Abstract
The SH2 domain containing inositol 5-phosphatase SHIP2 contains several interacting domains that are important for scaffolding properties. We and others have previously reported that SHIP2 interacts with the E3 ubiquitin ligase c-Cbl. Here, we identified human SHIP2 monoubiquitination on lysine 315. SHIP2 could also be polyubiquitinated but was not degraded by the 26 S proteasome. Furthermore, we identified a ubiquitin-interacting motif at the C-terminal end of SHIP2 that confers ubiquitin binding capacity. However, this ubiquitin-interacting motif is dispensable for its monoubiquitination. We showed that neither c-Cbl nor Nedd4-1 play the role of ubiquitin ligase for SHIP2. Strikingly, monoubiquitination of the ΔSH2-SHIP2 mutant (lacking the N-terminal SH2 domain) is strongly increased, suggesting an intrinsic inhibitory effect of the SHIP2 SH2 domain on its monoubiquitination. Moreover, SHIP2 monoubiquitination was increased upon 30 min of epidermal growth factor stimulation. This correlates with the loss of interaction between the SHIP2 SH2 domain and c-Cbl. In this model, c-Cbl could mask the monoubiquitination site and thereby prevent SHIP2 monoubiquitination. The present study thus reveals an unexpected and novel role of SHIP2 SH2 domain in the regulation of its newly identified monoubiquitination.
Introduction
SH2 domain-containing inositol phosphate 5-phosphatase 2 (SHIP2)3 is a type II lipid phosphatase that hydrolyzes the 5-phosphate of the phosphorylated inositol ring of inositol and phosphatidylinositol molecules (1, 2). Phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3), which is produced by class I phosphatidylinositol 3-kinase, is the major substrate of SHIP2 and is converted into PtdIns(3,4)P2 by its enzymatic activity (3, 4). In addition to its central catalytic domain, SHIP2 contains an N-terminal SH2 domain, and in its C-terminal part an NPXY motif, a proline-rich region, and a sterile α motif (SAM). Through these domains, SHIP2 interacts with numerous protein partners (the EGFR, p130(Cas), Cbl, Vinexin, Arap3, APS, JIP-1, and Intersectin) (5–12). Beside its phosphoinositide phosphatase activity, the scaffold properties of SHIP2 could also contribute to its function in signaling. For example, we showed that, through direct interaction with JIP1, SHIP2 was able to up-regulate JIP1-mediated JNK activation as well as to increase JIP1 tyrosine phosphorylation (12). SHIP2 mRNA and proteins are ubiquitously expressed in human, rat, and mouse (3, 4, 13). SHIP2 appears to be tyrosine phosphorylated by a very large number of extracellular ligands (EGF, platelet-derived growth factor, macrophage-colony stimulating factor, insulin, and human growth factor), but the biological significance of SHIP2 tyrosine phosphorylation remains controversial (5, 14–17). SHIP2 also functions in remodeling of actin structures, cell adhesion and spreading, and receptor endocytosis (18, 19). In vitro and in vivo studies in mice also showed that SHIP2 negatively regulates insulin signaling (20–23).
We and others have previously shown that SHIP2 is a c-Cbl partner in mammalian cells (8, 18). The mammalian Cbl family of proteins consists of three homologues known as c-Cbl, Cbl-b, and Cbl-c (also known as Cbl-3). All three Cbl proteins share highly conserved domains. c-Cbl and Cbl-b function as adaptor proteins by interacting with other signaling molecules through their various protein-protein interacting motifs. They share a C-terminal proline-rich region containing potential tyrosine phosphorylation sites and a ubiquitin-associated domain (UBA) (24). The three Cbl proteins also contain an N-terminal tyrosine kinase-binding domain and a catalytic RING finger domain responsible for their E3 ubiquitin ligase activity. Their E3 ubiquitin ligases activity mediates the ubiquitination of different activated substrates (e.g. EGFR, platelet-derived growth factor receptor, CSF-1, Met, Src, Abl, and Sprouty, among others) and targets them for degradation (25–30).
Polyubiquitinated proteins are subjected to proteolysis by the 26 S proteasome when lysine 48 of ubiquitin is the site of chain formation. Ubiquitin chains formed through lysine 63 are rather involved in a variety of processes, including DNA repair, translation, IκB kinase activation, endocytosis, and protein transport (31). Monoubiquitination has been associated with endocytosis, virus budding, chromatin remodeling, and transport of proteins in different cellular compartments (32–34). Moreover, many proteins (ubiquitin receptors) harbor ubiquitin binding domains/motifs (UBD), which interact with mono- and/or polyubiquitin chains (35). To date, eleven families of UBDs have been identified (36). Thus, ubiquitinated proteins and ubiquitin receptors constitute a signaling network within the cell.
In this study, we identified a ubiquitin-interacting motif (UIM) at the C-terminal extremity of SHIP2 that confers to SHIP2 the capacity to bind ubiquitin. We have observed that SHIP2 is monoubiquitinated by a process that is actively controlled by the ability of its SH2 domain to mask the monoubiquitination site.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
COS-7 and Hek293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), 2.5 μg/ml fungizone (Invitrogen), and 2 mm sodium pyruvate (Invitrogen). CHO-IR cells (Chinese hamster ovary cells stably overexpressing the human insulin receptor) were maintained in Ham's F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 2.5 μg/ml and 400 μg/ml Geneticin (G418, Invitrogen). Cells were grown at 37 °C in a 5% CO2 humidified atmosphere. For EGF and insulin stimulation, 16-h serum-starved cells were stimulated by 50 ng/ml EGF or 100 nm insulin for the indicated times.
Reagents and Antibodies
Lactacystin, cycloheximide, and Nonidet P-40 were purchased from Calbiochem. Mouse monoclonal anti-FLAG (M2), mouse monoclonal anti-Cyclin D1, rabbit polyclonal anti-β-actin, anti-β-catenin, and anti-Myc (9E10) antibodies were purchased from Sigma; mouse monoclonal anti-His was from BD Transduction Laboratories; mouse monoclonal anti-HA was from Roche Molecular Biochemicals; mouse monoclonal anti-phosphotyrosine (4G10) was from Upstate; mouse monoclonal anti-Ubiquitin (P4D1), rabbit polyclonal anti-EGFR (1005), and mouse monoclonal anti-Cbl (A-9) were from Santa Cruz Biotechnology; rabbit polyclonal anti-laminB1 was from Abcam; and mouse monoclonal anti-tubulin αAb2 was from NeoMarkers. Rabbit polyclonal SHIP2 and polyclonal anti-Type I 5-PPase I antibodies have been previously reported (13, 37).
Plasmids and Site-directed Mutagenesis
The human His-tagged SHIP2 and certain mutants (SHIP2-(18–935) or tSHIP2 mutant, SHIP2-(18–1194) or ΔSAM mutant, SHIP2-(885–1184) or PRO mutant, and ΔSH2-SHIP2 mutant) have been described before (3, 5, 8, 38). The human c-Cbl and FLAG-tagged ubiquitin cDNAs were kind gifts from Dr. Wallace Y. Langdon (University of Western Australia). The human Cbl-b cDNA was kindly provided by Dr. Stanley Lipkowitz (National Naval Medical Center, Bethesda, MD). The full-length c-Cbl and Cbl-b cDNAs were subcloned into a pcDNA3-HA vector. The human tagged-HA SHIP2 cDNA was a kind gift from Dr. Mitchell (Monash University, Clayton, Australia). The mouse FLAG-tagged Eps15 cDNA was kindly provided from Dr. S. Sigismund (Institute for Molecular Oncology, Milan, Italy). The human HA-tagged coiled-coil activator cDNA was kindly provided by Dr. Yang (University of Southern California, Los Angeles, CA). Ubc9 cDNA was a kind gift from Dr. Niedenthal (Hanover Medical School, Germany). FLAG-tagged SUMO-1 and SUMO-2 cDNA were kindly provided by Dr. LeGoff (University of Rennes, France). SHIP2 constructs corresponding to amino acids 935–1258, 18–650, and 651–1258 were prepared with SHIP2 as template and a 5′-primer containing an EcoRI restriction site (underlined) and a 3′-primer containing an XbaI site (underlined). The primers were: 5′-primer (5′-GGAATTCGTGGAGAGAAACCGCCACCAACGGGGAG-3′) and 3′-primer (5′-GCTCTAGAGTCACTTGCTGAGCTGCAGGGTG-3′) for the SHIP2-(935–1258); 5′-primer (5′-GGAATTCGTGGAGCCCCCTCCTGGTACCACCGCGAC-3′) and 3′-primer (5′-GCTCTAGATGAATCGAAGGAAGACCTTGTG-3′) for the SHIP2-(18–650); and 5′-primer (5′-GGAATTCGTGGAAGTGAGGAGGAGATCTCCTTC-3′) and 3′-primer (5′-GCTCTAGAGTCACTTGCTGAGCTGCAGGGTG-3′) for the SHIP2-(651–1258). The PCR products were subcloned in a pcDNA3-HisC vector. Site-directed mutagenesis was conducted by using the QuikChange site-directed mutagenesis kit according to the manufacturer's instruction (Stratagene). The mutants K315R, K515R, and K962R were generated from the full-length His-tagged SHIP2 construct by replacing the target lysines by arginine. The ΔUIM mutant was generated from the full-length His-tagged SHIP2 construct by replacing aspartate 1117, 1118, and 1134, leucine 1124, serine 1131, and glutamate 1132 by alanine, and alanine 1127 by glycine. The HA-tagged C381A mutant was generated from the full-length HA-c-Cbl construct by replacing cysteine 381 by alanine. All mutations were confirmed by DNA sequencing.
Transfections
COS-7 cells were plated at 1.5 × 106 cells/10-cm dish and CHO-IR cells at 2 × 106 cells/10-cm dish the day before transfection. To transfect the cells, we alternatively used 3 μg of FLAG-tagged ubiquitin, 4 μg of His-tagged SHIP2 and mutants, 3 μg of HA-tagged Cbl and mutants, 2 μg of Type I 5-PPase, 3 μg of Myc-tagged SHIP1, 3 μg of FLAG-tagged SUMO, 1 μg of HA-tagged coiled-coil activator, 3 μg of Ubc9, or 2 μg of FLAG-tagged Eps15 expressing plasmids using FuGENE 6 according to the manufacturer's instructions (Roche Diagnostics). Empty vector was used to equilibrate transfected DNA amounts. Transfection of HEK293T was performed using calcium phosphate based on the procedure described previously (39).
