Abstract
Ubiquitin ligases, together with their cognate ubiquitin-conjugating enzymes, are responsible for the ubiquitylation of proteins, a process that regulates a myriad of eukaryotic cellular functions. The first cullin-RING ligase discovered, yeast SCFCdc4, functions with the conjugating enzyme Cdc34 to regulate the cell cycle. Cdc34 orthologs are notable for their highly acidic C-terminal extension. Here we confirm that the Cdc34 acidic C-terminal tail has a role in Cdc34 binding to SCFCdc4 and makes a major contribution to the submicromolar Km of Cdc34 for SCFCdc4. Moreover, we demonstrate that a key functional property of the tail is its acidity. Our analysis also uncovers an unexpected new function for the acidic tail in promoting catalysis. We demonstrate that SCF is functional when Cdc34 is fused to the C terminus of Cul1 and that this fusion retains partial function even when the acidic tail has been deleted. The Cdc34-SCF fusion proteins that lack the acidic tail must interact in a fundamentally different manner than unfused SCF and wild type Cdc34, demonstrating that distinct mechanisms of E2 recruitment to E3, as is seen in nature, can sustain substrate ubiquitylation. Finally, a search of the yeast proteome uncovered scores of proteins containing highly acidic stretches of amino acids, hinting that electrostatic interactions may be a common mechanism for facilitating protein assembly.
Introduction
Regulation of protein degradation by the ubiquitin proteasome system is critical for cellular homeostasis (1). Ubiquitylation is governed by a transfer cascade involving three enzyme families (2). The E12 enzyme activates ubiquitin and transfers it to a ubiquitin-conjugating enzyme (E2). The ubiquitin-charged E2s engage ubiquitin ligases (E3) that recognize and bind to protein substrates targeted for ubiquitylation. This results in the transfer of ubiquitin from the E2 to substrate, either directly or via a covalent E3-ubiquitin intermediate. Subsequent ubiquitins can be attached to the ubiquitin conjugated to substrate to initiate formation of an ubiquitin chain. Transfer of at least four ubiquitins in the form of a polyubiquitin chain constitutes a signal for recognition by proteasomes and ensuing degradation (3).
Thanks to the advances of genome sequencing and powerful algorithms for comparing distantly related protein sequences, it is now understood that there are perhaps as many as 650 distinct ubiquitin ligases encoded by the human genome (4), a seemingly daunting statistic for those who wish to understand molecular mechanisms of ubiquitylation. However, up to 350 of these ligases may belong to a single family called cullin-RING-ligases (CRLs) (4), such that mechanistic insight gained by the analysis of one member of the CRL family may serve as a template for understanding up to half of all E3s. Moreover, mutations in or the misregulation of several CRL subunits have been shown to promote tumorigenesis in numerous forms of cancer (5, 6), and thus a mechanistic understanding of how these ligases function may aid in the process of drug discovery.
The founding member of the CRLs is the yeast ubiquitin ligase SCFCdc4, named after the Skp1, Cul1 (Cdc53 in yeast), and F-box proteins that form this multi-subunit enzyme complex (7, 8). F-box proteins specifically recognize protein substrates that are to be ubiquitylated by SCF. For instance, the F-box protein Cdc4 binds to the SCFCdc4 substrate Sic1, a cyclin-CDK inhibitor and key regulator of the cell cycle in yeast. A fourth subunit of SCF, Hrt1/Rbx1/Roc1, belongs to the RING family of proteins and binds to E2s (9–13). The C-terminal domain of Cdc53 binds Hrt1, whereas the N-terminal end of Cdc53 binds to Skp1, which binds to an array of different F-box proteins such as Cdc4. The x-ray structure of the cullin protein reveals an elongated, platform-like shape, such that the cullin acts as a scaffold that brings the F-box-substrate complex and the E2 into proximity (14). There are several excellent reviews that detail structural, functional, and biological aspects of the CRLs (15–17).
The E2-conjugating enzyme for SCFCdc4 is a protein named Cdc34 (18). Although all of the E2s share a common catalytic domain that has a conserved sequence and structure, some of them have additional domains appended to either their N or C termini. In budding yeast, there are three E2s that have unique C-terminal extensions that are remarkable for the abundance of acidic residues in the amino acid sequence: Cdc34, Ubc8, and Rad6. However, the acidic tail of Cdc34 is much longer than the extension on Rad6 and is far more conserved throughout eukaryotic evolution than is the tail on Ubc8. For these reasons, Cdc34 stands alone in the E2 family for its unique acidic tail (supplemental Fig. S1).
Mutants lacking all or part of the acidic tail have profound phenotypes by biochemical and genetic analysis. For instance, deletion of the acidic tail is lethal in yeast (19, 20). Interestingly, a Cdc34 null mutant can be rescued by grafting the acidic tail onto the E2 Rad6, suggesting that the function of the tail is portable (19, 20). GST pulldown assays with both wild type and tail-truncated yeast or human Cdc34 demonstrated that the acidic tail is likely involved in binding to SCF (21, 22). However, a different study found no defect in SCF binding for a tail-truncated human Cdc34 mutant (23), but nevertheless tail deletion mutants of human Cdc34 have been reported to be extremely defective in an in vitro ubiquitylation assay (22).
SCFCdc4 was the first CRL to be fully reconstituted in an in vitro ubiquitylation system, and SCF ligases remain the best understood members of the CRL family. Nevertheless, we still have only rudimentary insight into how the essential acidic tail of Cdc34 enables ubiquitylation of SCF substrates. To address this gap in our knowledge, we have taken an enzymological approach to understand the role of the acidic tail in Cdc34-SCF function. Our work extends previous observations that the Cdc34 tail is important for SCF binding and establishes that the acidity of the tail is its key functional attribute. We also provide evidence that the tail promotes ubiquitin transfer within the substrate-SCF-Cdc34·Ub Michaelis complex. Finally, we show that the acidic tail is not the only means by which Cdc34 can be productively tethered to SCF, a finding that has important implications for understanding the evolution of ubiquitylation mechanisms. Our results illustrate how kinetic analysis can provide important insights into the function of E2-E3 complexes.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Cdc34
All of the Cdc34 coding sequences were amplified by PCR to encode a six-histidine tag at the C terminus and expressed in Escherichia coli cells. Bacterial cell pellets were sonicated in 30 mm Hepes, pH 7.5, 250 mm NaCl, 0.1% detergent (Igepal CA-630 or Triton X-100), 10% glycerol, and proteasome inhibitor mixture (Roche Applied Science), and lysates were cleared by centrifugation. Cleared lysates were incubated with nickel-nitrilotriacetic acid beads (Qiagen) for 1 h at 4 °C, and afterward the beads were briefly washed with lysis buffer. The beads were loaded into a disposable column, which was then washed with at least 10 column volumes of lysis buffer. His-tagged proteins were eluted in a buffer containing 50 mm Hepes, pH 7.5, 100 mm NaCl, and 500 mm imidazole. This sample was then concentrated and loaded onto a Superdex 75 gel filtration column that had been equilibrated with 30 mm Tris, pH 7.5, 100 mm NaCl, 1 mm dithiothreitol, and 10% glycerol (storage buffer). Fractions containing Cdc34 were pooled, concentrated, and then aliquoted before storage at −80 °C.
