Abstract
Corticosteroids are potent anti-inflammatory agents, but corticosteroid insensitivity is a major barrier for the treatment of some chronic inflammatory diseases. Here, we show that hypoxia induces corticosteroid-insensitive inflammation via reduced transcription of histone deacetylase-2 (HDAC2) in lung epithelial and macrophage cells. HDAC2 mRNA and protein expression was reduced under hypoxic conditions (1% O2). Hypoxia enhanced interleukin-1β-induced interleukin-8 (CXCL8) production in A549 cells and decreased the ability of dexamethasone to suppress the CXCL8 production. Deletion or point mutation studies revealed that binding of the transcription factor hypoxia-inducible factor (HIF) 1α to a HIF response element at position −320, but not HIF-1β or HIF-2α, results in reduced polymerase II binding at the site, leading to reduced promoter activity of HDAC2. Our results suggest that activation of HIF-1α by hypoxia decreases HDAC2 levels, resulting in amplified inflammation and corticosteroid resistance.
Introduction
Hypoxia is defined as a decreased availability of oxygen in tissues and is linked to many diseases such as cancer, cardiovascular diseases, pulmonary diseases, and stroke (1). At the cellular level, hypoxia leads to the rapid stabilization of the transcription factor hypoxia-inducible factor (HIF)2 1α. HIF-1α is expressed in a wide range of cells, including those of the lung, and along with its dimerization partner HIF-1β, it regulates a cell's response to hypoxia. Under normal oxygen tension, HIF-1α is steadily transcribed and translated, but it is immediately targeted for degradation via the proteasome after hydroxylation by prolyl hydroxylases. Under low oxygen tension, the prolyl hydroxylases are inhibited, and HIF-1α is rapidly stabilized. HIF-1 regulates >100 genes controlling cellular responses such as angiogenesis, oxygen transport, and glucose metabolism as well as cell proliferation and survival (2). Recently, it has been shown that an increase in reactive oxygen species inhibits prolyl hydroxylases and thus stabilizes HIF-1α (3). Other findings have shown that inflammatory molecules such as lipopolysaccharide, interleukin-1, and NF-κβ can also enhance the transcription of HIF-1α (4–6). These findings highlight the stabilization of HIF-1α under non-hypoxic and mainly inflammatory conditions and may have important implications in chronic inflammatory diseases.
Histone deacetylases (HDACs) and histone acetyltransferases regulate gene transcription by modifying histones, which leads to remodeling of the chromatin complex. Non-histone-related functions and targets for HDACs and histone acetyltransferases have recently been uncovered, including their role in the inflammatory response (7, 8). Histone acetyltransferase activity is increased in bronchial biopsies of asthma patients (9), creating an imbalance in the ratio of histone acetyltransferase/HDAC activity in asthmatic lungs and a shift toward higher levels of transcriptional activity. More specifically, HDAC2 is recruited by the activated glucocorticoid receptor to repress NF-κB transcription in airway epithelial cells (10). Measurements of specific HDACs at the RNA and protein levels have shown that HDAC2 is reduced in macrophages and peripheral lung in the more severe stages of chronic obstructive pulmonary disease (COPD) (11). More importantly, most severe COPD patients suffer from hypoxia. A reduction in HDACs as observed in COPD may cause amplified inflammation as well as reduce the anti-inflammatory effects of corticosteroids.
In this study, we examined the role of HIF-1α in the reduction of HDAC2 observed in airway cells. We show that the transcription factor HIF-1α, induced by hypoxia or cobalt chloride (CoCl2), is necessary and sufficient to reduce the transcription of HDAC2 and that this reduction leads to increased inflammation as well as corticosteroid resistance.
EXPERIMENTAL PROCEDURES
Cell Culture
A549 cells were cultured in Dulbecco's modified Eagle's medium (Sigma, Gillingham, UK) with 10% fetal calf serum (FCS). Cells were exposed to 0% FCS medium overnight before treatment with fresh 0% FCS medium for protein and RNA extractions. U937 cells were cultured similarly but with RPMI 1640 medium (Sigma). Primary macrophages were obtained following lung resection surgery. The macrophage isolation procedure was published previously (12). Macrophages were kept in RPMI 1640 medium with 10% FCS, 2 mm l-glutamine, 0.1 mg/ml penicillin/streptomycin, and 2.5 μg/ml amphotericin B. Cells were counted 24 h after isolation and exposed to hypoxia (1% oxygen, 5% CO2/N2 balance) or CoCl2 for 24 h. Small airway epithelial cells were obtained from Cambrex (East Rutherford, NJ) and cultured in BulletKit medium with supplements according to the manufacturer's instructions. Cells were exposed to hypoxia or CoCl2 for 24 h in normal medium.
Nuclear Extraction and Western Blotting
Nuclear extractions were performed with a nuclear extraction kit (Active Motif, Rixensart, Belgium) following the manufacturer's instructions. 20–30 μg of nuclear extracts was separated on 4–20% Tris-HCl gels (Invitrogen) and transferred to nitrocellulose using iBlotTM dry transfer (Invitrogen). Membranes were blocked in 5% milk/Tris-buffered saline/Tween solutions for 30 min at room temperature prior to probing with primary antibodies overnight at 4 °C. Secondary antibodies were applied at room temperature for 1 h, and detection was performed with ECL solutions (GE Healthcare, Little Chalfont, UK). Densitometry was done using a Gel-Doc-It imaging system (UVP, Cambridge, UK).
