Abstract
Innate defense regulator-1 (IDR-1) is a synthetic peptide with no antimicrobial activity that enhances microbial infection control while suppressing inflammation. Previously, the effects of IDR-1 were postulated to impact several regulatory pathways including mitogen-activated protein kinase (MAPK) p38 and CCAAT-enhancer-binding protein, but how this was mediated was unknown. Using a combined stable isotope labeling by amino acids in cell culture-proteomics methodology, we identified the cytoplasmic scaffold protein p62 as the molecular target of IDR-1. Direct IDR-1 binding to p62 was confirmed by several biochemical binding experiments, and the p62 ZZ-type zinc finger domain was identified as the IDR-1 binding site. Co-immunoprecipitation analysis of p62 molecular complexes demonstrated that IDR-1 enhanced the tumor necrosis factor α-induced p62 receptor-interacting protein 1 (RIP1) complex formation but did not affect tumor necrosis factor α-induced p62-protein kinase ζ complex formation. In addition, IDR-1 induced p38 MAPK activity in a p62-dependent manner and increased CCAAT-enhancer-binding protein β activity, whereas NF-κB activity was unaffected. Collectively, these results demonstrate that IDR-1 binding to p62 specifically affects protein-protein interactions and subsequent downstream events. Our results implicate p62 in the molecular mechanisms governing innate immunity and identify p62 as a potential therapeutic target in both infectious and inflammatory diseases.
Introduction
Innate defense regulator-1 (IDR-1)4 (KSRIVPAIPVSLL-NH2) is a synthetic peptide with no antimicrobial activity that enhances host bacterial infection control while suppressing harmful inflammation. Treatment of mice with IDR-1 provides protection from otherwise lethal infections with Gram-positive and Gram-negative pathogens, including methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enteroccocus, and Salmonella enterica (1). In addition, IDR-1 enhances production of some monocyte-produced chemokines, including MCP-1 and RANTES (regulated on activation normal T cell expressed and secreted) and the anti-inflammatory cytokine interleukin-10. IDR-1 also suppresses production of Toll-like receptor-induced proinflammatory cytokines, including interleukin-6 and TNFα (1). Previously, the effects of IDR-1 were postulated to impact several intracellular pathways, including MAPK p38 and C/EBP, but the preceding molecular events remained unknown.
In this study, the cytoplasmic protein sequestosome-1 (p62) was identified as a molecular target of IDR-1. p62 is a multidomain scaffold (adaptor) protein, with many known interacting partners, including PKCζ (2, 3), p38 (4), RIP1 (5), and TRAF6 (6). p62 comprises an N-terminal PB1 domain that is primarily important for atypical PKC binding (2), a ZZ-type zinc finger (ZZ) domain that interacts with RIP1 (5), and a TRAF6 binding sequence domain recognized by TRAF6 (6). Additionally, a C-terminal ubiquitin-associated domain binds to polyubiquitin (7), a function recently demonstrated to facilitate the efficient activation of prosurvival and proapoptotic pathways by binding polyubiquitinated signaling proteins (8). The ubiquitin-associated domain is also considered the basis for the association between p62 and protein trafficking to the proteasome (9, 10). Thus, p62 functions as a nodal point in cellular signaling pathways, particularly in the regulation of NF-κB (11, 12) and cellular differentiation (13).
Here we provide evidence that IDR-1 specifically binds to the ZZ domain of p62. This binding event selectively stabilized TNFα-induced p62-RIP1 complex formation, but not TNFα-induced p62-PKCξ complex formation, and specifically modulated the downstream signaling pathways by activating MAPK p38 and C/EBPβ but not NF-κB. These studies provide new evidence for p62 as an important component of the immune system and demonstrate that IDR-1 can be used for the interrogation of the molecular events governing these innate responses.
EXPERIMENTAL PROCEDURES
Experimental Procedures are detailed in the supplemental material.