Note that yeast Cdc34-Δ271–295 (RDB929), which lacks the distal 25 residues from the tail (for simplicity, we now refer to all Cdc34 truncation proteins by the final C-terminal position (i.e. Cdc34-Δ270)), has been shown to have activity comparable with full-length yeast Cdc34 (24) (in the prior work Cdc34-Δ270 was referred to as Cdc34-ΔC). We initially chose to work with Cdc34-Δ270 because it is easier to purify and work with than the full-length protein. Cdc34-Δ230 (RDB2322), which was used as the starting point for analysis of a series of progressive tail truncations, has nearly the same activity as Cdc34-Δ270 (supplemental Fig. S2). Other yeast Cdc34 truncation proteins used in this study include: Cdc34-Δ225 (RDB2340), Cdc34-Δ220 (RDB2341), Cdc34-Δ210 (RDB2323), Cdc34-Δ205 (RDB2342), and Cdc34-Δ190 (RDB2343).
GST Fusion Proteins
Sequences encoding yeast acidic tail residues 171–270 were fused downstream of the GST coding region to generate RDB2344 (GST-YACT). GST fusion to the human acidic tail sequence comprised residues 175–238 (RDB2345) (GST-HACT). These proteins were expressed in and purified from E. coli cells. Briefly, bacterial cell pellets were sonicated in lysis buffer containing 30 mm Tris, pH 7.5, 250 mm NaCl, 5 mm dithiothreitol, 10% glycerol, and protease inhibitor mixture. Clarified lysates were incubated with glutathione-Sepharose 4B (GE Healthcare) resin for 2 h. The resin was washed with lysis buffer, followed by elution with 20 mm Tris, pH 8.0, 100 mm NaCl, and 10 mm glutathione. The purified protein was then equilibrated into storage buffer by gel filtration and then aliquoted for long term storage at −80 °C.
Human Cul1-Rbx1
Recombinant Cul1-Rbx1 was purified as previously described (25). Briefly, we used the split-n-co-express method for expression in E. coli, where the Cul1 subunit is divided and expressed as two fragments, the N-terminal domain (NTD) and C-terminal domain (CTD) (26). The NTD is expressed from a plasmid with chloramphenicol resistance (RDB2080), and the CTD is expressed along with GST-Rbx1 from a different plasmid with ampicillin resistance (RDB2081). These two plasmids were co-transformed into BL21 (DE3) bacterial cells. Transformants were cultured to an OD of ∼1.0, and expression was induced with isopropyl β-d-thiogalactopyranoside. Induction was carried out at 16 °C overnight. Bacterial pellets were lysed, and the lysates were clarified by centrifugation and incubated with glutathione-Sepharose 4B (GE Healthcare) resin for 2 h. The resin was washed with lysis buffer, followed by elution with 20 mm Tris, pH 8.0, 100 mm NaCl, and 10 mm glutathione. Thrombin was added to the eluate, and the proteolysis reaction was incubated at 4 °C overnight. This mixture was then loaded onto a HiTrap S HP ion exchange column and eluted by salt gradient (note that GST does not bind to this column). Fractions containing Cul1-Rbx1 were concentrated and loaded onto a Superdex-200 gel filtration column equilibrated in storage buffer. Fractions with Cul1-Rbx1 were concentrated, and the purified protein was frozen at −80 °C for long term storage.
Cul1-Cdc34 Fusions
Cul1-Cdc34 fusion constructs were prepared using an E. coli expression system similar to the one described above. Sequences used for fusion assembly were derived from the human proteins. Wild type Cdc34 and Cdc34-Δ190 were fused to the C-terminal end of the CTD sequence using the amino acid sequence KGTREGKGSPEGKGTR as a linker between CTD and Cdc34. CTD-wild type and CTD-Cdc34-Δ190 were cloned into plasmids with chloramphenicol resistance (RDB2346 and RDB2347, respectively), whereas NTD and GST-Rbx1 were cloned into a plasmid with ampicillin resistance (RDB2348). Either the CTD-Cdc34 or the CTD-Cdc34-Δ190 plasmid was co-transformed with the plasmid encoding NTD and GST-Rbx1 into BL21(DE3) cells. The resulting transformants were used for protein expression. The fusion proteins were purified in an identical manner as for the CTD-NTD-Rbx1 complex, with the exception that anion exchange using a HiTrap Q HP column (GE Healthcare) was used for the fusion constructs. Purified proteins were equilibrated into storage buffer by gel filtration and frozen at −80 °C for long term storage. Note that the activities of the fusion proteins, as well as the unfused human Cdc34 proteins, were negatively affected by multiple freeze-thaw cycles as well as long term storage (more than 1 month) at concentrations higher than 100 μm.
G1-CDK, SCF, and Sic1 Expression and Purification
G1-CDK and yeast SCF were expressed in insect cells by baculovirus infection as previously described (27). Wild type Sic1 (RDB1706) was expressed in E. coli and purified using nickel-nitrilotriacetic acid affinity chromatography. Stocks of purified 100 μm wild type Sic1 were equilibrated into storage buffer using a PD-10 column (GE Healthcare) and frozen at −80 °C.
Phosphorylation of Sic1
Phosphorylation of Sic1 was performed in a similar manner as was previously described (27), with the exception that the final concentration of 32P labeled wild type Sic1 was ∼12 μm.
Phosphorylation of K48R Ubiquitin and Purification of D77 Ubiquitin
K48R ubiquitin was purified from E. coli expressing K48R ubiquitin coding sequences fused to GST coding sequences (RDB1882). The fusion protein contained a protein kinase A phosphorylation site immediately upstream of the ubiquitin N terminus and downstream of a TEV protease recognition site. Phosphorylation was performed in the presence of [γ-32P]ATP and protein kinase A for 1 h at 30 °C. Unincorporated nucleotides were removed by desalting with a G25 column (GE Healthcare). Afterward, TEV protease and 0.5 mm dithiothreitol were added, and the sample was incubated for 2 h at 30 °C, followed by heating at 65 °C for 15 min to inactivate TEV protease. The reaction was then cooled on ice for 5 min, followed by centrifugation at maximum speed for 15 min in an Eppendorf 5415D bench top centrifuge (this step pellets the denatured GST protein). The supernatant containing labeled K48R ubiquitin was collected and frozen at −20 °C. D77 ubiquitin (RDB2226) was purified by virtue of its histidine tag at the N terminus as previously described (25).
Ubiquitylation Reactions
All of the ubiquitylation reactions were carried out in 30 mm Tris, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 2 mm ATP, and 2 mm dithiothreitol (see figure legends for the time of reaction and the concentrations of the reaction components). Sic1 ubiquitylation reactions were quenched with reducing SDS-PAGE buffer and separated on 10% Tris-glycine SDS-PAGE gels. The gels were dried and exposed to a phosphor screen for analysis. Quantification was performed with ImageQuant (GE Healthcare) and plotted using Prism. The Km of Cdc34 for SCF reported in this study was substantially lower than that reported in a prior publication from this laboratory (25). This discrepancy is due to differing salt concentrations in the reactions. We have recently discovered that binding of Cdc34 to SCF (and thus the Km of Cdc34 for SCF) was exquisitely sensitive to salt (45). In addition, the maximal rates of Sic1 ubiquitylation reported here (2 min−1) are substantially slower than what we reported previously (12 min−1) (24). This is due to inadvertent overestimation of Sic1 concentration in the prior study.