Immunoprecipitation and Immunoblotting
36 h after transfection, cells cultured in 100-mm dishes were washed twice with ice-cold phosphate-buffered saline, solubilized in 500 μl of lysis buffer (80 mm Tris-HCl, pH 7.5, 150 mm NaCl, 20 mm EDTA, 200 mm NaF, 1% Brij, 4 mm sodium vanadate, 5 mm Na4P2O7, 1 μm okadaic acid, and a mix of protease inhibitors (Complete, Roche Molecular Biochemicals)) by constant agitation for 30 min at 4 °C, and centrifuged at 14,000 × g for 20 min at 4 °C. 50 μg of total cell extracts was tested by Western blot to verify proteins expression. The supernatant (of one-half cell extract = 625 μg of total proteins) was incubated with the indicated antibodies by constant agitation for 2 h at 4 °C. Immune complexes were precipitated with protein G-Sepharose beads (Amersham Biosciences) by constant rotation for 2 h at 4 °C. Immunoprecipitates were collected by centrifugation, washed three times with lysis buffer, and boiled for 5 min in Laemmli buffer (125 mm Tris, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.01% bromphenol blue). The solubilized proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (Amersham Biosciences). The membranes were blocked with 5% nonfat dry milk in TBSN (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Nonidet P-40) for 40 min and subsequently incubated with primary antibodies in TBSN, washed three times with TBSN, and incubated with horseradish peroxidase-conjugated secondary antibody (1/25,000 dilution). After extensive washing, the membranes were incubated with enhanced chemiluminescence reagent (ECL, PerkinElmer Life Sciences). Sumoylation experiments were done in lysis buffer supplemented with 20 mm N-ethylmaleimide (Sigma). Quantification of the data was performed by using densitometric analysis of the autoradiography (Scion Image Software) or through Odyssey method. Statistical significance was performed by using Student's t test. A p value of <0.5 was considered to be statistically significant.
Pulldown Assay with Ubiquitin Beads
Two days after transfection of the expression constructs, COS-7 cells in 100-mm dishes were lysed in 500 μl of lysis buffer. After centrifugation at 12,000 × g for 15 min at 4 °C, the supernatants (of one-fourth cell extracts, 250 μg) were incubated with 30 μl of ubiquitin-agarose beads (Boston Biochem, Cambridge, MA) or with protein A-agarose beads (Sigma-Aldrich) for 3 h at 4 °C. The beads were then washed three times with washing buffer (10 mm Tris-HCl, pH 7.4, 100 mm NaCl, and 0.1% Nonidet P-40), and the bound proteins were eluted by boiling with 5× Laemmli sample buffer. Eluted proteins were separated by SDS-PAGE, and proteins were detected by immunoblotting and the enhanced chemiluminescence reagent (Amersham Biosciences).
Mass Spectrometry Analysis
COS-7 cells were transfected with plasmids encoding His-tagged SHIP2 and FLAG-tagged ubiquitin and cultured for 2 days. His-tagged SHIP2 was immunoprecipitated from 15 mg of proteins with anti-His monoclonal antibodies and separated by 7.5% SDS-PAGE. The two bands corresponding to immunoprecipitated SHIP2 were excised from the silver nitrate-stained gel and subjected to tryptic digestion with a MassPrep robot (Micromass), as described before (Shevshenko et al. (72)). Mass spectrometry analysis of the digested fragments was performed on a Q-TOF Ultima Global mass spectrometer equipped with a MALDI source. Microsequencing was performed by argon-induced fragmentation after selection of the parent ion.
Malachite Phosphatase Assay
PtdIns(3,4,5)P3 5-phosphatase activity was determined with the phosphate release assay using an acidic malachite green dye (40). Dioctanoylglycerol PtdIns(3,4,5)P3 was diluted in 30 μl of assay buffer (50 mm Hepes, pH 7.4, 2 mm MgCl2, 1 mg/ml bovine serum albumin). The phosphatase reaction was initiated by adding immunoprecipitated SHIP2 diluted in assay buffer (15 μl) to the substrate. Samples were incubated at 37 °C. After 7 min, reactions were stopped by the addition of 15 μl of 0.1 m EDTA. 75 μl of malachite green reagent (41) was added to 50 μl of the reaction solution. Samples were allowed to stay for 10 min for color development before measuring absorbance at 650 nm. Inorganic phosphate release was quantified by comparison to a standard curve of KH2PO4 in dH2O. We checked the linearity of the assay with respect of time and protein concentration. The enzymatic blank was made by adding the EDTA solution to the substrate before the enzyme. Each value results from triplicate determinations ± S.E.
Subcellular Fractionation
COS-7 cells (3 × 106 in 90-mm Petri dishes) were transiently transfected using FuGENE 6 (Roche Applied Science). Cells from 10 dishes were harvested at 48 h after transfection, on ice, in 2 ml of a homogenization medium containing 0.25 m sucrose, 3 mm imidazole, pH 7.0, 1 mm EDTA, and complete protease inhibitor mixture (Roche Applied Science). After a first homogenization by 10 passages through a tight Dounce homogenizer (Wheaton), the material was centrifuged at 1,200 × g for 10 min. The upper third of the supernatant was collected as a postnuclear supernatant (E), and the remainder was homogenized again by two Dounce passages and centrifugations at 925 × g for 7 and 5 min. Each time, the upper third was removed and added in fraction E before repeating Dounce homogenization and centrifugation. The final supernatant was entirely removed and combined with E fraction. To maximize subcellular particle extraction, 1 ml of the homogenization medium was again added to the pellet that was passed through the Dounce homogenizer and pelleted again at 765 × g for 2 min, a procedure repeated several times until reaching a final volume of 10 ml in fraction E. The final pellet, denoted the N fraction, was resuspended in 0.5 ml of homogenization medium. Pooled supernatants (denoted the E fraction) were submitted to high speed sedimentation in a Ti-75 rotor (Beckman) to yield postnuclear particles (referred to as the MLP pellet) at 110,000 × g for 60 min. The high speed supernatant is denoted the S fraction. Postnuclear particles were resuspended in 1 ml of homogenization medium and homogenized by ten passages through a loose Dounce homogenizer. All the homogenization and extraction procedures were efficient, because between 75 and 100% of the total proteins were recovered in the S and MLP fractions. Protein concentrations were determined in fractions E, N, S, and MLP, equivalent quantities were separated by SDS-PAGE and analyzed by immunoblotting.
RESULTS
SHIP2 Associates with c-Cbl and Cbl-b Proteins
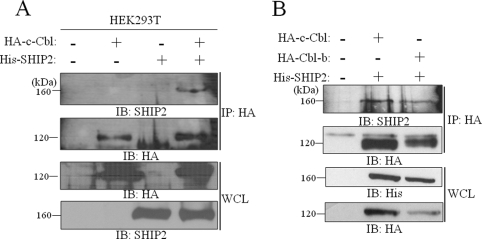
We previously reported that SHIP2 interacts with endogenous c-Cbl in CHO-IR cells in coimmunoprecipitation experiments (8). We confirmed this association in human HEK293T cells (Fig. 1A). Because Cbl-b presents a large sequence identity with c-Cbl, we tested whether SHIP2 could also interact with Cbl-b in COS-7 cells. As shown in Fig. 1B, His-tagged SHIP2 coimmunoprecipitated with HA-tagged Cbl-b indicating an association of SHIP2 with both forms of Cbl in intact cells. It was controlled that SHIP2 alone did not precipitate with anti-HA antibody (Fig. 1A, lane 3).
FIGURE 1.
SHIP2 associates with c-Cbl and Cbl-b proteins. A, cell lysates were prepared from HEK293T cells transfected with His-tagged SHIP2 and HA-tagged c-Cbl. Appropriate expression of c-Cbl and SHIP2 was confirmed with anti-HA and anti-SHIP2 antibodies, respectively (WCL) IB, immunoblotted; IP, immunoprecipitated. B, cell lysates prepared from COS-7 cells transfected with His-tagged SHIP2 and HA-tagged c-Cbl or HA-tagged Cbl-b were immunoblotted with anti-HA or anti-His antibodies. In both A and B, anti-HA immunoprecipitates were separated by SDS-PAGE and immunoblotted with anti-SHIP2 or anti-HA antibodies.
SHIP2 Is Monoubiquitinated
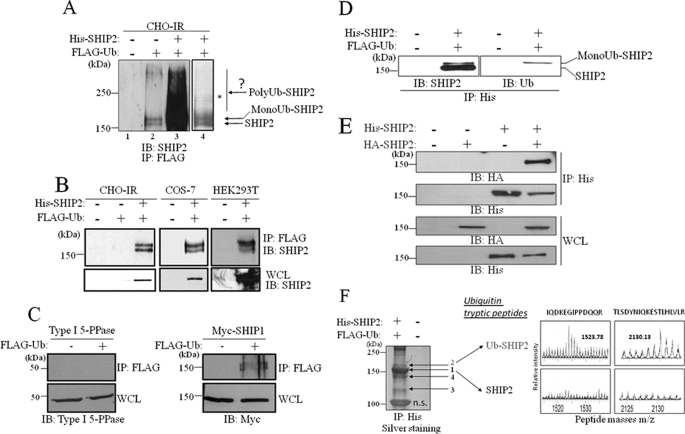
Given the association of SHIP2 with Cbl proteins, which presents an E3 ubiquitin ligase activity, we questioned if SHIP2 could be a target for Cbl-mediated ubiquitination. First, we examined whether SHIP2 could be ubiquitinated in intact cells by an in vivo ubiquitination assay. FLAG-tagged ubiquitin was transfected in CHO-IR cells, and ubiquitinated proteins were immunoprecipitated from cell lysates with an anti-FLAG antibody. Endogenous SHIP2 protein was immunodetected with anti-SHIP2 antibody (Fig. 2A, lane 2). We detected the ubiquitinated form of SHIP2 at 170 kDa showing that SHIP2 can be ubiquitinated when expressed at physiological levels.
FIGURE 2.
Monoubiquitination of SHIP2. A, CHO-IR cells were transiently transfected with His-tagged SHIP2 and FLAG-tagged ubiquitin. Anti-FLAG immunoprecipitates were resolved by SDS-PAGE, and endogenous or transfected SHIP2 were both revealed by immunoblotting with anti-SHIP2 antibodies. B, lysates of CHO-IR, COS-7, and HEK293T cells transiently transfected with FLAG-tagged ubiquitin, and His-tagged SHIP2 were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted (IB) with anti-SHIP2 antibodies. C, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and Type I 5-PPase or Myc-tagged SHIP1. Anti-FLAG immunoprecipitates with anti-FLAG antibodies were immunoblotted with anti-Type I 5-PPase or anti-Myc antibodies, respectively. D, CHO-IR cells were transiently transfected with His-tagged SHIP2 and FLAG-tagged ubiquitin. Anti-His immunoprecipitates were resolved by SDS-PAGE and detected by immunoblotting with anti-SHIP2 and anti-ubiquitin (anti-Ub) antibodies. E, COS-7 cells were transiently transfected with HA-tagged SHIP2 and/or His-tagged SHIP2. Anti-His immunoprecipitates were immunoblotted with anti-HA antibodies. F, COS-7 cells were transfected with His-tagged SHIP2 and FLAG-tagged ubiquitin. Anti-His immunoprecipitated proteins were separated by SDS-PAGE and silver stained. Arrows indicate bands of stained proteins excised for enzymatic digestion by trypsin and subsequent analysis by MALDI-Q-TOF mass spectrometry. In-gel tryptic digests of the control SHIP2 (band 1) and the monoubiquitinated SHIP2 (band 2) were analyzed by MALDI-Q-TOF mass spectrometry. Mass spectrometry spectra (enlarged) of the ubiquitin peptides isolated in monoubiquitinated SHIP2 (band 2) are shown. n.s., band is unspecific.