Quantitative Reverse Transcription (RT)-PCR
Cell samples were collected after washing with Hanks' balanced salt solution and centrifuged at 4 °C, and the cell pellets were stored at −80 °C. RNA extractions were done with an RNeasy kit (Qiagen, Hilden, Germany). 10 μl of RNA was used in a RT reaction using a cDNA Archive kit (Applied Biosystems, Foster City, CA). Real-time PCR for HDAC2 or GNB2L for normalization was run in a Corbett 3000 RotorGene (Corbett Research, Cambridge, UK) as described previously (10).
HDAC2 Promoter Amplification and Luciferase Plasmid Construction
pGL4.10-Luc2 (pLuc2) was purchased from Promega (Madison, WI). Human genomic DNA was extracted from A549 cells and used to amplify the HDAC2 promoter from positions −465 to +114 relative to the transcription start site (5′-primer, 5′-CTAGCTACCAGGCACTGGGGCGAT-3′; and 3′-primer, 5′-ATCGTGGGAGGAGAGGAGGGGG-3′). This product was ligated into pLuc2 and renamed pLuc2-(−465/+114). A second amplification from positions −1341 to −300 was performed, followed by digestion with EcoRV and BsaAI and ligation into the first construct (5′-primer, 5′-CTCACTTTACCTCTAGGCCAGTG-3′; and 3′-primer, 5′-TAGCCTTGGCGGTCTGGA-3′). Other deletion constructs were made by the following paired digests of pLuc2-(−1341/+114): EcoRI and HindIII, BsaAI, or MfeI, respectively, followed by end blunting with Klenow polymerase and ligation into pLuc2. All constructs were sequenced and matched with the published human chromosome 6 sequence in the NCBI Database (Build 36.2).
Transfection and Luciferase Assays
Transfections were done with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. 0.8–1 μg of DNA/plasmid (pLuc2-x and pSV-β-galactosidase, Promega) was transfected in 80–90% confluent cells. For small interfering RNAs (siRNAs), 100 nm siRNA was transfected into 50–60% confluent cells using Lipofectamine 2000. 4 h after the transfection, the medium was changed to the appropriate treatment in 0% FCS medium and incubated overnight. Cells were collected after washing with Hanks' balanced salt solution in Reporter Lysis Buffer (Promega). 25 μl of each extract was dispensed in an opaque white 96-well plate in duplicate. Luciferase assay reagent (50 μl) was added, samples were diluted 1:100 in water, and plates were read on a Synergy HT luminometer (BioTek, Winooski, VT) at 595/360 nm. Luminescence was standardized with a β-galactosidase assay (Promega) performed in duplicate with 30 μl of cell extract following the manufacturer's instructions.
Enzyme-linked Immunosorbent Assays
CXCL8 concentrations were determined by sandwich enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, MN). Cells were plated in 96-well plates on day 1; on day 2, the cells were starved overnight, followed by a 24-h exposure to normoxia or hypoxia. On day 4, cells were stimulated with interleukin-1β (IL-1β) after a 10-min incubation with the indicated dexamethasone dose and placed back under hypoxia as relevant. CXCL8 levels were normalized to cell number as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
HIF-1α Competitive Binding Assay
The TransAM HIF-1 kit (Active Motif) was used for the competitive binding assay. The sequences surrounding each HIF response element (HRE) were ordered as high pressure liquid chromatography-purified DNA oligonucleotides. A 10 μm solution of each pair of oligonucleotides (positive and negative strands) was annealed by heating to 100 °C for 15 min, followed by slow cooling to room temperature. This solution was then used for each site in the competitive binding assay in accordance with the manufacturer's recommendations. We used positive strand sequence for −817 HRE (cagctagggtgcgtggagtttgca), positive strand sequence for −368 HRE (tcgcggcacgtggtcggcttg), and positive strand sequence for −320 HRE (aggggagcagcgtgggacgccga). Matching negative strand sequences were ordered.
Chromatin Immunoprecipitation (ChIP) and Quantitative PCR
ChIP was performed after 24 h of hypoxia (or normoxia control) as described previously (8). 1 μg of rabbit polyclonal antibody to polymerase II (pol II; Santa Cruz Biotechnology, Santa Cruz, CA) was utilized for immunoprecipitation. After standard elution, proteinase K digestion, and DNA precipitation, 3 μl of diluted DNA was used for quantitative PCR with SYBR green (Qiagen). Primer sequences were as follows: −320, 5′-AGCTTCGCGACAGTGGGGG-3′ (forward) and 5′-AGCCTTGGCGGTCTGGACC-3′ (reverse); −368, 5′-CGGAAGGGGCGCGGGAGTGGG-3′ (forward) and 5′-CCGACCACGTGCCGCGAGCC-3′ (reverse); and −817, 5′-AATCTTCCAGTGTCTAGCCAGCAG-3′ (forward) and 5′-CACGCACCCTAGCTGTTCCA-3′ (reverse). PCRs were run in the Corbett 3000 RotorGene, with primer annealing temperatures used for the amplification step of each cycle. Standard curves were generated for each primer pair, and data were normalized to input control samples.
Statistics
Student's unpaired t test was calculated when three or more samples were present in each treatment group. p < 0.05 was considered significant.
