Abstract
The process of wing patterning involves precise molecular mechanisms to establish an organizing center at the dorsal–ventral boundary, which functions to direct the development of the Drosophila wing. We report that misexpression of dLMO, a Drosophila LIM-only protein, in specific patterns in the developing wing imaginal disc, disrupts the dorsal–ventral (D-V) boundary and causes errors in wing patterning. When dLMO is misexpressed along the anterior–posterior boundary, extra wing outgrowth occurs, similar to the phenotype seen when mutant clones lacking Apterous, a LIM homeodomain protein known to be essential for normal D-V patterning of the wing, are made in the wing disc. When dLMO is misexpressed along the D-V boundary in third instar larvae, loss of the wing margin is observed. This phenotype is very similar to the phenotype of Beadex, a long-studied dominant mutation that we show disrupts the dLMO transcript in the 3′ untranslated region. dLMO normally is expressed in the wing pouch of the third instar wing imaginal disc during patterning. A mammalian homolog of dLMO is expressed in the developing limb bud of the mouse. This indicates that LMO proteins might function in an evolutionarily conserved mechanism involved in patterning the appendages.
Keywords: wing development/dorsal–ventral patterning
The Drosophila wing and notum are derived from the wing imaginal disc (dorsal mesothoracic disc), which is subdivided first into anterior and posterior compartments and later into dorsal and ventral compartments during development (1, 2). The Drosophila LIM-homeodomain protein Apterous (Ap) is expressed only in the dorsal compartment, where it specifies dorsal cell fates (3, 4). Ap activates expression of fringe (fng), which modulates the activation of the Notch (N) receptor by its ligands Serrate (Ser) and Delta (Dl) across the dorsal–ventral (D-V) boundary (1, 5–7). Notch activation leads to the expression of wingless (wg) and cut (ct) at the D-V boundary (8–10). Cells along the D-V boundary eventually form the wing margin, the edge of the wing connecting apposed dorsal and ventral cell layers of the wing blade (1).
Loss of tissue at the wing margin occurs in Beadex (Bx) mutations. First identified by C. Bridges in 1925, Bx acts as a dominant mutation that maps to region 17C on the X chromosome (11). The Bx phenotype appears to be caused by excess activity of the wild-type heldup-a (hdp-a) gene, which also maps to 17C (12–14). The observation that a deletion that not only removes the Bx locus but also adjacent DNA results in a hdp-a phenotype without the Bx phenotype indicates that wild-type hdp-a gene function is necessary for the expression of the Bx phenotype (15). In this paper, we report results strongly suggesting that Bx is a regulatory mutation in dLMO, encoding a Drosophila LIM-only protein (16) that most likely corresponds to the activity associated with hdp-a.
The dLMO protein contains two LIM domains without any other recognizable domains. LIM domains are characterized by seven precisely spaced cysteine residues. This domain was identified originally in three transcription factors, Lin-11, Isl-1, and Mec-3, for which it was named (17, 18). These proteins, like Ap, contain two tandem LIM domains, a homeodomain (HD) and a transcriptional activation domain, placing them in the LIM-HD subfamily. In contrast, proteins in the LIM only (LMO) subfamily, such as dLMO, contain only LIM domains without homeodomains (17). dLMO was isolated by virtue of its sequence similarity with the human LMO genes (16), which are protooncogenes associated with forms of acute T cell leukemia (19, 20).
To determine the likely sites of dLMO function in Drosophila, we examined the expression pattern of dLMO RNA and found that dLMO is expressed in many tissues including the wing disc. We studied the effect of dLMO on wing development by misexpressing dLMO in different patterns in the developing wing disc. Misexpression of dLMO along the anterior–posterior (A-P) boundary induces ectopic wing margin formation and ectopic wing outgrowth, similar to the phenotype generated by clones of ap mutant cells (3, 4). Additionally, misexpression of dLMO along the D-V boundary results in a scalloping phenotype similar to that seen in Bx. Molecular analysis of Bx reveals that many alleles contain transposon insertions in the dLMO transcript.