Analysis of the Yeast Proteome
The yeast proteome was scanned for protein sequences with acidic stretches using the following algorithm. A defined length of residues (between 15 and 25) was used as a sliding window over each protein sequence in the yeast proteome. If an acidic stretch of residues (defined as containing aspartate or glutamate residues in greater than 60, 70, or 80% of the positions within the window) was encountered at least once during this process, this protein is counted once, and the next sequence was analyzed. Therefore, the total possible number of hits can be no greater than the number of protein sequences in the data base (5,862 protein sequences).
To determine the statistical significance of the results, the sequence of each protein was randomized and then analyzed for the presence of acidic stretches using identical criteria as above. This process was repeated 10 times for each sequence in the proteome. This was used to estimate the expected value of acidic stretches for each protein sequence where the expectation is the average number of hits for all 10 randomized sequences. Therefore, the expectation for each sequence will have a range between 0 and 1. The expected value of the number of proteins from the proteome is the total sum of the individual expectations. The entire process was repeated three times. These estimates were then used to calculate the means and standard deviations of the expected values (supplemental Table S1). The proteomic search was done using custom PERL software.
RESULTS
Deletion of the acidic tail of human Cdc34 results in a severe loss of Cdc34-SCF activity in an “autoubiquitylation” assay (22). This loss of activity can be explained by one of three possible hypotheses. First, deletion of the tail may affect binding of Cdc34 to SCF. Second, tail deletion may result in a loss of catalytic activity of the Cdc34-SCF complex. Finally, the acidic tail of Cdc34 may confer both binding and catalytic properties to the conjugating enzyme. Prior pulldown studies in yeast have pointed to a role for the acidic tail in binding to SCF (21, 22), and this conclusion was supported by a biochemical study of human Cdc34 that utilized autoubiquitylation as a functional read-out (22). However, a subsequent analysis of p27 ubiquitylation by human Cdc34 concluded that the acidic tail is dispensable for binding to SCF and instead appears to play a catalytic function (23). To deconvolve the functions of the acidic tail, we sought to measure its quantitative contribution to the kinetic parameters of ubiquitylation with an authentic substrate.
We began our investigation by comparing the activities of yeast Cdc34-Δ270 and Cdc34-Δ190. Cdc34-Δ270 lacks the distal 25 amino acids of the tail but retains multiple highly acidic stretches, is fully active in vitro (24), and supplies CDC34 function in vivo. Cdc34-Δ190 lacks ∼85% of the tail (which starts approximately at residue 171), including the major acidic stretches, and is unable to supply Cdc34 function in vivo. The assay we employed was a steady-state di-ubiquitin synthesis assay (28), a key advantage of which is that it can be used to assess both SCF-independent and SCF-dependent activities for Cdc34. Radiolabeled K48R ubiquitin was added to reactions containing E1, Cdc34, and ATP in either the presence or the absence of SCF. Because Cdc34 forms Lys48 ubiquitin chains with high specificity, K48R ubiquitin can form thioesters with Cdc34 but will not attack Cdc34·ubiquitin thioesters. Once Cdc34 was charged fully with the K48R ubiquitin, the reactions were initiated by the addition of unlabeled D77 ubiquitin, which cannot form thioesters with E2 because the terminal glycine residue is blocked but is nevertheless able to attack Cdc34·ubiquitin thioesters because Lys48 remains intact. The product appears in the form of radiolabeled D77-K48R di-ubiquitin.
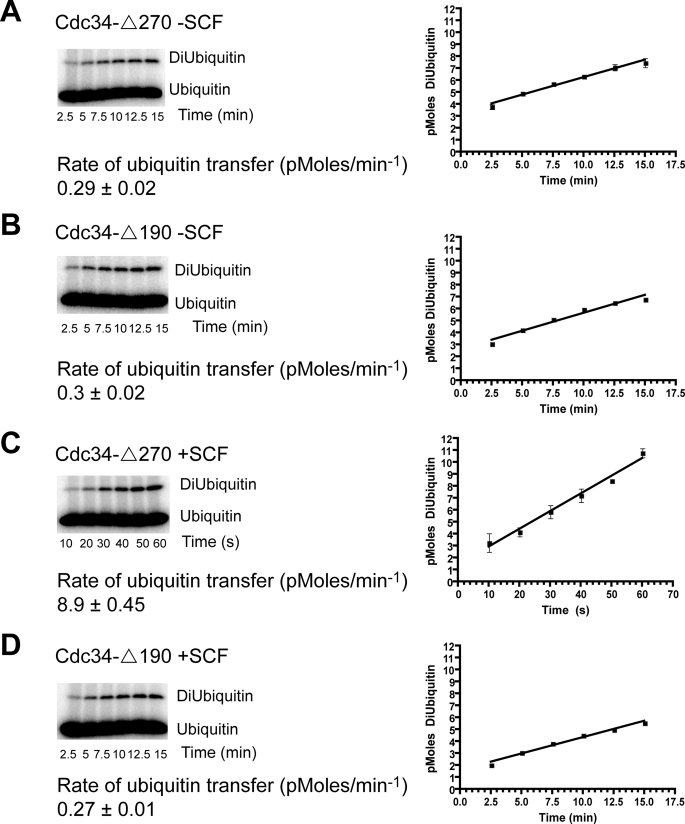
Using a concentration of both Cdc34-Δ270 and Cdc34-Δ190 (2 μm) that was sufficient for Δ270 to saturate SCF, we measured the rate of di-ubiquitin synthesis in both the presence and the absence of SCFCdc4 (for simplicity, we will refer to SCFCdc4 hereafter as SCF unless noted otherwise). Both Cdc34-Δ270 and Cdc34-Δ190 formed di-ubiquitin with nearly identical rates (Fig. 1, A and B). The addition of SCF stimulated Cdc34-Δ270 activity by ∼30-fold (Fig. 1C). In stark contrast, the rate of di-ubiquitin formed by Cdc34-Δ190 was not enhanced at all by SCF (Fig. 1D). These results demonstrate that although deletion of the acidic tail had no effect on the SCF-independent catalytic activity of Cdc34, it completely vitiated the SCF-enhanced boost in activity that was observed for Cdc34-Δ270.
FIGURE 1.
Yeast Cdc34-Δ190 is functional but cannot be activated by SCF. Di-ubiquitin synthesis assays containing 250 nm E1, 2 μm Cdc34-Δ270 or Cdc34-Δ190, 6 μm 32P-labeled K48R ubiquitin, and 50 μm D77 ubiquitin were performed for the specified times both in the absence and the presence of 100 nm SCF. A, Cdc34-Δ270; B, Cdc34-Δ190; C, Cdc34-Δ270 with SCF; D, Cdc34-Δ190 with SCF. Notice that Cdc34-Δ270 and Cdc34-Δ190 form product at similar rates in the absence of SCF, demonstrating that tail deletion does not disturb the structure and catalytic function of Cdc34-Δ190. Each graphical data point represents the mean of triplicate data values from three independent experiments, and the error bars are the standard deviations.