RESULTS
HDAC2 Protein Is Reduced by Hypoxia and Causes Corticosteroid Insensitivity
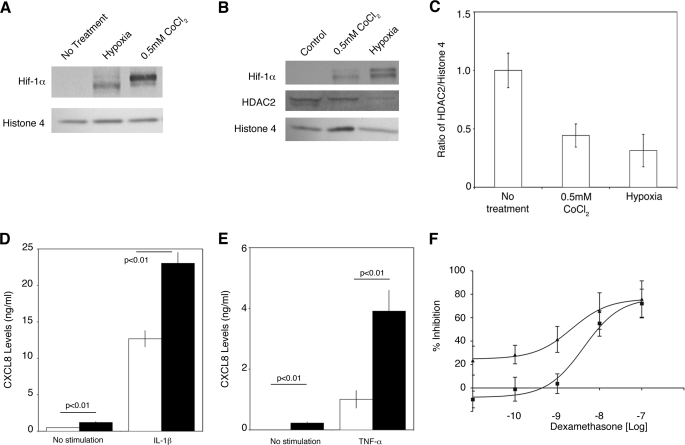
We first examined whether A549 cells, a human alveolar epithelial cell line, have a normal hypoxic response. The cells were placed under hypoxia (1% oxygen, 5% CO2/N2 balance) or treated with CoCl2 for 24 h, followed by nuclear extraction. CoCl2 mimics hypoxia by interfering with the prolyl hydroxylases (13). HIF-1α protein was clearly stabilized in the cells (Fig. 1A). After 24 h in 1% oxygen, nuclear extracts showed that although HIF-1α protein was stabilized, HDAC2 protein levels were unchanged compared with cells under normoxic conditions (data not shown). Cells were subsequently exposed to 48 h of hypoxia, and HDAC2 levels, relative to histone 4 as a housekeeping nuclear protein, were decreased in nuclear extracts (Fig. 1, B and C). Hypoxia reduced the protein levels by 69% and CoCl2 by 56%. In U937 cells (a human monocytic lymphoma cell line) exposed to 48 h of hypoxia, HDAC2 was also reduced by 59% (data not shown).
FIGURE 1.
HDAC2 protein reduction causes corticosteroid insensitivity. A, Western blot analysis demonstrating that HIF-1α protein is stabilized in nuclear extracts of A549 cells after a 24-h exposure to 0. 5 mm CoCl2 or hypoxia. The results are representative of four independent experiments. B, Western blot analysis of HDAC2 protein levels after a 48-h exposure to hypoxia or CoCl2. The results are representative of three independent experiments. C, graphical analysis of HDAC2 protein expression as reported in B 48 h after treatment with CoCl2 (0.5 mm) or hypoxia (means ± S.E., n = 3). D, hypoxia pretreatment (black bars) of A549 cells for 24 h enhances the ability of IL-1β to induce CXCL8 expression after 12 h (means ± S.D., n = 3). E, hypoxia pretreatment (black bars) of U937 cells for 24 h enhances the ability of tumor necrosis factor-α (TNF-α; 10 ng/ml) to induce CXCL8 expression after 12 h (means ± S.D., n = 3). F, the concentration-dependent ability of dexamethasone to suppress IL-1β-induced CXCL8 expression (■) is reduced by pre-exposure to hypoxia for 24 h (n = 3; p < 0.05). ▲, normoxia.
The effect of corticosteroids on cells under hypoxic conditions for 48 h was examined. Application of corticosteroids to cells quickly activates the glucocorticoid receptor, which translocates into the nucleus to activate anti-inflammatory gene transcription. As HDAC2 is recruited by the activated glucocorticoid receptor to repress NF-κB-mediated transcription of pro-inflammatory genes (10), we measured CXCL8 release after stimulating A549 cells with IL-1β (Fig. 1D) and U937 cells with tumor necrosis factor-α (Fig. 1E). Overall, CXCL8 levels were higher in hypoxic stimulated cells than in cells under normoxic conditions (Fig. 1, D and E), in agreement with findings in endothelial cells (14). A concentration-response curve for dexamethasone after IL-1β stimulation was calculated in A549 cells and was shifted to the right (normoxia IC50 = 12 nm and hypoxia IC50 = 36 nm; p < 0.05), showing a 3-fold decrease in the effectiveness of dexamethasone to repress CXCL8 release in hypoxic cells (Fig. 1F).
Hypoxia Reduces HDAC2 mRNA by a Reduction in Its Promoter Activity
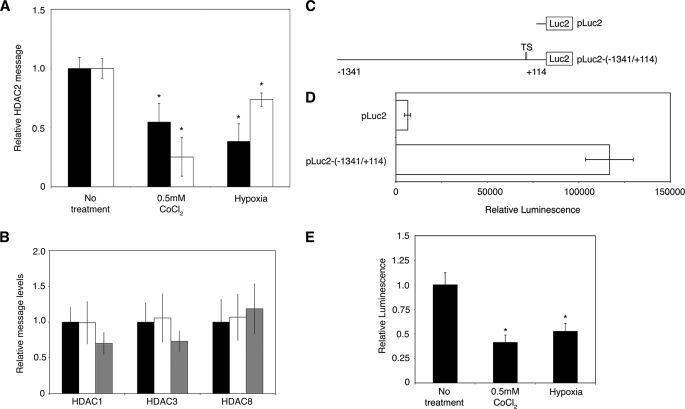
HDAC2 transcript levels were measured after exposure to hypoxia or CoCl2 by quantitative RT-PCR after 24 h. Hypoxia and 0.5 mm CoCl2 reduced RNA levels by 61.7 ± 14.9 and 45.3 ± 15.7%, respectively, in A549 cells and by 26.3 ± 5.6 and 74.8 ± 16.2%, respectively, in U937 cells (Fig. 2A). The levels of HDAC1, HDAC3, and HDAC8 were also examined in A549 cells after 24 h under hypoxia or 0.5 mm CoCl2, and no significant changes in message levels were detected (Fig. 2B).