Drosophila wing development and vertebrate limb development have intriguing parallels. Analogous to the margin in the Drosophila wing, the apical ectodermal ridge is formed at the D-V boundary of the developing vertebrate limb bud and serves as a signaling center for the outgrowth and patterning of the limb (21). In addition, vertebrate and Drosophila appendage formation involves many orthologous genes. We found that a mammalian homolog of dLMO, LMO-2, is expressed in the developing limb bud at a stage and location consistent with an involvement in limb development.
MATERIALS AND METHODS
Fly Strains.
Flies were grown on standard cornmeal/agar/yeast. Common stocks are described in ref. 11. scabrous-GAL4 (sca-G4) (22), wingless-lacZ (wg-lacZ) (23), apterous-lacZ (ap-lacZ) (24), and patched-GAL4 (ptc-G4) (25) are enhancer traps of the respective genes. Serrate-GAL4 (Ser-G4) is a construct containing GAL4 driven by a Ser enhancer (26). Bx1, Bx3, and BxJ were supplied by the Bloomington Drosophila Stock Center (Bloomington, IN).
Misexpression Screen.
Approximately 2,300 independent EP insertion lines, generated by P. Rørth (27, 28) and kindly provided by P. Rørth, T. Laverty, and G. M. Rubin, were crossed initially to sca-G4 at 25°C. The F1 of these crosses were shifted to 29°C 48–72 hr after egg laying. The emerging adults were scored for abnormal bristles. The lines that gave bristle phenotypes were crossed to sca-G4 and ptc-G4. Three lines, EP 1394, EP 1383, and EP 1306, were found to produce abnormal wings when crossed to ptc-G4, in addition to their bristle phenotypes. The DNA sequence flanking the EP insertion site was isolated by plasmid rescue and sequenced (27, 28).
Molecular Analysis of the Bx Locus.
Molecular techniques were performed according to Maniatis et al. (29). The dLMO cDNA was kindly provided by B. Royer-Pokora (Heinrich-Heine Universitaet, Duesseldorf). Genomic DNA was prepared from homozygous Bx flies (30). y w (yellow white) flies were used as a control.
Upstream Activation Sequence (UAS)-dLMO Transgenic Flies.
The plasmid containing dLMO cDNA was digested with XhoI and XbaI, and the released dLMO insert was subcloned into pUAST (31) to generate pUAS-dLMO. To add the myc tag, PCR was performed (Vent Polymerase, New England Biolabs) by using a primer over a PstI site within dLMO (5′-GCCTGCAGCAAGGTGATCCCAGCCTTCGAG-3′) and a primer at the stop codon (5′-GGCTCTAGACTACAGATCCTCCTCGGAGATCAGCTTCTGCTCCATGCTGGACGCGCCCAGTTGATTCTTCATATGGGC-3′), which added amino acids QGTEQKLISEEDLN (myc tag) in-frame with the dLMO ORF. The fragment generated was subcloned into pBluescript (Stratagene) containing the dLMO cDNA digested with PstI and XbaI. All cloned PCR fragments were sequenced to ensure no errors were introduced during PCR. The cDNA containing the myc tag and without the 3′ UTR was then subcloned into pUAST and used to create pUAS-dLMOmyc. A total of two UAS-dLMO and nine UAS-dLMOmyc transgenic lines were generated by using standard techniques (30).
Immunostaining, X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) Staining, and in situ Hybridization.
Immunostaining, X-gal staining, and in situ hybridization of Drosophila embryos and imaginal discs were performed as in ref. 30. The rabbit anti-β-gal antibody was purchased from Cappel and used at 1:500 dilution. The mouse anti-myc mAb (9E10) was purchased from Santa Cruz Biotechnology and used at 1:200 dilution. Rat anti-Cut antibody is described in ref. 32. In situ hybridization to whole-mount mouse embryos was done essentially as described (33). The mouse LMO-2 cDNA was obtained from Genome Systems (St. Louis). The sense RNA probe was used as a control (not shown).
RESULTS
Isolation of dLMO from a Misexpression Screen.