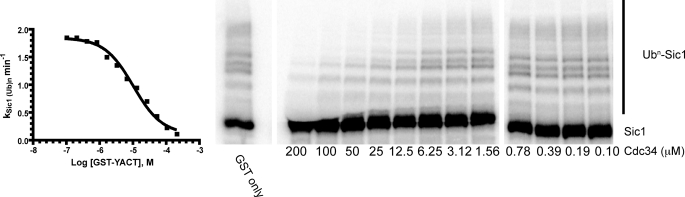
Prior work has demonstrated that the C-terminal tail of Cdc34 is both necessary and sufficient for binding SCF in a pulldown assay (21). We next sought to test whether this binding is sufficient to inhibit SCF activity. Titration of GST yeast tail into Sic1 ubiquitylation reactions resulted in the nearly complete inhibition of product formation at 200 μm GST-tail (Fig. 2) with an IC50 of 16 ± 6 μm. Note that inhibition was not an artifact of the high concentration of protein because 200 μm GST had no obvious effect on Sic1 ubiquitylation. This result is consistent with the observation that the acidic tail is important for Cdc34 function (21) and furthermore indicates that the function of the tail cannot be supplied in trans.
FIGURE 2.
The acidic tail inhibits Sic1 ubiquitylation by Cdc34-SCF. Reactions containing 1.6 μm yeast E1, 100 nm yeast SCF, 1 μm 32P-labeled Sic1, 2 μm yeast Cdc34-Δ270, and 150 μm ubiquitin were incubated for 1 min at 20–22 °C along with various concentrations of GST-tail (residues 171–270; GST-YACT) or GST prior to quenching with reducing SDS-PAGE buffer. Substrate conversion to product was quantified and plotted as the rate of Sic1 ubiquitylation versus the log of the GST fusion concentration. An IC50 of 16 ± 6 μm was estimated from nonlinear curve fitting the data to a sigmoidal dose-response curve with a fixed slope of 1 (Prism).
Kinetic Analysis of Cdc34 Tail Truncations Reveals Defects in Both Binding to and Catalysis with SCF
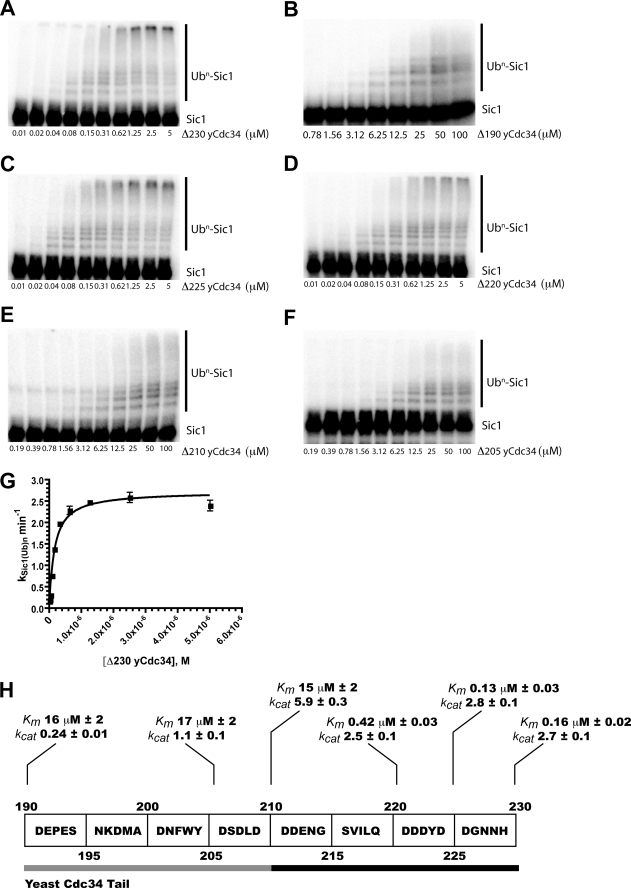
Our next goal was to characterize Cdc34-Δ270 and Cdc34-Δ190 by measuring the Michaelis-Menten kinetics for these E2s. To accomplish this, E2 was titrated into multi-turnover Sic1 ubiquitylation reactions. The initial rate of Sic1 ubiquitylation (defined as molecules of substrate modified with one or more ubiquitins as a function of time) was plotted against Cdc34 concentration, and data were fit to the Michaelis-Menten equation. If Cdc34 is in rapid equilibrium with SCF relative to the rate of ubiquitin transfer, the Km may be interpreted as a pseudo equilibrium binding constant between E2 and enzyme. Because numerous ubiquitin molecules may be transferred to substrate during the time course of a reaction, it is important to note that kcat, the maximum rate of Sic1 ubiquitylation, is a function of all the first order rate constants, including the ubiquitin transfer rate and product dissociation, and cannot be simply interpreted as the rate of the chemical step. However, it seems very unlikely that tail truncation would affect the dissociation of ubiquitylated product from SCF, and thus any reduction in kcat (as well as a reduction in the number of ubiquitins attached per substrate when E2 is saturating SCF) is likely to be indicative of a defect in catalysis.
We first measured Km and kcat for Cdc34-Δ230, which has nearly the same activity for Sic1 ubiquitylation as Cdc34-Δ270 (supplemental Fig. S2). Cdc34-Δ230 had a Km of ∼160 nm and a kcat of ∼3 min−1 (Fig. 3, A and G). Saturating concentrations of Cdc34-Δ230 resulted in highly ubiquitylated products that barely migrated into the SDS-PAGE gel. To complement the Km determination, we also measured the intracellular concentration of Cdc34 by a combination of quantitative immunoblotting and quantitative fluorescence (supplemental Fig. S3). Cdc34 was present at ∼10 μm in the nucleus and 2.9 μm in the cytoplasm. It should be noted that Km was measured at 50 mm NaCl, and interaction of human Cdc34 with human SCF is sensitive to salt (45). Nevertheless, it would appear that a significant fraction of SCF is bound to Cdc34 in vivo and in the nucleus is likely to be saturated.
FIGURE 3.
Progressive deletion of the Cdc34 acidic tail results in defects in both Cdc34 binding to SCF and catalysis. All of the reactions contained 1.6 μm yeast E1, 100 nm yeast SCF, 1.2 μm 32P-labeled Sic1, 300 μm ubiquitin, and Cdc34 (2-fold serial dilutions starting at 5 or 100 μm). A, various concentrations (see figure) of Cdc34-Δ230 were incubated with the above reactants at 20–22 °C for 1 min and quenched by SDS-PAGE buffer. B, same as above, except Cdc34-Δ190 was the E2, and reactions were incubated for 30 min prior to quenching. C, Cdc34-Δ225. The incubation period was 1 min. D, Cdc34-Δ220. The incubation period was 1 min. E, Cdc34-Δ210. The incubation period was 1 min. F, Cdc34-Δ205. The incubation period was 2 min. G, graph plotting the rate of Sic1 ubiquitylation against the concentration of Cdc34-Δ230. The data were fit to the Michaelis-Menten equation using nonlinear curve fitting (Prism). Each graphical data point represents the mean of duplicate data values from two independent experiments, and the error bars are the standard deviations. H, summary of the results from Fig. 3. Km and kcat values are given for each Cdc34 construct. The units for kcat are min−1. The black bar encompasses residues from the distal tail, and the gray bar encompasses residues from the proximal tail.