FIGURE 2.
HDAC2 transcription is reduced under hypoxia. A, A549 (black bars) and U937 (white bars) cells were treated for 24 h, and the effect of CoCl2 and hypoxia on HDAC2 mRNA was determined by quantitative RT-PCR (means ± S.E., n = 4). *, p < 0.01. B, the levels of HDAC1, HDAC3, and HDAC8 were measured by quantitative RT-PCR after a 24-h exposure to CoCl2 (gray bars) or hypoxia (white bars) in A549 cells (means ± S.E., n = 3). Black bars, no treatment. C, a HDAC2 promoter-luciferase reporter plasmid was designed (pLuc2-(−1341/+114) that includes the region from positions −1341 to +114 from the transcription start site (TS) of HDAC2. D, luciferase activity shows constitutive activation of the HDAC2 promoter after 24 h, with little background from the empty reporter vector (means ± S.D., n = 4). E, the effect of CoCl2 and hypoxia on pLuc2-(−1341/+114) luciferase activity after 24 h is shown (means ± S.D., n = 4). *, p < 0.001.
To study HDAC2 promoter activity under various conditions, we cloned a 1455-bp region of the human promoter into a luciferase reporter vector, named pLuc2-(−1341/+114) (Fig. 2C). Transfection of the plasmid into A549 cells resulted in high levels of luciferase activity after 24 h (Fig. 2D). The results indicate endogenous transcription of the HDAC2 promoter under normal conditions. Transfections with pLuc2-(−1341/+114) were repeated, and the cells were exposed to hypoxia or 0.5 mm CoCl2 for 24 h. The luciferase assay results show that the promoter activity was reduced 58.7 ± 5.7% by CoCl2 and 47.4 ± 6.1% by hypoxia (Fig. 2E). The observed reduction in HDAC2 transcription is therefore due to reduced promoter activity under low oxygen or in the presence of CoCl2.
HIF-1α Binds to the HDAC2 Promoter and Prevents Transcription
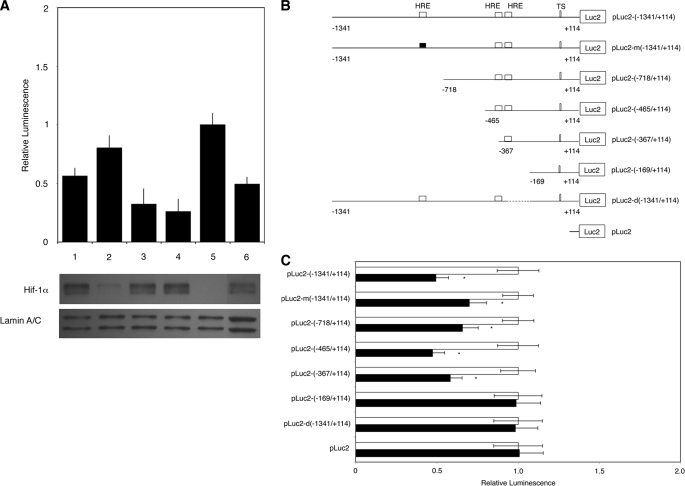
To determine whether HIF-1α plays a direct role in the reduction of HDAC2 transcription, transfection experiments with pLuc2-(−1341/+114) were repeated in the presence of a siRNA to HIF-1α (siHIF-1α). The siRNA was tested independently by transfection into A549 cells for 24 h and reduced HIF-1α RNA levels by 80–90% as determined by quantitative RT-PCR (data not shown). When siHIF-1α was cotransfected with pLuc2-(−1341/+114) and the cells were exposed to hypoxia, promoter activity (0.80 ± 0.11 versus 0.49 ± 0.06) was restored to near normal levels (set to 1.0) (Fig. 3A). The use of a nonspecific siRNA (siNeg), siHIF-2α, or siHIF-1β did not prevent the decrease in luciferase activity (0.56 ± 0.07, 0.32 ± 0.13, and 0.26 ± 0.11, respectively). HIF-1α levels were checked in transfected samples; we observed very low levels of HIF-1α protein after 24 h of hypoxia when siHIF-1α was present (Fig. 3A).
FIGURE 3.
HIF-1α and HREs reduce HDAC2 promoter activity under hypoxia. A, knockdown of HIF-1α prevents the decrease in pLuc2-(−1341/+114) promoter activity under hypoxia. siNeg, siHIF-2α, and siHIF-1β had no effect on promoter activity. Bar 1, siNeg; bar 2, siHIF-1α; bar 3, siHIF-2α; bar 4, siHIF-1β; bar 5, no treatment; bar 6, hypoxia (means ± S.D., n = 4; p < 0.05). Western blot analysis of transfected cells showed that only siHIF-1α reduced HIF-1α protein after a 24-h exposure to hypoxia. B, a schematic diagram of pLuc2-(−1341/+114) is shown with the position of each HRE indicated. The mutated HRE construct is shown with a black box. All five shorter promoter plasmids are shown with respective sizes and HRE sites. TS, transcription start site. C, the effect of removal of each HRE site in the HDAC2 promoter construct on the ability of hypoxia (black bars) to attenuate luciferase activity was investigated. Only pLuc2-(−169/+114) and the deletion mutant pLuc2-d(−1341/+114) failed to reduce reporter gene activity under hypoxic conditions. Results are presented as means ± S.D. of four independent experiments. *, p < 0.05.