RESULTS
p62 Is an Intracellular Target of IDR-1
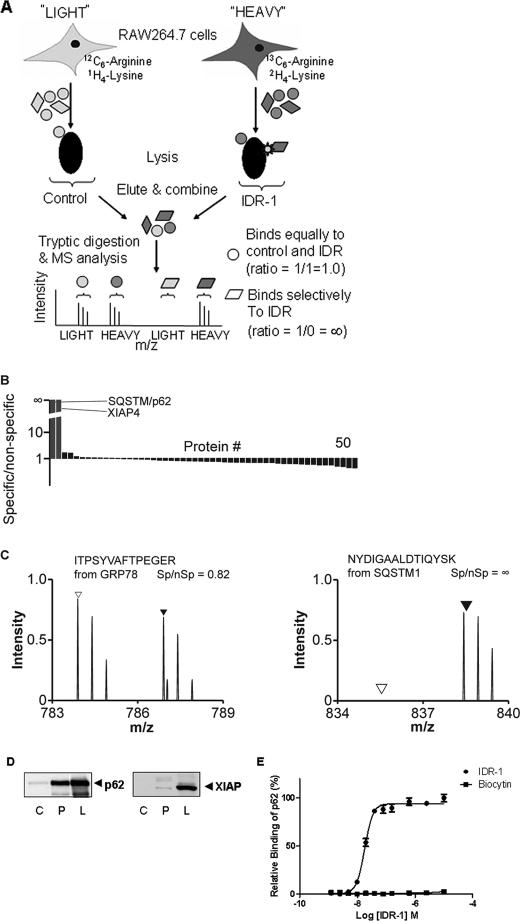
We utilized stable isotope labeling by amino acids in cell culture (SILAC) and quantitative mass spectrometry-based proteomics to identify the major intracellular binding target of IDR-1. Murine leukemic RAW264.7 cells were grown in isotopically labeled medium (SILAC-heavy condition containing [13C6]arginine and [2H4]lysine) and in isotopically unlabeled medium (SILAC-light condition containing normal isotopic arginine and lysine) (14). The cells from both conditions were harvested, lysed, and subjected to affinity enrichment with desthiobiotinylated IDR-1 (heavy condition) or desthiobiotin alone (light, control condition) (Fig. 1A). Proteins bound to the matrix after washing were eluted using free biotin, digested to peptides, and analyzed by liquid chromatography-tandem mass spectrometry. Quantitative SILAC ratios were then extracted after acquisition. Only two cellular proteins, p62 and XIAP-4, showed a marked and consistent differential binding to IDR-1 when compared with control in three repeat studies (Fig. 1B). p62 was present with a specific/nonspecific (Sp:nSp) ratio approaching infinity indicating selective binding (Fig. 1C, right panel). This analysis was confirmed using other, related IDR compounds, which detected only p62, but not XIAP-4, as the binding partner (not shown).
FIGURE 1.
IDR-1 binds to sequestosome-1/p62. A, schematic representation of the SILAC-proteomics approach utilized for the identification of the IDR-1 target. Streptavidin beads coated with desthiobiotinylated IDR-1 or desthiobiotin alone (negative control) were incubated with lysates prepared from SILAC-labeled and unlabeled RAW264.7 cells, respectively, followed by liquid chromatography-tandem mass spectrometry analysis of the combined bead eluent. Spectra derived from SILAC-labeled proteins and unlabeled proteins are presented as Sp and nSp. IDR-1 binding partners are identified by having an Sp:nSp ratio greater than 1. Proteins bound to the desthiobiotin control will produce Sp:nSp ratios close to or less than 1 and constitute background binders. B, proteins detected in the IDR-1 pulldown. Only proteins that were detected in more than one experiment are presented. p62 and XIAP-4 are highlighted by lines. C, IDR-1 interacts with p62. In the left panel, spectra from representative peptides of a non-specifically binding protein (GRP78) showed similar signals of the labeled (▲) and unlabeled (△) protein, i.e. the ratio of peak intensities of Sp over nSp is close to 1.0. In the right panel, spectra from representative peptides of p62 (SQSTM1) are observed only in the labeled condition, indicating the specific binding of p62 to IDR-1, i.e. the ratio of peak intensities of Sp over nSp is close to infinity. These data are representative of three replicate experiments. D, IDR-1 binds to human p62, but only weakly to XIAP. Lysates from HEK293T cells were pulled down against biocytin control (C) or biotinylated IDR-1 (P). Cell lysates are designated as (L) and were used as control. Proteins pulled down were immunoblotted with anti-p62 antibody (left panel) or with anti-XIAP antibody (right panel). E, IDR-1 binds to recombinant human p62 in vitro. Direct p62 binding by IDR-1 (●) or biocytin control (■) was determined using a plate coated with various concentrations of IDR-1. Error bars represent S.D.