By contrast with the efficient ubiquitylation of Sic1 by Cdc34-Δ230, titration of Cdc34-Δ190 into Sic1 ubiquitylation reactions yielded dramatically different kinetic parameters. The Km for Cdc34-Δ190 (16 μm) was ∼100-fold greater, which together with the GST tail inhibition experiment solidifies the notion that the acidic tail of Cdc34 is an important determinant of functional E2 binding to SCF (Fig. 3, B and H, and supplemental Fig. S4). We note that Km for Cdc34-Δ190 and the IC50 for the GST tail were nearly identical, suggesting that the catalytic domain and the acidic tail of Cdc34 alone have comparable weak affinities for SCF. Direct binding measurements confirmed the kinetic observations reported here that the acidic tail stabilized association of the catalytic domain with SCF (45).
In addition to the increase in Km, we also observed that the kcat for Cdc34-Δ190 was ∼10-fold less than kcat for Cdc34-Δ230. Even when Cdc34-Δ190 was close to saturating SCF, chain elongation on Sic1 was substantially attenuated (Fig. 3B). For example, notice that unlike Cdc34-Δ230, there were no high molecular weight products formed with Cdc34-Δ190 that migrated at the top of the gel. Thus, in addition to promoting binding to SCF, the acidic tail also promotes ubiquitin transfer within the Cdc34·Ub-SCF-substrate complex.
To dissect more finely the different activities of the acidic tail, we created a truncation series in which 5 or 10 residue blocks were progressively deleted from the C terminus of the Cdc34-Δ230 sequence. The first mutant in the truncation series, Cdc34-Δ225, resulted in an activity that was indistinguishable from Cdc34-Δ230 (Fig. 3, C and H, and supplemental Fig. S4). The next mutant, Cdc34-Δ220, exhibited a modest 3-fold increase in Km but no change in either kcat or ubiquitin chain elongation on substrate (Fig. 3, D and H, and supplemental Fig. S4). Deletion of an additional 10 residues created a mutant (Cdc34-Δ210) that was severely defective in Km (Fig. 3, E and H, and supplemental Fig. S4). The Km for Cdc34-Δ210 was comparable with the Km for Cdc34-Δ190, indicating that residues in between amino acids 211 and 220, in the sequence DDENGSVILQ, can mediate binding of Cdc34 to SCF. In addition to the Km defect, there was a noticeable decrease in ubiquitin chain elongation. Finally, Cdc34-Δ205 exhibited a Km similar to Cdc34-Δ210, but unlike Cdc34-Δ210, Cdc34-Δ205 showed a decrease in kcat in addition to a noticeable defect in ubiquitin chain elongation (Fig. 3, F and H, and supplemental Fig. S4). In conclusion, we found that residues 211–220 in the Cdc34 acidic tail contribute to binding to SCF, whereas the more proximal segment 191–210 appears to affect catalysis.
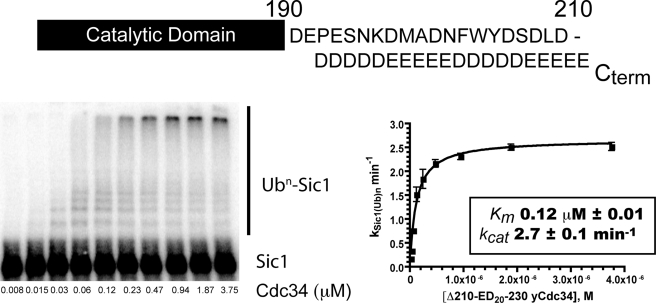
Although it is clear that the tail is essential for Cdc34 function and interaction with SCF, it is largely unknown exactly what property of the tail comprises its essential function. Our fine scale deletion mapping data cast doubt on the assumption that the acidity of the tail is its critical feature, because the sequence in the region spanning amino acids 211–220 (DDENGSVILQ) contains only three acidic residues, but its deletion increases Km by 36-fold. Meanwhile, both the segments immediately upstream (residues 201–210) and downstream (residues 221–230) are more enriched for Glu and Asp, but deletion of either segment has only a modest effect on Km. To resolve this apparent discrepancy, we sought to address directly the issue of whether the acidity of the tail is in fact relevant to its biological function by replacing the entire stretch of residues from 211 to 230 with acidic amino acids. Remarkably, this mutant, Cdc34-Δ210ED-230, was capable of ubiquitylating Sic1 with essentially identical kinetics as compared with Cdc34-Δ230 (Fig. 4). In addition to the highly similar values of Km and kcat for the two proteins, chain elongation on Sic1 was also very similar. This result provides compelling evidence that electrostatic interactions involving tail residues C-terminal to residue Asp210 play a key role in mediating contacts between Cdc34 and SCF.
FIGURE 4.
The distal segment of the acidic tail can be replaced entirely by poly(Glu-Asp). Reactions containing 1.6 μm yeast E1, 100 nm yeast SCF, 1.2 μm 32P-labeled Sic1, 300 μm ubiquitin, and a 2-fold linearly decreasing titration of Cdc34-Δ210ED-230 were incubated for 1 min and quenched by SDS-PAGE buffer. Each data point was measured in duplicate. The kinetic parameters obtained should be compared with those reported in Fig. 3 for Cdc34-Δ230 and Cdc34-Δ210.
Fusion of Cdc34 to Cul1 Partially Rescues Cdc34-Δ190 Activity
If a key function of the acidic tail is indeed to help tether Cdc34 to SCF, then it should be possible to at least partially compensate for deletion of the tail by directly fusing Cdc34 to SCF. To test this hypothesis, we sought to construct fusions between human Cul1 and human Cdc34. The rationale for using the human proteins for this experiment is 2-fold. First, there are crystal structures for human Cul1 and Cdc34 but not the yeast proteins. Second, the human Cul1 can be expressed at high levels in E. coli as two separate fragments, which would facilitate the expression and analysis of chimeric constructs. It is important to note that the acidic tail of human Cdc34 is important for its function in SCF-dependent substrate ubiquitylation (22). Moreover, we confirmed that the tail domain of human Cdc34, like that of yeast Cdc34 (21) bound Cul1-Rbx1 in a GST pulldown assay (supplemental Fig. S5).
To construct an SCF-Cdc34 chimera, we settled on a design in which the N terminus of Cdc34 was fused to the C terminus of Cul1 using a flexible 16-residue glycine-rich linker that was predicted by computational analysis to be long enough to span the two termini in an E2-SCF complex. We anticipated that the fusion should keep SCF and Cdc34 in close proximity, such that SCF would be continuously saturated with Cdc34.