The promoter region was next examined for the presence of hypoxia response elements (2). Three potential binding sites were found, one at position −817 and two in tandem around position −344 (Fig. 3B). Serial deletions in the HDAC2 promoter were made (pLuc2-(−718/+114), pLuc2-(−465/+114), pLuc2-(−367/+114), and pLuc2-(−169/+114)) as well as one HRE mutant (pLuc2-m(−1341/+114)) and one construct with an internal deletion of bases −383 to −192 (pLuc2-d(−1341/+114)). The engineered mutation changed the HRE site from CGTG to AATG at position −817.
Each plasmid was transfected into A549 cells and exposed to hypoxia for a total of 24 h. The presence of the HRE site at position −817 was found to be unnecessary for the decrease in promoter activity under hypoxia: neither the mutation of the site nor its deletion affected the reduction in promoter activity under hypoxia (Fig. 3C). The presence of the tandem pair of HREs is necessary for the reduction of promoter activity, but the closest site to the start position was able to reduce activity when present on its own, as demonstrated by clone pLuc2-(−367/+114).
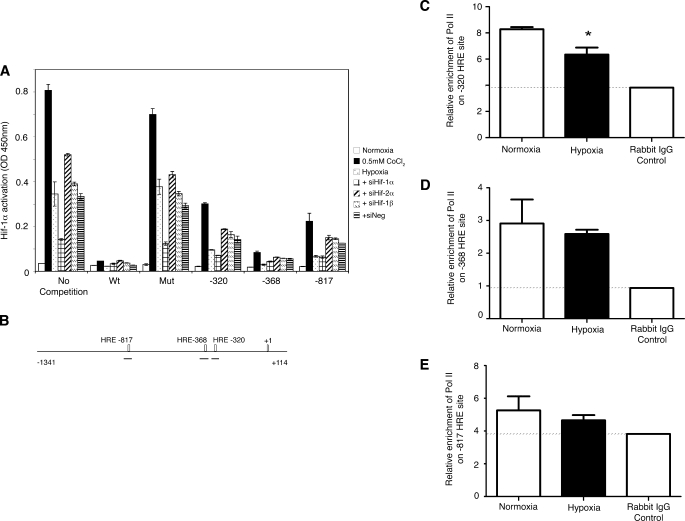
To confirm the binding of the HIF-1α to the promoter at that site, we performed a HIF-1α competitive binding assay (Fig. 4A). The assay provides double-stranded oligonucleotides of a wild-type HRE and a mutant HRE, and we added oligonucleotides corresponding to the three HRE sites from the HDAC2 promoter. Nuclear extracts of A549 cells treated with normal medium, hypoxia, or 0.5 mm CoCl2 or transfected with siNeg, siHIF-1α, siHIF-2α, or siHIF-1β followed by exposure to hypoxia were obtained, and each was assayed in duplicate in each competitive binding assay. HIF-1α was activated by hypoxia and CoCl2 treatment, and competition with the wild-type sequence reduced the detection signal to background levels, whereas transfection with siHIF-1α also reduced the activation levels of HIF-1α (Fig. 4A). All three sequences from the HDAC2 promoter resulted in reduction of HIF-1α detection, but the strongest competition was observed with the oligonucleotides for the −368 sequence.
FIGURE 4.
HIF-1α inhibits HDAC2 transcription. A, the effect of hypoxia, CoCl2 (0.5 mm), and HIF isoform knockdown on the ability of HIF-1α to bind to wild-type and truncated HDAC2 promoter oligonucleotides as determined by competitive HIF-1α binding assay shows a complete lack of HIF-1α signal with the wild-type sequence (Wt) compared with normal cells. The highest binding was obtained with the −368 oligonucleotides. Both the −320 and −817 oligonucleotides showed strong HIF-1α binding. The hypoxia, +siHIF-2α, +siHIF-1β, and +siNeg samples had similar levels of HIF-1α, whereas the siHIF-1α samples showed reduced binding capacity throughout. All hypoxia- or CoCl2-treated samples showed the same relative response to the five HRE sequences. Data are representative of two independent experiments (means ± S.E.). Mut, mutant. B, shown is a schematic diagram of the HDAC2 promoter with the three HRE sites indicated. Black lines represent each amplification region used in the quantitative PCR following ChIP assays. C, hypoxia treatment (black bars) for 24 h resulted in a decrease of pol II binding on the −320 HRE site as determined by quantitative PCR following pol II ChIP. Data are normalized to input control DNA, and the rabbit IgG control is shown (means ± S.E., n = 3). *, p < 0.05. D, the results of the ChIP experiment at the −368 HRE site show a small reduction of pol II binding relative to control samples (means ± S.E., n = 3). E, low detection levels were obtained for the −817 HRE site following pol II ChIP (means ± S.E., n = 3).