We performed a modular misexpression screen using approximately 2,300 EP lines, each containing an independent P element insertion, that causes misexpression of a downstream gene in the presence of GAL4 (27, 28). Our initial screen was designed to identify lines that yielded bristle phenotypes upon ectopic expression driven by GAL4 in neural precursors. Positives from the initial screen were subjected to a secondary screen, in which we used patched-GAL4 (ptc-G4) to drive misexpression along the A-P boundary including a subset of sensory organ precursors in the developing wing disc. Three EP lines (1306, 1383, 1394) were identified that displayed ectopic wing formation on the wing or notum when crossed to ptc-G4. The DNA flanking the P element in each of these EP lines was cloned and sequenced. After comparison with the GenBank database, the three lines were identified as independent genomic insertions approximately 310, 280, and 250 bp upstream of the reported dLMOa transcript (Fig. 1A) (16). To confirm that dLMO was, in fact, responsible for the ectopic wing outgrowth, we made transgenic flies containing a UAS-dLMO cDNA construct. Misexpression of UAS-dLMO with ptc-G4 results in ectopic wing outgrowth from the dorsal wing blade (data not shown), the same phenotype seen when EP 1394 is crossed to ptc-G4 (see below).
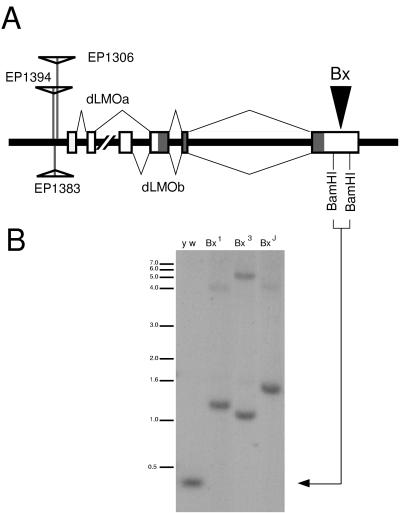
Figure 1.
(A) The genomic locus containing the dLMO gene. Three EP lines (1306, 1383, and 1394) contain insertions upstream of the first exon of dLMO. Two alternatively spliced transcripts, dLMOa and dLMOb, are reported (16). Beadex alleles Bx1, Bx3, and BxJ contain insertions (arrowhead) in the last exon of dLMO, within the 3′ UTR, between two BamHI sites. (B) Genomic Southern analysis of Beadex alleles. Genomic DNA from y w, Bx1, Bx3, and BxJ adult flies was digested with BamHI. The Southern was probed with a 403-bp BamHI fragment from the 3′ UTR of the dLMO cDNA. The 403-bp fragment is detected in DNA from y w flies (arrow) while larger fragments are detected in the DNA from Bx flies.
Genomic Southern Analysis of Beadex Mutants Detects Insertions in the Gene Encoding dLMO.
The dLMO gene is located in the 17C region of the X chromosome (16). Beadex (Bx) also maps to this region (11). Because of the wing phenotype associated with Bx alleles, we suspected that dLMO could be the gene mutated to cause the Bx phenotype. Many Bx alleles contain transposon insertions within a 500-bp region (15). Comparing the restriction map of the Bx alleles (15) with the restriction map of the dLMO genomic region (16), we suspected this 500-bp region corresponds to a BamHI fragment in the 3′ UTR of dLMO (Fig. 1A). To test this idea, we used this 403-bp BamHI fragment from dLMO cDNA to probe a genomic Southern blot of wild type and Bx mutant genomic DNA digested with BamHI. In each of the Bx alleles tested (Bx1, BxJ, Bx3), we found that the BamHI fragment thus identified was longer than the 403-bp BamHI fragment from wild-type DNA (Fig. 1B). This strongly suggests that the insertions in these previously studied Bx alleles (15) lie within the DNA encoding the 3′ UTR of the dLMO transcript.
The Expression Pattern of dLMO in Wild-Type Drosophila During Development.
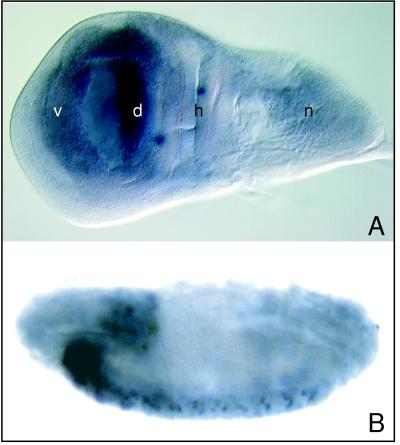
To determine the expression pattern of dLMO, we performed in situ hybridization using dLMO cDNA as a probe. In third instar wing imaginal discs, dLMO is expressed at high levels in the dorsal compartment and at lower levels in the ventral compartment (Fig. 2A). dLMO also is expressed in the leg and eye discs (not shown). In the embryo, dLMO is expressed in the brain and in a subset of cells in the developing central nervous system (Fig. 2B). The presence of dLMO in many different structures suggests that perhaps dLMO serves multiple functions during development.