For the experiments shown in Fig. 5, wild type Cdc34 and Cdc34-Δ190 fused to Cul1 were co-expressed with Rbx1 in E. coli and purified. To avoid confusion, the noncovalent complex formed by co-expressed Cul1 and Rbx1 is referred to as Rbx1+Cul1. Fused complexes were either assayed as is or assembled with Skp1-βTrCP expressed in baculovirus-infected insect cells. In a direct comparison, unfused wild type Cdc34 and Rbx1+Cul1 (Fig. 5A) and wild type Rbx1+Cul1-Cdc34 fusion (Fig. 5B) yielded similar activities in the di-ubiquitin synthesis assay. Because it is known that the same interface on E2 binds to both RING and E1 (i.e. E2 cannot bind to both E1 and RING simultaneously) (29), this result indicated that fusion of Cul1 to Cdc34 did not impede the access of Cdc34 to either Rbx1 or E1 enzyme and that the catalytic domain of the fused Cdc34 can dissociate from and reassociate with SCF to engage in multiple rounds of di-ubiquitin synthesis. As expected, assaying unfused human Cdc34-Δ190 (using the same concentration as wild type Cdc34) with Rbx1+Cul1 produced no detectable product during the reaction time course (Fig. 5A). However, the Rbx1+Cul1-Cdc34-Δ190 fusion was active and produced di-ubiquitin, albeit at a reduced rate compared with the wild type fusion (Fig. 5B).
FIGURE 5.
Fusion of human Cdc34-Δ190 to Cul1 can partially rescue the defect of Cdc34 tail deletion in ubiquitylation reactions. A, di-ubiquitin synthesis assay comparing wild type and Cdc34-Δ190 in the presence of Rbx1+Cul1. B, comparison of wild type (WT) and Cdc34-Δ190 fusions to Cul1. Reactions containing either 300 nm Rbx1+Cul1 and 300 nm Cdc34 or containing 300 nm Rbx1+Cul1-Cdc34, 0.7 μm human E1, 6 μm 32P-labeled K48R ubiquitin, and 50 μm D77 ubiquitin were incubated at 20–22 °C for the specified times and quenched with SDS-PAGE buffer. No products were observed when Cdc34-Δ190 was assayed as an unfused species. C and D, di-ubiquitin synthesis assay comparing Cul1 fusions to either wild type Cdc34 (C) or Δ190 Cdc34 (D). The reactions containing 300 nm Rbx1+Cul1-Cdc34, 0.7 μm human E1, 6 μm 32P-labeled K48R ubiquitin, and 50 μm D77 ubiquitin were incubated at 20–22 °C for the specified times and quenched with SDS-PAGE buffer. E, Ub-β-catenin (25) ubiquitylation reactions comparing unfused wild type and Cdc34-Δ190 (300 nm) in the presence of Rbx1+Cul1 (300 nm) or of wild type and Cdc34-Δ190 fusions (1.2 μm). SCFβTrCP complex formation was accomplished by briefly mixing equimolar amounts of either Rbx1+Cul1 or the Rbx1+Cul1-Cdc34 fusions with βTrCP prior to initiation of the reaction by the addition of substrate. The reactions also contained 0.7 μm E1, 75 μm Ub, and 3 μm 32P-labeled Ub-β-catenin peptide. The reactions were incubated at 20–22 °C for the specified times. All of the experiments were done in duplicate.
Human SCF activity is stimulated by covalent modification of a conserved lysine residue on Cul1 with the ubiquitin-like protein Nedd8 (30). We tested whether neddylation of the Cul1 fusion proteins would stimulate activity in a manner similar to the unfused proteins. Indeed, neddylation of both the wild type and Cdc34-Δ190 fusions resulted in a substantial increase in activity (supplemental Fig. S6), further demonstrating that fusion of Cul1 and Cdc34 did not corrupt normal SCF function. Given that the stimulatory effect of neddylation was modest under the multi-turnover reaction conditions used here, we chose to work with unmodified SCF for the remainder of the experiments with the fusion constructs.
Given the qualitative rescue effect of the fusion, the time courses for wild type and Δ190 fusion were repeated with more time points and longer reaction times to quantify the difference in activity, which was ∼10-fold (Fig. 5, C and D). Because the Cul1+Rbx1 module within the fusion complex should be saturated with Cdc34, the 10-fold reduction in the rate of di-ubiquitin formation most likely represents a defect in catalysis, which compares favorably with our kinetic analysis of unfused yeast Cdc34-Δ190 (Fig. 3B).
We next compared the activities of the wild type and Cdc34-Δ190 fusion SCFβTrcp complexes using the SCFβTrcp substrate, mono-ubiquitylated β-catenin peptide (Ub-β-catenin) (25). We chose to use Ub-β-catenin as substrate because it is conjugated with ubiquitin more rapidly than the unmodified peptide, thereby simplifying quantitative analysis. Remarkably, the activity of the Cdc34-Δ190 fusion was within 2-fold of the wild type fusion (Fig. 5E). Thus, the acidic tail of Cdc34 was almost entirely dispensable for ubiquitylation of Ub-β-catenin when Cdc34 was fused to Cul1. Interestingly, the activity of the wild type Cul1-Cdc34 fusion toward an authentic substrate was substantially reduced compared with unfused proteins (∼15-fold reduction of substrate consumption). Recall that the fused proteins had approximately normal activity in a di-ubiquitin synthesis assay (Fig. 5, A and B). We suggest that Cdc34 in the fusion complex is overconstrained by having three separate attachments to SCF (the fusion joint, the catalytic domain-RING interaction, and the acidic tail-SCF interaction). Together, these interactions may restrict rotational and conformational flexibility required for juxtaposition of Cdc34∼Ub and substrate within the SCF complex.
Acidic stretches in protein sequences may be a general strategy for promoting protein-protein interactions. Given the critical role of the acidic tail in Cdc34 function, we wondered whether there are additional proteins in the yeast proteome whose function might also depend on highly acidic peptides. To gain insight into this question, the yeast proteome was systematically searched for proteins that contain acidic stretches. Surprisingly there were numerous proteins that contained at least one highly acidic stretch of residues of significant length.
For example, consider that there is an acidic stretch of 25 contiguous residues in the yeast Cdc34 amino acid sequence (residues 208–232) where 72% of those residues are aspartates or glutamates. We found 83 protein sequences in the yeast proteome that each contain at least one acidic stretch spanning 25 contiguous residues in which at least 70% of the residues were Glu or Asp (Fig. 6B). When protein sequences from the entire yeast proteome were shuffled and reanalyzed for the presence of equivalent acidic stretches, essentially no proteins were identified (supplemental Table S1), eliminating the trivial possibility that our analysis revealed protein sequences loaded with acidic residues.
FIGURE 6.
Proteins that contain long acidic stretches are abundant in the yeast proteome, suggesting that the paradigm of Cdc34-SCF represents a common mechanistic solution for facilitating protein assembly. Each protein sequence in the yeast proteome was searched for peptides of lengths between 15 and 25 residues that contain at least 60% acidic residues (A), 70% acidic residues (B), or 80% acidic residues (C).
Careful inspection of the Cdc34-Δ230 acidic tail sequence (as well as the human ones) identifies a smaller stretch of residues that is even more acidic. These concentrated stretches may be important for biological function. Searching the yeast proteome for acidic stretches of 15 residues where at least 80% of the residues are acidic yielded 146 proteins (Fig. 6C), hinting that the binding mechanism utilized by Cdc34 and SCF may be a general phenomenon.