To verify that the binding of HIF-1α to the HDAC2 promoter prevents transcription, we performed a ChIP/quantitative PCR experiment using an antibody to pol II and three sets of primers to amplify the three HRE sites on the promoter. The hypoxic samples showed a decrease in pol II binding at all three amplification sites compared with the normoxic samples (Fig. 4, C–E). The primers for the HRE at position −817 showed the least binding overall relative to the rabbit IgG control (Fig. 4E), whereas the primers at positions −368 (Fig. 4D) and −320 (Fig. 4C) had much greater signals. Although all three sites were bound by HIF-1α in the competitive binding assay, the HRE at position −320 is the only site that is necessary to cause the decrease in HDAC2 transcription.
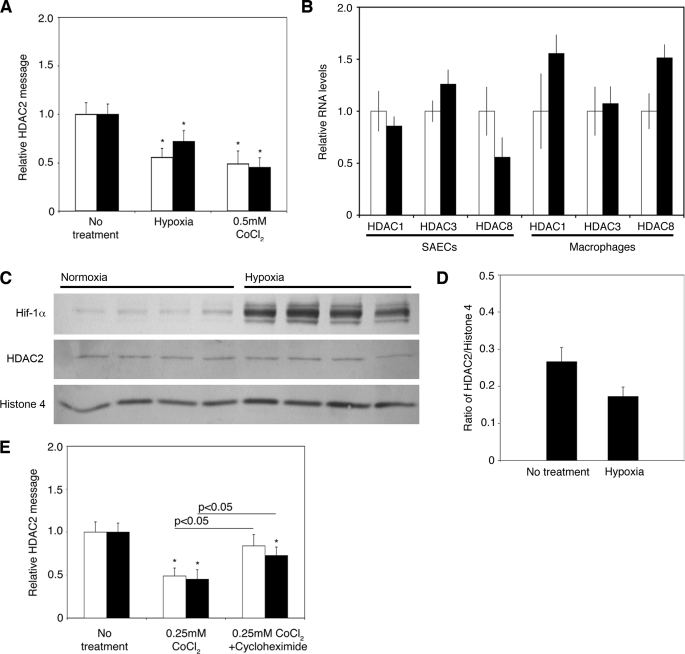
Translation Inhibition Prevents the Decrease of HDAC2 in Primary Pulmonary Cells
To confirm the HIF-1α-mediated molecular mechanism of HDAC2 reduction, we examined primary small airway epithelial cells (SAECs) and alveolar macrophages for HDAC2 RNA levels after a 24-h exposure to hypoxia or 0.25 mm CoCl2. HDAC2 mRNA was reduced by exposure to hypoxia by 45 ± 9% (p < 0.05) in alveolar macrophages and by 28 ± 11% (p < 0.05) in SAECs and by 51 ± 13 and 55 ± 10% (p < 0.05), respectively, in the presence of CoCl2 compared with cells exposed to normoxic conditions (Fig. 5A). A lower concentration of CoCl2 was used in the primary cells (0.25 mm) to prevent loss of cell viability after the 24-h exposure. The levels of three other class I HDACs (HDAC1, HDAC3, and HDAC8) were examined, and no significant changes between hypoxia and normoxia were detected in either cell type (Fig. 5B). Protein levels were examined in SAECs after a 24 h-exposure to hypoxia (Fig. 5, C and D). HDAC2 protein was reduced by 35.1 ± 2.5% (p < 0.05) in nuclear extracts of hypoxic cells. These results confirm that the mechanism is also present in primary cells that are exposed to hypoxia and is not restricted to culture cell lines.
FIGURE 5.
HIF-1α mediates HDAC2 reduction in primary cells. A, shown is the effect of hypoxia and CoCl2 (0.25 mm) on HDAC2 mRNA expression measured by quantitative RT-PCR after 24 h in human bronchoalveolar lavage macrophages (white bars) and SAECs (black bars). The results are presented as means ± S.D. of three independent experiments. *, p < 0.05 to no treatment. B, the levels of HDAC1, HDAC3, and HDAC8 were also measured by quantitative RT-PCR in the same samples. White bars, no treatment; black bars, hypoxia. t test results showed no significant differences. C, shown are the results from representative Western blot analysis of nuclear extracts of SAECs exposed to 24 h of hypoxia. HDAC2 levels were reduced relative to histone 4. D, data shown in C (and data not shown) were quantified. The results are presented as means ± S.E. of six independent experiments. E, the effect of cycloheximide (20 μg/ml) on the ability of CoCl2 (0.25 mm) to attenuate HDAC2 mRNA in bronchoalveolar macrophages (white bars) and SAECs (black bars) is compared with CoCl2-only treated cells. The results are expressed as means ± S.D. of three independent experiments. *, p < 0.05 compared with 0.25 mm CoCl2 as indicated.
The primary cells were treated with 0.25 mm CoCl2 for 1 h, and then 20 μg/ml cycloheximide was added for 23 h to prevent further translation of the oxygen-regulated protein. This protocol allowed a proportion of HIF-1α to be stabilized, and the effect on HDAC2 RNA levels was examined. HDAC2 RNA levels were found to be reduced compared with non-treated cells, but not by as much as in CoCl2-exposed cells (27 ± 13% versus 55 ± 10% (p < 0.05) in SAECs and 16 ± 13% versus 51 ± 13% (p < 0.05) in alveolar macrophages) (Fig. 5E).