This result was confirmed overexpressing His-SHIP2 in CHO-IR cells (Fig. 2A, lanes 3 and 4; Fig. 2B, left panel (intermediate exposure)). Similar data were obtained in COS-7 and HEK293T cells transfected by the same constructs (Fig. 2B, middle and right panels).
We then tested whether other inositol phosphate 5-phosphatases, Type I 5-PPase and SHIP1, could also be ubiquitinated. Type I 5-PPase was not ubiquitinated under the same experimental conditions (Fig. 2C, left panel), whereas SHIP1 was monoubiquitinated (Fig. 2C, right panel).
Interestingly, for both SHIP1 and SHIP2, the antibody revealed two bands with a molecular mass that differs by ∼10 kDa. We detected the ubiquitinated form of SHIP2 at 170 kDa, as well as, surprisingly, a band at 160 kDa corresponding to the molecular mass of non-ubiquitinated SHIP2 (160 kDa). The presence of the lower band suggested that ubiquitinated SHIP2 associated with non-ubiquitinated SHIP2, directly or indirectly. The same observation was made for SHIP1 with molecular masses of ∼145 and 155 kDa. To identify the ubiquitinated form of SHIP2 in the doublet, we specifically immunoprecipitated SHIP2 with anti-His antibody and revealed ubiquitin (Fig. 2D, right panel). Only the 170-kDa form of SHIP2 was detected, whereas anti-His antibody revealed both bands as SHIP2 protein (Fig. 2D, left panel). The hypothesis that the interaction between SHIP2 and itself is the consequence of ubiquitination was discarded. Indeed, differentially His- and HA-tagged SHIP2 were coimmunoprecipitated in COS-7 cells where ubiquitin was not overexpressed (Fig. 2E) confirming that two molecules of SHIP2 can be associated.
As the molecular mass of one ubiquitin molecule is 10 kDa, the observed shift in molecular mass of SHIP2 could be caused by the attachment of a single ubiquitin molecule. This was assessed by mass spectrometry analysis. FLAG-tagged ubiquitin and His-tagged SHIP2 or empty vector were transfected in COS-7 cells. SHIP2 was immunoprecipitated with anti-His. Precipitated proteins were separated by SDS-PAGE and stained with silver (Fig. 2F). Both bands at 160 and 170 kDa (respectively, bands 1 and 2 in Fig. 2F) were excised, subjected to tryptic digestion, and analyzed by MALDI-Q-TOF mass spectrometry to identify SHIP2 and ubiquitin peptide fragments. Both bands proved to be derived from SHIP2 as the digested peptides corresponded with the expected sequences of SHIP2 (supplemental Fig. S1, A and B). The upper band generated additional peptides with m/z values of 1523.78 and 2130.13, corresponding to ubiquitin tryptic peptides, whereas the lower band did not (Fig. 2F). Microsequencing of these peptides by fragmentation confirmed that they corresponded to ubiquitin sequences (supplemental Fig. S2). In the same mass spectrometry experiment, the 120-kDa protein coimmunoprecipitated with SHIP2 (Fig. 2F, band 3) was also excised, subjected to tryptic digestion, and analyzed by MALDI-Q-TOF mass spectrometry. Two ions were selected for tandem mass spectrometry analysis, and peptide sequence information unequivocally identified this protein as c-Cbl (supplemental Fig. S3) by peptide mass fingerprint analysis using the SwissProt program. Band 4 on the Fig. 2F derived from degraded SHIP2 (supplemental Fig. S4).
SHIP2 Could Be Polyubiquitinated in Vivo but Is Not Degraded by the 26 S Proteasome
Fig. 2A (lanes 3 and 4) revealed a pattern above the monoubiquitinated SHIP2 suggesting that SHIP2 could also be polyubiquitinated. In most cases, polyubiquitination targets modified proteins for degradation by the 26 S proteasome. To determine whether SHIP2 could be degraded by the proteasomal system, CHO-IR cells were treated for various times with or without the 26 S proteasome inhibitor lactacystin. Lactacystin did not affect the protein level of endogenous SHIP2, while the positive control protein β-catenin, which is degraded via the ubiquitin-proteasome pathway (supplemental Fig. S5A) (42), clearly accumulated upon treatment with this inhibitor. Similar results were obtained after treatment of the cells by the aldehyde peptide MG132, another 26 S proteasome inhibitor (data not shown). These results suggest that the protein level of endogenous SHIP2 is not regulated by the 26 S proteasome degradation system in CHO-IR cells under basal conditions.
Simultaneously, we evaluated the stability of endogenous SHIP2 protein. CHO-IR cells were treated for various times with the protein synthesis inhibitor cycloheximide, and endogenous SHIP2 turnover was monitored during 24 h (supplemental Fig. S5B). No change in the level of endogenous SHIP2 was detected. We reprobed the membrane with anti-β-actin antibodies to confirm equal loading of proteins and with anti-cyclin D1 antibodies as positive control (43). Similar data were obtained in 3T3-L1 and C2C12 cells, with the mRNA synthesis inhibitor Actinomycin D and with the protein synthesis inhibitor Puromycin (data not shown). These results show that SHIP2 is a stable protein with a half-life longer than 24 h.
Mapping of the Ubiquitination Domain in SHIP2
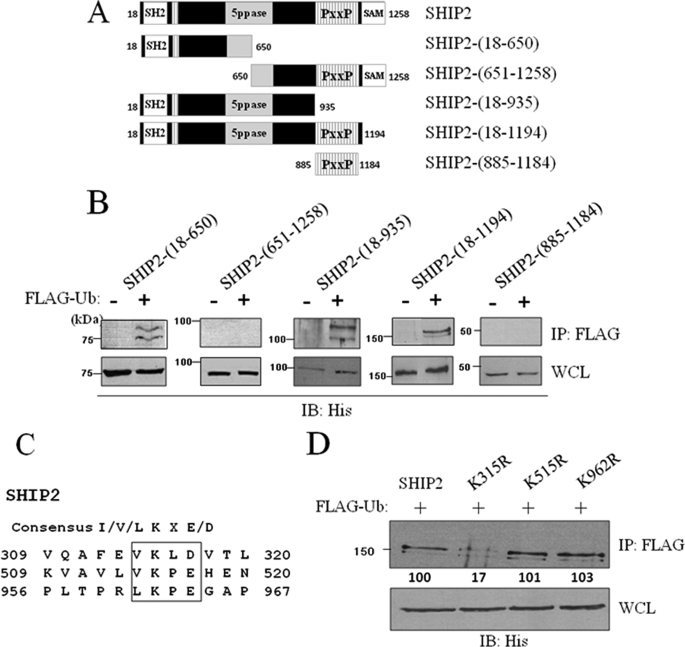
We have determined the regions of SHIP2 necessary for its monoubiquitination. We created two deletion mutants (SHIP2-(18–650) and SHIP2-(651–1258)) from wild-type SHIP2 (Fig. 3A) and verified their ubiquitination by an ubiquitination assay in COS-7 cells. The N-terminal SHIP2-(18–650) mutant (75 kDa) was monoubiquitinated, whereas the C-terminal SHIP2-(651–1258) mutant (85 kDa) was not (Fig. 3B). The monoubiquitination of the SHIP2-(18–650) mutant indicated that this N-terminal region of SHIP2 contains a monoubiquitination site and that amino acids 651–1258 are dispensable for this post-translational modification. We confirmed these results in COS-7 cells transiently transfected with three other deletion SHIP2 mutants (SHIP2-(18–935) (100kDa), SHIP2-(18–1194) (150 kDa), and SHIP2-(885–1184) (40 kDa)) (Fig. 3A). As shown in Fig. 3B, SHIP2 mutants, which contain amino acids 18–650, were monoubiquitinated unlike those that did not contain this region.
FIGURE 3.
Mapping of the lysine 315 as ubiquitination site in SHIP2. A, deletion mutants of wild-type SHIP2. B, COS-7 cells were transiently transfected with His-tagged SHIP2-(18–650), His-tagged SHIP2-(651–1258), His-tagged SHIP2-(18–935), His-tagged SHIP2-(885–1184), or His-tagged SHIP2-(18–1194) mutants of SHIP2. Cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted with an anti-His antibody. Expression of SHIP2 mutants was confirmed by immunoblotting of the cell lysates with an anti-His antibody. C, sequence analysis of SHIP2 showed that SHIP2 contains three consensus ubiquitination/sumoylation sites (lysine residues in position 315, 515, and 962). D, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and His-tagged SHIP2, His-tagged K315R, His-tagged K515R, or His-tagged K962R. Expression of SHIP2 or mutants was confirmed with anti-His antibody. Cell lysates were immunoprecipitated with an anti-FLAG antibody, immunoblotted with an anti-His antibody, and SHIP2 ubiquitination was quantified by densitometric analysis. Each value represents the mean ± S.E. of four independent experiments (*, p < 0.05 compared with control).
Lysine 315 Is the Major Site for SHIP2 Ubiquitination
To identify precisely the target lysine residue that could be implicated in the monoubiquitination of SHIP2, mass spectrometry was used. Analysis of ubiquitinated proteins by trypsin digestion cleaves the original ubiquitin molecule leaving a di-peptide (-GG) that adds a monoisotopic mass of 114.04 Da to the targeted lysine residue; sometimes miscleavage in ubiquitin generates a longer tag (-LRGG) (supplemental Fig. S6A). In addition, the modified peptides carry a proteolytic miscleavage, because trypsin digestion cannot occur at the modified lysine site due to sterical hindrance by the ubiquitin molecule. Both modifications lead to unique mass spectrometry spectra for peptides modified by ubiquitination. His-tagged SHIP2 and FLAG-tagged ubiquitin or empty vector were transfected in COS-7 cells, and SHIP2 was immunoprecipitated with anti-His. Precipitated proteins were separated by SDS-PAGE and stained with silver (Fig. 2F). The band at 170 kDa (band 2 on the Fig. 2F) was excised and processed for mass spectrometry. This revealed the presence of a 2301.2 m/z value peptide, corresponding to the miscleaved peptide of SHIP2 with attached GG moieties derived from the C terminus of ubiquitin (supplemental Fig. S6B). The detected peptide mass corresponded to a monoubiquitinated branched peptide centered on lysine 315 of SHIP2. The purified peptide was in too low quantity to be sequenced.