DISCUSSION
The results of our quantitative biochemical characterization of the Cdc34 acidic tail rationalize prior discrepancies in the literature by providing strong evidence that the acidic tail has multiple effects on the ability of Cdc34 to engage productively the SCF ubiquitin ligase. The acidic tail can be dissected into two domains: a highly acidic distal portion and a less acidic more proximal segment that links the acidic segment to the catalytic domain. The more distal segment of the acidic tail promotes binding of Cdc34 to SCF. The key role of acidity in this interaction is underscored by the observation that a functionally critical segment of the tail (amino acids 211–230) can be replaced by poly(Glu-Asp). Strikingly, the role of the tail in recruitment of Cdc34 to SCF can be partially suppressed in vitro by direct fusion of Cdc34 to Cul1. Contrary to findings reported by Wu et al. (22), we found that the more proximal segment of the tail does not appear to affect the Km of Cdc34 for SCF but instead promotes ubiquitin transfer from E2 to substrate within the Cdc34·Ub-SCF-substrate complex. We note that this discrepancy may be due to the fact that the kinetic analysis presented here employed the yeast Cdc34 protein, whereas human Cdc34 was analyzed by Wu et al. (22). Taken together, our results provide a quantitative biochemical rationale for why deletion of the Cdc34 tail influences both recruitment to SCF (21, 22) and catalysis (23) and is a lethal mutation in budding yeast.
Acidic Tail Sequences Have a Role in Binding to SCF
Enzyme kinetic assays reported here indicate that the acidic tail of Cdc34 contributes to recruitment of Cdc34·Ub to SCF to form the Michaelis complex. Deletion of the tail increases Km for Cdc34 by ∼100-fold, testifying to its importance. This observation, combined with the surprising result that residues in the tail important for binding can be completely replaced with acidic residues, argues that the principle function of the tail is to mediate an electrostatically driven interaction with SCF. Further evidence that the tail plays an important role in recruitment of Cdc34 to SCF is provided by the observation that a tail deletion mutant can be partially suppressed by direct fusion of Cdc34 to the Cul1 subunit of SCF. These observations supplement prior work, which had shown that the Cdc34 tail can confer CDC34 genetic function when fused to the E2 Rad6 (19, 20) and that the Cdc34 tail binds SCF in a pulldown assay (21, 22). However, an important distinction is that pulldown assays yield correlative information on binding that may or may not be relevant to catalysis. For example, it is well known that some E2-RING E3 pairs exhibit strong biochemical activity even if direct binding cannot be detected in a pulldown assay, and conversely, it is possible to form E2-E3 complexes that lack catalytic activity (4). Indeed, our own analysis of Cdc34 truncation mutants revealed that deletion of the C-terminal 65 residues from yeast Cdc34 eliminated binding to SCF (as determined by pulldown assay) but had no effect on the Km of Cdc34 for SCF (this work).3 By contrast, the Km effects reported here provide direct confirmation that the acidic tail contributes to the formation of a functional Cdc34·Ub-SCF-substrate complex.
Our progressive deletion analysis revealed a particularly critical role for tail amino acids 211–220 in mediating Cdc34-SCF association. However, the essentially normal activity of a synthetic chimera in which amino acids 211–230 were replaced by poly(Glu-Asp) argues against the presence of any specific information in this segment other than acidic amino acids. Because every subsequent block of 10 amino acids on through the C terminus contains at least three acidic residues, we suggest that there is nothing unique about amino acids 211–220 other than that it contains the most proximal acidic segment that can engage SCF productively. We suggest that in the wild type Cdc34, multiple short acidic stretches in the distal half of the tail can bind SCF in a quasi-redundant manner.
Evaluating the role of the acidic tail in light of the structure of other E2s raises a question: why does Cdc34 use an acidic tail to interact with SCF, whereas other E2s appear to dock to their cognate E3 principally via the N-terminal α helix and loop regions flanked by β strands 3 and 4 (loop 1) and α helices 2 and 3 (loop 2)? Our data indicate that on their own, the catalytic domain and the acidic tail of Cdc34 each have very weak affinities for SCF (∼16 μm). We propose that the tail has weak affinity for SCF because charged residues must be desolvated before they can interact with each other (31), which renders electrostatically driven interactions inherently unstable because of the unfavorable energetics of desolvating charged side chains. However, upon binding SCF, the tail would tether the catalytic domain nearby and thereby drive its efficient docking to the RING and Cul1, despite the fact that the catalytic domain by itself has relatively poor affinity. Thus, instead of deriving favorable binding energy from a single interface, the Cdc34-SCF complex is stabilized by independent contributions of two distinct interfaces. In this way, each domain stabilizes binding of the other via an avidity effect, similar to how each Fab contributes to the tight binding of an intact IgG to polyvalent antigen. This strategy of achieving high affinity binding by brandishing two relatively weak interfaces as opposed to a single tight interface may provide a kinetic advantage for forming a functional complex, because there is evidence that electrostatic interactions can occur very rapidly (32, 33). Indeed, in other work we have shown that the acidic tail maintains a high affinity yet highly dynamic interaction between Cdc34 and SCF by binding to a basic region on Cul1 (45).
The Role of Proximal Tail Sequences in Promoting Catalysis
In addition to promoting functional interaction between Cdc34 and SCF, the proximal region of the acidic tail (i.e. residues upstream of residue 210) promotes ubiquitin transfer from Cdc34·Ub to substrate within the Cdc34·Ub-SCF-substrate Michaelis complex. The magnitude of the effect we measured was ∼10-fold, but this may be an underestimate, because we calculated kcat based on molecules of substrates modified per SCF per unit time, and comparisons of single- and multi-turnover reactions (25) suggest that some step other than the chemical step is rate-limiting under multi-turnover conditions. This is also evident from examining the pattern of conjugates produced by Cdc34 variants that either contain or lack the proximal region of the tail; mutants deleted for the proximal tail yield reaction products of lower average mass (i.e. bearing fewer ubiquitin modifications). In any event, this catalytic role of the tail reported here went undetected in prior studies, which relied primarily on binding assays. Although the acidic tail has a demonstrable kcat effect, the mechanism by which this occurs remains unknown and awaits further study.
Flirting with Evolution: Redesigning the Cdc34-SCF Interaction
Our results indicate that Cdc34 employs an acidic tail to stabilize its productive interaction with SCF. Other E2-E3 pairs (i.e. Rad6-Ubr1 and Ube2g2-gp78) also form contacts other than the canonical catalytic domain-RING interface to stabilize association (34–36). Indeed, there is even an extreme example in which E2 and E3 are expressed as a single polypeptide chain (37). This suggested to us that it may be possible to redesign the Cdc34-SCF pair such that productive interaction of the E2 catalytic domain with the RING subunit of SCF is facilitated by direct protein fusion rather than the distal segment of the acidic tail. Remarkably, fusion of the N terminus of Cdc34 to the C terminus of Cul1 largely negated the benefit of possessing an acidic tail when assays were carried out with a mono-ubiquitylated β-catenin peptide substrate. Taken together with the properties of other E2-E3 pairs noted above, this result suggests that it is important for E2 to be able to bind rapidly to its cognate E3 to effect ubiquitin transfer to substrate but that the exact mechanism by which rapid binding is achieved is not necessarily critical. Thus, we suggest that there will be diverse, convergent evolutionary solutions to facilitate rapid association of E2s and E3s.