Figure 2.
Localization of dLMO RNA. (A) dLMO RNA is present in the dorsal and ventral compartments of the third instar wing disc. The dorsal compartment contains higher levels of dLMO RNA than the ventral. n, notum precursor region; h, hinge precursor region; d, dorsal wing precursor compartment; v, ventral wing precursor compartment. (B) dLMO RNA is present in the brain and developing central nervous system of the stage 15 embryo. Each segment contains a repeated pattern of cells containing dLMO RNA. Anterior is to the left; dorsal is at the top.
Misexpression of dLMO at the D-V Boundary Disrupts Margin Formation Mimicking the Beadex Phenotype.
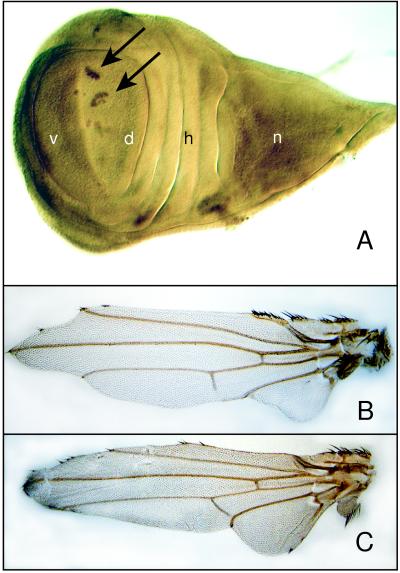
To test the effect of misexpression of dLMO along the boundary between the dorsal and ventral compartments of the wing disc, we used Serrate-Gal4 (Ser-G4), which drives expression in the developing wing margin during third instar (22). Wing margin formation in third instar wing discs then was assessed by using wg-lacZ and Ct as markers for the developing wing margin. We found that the margin is discontinuous in discs misexpressing dLMO along the D-V boundary (Fig. 3A). This disruption of margin formation results in wing scalloping (Fig. 3B) that resembles the Bx phenotype (Fig. 3C).
Figure 3.
Misexpression of dLMO using Ser-G4 to drive expression of EP 1394 at the D-V boundary during third instar. (A) Immunohistochemistry using an antibody against the Ct protein demonstrates loss of continuity in the developing wing margin of third instar wing discs. Anti-Ct antibody, similar to wg-lacZ (Fig. 4A), stains the D-V boundary in wild-type wing discs. The arrows point out the remaining Ct-positive cells forming islands along the D-V boundary. n, notum precursor region; h, hinge precursor region; d, dorsal wing precursor compartment; v, ventral wing precursor compartment. (B) The adult wing produced when dLMO is misexpressed at the D-V boundary. The wing margin shows a substantial loss of tissue. (C) The adult wing from a Bx3 mutant displays a similar phenotype of loss of wing margin tissue.
Misexpression of dLMO Along the A-P Boundary Leads to Ectopic Margin Formation and Extra Wing Outgrowth.