DISCUSSION
Collectively, our results present a novel mechanism of corticosteroid insensitivity via a HIF-1α-dependent negative regulation of gene expression. We have shown that HIF-1α stabilization leads to a reduction in transcription at the HDAC2 promoter due to the direct binding of the transcription factor (HIF-1α). The reduced transcriptional activity leads to lower levels of HDAC2 mRNA in lung epithelial and alveolar macrophages, resulting in reduced protein levels. Moreover, this work is one of the few reports of transcriptional regulation of a HDAC under a specific physical condition (15).
Negative regulation of gene expression by a transcription factor is not unique. The transcription factor p53 has been shown to repress the transcription of NF-κB-regulated genes in vivo, but no direct binding of p53 to the promoters was demonstrated (16), whereas SP1 was shown to bind to the cyclin D1 promoter and repress its transcription (17). Not all HREs on HDAC2 seem to have the same level of binding and negative effect from HIF-1α. Although the HRE at position −817 was bound by HIF-1α in the competitive assay, it showed the least pol II enrichment in the ChIP assay; thus, its greater distance from the transcription start site explains why it was shown to be unnecessary for the hypoxia-mediated effect in the deletion experiments. Tandemly arranged HREs are known to be more effective in activating transcription by HIF-1α than single HREs and perhaps may also be more effective in preventing transcription (2, 18). Interestingly, a cis-element present on TATA-less promoters of cell cycle and other essential genes is located around the same position on the HDAC2 promoter (19). The binding of HIF-1α to the tandem HREs prevents pol II and the transcription machinery from binding at or near that site, thus resulting in lower HDAC2 transcription levels in the presence of HIF-1α.
Other class I HDACs were not significantly affected at the messenger level by hypoxia in the cell lines examined. This result contrasts with other findings that HDAC1 is elevated after 16 h of hypoxia (20). Another study showed increased HDAC activity due to post-translational modifications of HDAC1 and HDAC2 after short-term exposures to hypoxia (21). We examined long-term exposure to hypoxia to evaluate the effect of the chronic presence of HIF-1α on HDAC2. The different times of exposure to hypoxia could explain the discrepancies between ours and other studies. At 24 h, protein levels in the cell lines were not affected by hypoxia, consistent with the study by Pluemsampant et al. (21). Our results reveal a new level of hypoxic regulation by using the HIF-1α transcription factor to down-regulate a class I HDAC. Interestingly, an increased protein interaction between HIF-1α and class II HDACs has been observed during hypoxia (22).
The incubation of the primary cells with cycloheximide resulted in reduced HDAC2 levels, but less pronounced than in CoCl2-only cells. Therefore, the levels of HIF-1α in primary cells mediate the reduction in HDAC2 transcription, just like the siRNA results in A549 cells, where the incomplete knockdown of HIF-1α resulted in a small but detectable decrease in promoter activity in the presence of the siRNA. Even small reductions in HDAC2 levels could have a clear impact on the effect of corticosteroids in various parts of a diseased tissue with high levels of inflammation, leading to non-hypoxic stabilization of HIF-1α.
Nitric oxide (NO) has been shown to impair the function of prolyl hydroxylases, thus causing the stabilization of HIF-1α under normoxic conditions (23). Exhaled NO is elevated in COPD patients and is correlated with disease severity (24). NO synthesis is also increased in rheumatoid arthritis in both macrophages and T lymphocytes (25). In cancer, where corticosteroids can be prescribed in combination with chemotherapy, endothelial nitric-oxide synthase is thought to be necessary for tumor cell maintenance (26). Increased levels of NO could stabilize HIF-1α and lead to decreased effectiveness of steroids in any of these diseases. Reactive oxygen species, especially H2O2, diminish the activity of prolyl hydroxylases by limiting the availability of Fe2+ (27), leading to the stabilization of HIF-1α. Alternatively, reactive oxygen species have been shown to activate HIF-1α transcription by activating NF-κB (28). The chronic inflammation in COPD and severe asthma patients, where high levels of oxidative stress are seen, could therefore lead to a chronic activation of HIF-1α in the lung, leading to the reduction in HDAC2, and contribute to the poor response to corticosteroids seen in these patients (29). The reduction in HDAC2 may also contribute to rheumatoid arthritis, where reactive oxygen species are also found to be elevated (30). Besides the use of CoCl2, we did not examine other non-hypoxic stabilizations of HIF-1α in our cells. Our results clearly indicate that the HDAC2 regulation is HIF-1α-specific and that a chronic non-hypoxic stabilization of HIF-1α should result in the same down-regulation of HDAC2. Additional work is needed to examine this effect under chronic inflammatory conditions to improve treatment options for patients.
Acknowledgments
We thank Misa Ito and Yasuo To for technical assistance.
This work was supported by Wellcome Trust Grant 076472/Z/05/Z and Medical Research Council Grant G0401662.
- HIF
- hypoxia-inducible factor
- HDAC
- histone deacetylase
- COPD
- chronic obstructive pulmonary disease
- FCS
- fetal calf serum
- RT
- reverse transcription
- siRNA
- small interfering RNA
- IL-1β
- interleukin-1β
- HRE
- HIF response element
- ChIP
- chromatin immunoprecipitation
- pol II
- polymerase II
- siHIF
- HIF siRNA
- siNeg
- nonspecific siRNA
- SAECs
- small airway epithelial cells.