To test direct binding of IDR-1 to human p62 and XIAP, lysates from HEK293T cells were incubated with streptavidin beads coated with biotinylated IDR-1 or biocytin as background control. Immunoblot analysis of the pulldown samples showed a strong band at ∼62 kDa that was detected with antibodies against p62 (Fig. 1D, left panel). Densitometric analysis of this band revealed that it constitutes 64% of the intensity of p62 present in the cell lysate control, suggesting that the majority of the p62 from the lysate bound to the peptide. In contrast, the IDR-1 pulldown of the XIAP protein was very faint (Fig. 1D, right panel) and constituted only 4% of the lysate control.
We next investigated direct IDR-1 binding to recombinant human p62. To test this, an in vitro protein binding assay using biotinylated IDR-1 and human recombinant glutathione S-transferase-labeled p62 was performed. As shown in Fig. 1E, a concentration-dependent binding of p62 by IDR-1 was observed, whereas the control biocytin did not bind to p62 over the same concentration range. This binding was confirmed using a competitive binding experiment with biotinylated IDR-1 and unlabeled IDR-1 and with other IDR analogs in the same class (data not shown). These results confirmed direct binding of IDR-1 to the human p62 protein.
IDR-1 Binds to the ZZ Domain of p62
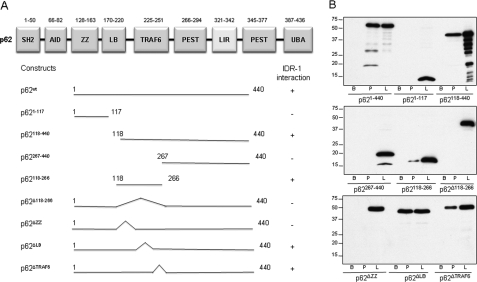
To determine which region of p62 is required for IDR-1 binding, FLAG-tagged human p62 deletion mutants (Fig. 2A) were transfected into HEK293T cells and tested in biotinylated IDR-1 pulldown assays, as described above. Initially, the analysis was focused on the N-terminal, central, and C-terminal regions (constructs p621–117, p62118–440, and p62267–440). IDR-1 did not bind to constructs p621–117 and p62267–440, whereas it bound to p62118–440 (Fig. 2B). Further, a p62 construct composed only of residues 118–266 (p62118–266) was also able to bind IDR-1. This central region of p62 (residues 118–266) is made up of three subdomains, including a ZZ (zinc finger) structural motif (residues 128–163), an intermediate domain LB (residues 170–220), and a TRAF6 binding region (residues 225–251). Successive deletions of each of these subdomains revealed that removal of residues 128–163 was sufficient to abolish IDR-1 binding, whereas deletion of LB or TRAF6 domains did not affect binding (Fig. 2). These findings demonstrated that the ZZ domain of p62 is required for IDR-1 binding.
FIGURE 2.
IDR-1 binds to the ZZ domain of p62. A, schematic representation of the constructs used for mapping of the p62 domains that interact with IDR-1. SH2, an SH2 binding domain; AID, an acidic interaction domain (PB1 domain comprises the SH2 and AID); LB, LIM protein Ajuba binding domain; PEST, amino acids Pro, Glu, Ser, Thr-enriched domain; LIR, LC3-interacting region; UBA, ubiquitin-associated domain. B, analysis of IDR-1 pulldown assays with constructs presented in A. Lysates from HEK293T cells overexpressing FLAG-tagged deletion mutants of human p62 were pulled down against biocytin (B) or biotinylated IDR-1 (P). Cell lysates (L) were used as control. Proteins pulled down were immunoblotted with anti-FLAG antibody.