Interestingly, lysine 315 is fully conserved among all members of the SHIP2 family in various species and located in a sumoylation/ubiquitination consensus sequence ΦKXe (Φ, hydrophobic residue; K, lysine; X, amino acid; and e, acidic residue) (Fig. 3C) (44, 45). Two additional sumoylation/ubiquitination consensus sequences are present in SHIP2: one located in the N-terminal 18–650 region (lysine 515), and a second one is located in the proline-rich domain of the C-terminal region of SHIP2 (lysine 962). To evaluate the respective implication of these three lysine residues in SHIP2 monoubiquitination, we mutated separately each lysine residue to arginine and performed an ubiquitination assay in COS-7 cells. Mutation of lysine 515 or lysine 962 did not affect SHIP2 monoubiquitination, whereas with the K315R mutant, a loss of SHIP2 monoubiquitination as compared with the wild type was observed when lysine 315 was mutated (Fig. 3D). Our results thus demonstrate that lysine 315 is the main target site for SHIP2 monoubiquitination.
In addition, to test whether SHIP2 could be covalently modified by SUMO, COS-7 cells were transiently transfected with His-tagged SHIP2, Ubc9, and FLAG-tagged SUMO-1 or SUMO-2. Cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-His antibody. No sumoylated SHIP2 could be observed in COS-7 cells that co-express SUMO-1 or SUMO-2 with SHIP2 (supplemental Fig. S7A), whereas we could reproduce in our experimental conditions the already described sumoylation of coiled-coil activator (supplemental Fig. S7B) (46).
SHIP2 Binds Ubiquitin by a UIM Motif
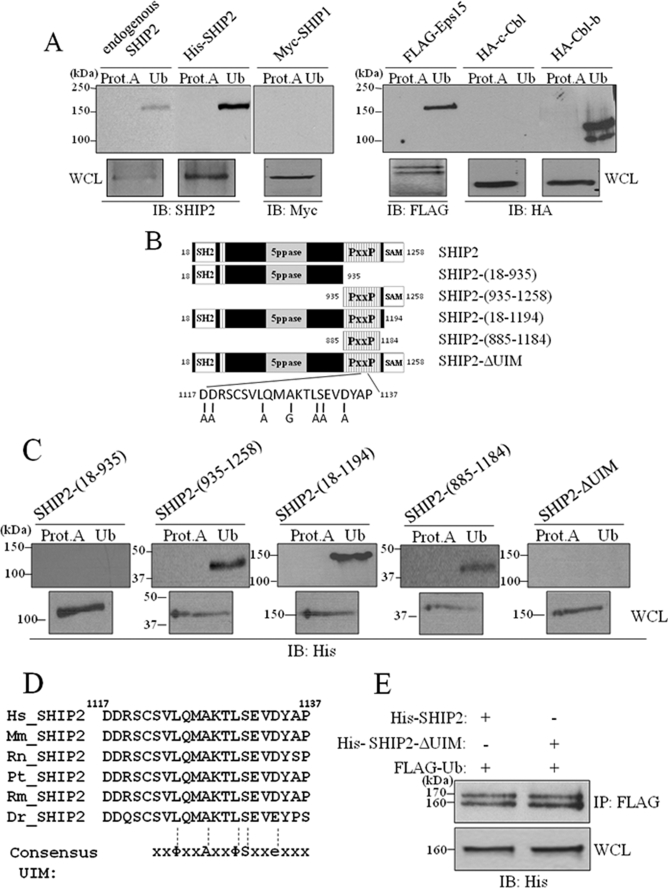
It is well documented that proteins that undergo monoubiquitination have an intrinsic propensity to bind to monoubiquitin (35, 47–49). Therefore, we investigated whether SHIP2 can interact with ubiquitin. Lysates were prepared from intact COS-7 cells or COS-7 cells overexpressing His-tagged SHIP2 (whole cell lysate corresponds to 50 μg of cell extract). 300 μg of the extract were incubated with agarose beads covalently bound to ubiquitin. By SDS-PAGE and immunoblotting analysis with anti-SHIP2 antibodies, we found that endogenous and overexpressed SHIP2 bound to ubiquitin-agarose beads, but not to our control beads of protein-A-agarose (Fig. 4A). The same results were obtained for Eps15 and Cbl-b, known to bind ubiquitin (50, 51) (Fig. 4A). c-Cbl was used as a negative control (51). These data suggest that SHIP2 is able to bind to ubiquitin.
FIGURE 4.
SHIP2 binds monoubiquitin by a UIM motif. A, one-fourth lysates of non-transfected COS-7 cells or of COS-7 cells transfected with His-tagged SHIP2, FLAG-tagged Eps15, HA-tagged c-Cbl, HA-tagged Cbl-b, or Myc-tagged SHIP1 were incubated with ubiquitin-agarose (Ub) or protein A-agarose (Prot.A) for 4 h at 4 °C. Bound proteins were analyzed by immunoblotting (IB) with anti-SHIP2, anti-FLAG, anti-HA, or anti-Myc antibodies for the indicated proteins. Expression of transfected proteins was confirmed by immunoblotting of 1/25 lysates with corresponding antibodies. B, schematic representation of the deletion mutants of wild-type SHIP2 we used. C, COS-7 cell lysates transfected with indicated mutants of SHIP2 were incubated with ubiquitin-agarose (Ub) or protein A-agarose (Prot.A) for 4 h at 4 °C. Bound proteins were analyzed by immunoblotting with an anti-His antibody. Expression of transfected proteins was confirmed by immunoblotting of 1/25 lysates with an anti-His antibody. D, alignment of human SHIP2 (amino acids 1117–1137) with its orthologs from different organisms. Consensus UIM is aligned with SHIP2 sequences: Φ, hydrophobic residue; A, alanine; S, serine; and e, acidic residue. E, COS-7 cells were transiently transfected with His-tagged SHIP2 or His-tagged SHIP2-ΔUIM mutant and FLAG-tagged ubiquitin. Cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted with an anti-His antibody. Expression of transfected SHIP2 proteins was confirmed with an anti-His antibody.
To identify the ubiquitin-binding region of SHIP2, several truncated mutants of SHIP2, described in Fig. 4B, were tested for their ability to bind to ubiquitin-agarose beads. As shown in Fig. 4C, the SHIP2-(18–935) mutant failed to interact with ubiquitin, whereas the three other mutants were still able to bind to ubiquitin. Thus, only the proline-rich domain of SHIP2 (residues 885–1184) has a strong ability to bind to ubiquitin, which suggests that the ubiquitin-binding region of SHIP2 resides within the C terminus, between amino acids 885 and 1184. In this region we identified a putative UIM domain (residing between amino acid residues 1117 and 1137) (Fig. 4D). The UIM domain is a single α-helix that binds to monoubiquitin (36, 52). The peptide sequence of the UIM domain consists of a highly conserved ΦXXAXXΦSXXe core (Φ, hydrophobic residue; A, alanine; S, serine; X, amino acid; and e, acidic residue), which is preceded by a block of acidic residues (51, 53). Sequence alignment of the region comprised between amino acid 1117 and 1137 of different species of SHIP2 showed a strong conservation (Fig. 4D).
To further confirm that the UIM domain is responsible for binding to ubiquitin, we replaced each conserved residue of the putative UIM domain with alanine, or with glycine if alanine was already present, in the full-length His-tagged SHIP2. The ubiquitin interaction ability of the UIM mutant (SHIP2-ΔUIM) was tested and is shown in Fig. 4C. In SHIP2-ΔUIM the critical amino acids of the domain were mutated in SHIP2, and this abolished the interaction with ubiquitin. A partial inhibition was also obtained for L1124A-S1131A, D1117A-D1118A, and L1124A/S1131A/D1134A mutants (data not shown). Together, these findings indicate that SHIP2 contains a UIM domain necessary for interaction with ubiquitin. We also demonstrated that SHIP1 cannot bind to ubiquitin (Fig. 4A), which is consistent with the fact that SHIP1 does not contain such a UIM domain.
Woelk et al. showed that covalent monoubiquitination of several proteins harboring UBDs depends on the ability of the UBD to engage ubiquitin noncovalently, a process termed “coupled monoubiquitination” (54). To test if SHIP2 monoubiquitination depends on its UIM domain, we performed a ubiquitination assay in COS-7 cells transiently transfected with SHIP2 or the UIM mutant of SHIP2 and ubiquitin. As shown in Fig. 4E, mutation of the UIM motif did not affect SHIP2 monoubiquitination. Moreover, in Fig. 3B, we showed that the C-terminal mutant of SHIP2-(18–650) and SHIP2-(18–935), which lacks the UIM domain, can still be monoubiquitinated. SHIP2 monoubiquitination is thus independent of its UIM.
Cbl and Nedd4-1 Do Not Play the Role of Ubiquitin Ligase for SHIP2 Monoubiquitination
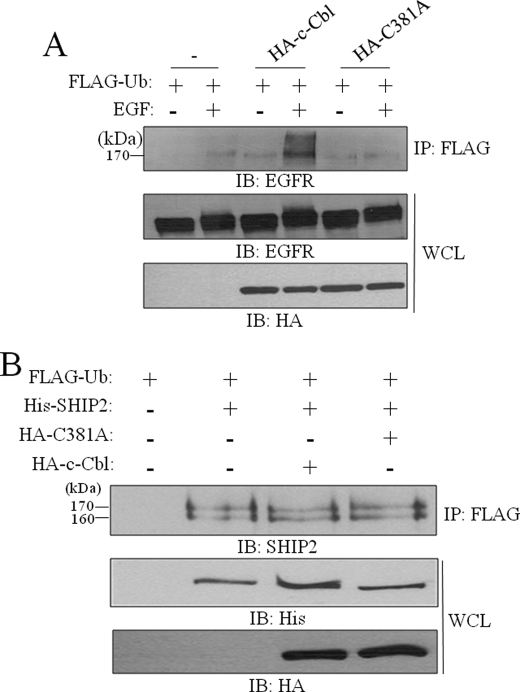
Given the association of SHIP2 with Cbl, which presents an E3 ubiquitin ligase activity, we then examined whether Cbl could play the role of the E3 ligase in SHIP2 monoubiquitination. For this purpose, we used the HA-tagged c-Cbl RING finger mutant C381A, which contains a mutation of the conserved cysteine 381 to alanine, which disrupts the E2 binding and ligase activity of c-Cbl (24). The loss of activity of the C381A mutant is confirmed in Fig. 5A, by comparing the effect of c-Cbl and C381A mutant on EGFR ubiquitination. We thus coexpressed HA-tagged c-Cbl or HA-tagged C381A mutant with His-tagged SHIP2 and FLAG-tagged ubiquitin in COS-7 cells. Equal amounts of cell lysates were immunoprecipitated with anti-FLAG and immunoblotted with anti-SHIP2 to detect monoubiquitinated SHIP2. As shown in Fig. 5B, overexpression of c-Cbl or C381A mutant did not affect SHIP2 monoubiquitination. Similar results were obtained in COS-7 cells stimulated or not with EGF (supplemental Fig. S8). This result indicates that c-Cbl does not act as E3 ligase for SHIP2 monoubiquitination.