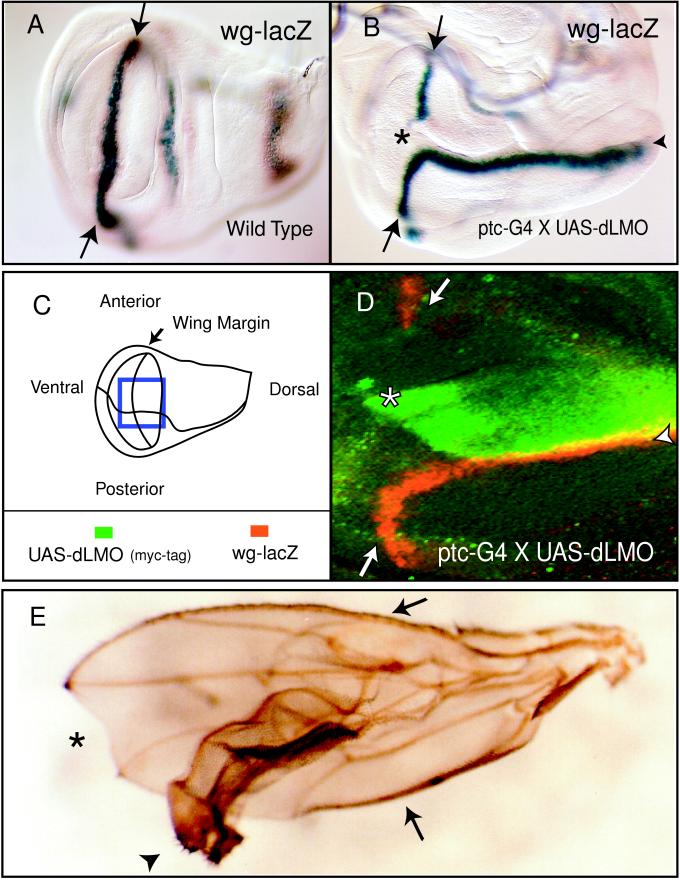
ptc-G4 was used to drive misexpression of dLMO along the A-P boundary. A gap in wg-lacZ staining appears in the third instar wing disc where dLMO misexpression intersects the D-V boundary (Fig. 4B). Additionally, an ectopic wg-lacZ stripe appears along the posterior edge of the ptc-G4 expression domain in the dorsal compartment, perpendicular to the endogenous wing margin (Fig. 4B). Double-staining for myc-tagged UAS-dLMO and wg-lacZ reveals that wg-lacZ is being expressed in cells both in the dLMO expression domain and in the adjacent cells (Fig. 4D), similar to wg expression in both the dorsal and ventral cells of the endogenous wing margin (1). Ct is also expressed along the posterior edge of the dLMO misexpression domain (data not shown). These markers signify the formation of an ectopic wing margin in the dorsal compartment. Outgrowth organized by this ectopic margin leads to extra wing formation from the dorsal wing blade as originally observed (Fig. 4E).
Figure 4.
Misexpression of dLMO using ptc-G4 to drive expression of a UAS-dLMO transgene or EP 1394 along the A-P boundary. The wg-lacZ enhancer trap marks wg expression in late third instar wing discs. (A) In a wild-type disc, wg-lacZ is found along the D-V boundary, indicated by the arrows. (B) dLMO misexpression prevents expression of wg-lacZ where the ptc-G4 expression domain intersects the D-V boundary, at the A-P boundary (asterisk). wg-lacZ is ectopically expressed at the posterior edge of the ptc-G4 stripe only in the dorsal compartment, perpendicular to the D-V boundary (arrowhead). Note the malformation of the wing disc at this stage of outgrowth. (C) Diagram of the third instar wing disc showing the compartments of the wing pouch. The box indicates the area shown in D. (D) Immunohistochemistry using antibody to β-gal (wg-lacZ) in red and myc epitope-tagged dLMO in green on third instar wing discs. UAS-dLMOmyc is being driven with ptc-G4. Formation of an ectopic wg-lacZ stripe (arrowhead) occurs at the posterior edge of the ptc-G4 expression domain in the dorsal compartment. wg-lacZ is expressed both in the dLMO misexpression domain (double-staining appears yellow) and in the cells lying adjacent to it along the posterior edge. (E) The adult wing produced when dLMO is overexpressed with ptc-G4 and EP 1394 contains an ectopic wing outgrowth (arrowhead). The endogenous anterior and posterior wing margins are indicated by the arrows. Note the notch in the margin at the A-P boundary because of disruption of endogenous D-V boundary signaling (asterisk).
The Vertebrate dLMO Homolog LMO-2 Is Expressed in the Developing Mouse Limb Buds.
Many vertebrate homologs of Drosophila genes important for wing patterning have been found to play a role in limb development (21). To determine whether the LMO genes might also be involved, we performed in situ hybridization using the mouse LMO-2 cDNA on E10.5 mouse embryos. (LMO-1 and LMO-3 were not characterized.) We found that LMO-2 RNA is present in the developing limb bud at E10.5 (Fig. 5). Expression is seen in a band centered on the D-V boundary of the developing limb bud. In situ hybridization to sections of the limb bud reveals that LMO-2 RNA is present in a broad field of the mesenchyme underlying the apical ectodermal ridge (data not shown), a structure known to be an important organizing structure of the limb bud (21). Expression is also seen at the somite boundaries (data not shown).