IDR-1 Selectively Affects p62 Complex Formation
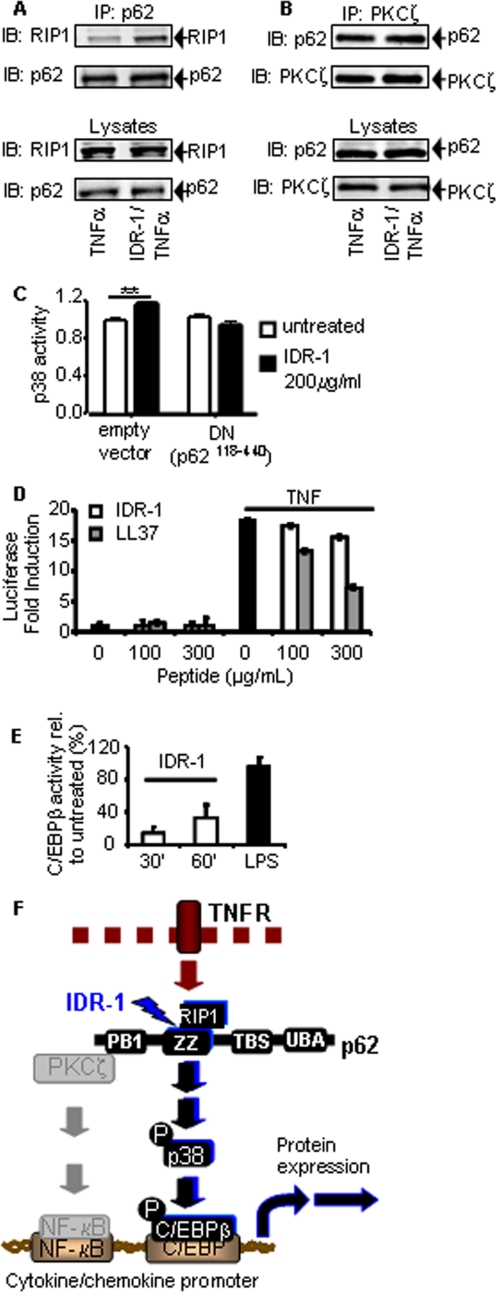
The p62 protein is a critical signaling component that functions as a scaffold by forming multiple intracellular protein signaling complexes. Because IDR-1 bound to p62 directly, we investigated the consequences of this binding on intracellular p62 protein complex formation. Sanz et al. (5, 6) previously reported that TNFα receptor ligation stimulates complex formation between RIP1 and p62 via the ZZ domain of p62. Thus, we hypothesized that the p62-RIP1 complex formation might be affected in cells treated with IDR-1. In HEK293T cells overexpressing RIP1, IDR-1 induced an increase in TNFα-stimulated complex formation between RIP1 and p62 proteins (Fig. 3A), suggesting that IDR-1 increased or stabilized this complex formation.
FIGURE 3.