FIGURE 5.
Cbl do not play the role of ubiquitin ligase for SHIP2 monoubiquitination. A, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and HA-tagged c-Cbl or HA-tagged C381A. Cells were serum-starved for 16 h and stimulated with 50 ng/ml EGF for 10 min. Equal amounts of cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted (IB) with anti-EGFR antibodies. Expression of EGFR, c-Cbl, and C381A was confirmed with anti-EGFR or anti-HA antibodies. B, COS-7 cells were transiently transfected with His-tagged SHIP2, FLAG-tagged ubiquitin, HA-tagged c-Cbl, HA-tagged C381A, or an empty vector. Cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-SHIP2 antibodies. Expression of SHIP2, c-Cbl, and c-Cbl-C381A was confirmed with anti-His or anti-HA antibodies.
Different studies reported that several UBD-containing proteins are monoubiquitinated by the E3 ligase Nedd4-1 (50, 54). Because SHIP2 contains a UIM domain, we tested also whether Nedd4-1 could be the E3 ligase for its monoubiquitination. By a ubiquitination assay, we showed that the coexpression of Nedd4-1 did not influence monoubiquitination of SHIP2, which is consistent with the fact that in addition SHIP2 cannot interact with Nedd4-1 (with or without EGF stimulation of the cells) (data not shown).
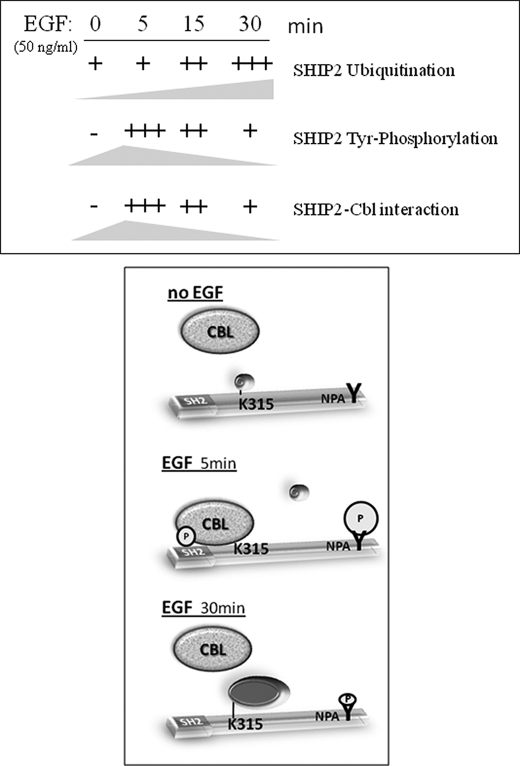
SHIP2 Monoubiquitination and SHIP2-Cbl Interaction Are Regulated by EGF
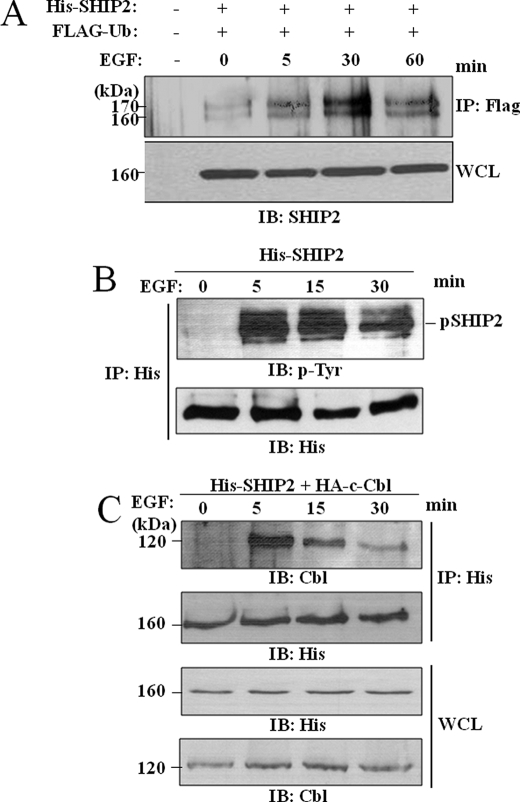
Many proteins containing a UBD as Hrs, Eps15, or Epsin, are also monoubiquitinated in response to EGF treatment (50). Therefore we tested whether EGF stimulation can influence SHIP2 monoubiquitination and if this correlates to phosphorylation. Serum-starved COS-7 cells transfected with FLAG-ubiquitin and His-SHIP2 were stimulated with EGF for 5, 30, and 60 min. Equal amounts of total cell lysates were immunoprecipitated with anti-FLAG antibody, and monoubiquitinated SHIP2 was detected with anti-SHIP2 antibodies. As shown in Fig. 6A, a small fraction of SHIP2 was monoubiquitinated without EGF stimulation, but this monoubiquitination was strongly increased upon EGF stimulation with a maximal level at 30 min.
FIGURE 6.
SHIP2 monoubiquitination is regulated by EGF stimulation. A, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and His-tagged SHIP2 or an empty vector, serum-starved for 16 h, and stimulated with 50 ng/ml EGF for 5, 30, and 60 min. Equal amounts of cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-SHIP2 antibodies. Expression of SHIP2 was confirmed by immunoblotting of the cell lysates with anti-SHIP2 antibodies. B and C, COS-7 cells were transfected with His-tagged SHIP2 (B) or with HA-tagged c-Cbl (C), serum-starved for 16 h, and stimulated with 50 ng/ml EGF at 5, 15, and 30 min. Equal amounts of cell lysates were immunoprecipitated (IP) with an anti-His antibody, and immunoprecipitates were separated by SDS-PAGE and immunoblotted (IB) with anti-4G10 (anti-phosphotyrosine) or anti-His antibodies (B) or with anti-Cbl or anti-His antibodies (C).
Ubiquitination of proteins often depends on the phosphorylation state of the target. We have shown before that SHIP2 tyrosine phosphorylation is increased after 5 min of EGF stimulation (3). Here we prolonged EGF stimulation until 30 min in serum-starved COS-7 cells transiently transfected with His-tagged SHIP2. Equal amounts of total cell lysates were immunoprecipitated with anti-His antibody, and tyrosine-phosphorylated SHIP2 was detected with anti-phosphotyrosine antibodies (Fig. 6B). We observed that tyrosine phosphorylation of SHIP2 was maximal at 5 min and strongly decreased after 30 min in contrast to its ubiquitination.
This phosphorylation time course also fits well with the association of SHIP2 with c-Cbl upon EGF stimulation in our COS-7 cells model. Serum-starved COS-7 cells transiently transfected with His-tagged SHIP2 and HA-tagged c-Cbl were stimulated with EGF for 5, 15, and 30 min. Equal amounts of total cell lysates were immunoprecipitated with an anti-His antibody, and immunoprecipitates were analyzed by immunoblotting with anti-Cbl and anti-His antibodies. As shown in Fig. 6C, SHIP2 is not associated with c-Cbl in non-stimulated cells. This interaction is clearly seen at 5 min of EGF stimulation and strongly decreased after 30 min. Taken together, these observations strongly suggest that the interaction between SHIP2 and c-Cbl is not constitutive but is regulated by EGF in COS-7 cells.
Loss of the SH2 Domain of SHIP2 Favors Its Monoubiquitination
To further address the relation between c-Cbl binding and SHIP2 phosphorylation on the ubiquitination, we analyzed the ubiquitination state of two important SHIP2 mutants, ΔSH2-SHIP2 (lacking the N-terminal SH2 domain) and NPAW-SHIP2 (lacking the tyrosine in the NPAY motif).
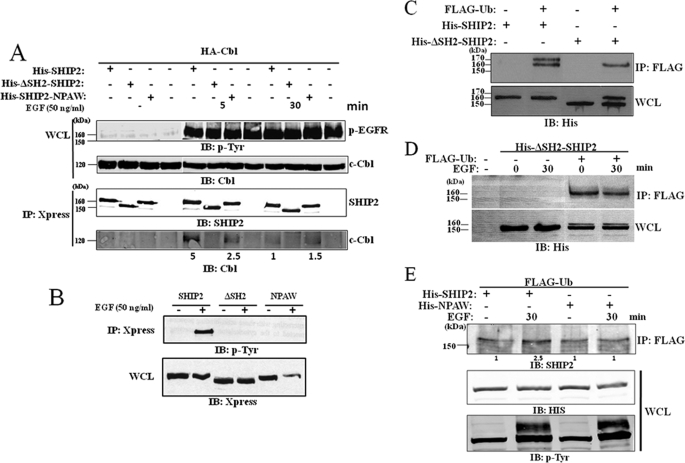
A difference in c-Cbl binding properties of the different mutants is shown as well in Fig. 7A in which we noticed that, if ΔSH2-SHIP2 did not bind c-Cbl at all, NPAW mutant binds to c-Cbl with much lower affinity than the wt-SHIP2. In addition this binding is less sensitive to EGF treatment. It is interesting to note that these mutants are not phosphorylated on tyrosine under EGF (Fig. 7B).
FIGURE 7.