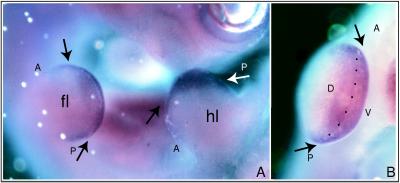
Figure 5.
Localization of LMO-2 RNA in E10.5 mouse embryos. (A) Expression of LMO-2 RNA is detected in the developing limb buds. This lateral view looks down on the dorsal surface of the forelimb (fl) and hindlimb (hl), with anterior to the left and posterior to the right. (B) Higher magnification looking at the distal edge of the forelimb in A reveals expression in a broad band centered on the D-V boundary (arrows and dotted line) and extending into the dorsal and ventral regions of the limb bud. A, anterior; P, posterior; D, dorsal; V, ventral.
DISCUSSION
In this report we provide evidence that improper regulation of dLMO, the Drosophila member of the evolutionarily conserved LIM-only gene family, can disrupt the development of the Drosophila wing. Misexpression or overexpression of dLMO in specific patterns in the developing wing imaginal disc causes errors in wing patterning such as ectopic wing outgrowth or loss of the wing margin. Additionally, we have found that the long-studied dominant mutation Beadex (Bx) disrupts the dLMO 3′ UTR. Below, we discuss possible explanations for the wing patterning effects of dLMO misexpression in the context of our current knowledge of wing development.
The Drosophila LIM-homeodomain protein Ap serves as the selector gene for the dorsal compartment (3, 4, 24). The juxtaposition of Ap-expressing dorsal cells and nonexpressing ventral cells establishes a D-V organizing center at their boundary (3). In the late third instar disc, wg and ct expression is induced in a three- to six-cell-wide stripe that straddles the D-V boundary, the location of the future wing margin (5, 8, 9). It is known that ap expression needs to be maintained continuously in the dorsal compartment, because loss of Ap function in mutant clones as late as third instar causes cell-autonomous fate transformation from dorsal to ventral (3). ap mutant clones in the interior of the dorsal compartment create a new boundary of dorsal versus ventral cells, thereby initiating the genetic program normally reserved for the endogenous D-V boundary. wg and ct are expressed in stripes of cells flanking the ectopic boundary, signifying the formation of an ectopic margin (3–5, 8, 9). Outgrowth organized by this ectopic margin eventually leads to the outgrowth of an ectopic wing from the dorsal surface of the wing blade (1).
We have generated an ectopic wing outgrowth phenotype similar to that produced by ap mutant clones by overexpression of dLMO in the wing disc. When dLMO expression is driven by ptc-G4, an ectopic margin is formed that, as with ap mutant clones, expresses wg-lacZ and Ct in the dorsal compartment of third instar wing discs. These similarities, along with the common LIM domain structure between dLMO and Ap, make it likely that misexpressed dLMO exerts its effect through interference with Ap function. Two not mutually exclusive models can be proposed for the possible molecular interactions between dLMO and Ap protein based on the extensive biochemical studies that have been carried out on LIM-HD and LMO class proteins. In one scenario, dLMO binds directly to Ap. High levels of dLMO could prevent Ap from binding DNA either by interaction with the homeodomain or the LIM domains of Ap. If dLMO binds the homeodomain, it could mask the DNA-binding portion of Ap, analogous to the ability of the LIM domains of other LIM-HD proteins to inhibit DNA-binding activity of the homeodomain (34). Alternatively, if dLMO binds the LIM domains of Ap, analogous to the observed LIM–LIM interactions between many LIM domain proteins (35, 36), it may negatively regulate Ap by forming nonfunctional heterodimers, reminiscent of the mechanism used by Id to prevent DNA binding by MyoD (37). A second model involves the sequestration of coactivators of Ap by dLMO rather than direct binding of dLMO to Ap. Recently, Drosophila Chip has been identified as a homolog of the mouse LIM domain-binding protein (Ldb-1/Nli/cLIM-2) (38–41). Chip binds the LIM domains of Ap protein, and mutations in Chip enhance ap mutations genetically (38). dLMO might be able to bind Chip, analogous to what is seen between mouse LMO proteins and Ldb-1 (42–44). High levels of dLMO might then interfere with Ap function by sequestering its putative coactivator, Chip. The expression of dLMO in the wing disc during patterning makes it possible that one of these mechanisms, or some other mechanism, is used by dLMO to regulate wing development.