IDR-1 selectively affects intracellular p62 complex formation and signaling pathways. A, IDR-1 enhances the p62 and RIP1 complex formation. HEK293T cells overexpressing RIP1 were stimulated with 100 ng/ml TNFα for 2 min with or without IDR-1 (200 μm) pretreatment for 30 min. Lysates were immunoprecipitated (IP) with anti-p62 antibody and immunoblotted (IB) with anti-RIP1 and anti-p62 antibodies. B, IDR-1 has no effect on the p62 and PKCζ complex formation. HEK293T cells were stimulated with 100 ng/ml TNFα for 2 min with or without IDR-1 (200 μm) pretreatment for 30 min. Lysates were immunoprecipitated with anti-PKCζ antibody and immunoblotted with anti-p62 and anti-PKCζ antibodies. C, IDR-1 treatment activates MAPK p38 in a p62-dependent manner. A549 cells were co-transfected with the PathDetect CHOP trans luciferase reporting system (Stratagene) for p38 activity and a p62 dominant-negative mutant (DN, p62118–440) or empty vector. 24 h after transfection, cells were untreated or treated with 200 μg/ml IDR-1, and p38 phosphorylation-dependent luciferase activity was analyzed. **, p < 0.01. The result is representative of an experiment repeated five times. Error bars represent S.D. in C–E. D, IDR-1 does not modulate NF-κB activity in a HEK293T NF-κB reporter cell line. Cells were pretreated with IDR-1 at 100 or 300 μg/ml, with or without 100 ng/ml TNFα stimulation. TNFα stimulation alone was used as control. Pretreatment with 100 or 300 μg/ml LL-37 prior to TNFα stimulation was used as a control for peptide modulation of NF-κB activity. E, treatment of A549 cells with IDR-1 induces C/EBPβ activity. Cells were treated with 200 μm IDR-1 or with 100 ng/ml lipopolysaccharide (LPS), and C/EBPβ activity was detected by enzyme-linked immunosorbent assay. F, proposed model of IDR-1 mechanism of action via p62. IDR-1 binds to the ZZ domain on the p62 protein. This event results in stabilization of the intracellular complex with RIP1. IDR-1 interaction with p62 activates the p38 MAPK pathway, which results in activation of the transcription factor C/EBPβ. In the presence of TNFα stimulation, the NF-κB pathway is activated, although IDR-1 does not change complex formation with PKCζ nor, consequently, modulate NF-κB activity. Activation of transcriptional complexes results in modulation of cytokine/chemokine production in anti-infectious and inflammatory immune responses.
p62 interacts with PKCζ via its PB1 domain (15), distinct from the IDR-1 binding ZZ domain, and has been postulated as a molecular link between the atypical PKC-dependent NF-κB activation and the TNFα-RIP1 signaling axes (5). Therefore, the effect of IDR-1 on p62-PKCζ complex formation was also investigated. IDR-1 pretreated or untreated parental HEK293T cells were stimulated with TNFα followed by PKCζ immunoprecipitation. The amount of p62 co-immunoprecipitated with PKCζ was unaffected by IDR-1 treatment (Fig. 3B), suggesting that IDR-1 does not alter the TNFα-induced complex formation between PKCζ and p62.
IDR-1 Selectively Affects Cellular Signaling Pathways
Complexes formed by p62 protein interactions regulate several intracellular signaling pathways (10, 16). Previously, independent studies have shown that p62 is involved in the p38 MAPK cascade (4) and that IDR-1-modulated cytokine production is linked to p38 (1). To investigate whether IDR-1 affects p38 MAPK signaling through p62, we utilized an approach based on a luciferase reporter of p38 activity and a dominant-negative mutant of p62 (p62118–440 cloned in pcDNA3.1) in the A549 epithelial cell line. IDR-1 treatment of these cells co-transfected with p38 activity reporter plasmids and an empty vector (pcDNA3.1) induced luciferase expression, indicating the activation of p38 (Fig. 3C). This effect was abolished in cells co-transfected with the dominant-negative mutant of p62, suggesting that p38 activation by IDR-1 is mediated by p62.
The p62 protein has been reported to play a role in NF-κB activation by serving as a scaffold for PKCζ (17). Given the central role of NF-κB in cell signaling, we evaluated the effect of IDR-1 on NF-κB activity using a HEK293T NF-κB luciferase reporter cell line. The treatment of unstimulated cells with IDR-1 did not induce NF-κB activation, in contrast to the high NF-κB activation induced by TNFα stimulation (Fig. 3D). Furthermore, pretreatment with IDR-1 did not alter the NF-κB activation by TNFα, whereas the cationic peptide LL-37 reduced NF-κB activity in a dose-dependent manner, as previously reported (18). These experiments were repeated in an A549 NF-κB luciferase reporter cell line with similar results (not shown).
Because NF-κB activity did not appear to be affected by IDR-1 treatment of cells, we investigated the IDR-1 effects on C/EBPβ given our previous findings of IDR-1 induction of this pathway (1) and the known role of p38 in the activation of the C/EBP family of transcription factors (19, 20). An induction of C/EBPβ activity at 30 and 60 min after IDR-1 treatment was observed in nuclear extracts of A549 cells assayed by C/EBPβ specific enzyme-linked immunosorbent assay (Fig. 3E). These experiments confirm earlier findings (1) and indicate that C/EBPβ activity, but not NF-κB activity, is modulated by IDR-1 treatment of cells.