Loss of the SH2 domain of SHIP2 favors its monoubiquitination. A, COS-7 cells were transiently transfected with HA-Cbl and Xpress-tagged SHIP2, ΔSH2-SHIP2, or NPAW-SHIP2 mutants. Cells are serum-starved for 16 h and stimulated with 50 ng/ml EGF for 5 and 30 min. Cell lysates were immunoprecipitated with an anti-Xpress antibody and immunoblotted (IB) with an anti-HA antibody. The amount of associated c-Cbl was quantified by the Odyssey method. B, COS-7 cells were transiently transfected with Xpress-tagged SHIP2, ΔSH2-SHIP2, or NPAW-SHIP2 mutants. Cells are serum-starved for 16 h and stimulated with 50 ng/ml EGF for 5 min. Phosphorylated SHIP2 was revealed using anti-phosphotyrosine antibody. C, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin, His-tagged SHIP2, or His-tagged ΔSH2-SHIP2 mutant. Cell lysates were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted with an anti-His antibody. Expression of His-tagged SHIP2 and ΔSH2-SHIP2 mutant was confirmed with an anti-His antibody. D, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and His-tagged ΔSH2-SHIP2 mutant as indicated, serum-starved for 16 h, and stimulated with 50 ng/ml EGF for 30 min. Equal amounts of cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-His antibody. E, COS-7 cells were transiently transfected with FLAG-tagged ubiquitin and His-tagged SHIP2 or NPAW SHIP2 mutant as indicated, serum-starved for 16 h, and stimulated with 50 ng/ml EGF for 5, 30, and 60 min. Equal amounts of cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-SHIP2 antibody. Expression of SHIP2 mutant was confirmed by immunoblotting of the cell lysates with an anti-His antibody, and EGF stimulation was verified using anti-phosphotyrosine antibody.
We further examined the monoubiquitination of these mutants by an ubiquitination assay. An interesting observation shown in the Fig. 7C was that the monoubiquitination of the ΔSH2-SHIP2 mutant was very much increased as compared with the wt-SHIP2. It was detectable in COS-7 whole cell lysates transfected with FLAG-tagged ubiquitin and His-tagged ΔSH2-SHIP2 mutant, which was not the case for SHIP2, for which monoubiquitination in whole cell extracts was almost undetectable (Fig. 7C, WCL). This result suggests that the ΔSH2-SHIP2 mutant is more subject to monoubiquitination than wild-type SHIP2 in these experimental conditions. The SH2 domain of SHIP2 thus negatively controls SHIP2 monoubiquitination. Therefore, we tested the EGF stimulation on ΔSH2-SHIP2 mutant monoubiquitination. Serum-starved COS-7 cells transiently transfected with FLAG-tagged ubiquitin and His-tagged ΔSH2-SHIP2 mutant were stimulated with EGF for 30 min. Equal amounts of total cell lysates were immunoprecipitated with an anti-FLAG antibody, and monoubiquitinated ΔSH2-SHIP2 mutant was detected with anti-His antibodies. No change in the level of the monoubiquitinated ΔSH2-SHIP2 mutant was observed with or without EGF (Fig. 7D, additional time in supplemental Fig. S9). We concluded that ΔSH2-SHIP2 monoubiquitination is abundant in control cells and not further increased by EGF. The monoubiquitination of the NPAW mutant was comparable to one of the wt-SHIP2 in unstimulated cells, and no increase was observed with EGF (Fig. 7E) suggesting that the tyrosine phosphorylation could be an important factor in EGF regulation of SHIP2 ubiquitination.
Monoubiquitinated SHIP2 Retains Its Catalytic Activity and Is Located in a Particular Fraction
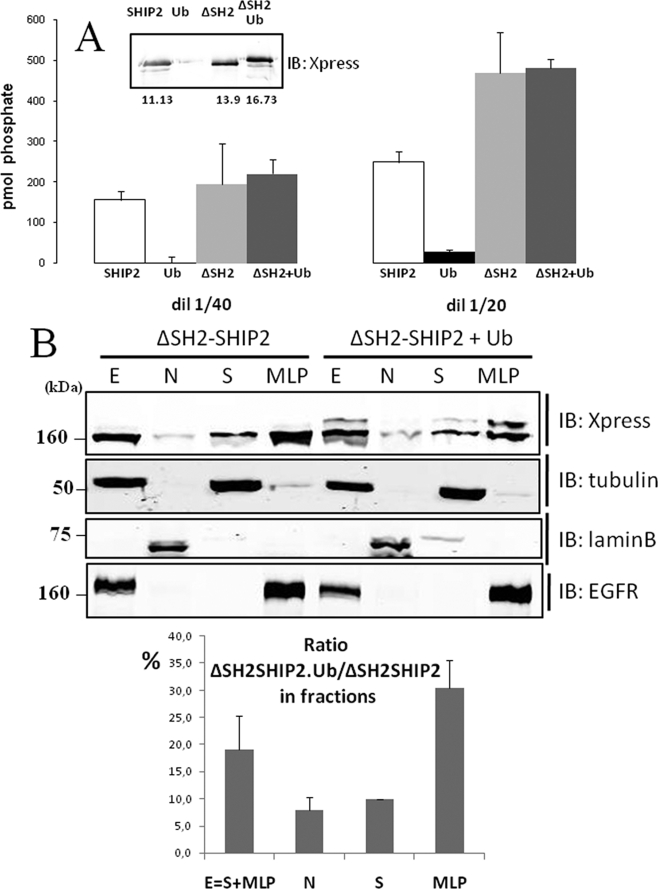
Surprisingly, we observed in Fig. 7C that monoubiquitinated SHIP2 (170 kDa) was found associated with SHIP2 (160 kDa), whereas monoubiquitinated ΔSH2-SHIP2 mutant (160 kDa) was not coimmunoprecipitated with non-ubiquitinated ΔSH2-SHIP2 mutant (150 kDa). This result suggests that the SHIP2 SH2 domain is needed for the SHIP2-SHIP2 association. This mutant was suitable, without any wt SHIP2 interference, to investigate if the catalytic activity of the monoubiquitinated ΔSH2-SHIP2 was modulated compared with the non-ubiquitinated ΔSH2-SHIP2 isoform. PtdIns(3,4,5)P3 5-phosphatase activity of ubiquitinated ΔSH2-SHIP2, Ub-ΔSH2-SHIP2, and SHIP2 was determined after immunoprecipitation. PtdIns(3,4,5)P3 5-phosphatase activity was determined into two different amounts of the immunoprecipitated beads (Fig. 8A). No significant difference was observed between the catalytic activity of monoubiquitinated ΔSH2-SHIP2 and that with the ΔSH2-SHIP2. This suggests that monoubiquitination does not modulate the catalytic activity of SHIP2 under these experimental conditions. We also showed that the catalytic activity of SHIP2 was not necessary for its monoubiquitination, because the catalytic mutant of SHIP2 (D607A) was still monoubiquitinated and able to dimerize (data not shown).
FIGURE 8.
Catalytic activity and subcellular localization of monoubiquitinated ΔSH2-SHIP2. A, COS-7 cells were transiently transfected with Xpress-tagged ΔSH2-SHIP2 alone or with FLAG-tagged ubiquitin, lysed, and immunoprecipitated overnight with Xpress or FLAG antibodies, respectively. Immunoblotting (IB) with anti-Xpress are shown in the inset. A fraction of the immunoprecipitation (50 and 25 μl) was used to assay phosphatase activity. The results are representative of four different experiments. In each experiments data are means of triplicates ± S.E. B, COS-7 cells were transiently transfected with His-tagged ΔSH2-SHIP2. SHIP2 in each fraction was probed by Western blotting with anti-Xpress antibodies. Tubulin α, lamin B1, and EGFR were used as controls to validate the fractionation. The percentage of ubiquitinated ΔSH2-SHIP2 over non-ubiquitinated SHIP2 in each fraction was estimated by quantitative Western blotting using anti-Xpress antibodies.
To investigate whether monoubiquitination of SHIP2 is related to its intracellular localization, the distribution of non-ubiquitinated and monoubiquitinated ΔSH2-SHIP2 was determined. COS-7 cells transiently transfected with ΔSH2-SHIP2 and with or without FLAG-tagged ubiquitin were fractionated into nuclear (N) and cytoplasmic (E) fractions (further subfractionated into high speed supernatant (S) and particles (MLP) fractions) by sequential centrifugations and homogenizations. The different fractions from COS-7 cells transfected with ΔSH2-SHIP2 alone or with FLAG-ubiquitin were immunoblotted with an anti-X-press antibody. As shown in Fig. 8B, non-ubiquitinated ΔSH2-SHIP2 was localized in both soluble (S) and particular (N and MLP) fractions. Interestingly ubiquitinated ΔSH2-SHIP2 is essentially associated with the MLP fraction (30%) and considerably less abundant in nuclear (7%) and cytosolic (10%) fractions. It is notable that ubiquitinated ΔSH2-SHIP2 and EGFR were immunodetected in the same MLP fraction. The same results were obtained with wt-SHIP2 but needed FLAG-immunoprecipitation because Ub-SHIP2 is not detectable by simple Western blot (data not shown).
DISCUSSION
In this study, based on cell ubiquitination assays and mass spectrometry, we identified SHIP2 monoubiquitination. We also showed that SHIP1 was also monoubiquitinated, albeit with lower intensity. In contrast, Type I 5-PPase was apparently not modified by ubiquitination. This suggests that overexpression of the protein is not sufficient to lead to ubiquitination. We consider this result as a significant negative control. It has been postulated that the tumor suppressor PTEN, is subjected to both poly- and monoubiquitination leading to PTEN degradation and nuclear localization respectively (34, 55). Controversial data on the implication of the ubiquitin ligase Nedd4-1 suggest that it is conceivable that PTEN mono- and polyubiquitination may be catalyzed by different ligases, possibly acting in cell- and tissue-specific manners (34, 55, 56). Our experiments also suggested that SHIP2 could be polyubiquitinated. Nevertheless, using 26 S proteasome inhibitors (lactacystin and MG132) we excluded that SHIP2 is degraded by the proteasome pathway and demonstrated that, in the experimental conditions tested, SHIP2 is a stable protein with a half-life greater than 24 h. In the case of polyubiquitin chain, the linkage via the lysine 48 targets polyubiquitinated proteins to the 26 S proteasomal degradation pathway. The potential SHIP2 polyubiquitination should thus be due to another ubiquitin linkage (35).
The major modification of SHIP2 thus consists of its monoubiquitination. We detected endogenous SHIP2 monoubiquitination in conditions where ubiquitin is overexpressed. Detecting monoubiquitination of endogenous proteins in native conditions is known to be rather difficult considering the low abundance of monoubiquitinated proteins and the lack of sensitive antibodies and methods to detect them. Accordingly, we speculate that SHIP2 monoubiquitination could be very rapid and transient in cells. Polyubiquitination of transfected PTEN was detected without adding ubiquitin in cells (34). Nevertheless to observe PTEN monoubiquitination, the authors needed to add tagged ubiquitin as well.
We have shown that Lys-315 is the major site for SHIP2 monoubiquitination. Lys-315 is in a sumoylation/ubiquitination consensus site (ΦKXe: Φ, hydrophobic residue; K, lysine; X, amino acid; and e, acidic residue) (44, 45, 57). Nevertheless SHIP2 was not covalently modified by sumoylation in our experimental model despite the presence of three different such consensus (around Lys-315, Lys-515, and Lys-962). This consensus site is present in other members of the 5-phosphatases family, two of them in SHIP1, SIP-110 and SIP-145, and INPP5B, which could be of potential interest in the regulation of these proteins.