REFERENCES
- 1.Lee K. A., Roth R. A., LaPres J. J. (2007) Pharmacol. Ther. 113, 229–246 [DOI] [PubMed] [Google Scholar]
- 2.Wenger R. H., Stiehl D. P., Camenisch G. (2005) Sci. STKE 2005, re12. [DOI] [PubMed] [Google Scholar]
- 3.Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., Schumacker P. T. (2005) Cell Metab. 1, 401–408 [DOI] [PubMed] [Google Scholar]
- 4.Blouin C. C., Pagé E. L., Soucy G. M., Richard D. E. (2004) Blood 103, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 5.Thornton R. D., Lane P., Borghaei R. C., Pease E. A., Caro J., Mochan E. (2000) Biochem. J. 350, 307–312 [PMC free article] [PubMed] [Google Scholar]
- 6.Tacchini L., De Ponti C., Matteucci E., Follis R., Desiderio M. A. (2004) Carcinogenesis 25, 2089–2100 [DOI] [PubMed] [Google Scholar]
- 7.Barnes P. J., Adcock I. M., Ito K. (2005) Eur. Respir. J. 25, 552–563 [DOI] [PubMed] [Google Scholar]
- 8.Ito K., Barnes P. J., Adcock I. M. (2000) Mol. Cell. Biol. 20, 6891–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K., Caramori G., Lim S., Oates T., Chung K. F., Barnes P. J., Adcock I. M. (2002) Am. J. Respir. Crit. Care Med. 166, 392–396 [DOI] [PubMed] [Google Scholar]
- 10.Ito K., Yamamura S., Essilfie-Quaye S., Cosio B., Ito M., Barnes P. J., Adcock I. M. (2006) J. Exp. Med. 203, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito K., Ito M., Elliott W. M., Cosio B., Caramori G., Kon O. M., Barczyk A., Hayashi S., Adcock I. M., Hogg J. C., Barnes P. J. (2005) N. Engl. J. Med. 352, 1967–1976 [DOI] [PubMed] [Google Scholar]
- 12.Smith S. J., Fenwick P. S., Nicholson A. G., Kirschenbaum F., Finney-Hayward T. K., Higgins L. S., Giembycz M. A., Barnes P. J., Donnelly L. E. (2006) Br. J. Pharmacol. 149, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonough M. A., Li V., Flashman E., Chowdhury R., Mohr C., Liénard B. M., Zondlo J., Oldham N. J., Clifton I. J., Lewis J., McNeill L. A., Kurzeja R. J., Hewitson K. S., Yang E., Jordan S., Syed R. S., Schofield C. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K. S., Rajagopal V., Gonsalves C., Johnson C., Kalra V. K. (2006) J. Immunol. 177, 7211–7224 [DOI] [PubMed] [Google Scholar]
- 15.Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 16.Komarova E. A., Krivokrysenko V., Wang K., Neznanov N., Chernov M. V., Komarov P. G., Brennan M. L., Golovkina T. V., Rokhlin O. W., Kuprash D. V., Nedospasov S. A., Hazen S. L., Feinstein E., Gudkov A. V. (2005) FASEB J. 19, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 17.Mejlvang J., Kriajevska M., Vandewalle C., Chernova T., Sayan A. E., Berx G., Mellon J. K., Tulchinsky E. (2007) Mol. Biol. Cell 18, 4615–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert B. L., Bunn H. F. (1998) Mol. Cell. Biol. 18, 4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyrwicz L. S., Gaj P., Hoffmann M., Rychlewski L., Ostrowski J. (2007) Acta Biochim. Pol. 54, 89–98 [PubMed] [Google Scholar]
- 20.Kim M. S., Kwon H. J., Lee Y. M., Baek J. H., Jang J. E., Lee S. W., Moon E. J., Kim H. S., Lee S. K., Chung H. Y., Kim C. W., Kim K. W. (2001) Nat. Med. 7, 437–443 [DOI] [PubMed] [Google Scholar]
- 21.Pluemsampant S., Safronova O. S., Nakahama K., Morita I. (2008) Int. J. Cancer 122, 333–341 [DOI] [PubMed] [Google Scholar]
- 22.Kato H., Tamamizu-Kato S., Shibasaki F. (2004) J. Biol. Chem. 279, 41966–41974 [DOI] [PubMed] [Google Scholar]
- 23.Metzen E., Zhou J., Jelkmann W., Fandrey J., Brüne B. (2003) Mol. Biol. Cell 14, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brindicci C., Ito K., Resta O., Pride N. B., Barnes P. J., Kharitonov S. A. (2005) Eur. Respir. J. 26, 52–59 [DOI] [PubMed] [Google Scholar]
- 25.Nagy G., Clark J. M., Buzas E., Gorman C., Pasztoi M., Koncz A., Falus A., Cope A. P. (2008) Immunol. Lett. 118, 55–58 [DOI] [PubMed] [Google Scholar]
- 26.Lim K. H., Ancrile B. B., Kashatus D. F., Counter C. M. (2008) Nature 452, 646–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerald D., Berra E., Frapart Y. M., Chan D. A., Giaccia A. J., Mansuy D., Pouysségur J., Yaniv M., Mechta-Grigoriou F. (2004) Cell 118, 781–794 [DOI] [PubMed] [Google Scholar]
- 28.Bonello S., Zähringer C., BelAiba R. S., Djordjevic T., Hess J., Michiels C., Kietzmann T., Görlach A. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 755–761 [DOI] [PubMed] [Google Scholar]
- 29.Barnes P. J. (2007) PLoS Med. 4, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippin L. I., Vercelino R., Marroni N. P., Xavier R. M. (2008) Clin. Exp. Immunol. 152, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]