DISCUSSION
The intracellular signaling network in host cells allows for efficient communication between the sensing of an incoming pathogen and the initiation of anti-infective responses. The p62 protein is emerging as one of the central signaling hubs of the cell, controlling this process (8–10, 21).
The data presented here demonstrate direct binding of IDR-1 to p62 and provide evidence that this binding has specific consequences on protein-protein interactions, as well as on downstream signaling pathways. p62 was discovered as the intracellular target of IDR-1 by an unbiased, non-hypothesis-driven proteomics approach. Due to high affinity of IDR-1 for p62, it is likely that p62 is the most physiologically relevant target. Although we identified XIAP-4 in the SILAC experiments, strong direct binding of IDR-1 to XIAP was not detected in human cell lysates, nor was binding to XIAP detected with other related IDR peptides that did bind p62. Another IDR-1 weak binding partner, glyceraldehyde 3-phosphate dehydrogenase, was also recently described (22).
Human p62 was confirmed as the IDR-1 target using binding assays and pulldown experiments from both endogenous and p62-overexpressing cell lysates. IDR-1 binding to both human and mouse p62, which share 99% amino acid sequence identity, was demonstrated. Through the investigation of p62 deletion mutants, we discovered that IDR-1 binds to the ZZ domain of p62, a zinc-binding protein interaction domain. Most compounds that act on protein-protein interactions antagonize or inhibit complex formation, but IDR-1 addition to cells led to a surprisingly specific effect; the TNFα-dependent receptor signaling complex of RIP1 and p62, which occurs at the ZZ domain, was stabilized, whereas the p62-PKCζ complex, which occurs at the PB1 domain proximal to the RIP1 binding region, was not affected.
The specific modulation of p62 signaling complexes by IDR-1 appears to have a corresponding effect on the downstream signaling. On one hand, p62 has been described as an activator of NF-κB due to its receptor-induced signaling complex formation with PKCζ (11, 12), but IDR-1 treatment did not modulate NF-κB activity downstream of TNFα, consistently with the p62-PKCζ complex formation being unaffected. On the other hand, p38 MAPK activation downstream of TNFα is regulated by RIP1 (23), and the increase of RIP1-p62 interaction by IDR-1 may explain the previously reported role of IDR-1 in p38-mediated signaling pathways (1). This is further strengthened by the observation that IDR-1-mediated activation of p38 is dependent on p62. The IDR-1-induced activation of C/EBPβ, downstream of p38, is also consistent with these and previous findings. Thus, we propose that the specific effects on intracellular signaling presented here contribute to the previously described IDR-1-modulated cytokine profile (Fig. 3F).
IDR-1 has been demonstrated to prevent and treat infections and reduce associated inflammation (1). In addition, recent studies report that p62 expression contributes to regulating macrophage-mediated (21) and cancer-associated inflammation (11, 16, 24), raising the question as to whether IDR-1 might also affect inflammatory responses in the absence of pathogen stimulation. The discovery that p62 is the target of IDR-1 highlights the importance of p62 in innate immunity and reveals p62 as a potential therapeutic target for anti-infective therapy without induction of harmful inflammation, and conversely, potential anti-inflammatory therapy with added anti-infective function.
Acknowledgment
We gratefully acknowledge the technical expertise of Mira Jovanovic.
This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) and the Foundation for the National Institute of Health through the Grand Challenges in Global Health Initiative (to B. B. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures.”
- IDR
- innate defense regulator
- C/EBP
- CCAAT-enhancer-binding protein
- NF-κB
- nuclear factor κ-light-chain-enhancer of activated B cells
- PKC
- protein kinase C
- SILAC
- stable isotope labeling of amino acids in cell culture
- TRAF6
- tumor necrosis factor receptor-associated factor 6
- RIP1
- receptor-interacting protein 1
- XIAP-4
- X-linked inhibitor-of-apoptosis proteins
- ZZ
- ZZ-type zinc finger
- MAPK
- mitogen-activated protein kinase
- TNF
- tumor necrosis factor
- Sp
- specific
- nSP
- nonspecific.