Interestingly, we also identified that SHIP2 contained, as other numerous proteins implicated in endocytosis, a functional UIM, very well conserved between species, which confers to SHIP2 the ability to bind ubiquitin. SHIP1 does not contain any UIM motif and does not bind ubiquitin. It was reported that several proteins harboring functional UBDs can undergo monoubiquitination by a process referred to as coupled monoubiquitination (54). Seven classes of UBDs (UIM, CUE, UBA, MIU, GAT, UBM, and UBZ) are currently known to sustain this process (35, 58, 59). However, we demonstrated that SHIP2 is not subjected to “coupled monoubiquitination,” because its UIM is dispensable for its monoubiquitination. Monoubiquitinated proteins that do not contain UBDs are also known (60).
The E3 ubiquitin ligase c-Cbl mediates ubiquitination of tyrosine kinases, receptors (e.g. EGFR, platelet-derived growth factor receptor, CSF-1, and Met) or non-receptors (e.g. Abl and Src) but also of cytoplasmic proteins (such as Sprouty) (25–30). We showed that c-Cbl, which associates with SHIP2 upon EGF stimulation, does not modify the level of SHIP2 monoubiquitination. We also tested Nedd4-1, which was shown to interact with and ubiquitinate PTEN (61). Nedd4-1 did not interact with or ubiquitinate SHIP2 in our experimental conditions. Hoeller et al. recently described that some UBD-containing proteins can be monoubiquitinated directly by E2-conjugating enzymes, associating with the UBD, bypassing the requirement of E3 ligase (62). It seems that it is not the case for SHIP2, because the UIM is dispensable for its monoubiquitination. However, both modes of ubiquitination of the same substrate, E3-independent and E3-dependent, can coexist (62). Thus, the identification of the physiological ubiquitin ligase for SHIP2 (among around 1000 ubiquitin ligases of the genome) remains to be identified.
We also observed that ubiquitinated SHIP2 was coimmunoprecipitated with unmodified SHIP2 suggesting that two SHIP2 molecules are able to associate. This association was not dependent on the ubiquitination of SHIP2, because we were able to coimmunoprecipitate differentially tagged SHIP2 in the absence of ubiquitin. Previous studies showed an association between SHIP1 and SHIP2 via the SHIP2 SH2-domain (63). The ΔSH2-SHIP2 mutant (which does not contain the N-terminal SH2 domain of SHIP2) is not able to coimmunoprecipitate with itself indicating the importance of the SH2 domain for the SHIP2-SHIP2 association.
Strikingly we also found that the ΔSH2-SHIP2 mutant was monoubiquitinated to a much more extent as compared with the wild-type SHIP2. This is interesting because SHIP2 SH2 domain is essentially known to interact with partners (e.g. c-Cbl) or immunoreceptors (FcγRIIB) (18, 64). Our data suggest that the N-terminal SH2 domain of SHIP2 possesses an intrinsic inhibitory effect on the monoubiquitination of SHIP2. Such an intramolecular inhibitory effect was also recently reported for PTEN. Its Nedd4-1-mediated ubiquitination and degradation is antagonized by its C-terminal region containing a C2 domain (61).
We also observed that EGF stimulation strongly increases SHIP2 monoubiquitination after 30 min. At the same time, tyrosine phosphorylation of SHIP2 and binding with c-Cbl strongly decreases. Both are maximal upon 5-min EGF stimulation. The fact that neither ΔSH2-SHIP2 nor NPAW-SHIP2 mutant monoubiquitination were regulated by EGF stimulation suggests that phosphorylation and SH2 target binding are crucial for this regulation.
To address the functional consequences of SHIP2 monoubiquitination, we tested whether this post-translational modification could modulate its catalytic activity. This question remains controversial in the case of phosphorylation and may be tissue-specific (17, 65, 66). In our experiments, monoubiquitinated SHIP2 remained fully active. We also showed by subcellular fractionation that monoubiquitinated SHIP2 was enriched in the particular fractions of the cells containing membranes, lysosomes, and vesicles (30% of the SHIP2 is monoubiquitinated) while not in the nuclear (7%) or in the soluble fractions (10%). SHIP2 monoubiquitination could thus take place in the particular subunits of the cell or the monoubiquitinated SHIP2 is further recruited in the particular fraction, perhaps targeting the PtdIns(3,4,5)P3 5-phosphatase activity to a specific cellular site. Previous studies have shown that monoubiquitination regulates cellular transport (33, 34, 67). PTEN monoubiquitination regulates its nuclear import, which represents a protective function of cytoplasmic proteasomal degradation (34). Monoubiquitination of p53 implies its nuclear export, but several nuclear localization signal and nuclear export signal sequences in p53 are also implicated in this transport (33, 67). An interesting fact resides in the colocalization of the monoubiquitinated SHIP2 with the EGFR in MLP fraction. As shown previously by Prasad and collaborators, SHIP2 regulates the ligand-dependent down-regulation of the EGFR (18).
On the basis of these data we propose the following model for regulation of SHIP2 monoubiquitination (Fig. 9): at the basal state or upon short times of EGF stimulation (5 min), SHIP2 is protected from monoubiquitination due to the integrity of its SH2 domain, which could be engaged in an intramolecular interaction with an internal phosphorylated tyrosine or in an interaction with a phosphorylated tyrosine of a proteic partner or both. This masks the potential interaction site for the E3 ligase or the target lysine 315, which prevents SHIP2 monoubiquitination. SHIP2 contains many tyrosine residues that are maximally phosphorylated after 5 min of EGF stimulation and could interact intramolecularly with its N-terminal SH2 domain (3). In the case of an interaction with a proteic partner, c-Cbl could play this role. Indeed, c-Cbl strongly interacts with SHIP2 upon 5-min EGF stimulation, and it does so via the SH2 domain of SHIP2 (18, 68). In this model, c-Cbl could be a bridge that assembles two SHIP2 molecules and competes for the addition of the ubiquitin moiety. This is consistent with our observation that SHIP2 is able to coimmunoprecipitate with itself and that the SH2 domain is also necessary for this interaction. After EGF stimulation for longer times (30 min), disruption of the inter- or intramolecular interaction, caused by a decrease in SHIP2 tyrosine phosphorylation, releases the monoubiquitination site and thus allows for SHIP2 monoubiquitination. Our findings showed SHIP2 is still weakly monoubiquitinated at the basal state (in the absence of serum). SHIP2 can thus also undergo this post-translational modification in the absence of EGF stimulation. This was also observed for Eps15, which is monoubiquitinated in Her14 cells upon EGF stimulation (69, 70), whereas in other cell lines endogenous Eps15 is constitutively monoubiquitinated (70). Thus, although monoubiquitination of some sorting proteins is EGF-dependent, other proteins are constitutively ubiquitinated.
FIGURE 9.
Proposed model for the regulation of SHIP2 monoubiquitination. At short times of EGF stimulation (5 min), SHIP2 would be protected from monoubiquitination due to the integrity of its SH2 domain, which could be engaged in an intramolecular interaction with an internal phosphorylated tyrosine or through interaction with a phosphorylated tyrosine of a proteic partner. After longer EGF stimulation times (30 min), disruption of the intramolecular interaction, caused by decreasing of SHIP2 tyrosine phosphorylation, or disruption of interaction with a partner would allow the accessibility of the monoubiquitination site and favor the SHIP2 monoubiquitination. The labels used are as follows: SH2, SHIP2 SH2 domain; K315, lysine on position 315 in SHIP2; and pY, phosphorylated tyrosine.
In summary, we demonstrate that SHIP2 is able to recognize other ubiquitinated proteins by a UIM domain and that its monoubiquitination is a process that is actively controlled by the ability of its SH2 domain to mask the monoubiquitination site. We also showed that monoubiquitination of SHIP2 could be implicated in its cellular translocation and does not modify its phosphatase activity. Recently, novel functions of SHIP2 were shown such as scaffold properties in the c-Jun N-terminal kinase cascade, negative regulation of EGFR internalization and degradation, EphA2 endocytosis, and angiotensin II signaling (12, 18, 19, 71). SHIP2 monoubiquitination is therefore a novel protein modification potentially involved in these molecular functions.
Acknowledgments
We thank Dr. Wallace Y. Langdon (University of Western Australia) for the generous gifts of c-Cbl and FLAG-tagged ubiquitin cDNAs, Dr. Stanley Lipkowitz (National Naval Medical Center, Bethesda, MD) for human Cbl-b cDNA, Dr. Mitchell (Monash University, Clayton, Australia) for HA-tagged SHIP2 cDNA, Dr. S. Sigismund (Institute for Molecular Oncology, Milan, Italy) for FLAG-tagged Eps15 cDNA, Dr. Yang (University of Southern California, Los Angeles, CA), for human HA-tagged coiled-coil activator, Dr. Niedenthal (Hanover Medical School, Germany), and Dr. Le Goff (University of Rennes, France) for FLAG-tagged SUMO-1 and SUMO-2. We thank Dr. Stéphane Martin, Sheela Onnockx, Jingwei Xie, Séverine Steuve, and Christine Jacobs for much experimental help and many fruitful discussions and Colette Moreau for careful technical assistance.
This work was supported by a grant from the National Fund for Scientific Research (FNRS), Fonds pour la formation à Recherche clans l'Industrie et dans l'Agriculture, and Télévie, Brussels, Belgium.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
- SHIP2
- SH2 domain-containing inositol phosphate 5-phosphatase 2
- EGF
- epidermal growth factor
- EGFR
- epidermal growth factor receptor
- HA
- hemagglutinin
- UBD
- ubiquitin-binding domain
- UIM
- ubiquitin-interacting motif
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- SAM
- sterile α motif
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- E2
- ubiquitin carrier protein
- E3
- ubiquitin-protein isopeptide ligase
- wt
- wild type
- PTEN
- phosphatase and tension homolog
- 5PPase
- inositol phosphate-5-phosphatase.
