-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegmund B, Lehr HA, Fantuzzi G. Leptin: a pivotal mediator of intestinal inflammation in mice. Gastroenterology. 2002;122:2011–2025. doi: 10.1053/gast.2002.33631. [DOI] [PubMed] [Google Scholar]
- 23.Murillo G, Choi JK, Pan O, Constantinou AI, Mehta RG. Efficacy of garbanzo and soybean flour in suppression of aberrant crypt foci in the colons of CF-1 mice. Anticancer Res. 2004;24:3049–3055. [PubMed] [Google Scholar]
- 24.Koratkar R, Rao AV. Effect of soya bean saponins on azoxymethane-induced preneoplastic lesions in the colon of mice. Nutr Cancer. 1997;27:206–209. doi: 10.1080/01635589709514526. [DOI] [PubMed] [Google Scholar]
- 25.Finley JW, Burrell JB, Reeves PG. Pinto bean consumption changes SCFA profiles in fecal fermentations, bacterial populations of the lower bowel, and lipid profiles in blood of humans. J Nutr. 2007;137:2391–2398. doi: 10.1093/jn/137.11.2391. [DOI] [PubMed] [Google Scholar]
- 26.Lin DW, Neuhouser ML, Schenk JM, et al. Low-fat, low-glycemic load diet and gene expression in human prostate epithelium: a feasibility study of using cDNA microarrays to assess the response to dietary intervention in target tissues. Cancer Epidemiol Biomarkers Prev. 2007;16:2150–2154. doi: 10.1158/1055-9965.EPI-07-0154. [DOI] [PubMed] [Google Scholar]
- 27.Powers HJ, Hill MH, Welfare M, et al. Responses of biomarkers of folate and riboflavin status to folate and riboflavin supplementation in healthy and colorectal polyp patients (the FAB2 Study) Cancer Epidemiol Biomarkers Prev. 2007;16:2128–2135. doi: 10.1158/1055-9965.EPI-07-0208. [DOI] [PubMed] [Google Scholar]
- 28.Roy N, Barnett M, Knoch B, Dommels Y, McNabb W. Nutrigenomics applied to an animal model of inflammatory bowel diseases: transcriptomic analysis of the effects of eicosapentaenoic acid- and arachidonic acid-enriched diets. Mutat Res. 2007;622:103–116. doi: 10.1016/j.mrfmmm.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Jang S, Kelley KW, Johnson RW. Luteolin reduces IL-6 production in microglia by inhibiting JNK phosphorylation and activation of AP-1. Proc Natl Acad Sci U S A. 2008;105:7534–7539. doi: 10.1073/pnas.0802865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cederroth CR, Vinciguerra M, Kuhne F, et al. A phytoestrogen-rich diet increases energy expenditure and decreases adiposity in mice. Environ Health Perspect. 2007;115:1467–1473. doi: 10.1289/ehp.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg AS, Obin MS. Obesity and the role of adipose tissue in inflammation and metabolism. Am J Clin Nutr. 2006;83:461S–465S. doi: 10.1093/ajcn/83.2.461S. [DOI] [PubMed] [Google Scholar]
- 32.Cinti S, Mitchell G, Barbatelli G, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Fried SK, Bunkin DA, Greenberg AS. Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J Clin Endocrinol Metab. 1998;83:847–850. doi: 10.1210/jcem.83.3.4660. [DOI] [PubMed] [Google Scholar]
- 34.Yu CY, Wang L, Khaletskiy A, et al. STAT3 activation is required for interleukin-6 induced transformation in tumor-promotion sensitive mouse skin epithelial cells. Oncogene. 2002;21:3949–3960. doi: 10.1038/sj.onc.1205499. [DOI] [PubMed] [Google Scholar]
- 35.Hodge DR, Peng B, Cherry JC, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promoter methylation. Cancer Res. 2005;65:4673–4682. doi: 10.1158/0008-5472.CAN-04-3589. [DOI] [PubMed] [Google Scholar]
- 36.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41:2502–2512. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen A, Mumick S, Zhang C, et al. Diet induction of monocyte chemoattractant protein-1 and its impact on obesity. Obes Res. 2005;13:1311–1320. doi: 10.1038/oby.2005.159. [DOI] [PubMed] [Google Scholar]
- 39.Zhou HR, Kim EK, Kim H, Claycombe KJ. Obesity-associated mouse adipose stem cell secretion of monocyte chemotactic protein-1. Am J Physiol Endocrinol Metab. 2007;293:E1153–E1158. doi: 10.1152/ajpendo.00186.2007. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto I, Wada J, Hida A, et al. Elevated serum monocyte chemoattractant protein-4 and chronic inflammation in overweight subjects. Obesity. 2006;14:799–811. doi: 10.1038/oby.2006.93. [DOI] [PubMed] [Google Scholar]
- 41.Kim CS, Park HS, Kawada T, et al. Circulating levels of MCP-1 and IL-8 are elevated in human obese subjects and associated with obesity-related parameters. Int J Obes (Lond) 2006;30:1347–1355. doi: 10.1038/sj.ijo.0803259. [DOI] [PubMed] [Google Scholar]
- 42.Qin S, Rottman JB, Myers P, et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musha H, Ohtani H, Mizoi T, et al. Selective infiltration of CCR5+CXCR3+ T lymphocytes in human colorectal carcinoma. Int J Cancer. 2005;116:949–956. doi: 10.1002/ijc.21135. [DOI] [PubMed] [Google Scholar]
- 44.Harkins JM, Moustaid-Moussa N, Chung YJ, et al. Expression of interleukin-6 is greater in preadipocytes than in adipocytes of 3T3-L1 cells and C57BL/6J and ob/ob mice. J Nutr. 2004;134:2673–2677. doi: 10.1093/jn/134.10.2673. [DOI] [PubMed] [Google Scholar]
- 45.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 46.Jager J, Gremeaux T, Cormont M, Le Marchand-Brustel Y, Tanti JF. Interleukin-1β-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology. 2007;148:241–251. doi: 10.1210/en.2006-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugita S, Kamei Y, Oka J, Suganami T, Ogawa Y. Macrophage-colony stimulating factor in obese adipose tissue: studies with heterozygous op/+ mice. Obesity (Silver Spring) 2007;15:1988–1995. doi: 10.1038/oby.2007.237. [DOI] [PubMed] [Google Scholar]
- 48.Dahlman I, Kaaman M, Olsson T, et al. A unique role of monocyte chemoattractant protein 1 among chemokines in adipose tissue of obese subjects. J Clin Endocrinol Metab. 2005;90:5834–5840. doi: 10.1210/jc.2005-0369. [DOI] [PubMed] [Google Scholar]
- 49.Harman-Boehm I, Bluher M, Redel H, et al. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92:2240–2247. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 50.Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]