REFERENCES
- 1.Scott M. G., Dullaghan E., Mookherjee N., Glavas N., Waldbrook M., Thompson A., Wang A., Lee K., Doria S., Hamill P., Yu J. J., Li Y., Donini O., Guarna M. M., Finlay B. B., North J. R., Hancock R. E. (2007) Nat. Biotechnol. 25, 465–472 [DOI] [PubMed] [Google Scholar]
- 2.Moscat J., Diaz-Meco M. T., Albert A., Campuzano S. (2006) Mol. Cell 23, 631–640 [DOI] [PubMed] [Google Scholar]
- 3.Puls A., Schmidt S., Grawe F., Stabel S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6191–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai K., Saito A., Sudo T., Osada H. (2008) J. Biochem. 143, 765–772 [DOI] [PubMed] [Google Scholar]
- 5.Sanz L., Sanchez P., Lallena M. J., Diaz-Meco M. T., Moscat J. (1999) EMBO J. 18, 3044–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vadlamudi R. K., Joung I., Strominger J. L., Shin J. (1996) J. Biol. Chem. 271, 20235–20237 [DOI] [PubMed] [Google Scholar]
- 8.Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 9.Seibenhener M. L., Geetha T., Wooten M. W. (2007) FEBS Lett. 581, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 11.Duran A., Linares J. F., Galvez A. S., Wikenheiser K., Flores J. M., Diaz-Meco M. T., Moscat J. (2008) Cancer Cell 13, 343–354 [DOI] [PubMed] [Google Scholar]
- 12.Durán A., Serrano M., Leitges M., Flores J. M., Picard S., Brown J. P., Moscat J., Diaz-Meco M. T. (2004) Dev. Cell 6, 303–309 [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez A., Durán A., Selloum M., Champy M. F., Diez-Guerra F. J., Flores J. M., Serrano M., Auwerx J., Diaz-Meco M. T., Moscat J. (2006) Cell Metab. 3, 211–222 [DOI] [PubMed] [Google Scholar]
- 14.Rogers L. D., Foster L. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18520–18525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moscat J., Diaz-Meco M. T. (2000) EMBO Rep. 1, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez P., De Carcer G., Sandoval I. V., Moscat J., Diaz-Meco M. T. (1998) Mol. Cell. Biol. 18, 3069–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mookherjee N., Brown K. L., Bowdish D. M., Doria S., Falsafi R., Hokamp K., Roche F. M., Mu R., Doho G. H., Pistolic J., Powers J. P., Bryan J., Brinkman F. S., Hancock R. E. (2006) J. Immunol. 176, 2455–2464 [DOI] [PubMed] [Google Scholar]
- 19.Csóka B., Németh Z. H., Virág L., Gergely P., Leibovich S. J., Pacher P., Sun C. X., Blackburn M. R., Vizi E. S., Deitch E. A., Haskó G. (2007) Blood 110, 2685–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poli V. (1998) J. Biol. Chem. 273, 29279–29282 [DOI] [PubMed] [Google Scholar]
- 21.Kim J. Y., Ozato K. (2009) J. Immunol. 182, 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mookherjee N., Lippert D. N., Hamill P., Falsafi R., Nijnik A., Kindrachuk J., Pistolic J., Gardy J., Miri P., Naseer M., Foster L. J., Hancock R. E. (2009) J. Immunol. 183, 2688–2696 [DOI] [PubMed] [Google Scholar]
- 23.Lee T. H., Huang Q., Oikemus S., Shank J., Ventura J. J., Cusson N., Vaillancourt R. R., Su B., Davis R. J., Kelliher M. A. (2003) Mol. Cell. Biol. 23, 8377–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiruma Y., Honjo T., Jelinek D. F., Windle J. J., Shin J., Roodman G. D., Kurihara N. (2009) Blood 113, 4894–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]