REFERENCES
- 1.Ooms L. M., Horan K. A., Rahman P., Seaton G., Gurung R., Kethesparan D. S., Mitchell C. A. (2009) Biochem. J. 419, 29–49 [DOI] [PubMed] [Google Scholar]
- 2.McCrea H. J., De Camilli P. (2009) Physiology 24, 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pesesse X., Deleu S., De Smedt F., Drayer L., Erneux C. (1997) Biochem. Biophys. Res. Commun. 239, 697–700 [DOI] [PubMed] [Google Scholar]
- 4.Ishihara H., Sasaoka T., Hori H., Wada T., Hirai H., Haruta T., Langlois W. J., Kobayashi M. (1999) Biochem. Biophys. Res. Commun. 260, 265–272 [DOI] [PubMed] [Google Scholar]
- 5.Pesesse X., Dewaste V., De Smedt F., Laffargue M., Giuriato S., Moreau C., Payrastre B., Erneux C. (2001) J. Biol. Chem. 276, 28348–28355 [DOI] [PubMed] [Google Scholar]
- 6.Dyson J. M., O'Malley C. J., Becanovic J., Munday A. D., Berndt M. C., Coghill I. D., Nandurkar H. H., Ooms L. M., Mitchell C. A. (2001) J. Cell Biol. 155, 1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad N., Topping R. S., Decker S. J. (2001) Mol. Cell. Biol. 21, 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandenbroere I., Paternotte N., Dumont J. E., Erneux C., Pirson I. (2003) Biochem. Biophys. Res. Commun. 300, 494–500 [DOI] [PubMed] [Google Scholar]
- 9.Paternotte N., Zhang J., Vandenbroere I., Backers K., Blero D., Kioka N., Vanderwinden J. M., Pirson I., Erneux C. (2005) FEBS. J. 272, 6052–6066 [DOI] [PubMed] [Google Scholar]
- 10.Raaijmakers J. H., Deneubourg L., Rehmann H., de Koning J., Zhang Z., Krugmann S., Erneux C., Bos J. L. (2007) Cell. Signal. 19, 1249–1257 [DOI] [PubMed] [Google Scholar]
- 11.Onnockx S., De Schutter J., Blockmans M., Xie J., Jacobs C., Vanderwinden J. M., Erneux C., Pirson I. (2008) J. Cell. Physiol. 214, 260–272 [DOI] [PubMed] [Google Scholar]
- 12.Xie J., Onnockx S., Vandenbroere I., Degraef C., Erneux C., Pirson I. (2008) Cell. Signal. 20, 1432–1441 [DOI] [PubMed] [Google Scholar]
- 13.Muraille E., Pesesse X., Kuntz C., Erneux C. (1999) Biochem. J. 342, 697–705 [PMC free article] [PubMed] [Google Scholar]
- 14.Habib T., Hejna J. A., Moses R. E., Decker S. J. (1998) J. Biol. Chem. 273, 18605–18609 [DOI] [PubMed] [Google Scholar]
- 15.Kalesnikoff J., Sly L. M., Hughes M. R., Büchse T., Rauh M. J., Cao L. P., Lam V., Mui A., Huber M., Krystal G. (2003) Rev. Phys. Biochem. Pharmacol. 149, 87–103 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y. J., Keogh R. J., Hunter M. G., Mitchell C. A., Frey R. S., Javaid K., Malik A. B., Schurmans S., Tridandapani S., Marsh C. B. (2004) J. Immunol. 173, 6820–6830 [DOI] [PubMed] [Google Scholar]
- 17.Batty I. H., van der Kaay J., Gray A., Telfer J. F., Dixon M. J., Downes C. P. (2007) Biochem. J. 407, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad N. K., Decker S. J. (2005) J. Biol. Chem. 280, 13129–13136 [DOI] [PubMed] [Google Scholar]
- 19.Zhuang G., Hunter S., Hwang Y., Chen J. (2007) J. Biol. Chem. 282, 2683–2694 [DOI] [PubMed] [Google Scholar]
- 20.Blero D., De Smedt F., Pesesse X., Paternotte N., Moreau C., Payrastre B., Erneux C. (2001) Biochem. Biophys. Res. Commun. 282, 839–843 [DOI] [PubMed] [Google Scholar]
- 21.Wada T., Sasaoka T., Funaki M., Hori H., Murakami S., Ishiki M., Haruta T., Asano T., Ogawa W., Ishihara H., Kobayashi M. (2001) Mol. Cell. Biol. 21, 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clément S., Krause U., Desmedt F., Tanti J. F., Behrends J., Pesesse X., Sasaki T., Penninger J., Doherty M., Malaisse W., Dumont J. E., Le Marchand-Brustel Y., Erneux C., Hue L., Schurmans S. (2001) Nature 409, 92–97 [DOI] [PubMed] [Google Scholar]
- 23.Sleeman M. W., Wortley K. E., Lai K. M., Gowen L. C., Kintner J., Kline W. O., Garcia K., Stitt T. N., Yancopoulos G. D., Wiegand S. J., Glass D. J. (2005) Nat. Med. 11, 199–205 [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro C. A., Wing S. S., Huang H., Leverson J. D., Hunter T., Liu Y. C. (1999) Science 286, 309–312 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Yeung Y. G., Langdon W. Y., Stanley E. R. (1996) J. Biol. Chem. 271, 17–20 [DOI] [PubMed] [Google Scholar]
- 26.Lee P. S., Wang Y., Dominguez M. G., Yeung Y. G., Murphy M. A., Bowtell D. D. L., Stanley E. R. (1999) EMBO J. 18, 3616–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. (1998) Genes Dev. 12, 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyake S., Lupher M. L., Jr., Druker B., Band H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soubeyran P., Barac A., Szymkiewicz I., Dikic I. (2003) Biochem. J. 370, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall A. B., Jura N., DaSilva J., Jang Y. J., Gong D., Bar-Sagi D. (2003) Curr. Biol. 13, 308–314 [DOI] [PubMed] [Google Scholar]
- 31.Weissman A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178 [DOI] [PubMed] [Google Scholar]
- 32.Hicke L. (2001) Cell 106, 527–530 [DOI] [PubMed] [Google Scholar]
- 33.Li M., Brooks C. L., Wu-Baer F., Chen D. L., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 34.Trotman L. C., Wang X. J., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N. P., Carver B. S., Cordon-Cardo C., Erdjument-Bromage H., Tempst P., Chi S. G., Kim H. J., Misteli T., Jiang X., Pandolfi P. P. (2007) Cell 128, 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 36.Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Communi D., Gevaert K., Demol H., Vandekerckhove J., Erneux C. (2001) J. Biol. Chem. 276, 38738–38747 [DOI] [PubMed] [Google Scholar]
- 38.Drayer A. L., Pesesse X., De Smedt F., Communi D., Moreau C., Emeux C. (1996) Biochem. Soc. Trans. 24, 1001–1005 [DOI] [PubMed] [Google Scholar]
- 39.Shiraki K., Hayakawa Y., Mori H., Namazue J., Takamizawa A., Yoshida I., Yamanishi K., Takahashi M. (1991) J. Gen. Virol. 72, 1393–1399 [DOI] [PubMed] [Google Scholar]
- 40.Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 41.Vandeput F., Backers K., Villeret V., Pesesse X., Erneux C. (2006) Cell. Signal. 18, 2193–2199 [DOI] [PubMed] [Google Scholar]
- 42.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. (1997) EMBO J. 16, 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campo P. A., Das S., Hsiang C. H., Bui T., Samuel C. E., Straus D. S. (2002) Cell Growth Differ. 13, 409–420 [PubMed] [Google Scholar]
- 44.Desterro J. M., Rodriguez M. S., Hay R. T. (1998) Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 45.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 46.Yang C. K., Kim J. H., Ann D. K., Stallcup M. R. (2008) BMC Mol. Biol. 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oldham C. E., Mohney R. P., Miller S. L., Hanes R. N., O'Bryan J. P. (2002) Curr. Biol. 12, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 48.Di Fiore P. P., Polo S., Hofmann K. (2003) Nat. Rev. Mol. Cell Biol. 4, 491–497 [DOI] [PubMed] [Google Scholar]
- 49.Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Nat. Cell Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 50.Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 51.Miller S. L., Malotky E., O'Bryan J. P. (2004) J. Biol. Chem. 279, 33528–33537 [DOI] [PubMed] [Google Scholar]
- 52.Fisher R. D., Wang B., Alam S. L., Higginson D. S., Robinson H., Sundquist W. I., Hill C. P. (2003) J. Biol. Chem. 278, 28976–28984 [DOI] [PubMed] [Google Scholar]
- 53.Hofmann K., Falquet L. (2001) Trends Biochem. Sci. 26, 347–350 [DOI] [PubMed] [Google Scholar]
- 54.Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Nat. Cell Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 55.Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z. H., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., Jiang X. J. (2007) Cell 128, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., Rotin D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 58.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 59.Penengo L., Mapelli M., Murachelli A. G., Confalonieri S., Magri L., Musacchio A., Di Fiore P. P., Polo S., Schneider T. R. (2006) Cell 124, 1183–1195 [DOI] [PubMed] [Google Scholar]
- 60.Polo S., Confalonieri S., Salcini A. E., Di Fiore P. P. (2003) Sci. STKE 2003, re17. [DOI] [PubMed] [Google Scholar]
- 61.Wang X., Shi Y., Wang J., Huang G., Jiang X. (2008) Biochem. J. 414, 221–229 [DOI] [PubMed] [Google Scholar]
- 62.Hoeller D., Hecker C. M., Wagner S., Rogov V., Döetsch V., Dikic I. (2007) Mol. Cell 26, 891–898 [DOI] [PubMed] [Google Scholar]
- 63.Giuriato S., Pesesse X., Bodin S., Sasaki T., Viala C., Marion E., Penninger J., Schurmans S., Erneux C., Payrastre B. (2003) Biochem. J. 376, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muraille E., Bruhns P., Pesesse X., Daëron M., Erneux C. (2000) Immunol. Lett. 72, 7–15 [DOI] [PubMed] [Google Scholar]
- 65.Taylor V., Wong M., Brandts C., Reilly L., Dean N. M., Cowsert L. M., Moodie S., Stokoe D. (2000) Mol. Cell. Biol. 20, 6860–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prasad N. K., Werner M. E., Decker S. J. (2009) Biochemistry 48, 6285–6287 [DOI] [PubMed] [Google Scholar]
- 67.Lohrum M. A., Woods D. B., Ludwig R. L., Bálint E., Vousden K. H. (2001) Mol. Cell. Biol. 21, 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prasad N. K., Tandon M., Badve S., Snyder P. W., Nakshatri H. (2008) Carcinogenesis 29, 25–34 [DOI] [PubMed] [Google Scholar]
- 69.van Delft S., Schumacher C., Hage W., Verkleij A. J., van Bergen E., Henegouwen P. M. (1997) J. Cell Biol. 136, 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klapisz E., Sorokina I., Lemeer S., Pijnenburg M., Verkleij A. J., van Bergen E., Henegouwen P. M. (2002) J. Biol. Chem. 277, 30746–30753 [DOI] [PubMed] [Google Scholar]
- 71.Sanada F., Taniyama Y., Azuma J., Iekushi K., Dosaka N., Yokoi T., Koibuchi N., Kusunoki H., Aizawa Y., Morishita R. (2009) Hypertension 53, 77–82 [DOI] [PubMed] [Google Scholar]
- 72.Shevshenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]