We demonstrate in this report that Bx mutations most likely disrupt dLMO, which was cloned previously based on sequence similarity with a human oncogene (16). The Bx phenotype appears to be associated with elevated activity of the hdp-a gene because this phenotype can be produced simply by increasing the copy number of the wild-type hdp-a gene (12–14). We believe the hdp-a gene encodes dLMO. In support of this idea, a phenotype very similar to that of Bx can be produced when Ser-G4 is used to drive overexpression from UAS-dLMO transgenes. Several independently derived Bx alleles contain transposable element insertions in the 3′ UTR of the dLMO gene. We suspect that at least part of the effect of Bx mutations is because of interference with the postulated mRNA destabilizing function of the dLMO 3′ UTR (16), possibly leading to abnormally high levels of dLMO protein. Having cloned dLMO, Zhu et al. (16) noted that the 3′ UTR contains 9 copies of an AT-rich motif common to short-lived mRNAs. Drosophila dLMO and human LMO-2 contain a 25-bp sequence in their 3′ UTRs that is 83% identical and contains one of these AT-rich motifs (16). This conservation might indicate an important role for the 3′ UTR in regulating the activity of LMO genes.
The expression of LMO-2, a mammalian homolog of dLMO, in the mesenchyme of the developing mouse limb bud raises the interesting possibility that the function of the LMO genes has been conserved evolutionarily between insects and mammals. Lmx-1, a LIM-HD protein like Ap, is expressed in the mesenchyme of the dorsal limb bud during development (21). Loss of Lmx-1b function causes a biventral phenotype, implicating Lmx-1b as a primary dorsalizing activity in the mouse limb (45). As with dLMO, LMO-2 is expressed in both the dorsal and ventral compartments during limb patterning. LMO-2 or other LMO proteins could interact with Lmx-1 in a manner similar to the proposed interaction between Ap and dLMO in Drosophila wing. LMO-2 also is expressed at the somite boundaries (data not shown). Many genes required for forming the D-V boundary in the developing limb, such as members of the fringe, Wnt, and Notch gene families, also play an important role during somitogenesis (46). The presence of LMO-2 RNA at the somite boundaries might indicate a conserved role of LMO gene family members in the context of boundary formation, including the limb bud, somite, and insect wing disc. Future work with both dLMO and vertebrate LMOs should further our understanding of the complex molecular interactions involved in patterning during insect and vertebrate development.
Acknowledgments
We thank M.-M. Jiang, S. Barbel, S. Younger-Shepherd, and L. Sharp for technical assistance and M. Lewandoski, S. Younger-Shepherd, and L. Ma for discussion and comments on the manuscript. We are indebted to B. Royer-Pokora for dLMO cDNA clone, to P. Rørth, T. Laverty, and G. M. Rubin for the EP lines, and to N. Perrimon, F. M. Hoffmann, K. Irvine, R. Fleming, S. Carroll, G. Struhl, J. Thomas, W. Janning, and Bloomington Drosophila Stock Center for fly strains. S.A. is supported by a fellowship from the Deutsche Forschungsgemeinschaft. Y.-M.C. currently is supported by the Program in Biological Sciences Markey Grant and the Herb Boyer Fund. This study was supported in part by the National Institute of Mental Health Silvio Conte Center for Neuroscience at University of California at San Francisco. C.Z. is a postdoctoral associate, N.J.J. is a predoctoral fellow, and L.Y.J. and Y.N.J. are investigators of Howard Hughes Medical Institute.