Other Examples of Acidic Tail-containing Proteins and Their Functional Similarity to Cdc34-SCF
Our observation that a synthetic distal tail with very low sequence complexity (poly(Glu-Asp)) is nevertheless capable of mediating functional interaction of Cdc34 with SCF suggested the possibility that polyacidic (or polybasic) stretches might be used more generally to drive protein-protein interactions. Searching the yeast proteome for other proteins that contain acidic regions produced a bolus of hits. Unlike Cdc34, acidic stretches were not confined to the C-terminal regions of protein sequences but were also found appended at the N terminus or sometimes even in the middle of these proteins, likely in the form of loops. Although it is beyond the scope of this manuscript to delve deeply into a functional analysis of all of these proteins, a cursory glance revealed a couple of striking examples, as detailed below.
One protein, Qcr6, contains an extraordinary acidic tail of 48 residues near its N terminus. Almost 80% of these residues are either glutamates or aspartates. Qcr6 is a subunit of the cytochrome bc1 complex, a highly conserved multi-subunit enzyme involved in cytochrome c maturation (38, 39). It has been known for some time that cytochrome c maintains a large positive dipole moment (40). Although no direct biochemical studies have been done, it has been shown that the reaction rate decreases steadily with increasing ionic strength of the reaction solution (41), a result that is typically interpreted as evidence for electrostatic interactions between two proteins. It is tempting to speculate that the tremendous rate of electron transfer between cytochrome c and the bc1 enzyme is enabled by an electrostatic interaction between Qcr6 and cytochrome c, where the acidic tail of Qcr6 helps align cytochrome c to promote productive binding to the cytochrome bc1 complex.
In another example, we found a ∼60-residue-long acidic tail at the C terminus of Vhs3, a functionally redundant inhibitory subunit of Ppz1p, a Ser/Thr protein phosphatase (42). Interestingly, Ppz1 has a basic N-terminal domain with a theoretical pI of 9.7 and an estimated charge (at pH 7) of 17. Furthermore, the functionally redundant inhibitory subunit of Ppz1, Sis2/Hal3, also maintains an acidic tail at its C terminus.
As a final note, it has long been understood from the work of Ptashne and co-workers (43, 44) that some transcriptional regulatory proteins have highly acidic stretches of amino acids in their activator domains. In fact, these acidic sequences can be transferred from one regulator to another, reminiscent of the ability of the Cdc34 tail to function with the catalytic domain of the E2 Rad6 (19, 20).
Acknowledgments
We thank Jost Vielmetter and members of the Caltech PEC for protein expression. We thank Shu-ou Shan for expert advice on the kinetic analysis of the data and members of the Deshaies lab for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Ruth Kirschstein Postdoctoral Fellowship F32 GM074471-01.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Table S1, and Figs. S1–S6.
M. Petroski, unpublished data.
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin conjugating enzyme
- E3
- ubiquitin-protein isopeptide ligase
- CRL
- cullin-RING-ligase
- GST
- glutathione S-transferase
- Ub
- ubiquitin
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- D77 ubiquitin
- ubiquitin with an aspartate residue appended to the C terminus.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Dye B. T., Schulman B. A. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 [DOI] [PubMed] [Google Scholar]
- 3.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 5.Frescas D., Pagano M. (2008) Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 7.Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J. (1997) Cell 91, 221–230 [DOI] [PubMed] [Google Scholar]
- 8.Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 9.Ohta T., Michel J. J., Schottelius A. J., Xiong Y. (1999) Mol. Cell 3, 535–541 [DOI] [PubMed] [Google Scholar]
- 10.Seol J. H., Feldman R. M., Zachariae W., Shevchenko A., Correll C. C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Deshaies R. J., Shevchenko A., Deshaies R. J. (1999) Genes Dev. 13, 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skowyra D., Koepp D. M., Kamura T., Conrad M. N., Conaway R. C., Conaway J. W., Elledge S. J., Harper J. W. (1999) Science 284, 662–665 [DOI] [PubMed] [Google Scholar]
- 12.Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999) Mol. Cell 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 13.Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G., Jr., Elledge S. J., Conaway R. C., Harper J. W., Conaway J. W. (1999) Science 284, 657–661 [DOI] [PubMed] [Google Scholar]
- 14.Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 15.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 16.Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 17.Willems A. R., Schwab M., Tyers M. (2004) Biochim. Biophys. Acta 1695, 133–170 [DOI] [PubMed] [Google Scholar]
- 18.Schwob E., Böhm T., Mendenhall M. D., Nasmyth K. (1994) Cell 79, 233–244 [DOI] [PubMed] [Google Scholar]
- 19.Kolman C. J., Toth J., Gonda D. K. (1992) EMBO J. 11, 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. (1992) EMBO J. 11, 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathias N., Steussy C. N., Goebl M. G. (1998) J. Biol. Chem. 273, 4040–4045 [DOI] [PubMed] [Google Scholar]
- 22.Wu K., Chen A., Tan P., Pan Z. Q. (2002) J. Biol. Chem. 277, 516–527 [DOI] [PubMed] [Google Scholar]
- 23.Block K., Appikonda S., Lin H. R., Bloom J., Pagano M., Yew P. R. (2005) Cell Cycle 4, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 24.Petroski M. D., Deshaies R. J. (2005) Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 25.Saha A., Deshaies R. J. (2008) Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Pavletich N. P., Schulman B. A., Zheng N. (2005) Methods Enzymol. 398, 125–142 [DOI] [PubMed] [Google Scholar]
- 27.Petroski M. D., Deshaies R. J. (2005) Methods Enzymol. 398, 143–158 [DOI] [PubMed] [Google Scholar]
- 28.Hofmann R. M., Pickart C. M. (2001) J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 29.Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 30.Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K. (2004) Oncogene 23, 1985–1997 [DOI] [PubMed] [Google Scholar]
- 31.Creighton T. E. (1993) Proteins: Structures and Molecular Properties, 2nd Ed., pp. 272–273, W. H. Freeman, New York [Google Scholar]
- 32.Schreiber G., Fersht A. R. (1993) Biochemistry 32, 5145–5150 [DOI] [PubMed] [Google Scholar]
- 33.Schreiber G., Fersht A. R. (1996) Nat. Struct. Biol. 3, 427–431 [DOI] [PubMed] [Google Scholar]
- 34.Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Tu D., Li L., Wollert T., Ghirlando R., Brunger A. T., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y., Varshavsky A. (1999) EMBO J. 18, 6832–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pohl C., Jentsch S. (2008) Ernst Schering Found. Symp. Proc. 1, 115–126 [DOI] [PubMed] [Google Scholar]
- 38.Van Loon A. P., De Groot R. J., De Haan M., Dekker A., Grivell L. A. (1984) EMBO J. 3, 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M., Trumpower B. L. (1994) J. Biol. Chem. 269, 1270–1275 [PubMed] [Google Scholar]
- 40.Koppenol W. H., Margoliash E. (1982) J. Biol. Chem. 257, 4426–4437 [PubMed] [Google Scholar]
- 41.Schoppink P. J., Hemrika W., Berden J. A. (1989) Biochim. Biophys. Acta 974, 192–201 [DOI] [PubMed] [Google Scholar]
- 42.Ruiz A., Muñoz I., Serrano R., González A., Simón E., Ariño J. (2004) J. Biol. Chem. 279, 34421–34430 [DOI] [PubMed] [Google Scholar]
- 43.Gill G., Sadowski I., Ptashne M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2127–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gill G., Ptashne M. (1987) Cell 51, 121–126 [DOI] [PubMed] [Google Scholar]
- 45.Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]