ABBREVIATIONS
- D-V
dorsal–ventral
- A-P
anterior–posterior
- Ap
Apterous
- HD
homeodomain
- wg
wingless
- ct
cut
- Bx
Beadex
- UAS
upstream activation sequence
References
- 1.Cohen S M. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 747–841. [Google Scholar]
- 2.Garcia-Bellido A. CIBA Found Symp. 1975;29:161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Benjumea F J, Cohen S M. Cell. 1993;75:741–752. doi: 10.1016/0092-8674(93)90494-b. [DOI] [PubMed] [Google Scholar]
- 4.Blair S S, Brower D L, Thomas J B, Zavortink M. Development. 1994;120:1747–1758. doi: 10.1242/dev.120.7.1805. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Irvine K D, Carroll S B. Cell. 1995;82:785–794. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- 6.Irvine K D, Wieschaus E. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 7.Doherty D, Feger G, Younger-Shepherd S, Jan L Y, Jan Y N. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- 8.Micchelli C A, Rulifson E J, Blair S S. Development. 1997;124:1497–1507. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- 9.Neumann C J, Cohen S M. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- 10.Couso J P, Knust E, Martinez Arias A. Curr Biol. 1995;5:1424–1436. doi: 10.1016/s0960-9822(95)00281-8. [DOI] [PubMed] [Google Scholar]
- 11.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 12.Green M M. Genetics. 1953;38:91–105. doi: 10.1093/genetics/38.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green M M. Z indukt Abstamm VerebLehre. 1953;85:435–449. doi: 10.1007/BF00308296. [DOI] [PubMed] [Google Scholar]
- 14.Lifschytz E, Green M M. Mol Gen Genet. 1979;171:153–159. doi: 10.1007/BF00270001. [DOI] [PubMed] [Google Scholar]
- 15.Mattox W W, Davidson N. Mol Cell Biol. 1984;4:1343–1353. doi: 10.1128/mcb.4.7.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu T H, Bodem J, Keppel E, Paro R, Royer-Pokora B. Oncogene. 1995;11:1283–1290. [PubMed] [Google Scholar]
- 17.Sanchez-Garcia I, Rabbitts T H. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 18.Curtiss R, Heilig J S. BioEssays. 1998;20:58–69. doi: 10.1002/(SICI)1521-1878(199801)20:1<58::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 19.Boehm T, Buluwela L, Williams D, White L, Rabbitts T H. EMBO J. 1988;7:2011–2017. doi: 10.1002/j.1460-2075.1988.tb03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Royer-Pokora B, Loos U, Ludwig W D. Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 21.Johnson R L, Tabin C J. Cell. 1997;90:979–990. doi: 10.1016/s0092-8674(00)80364-5. [DOI] [PubMed] [Google Scholar]
- 22.Nakao K, Campos-Ortega J A. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 23.Hays R, Gibori G B, Bejsovec A. Development. 1997;124:3727–3736. doi: 10.1242/dev.124.19.3727. [DOI] [PubMed] [Google Scholar]
- 24.Cohen B, McGuffin M E, Pfeifle C, Segal D, Cohen S M. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- 25.Hinz U, Giebel B, Campos-Ortega J A. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 26.Fleming R J, Gu Y, Hukriede N A. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- 27.Rørth P. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin G M, Weigmann K, Milán M, Benes V, Ansorge W, Cohen S M. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 30.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Brand A H, Perrimon N. Development. 1993;118:339–352. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 32.Blochlinger K, Bodmer R, Jan L Y, Jan Y N. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 33.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Garcia I, Osada H, Forster A, Rabbitts T H. EMBO J. 1993;12:4243–4250. doi: 10.1002/j.1460-2075.1993.tb06108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feuerstein R, Wang X, Song D, Cooke N E, Liebhaber S A. Proc Natl Acad Sci USA. 1994;91:10655–10659. doi: 10.1073/pnas.91.22.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arber S, Caroni P. Genes Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- 37.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 38.Morcillo P, Rosen C, Baylies M K, Dorsett D. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Nature (London) 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 40.Jurata L W, Kenny D A, Gill G N. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bach I, Carrière C, Ostendorff H P, Andersen B, Rosenfeld M G. Genes Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- 42.Wadman I A, Osada H, Gruetz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurata L W, Gill G N. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visvader J E, Mao X, Fujiwara Y, Hahm K, Orkin S H. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg K C, Pepicelli C V, Gan L, Lee B, Johnson R L. Nat Genet. 1998;19:51–55. doi: 10.1038/ng0598-51. [DOI] [PubMed] [Google Scholar]
- 46.Gossler A, Hrabe de Angelis M. Curr Top Dev Biol. 1998;38:225–287. [PubMed] [Google Scholar]