Abstract
Cryptococcus neoformans is a fungal pathogen that causes serious disease in immunocompromised individuals. The organism produces a distinctive polysaccharide capsule that is necessary for its virulence, a predominantly polysaccharide cell wall, and a variety of protein- and lipid-linked glycans. The glycan synthetic pathways of this pathogen are of great interest. Here we report the detection of a novel glycosylphosphotransferase activity in C. neoformans, identification of the corresponding gene, and characterization of the encoded protein. The observed activity is specific for UDP-xylose as a donor and for mannose acceptors and forms a xylose-α-1-phosphate-6-mannose linkage. This is the first report of a xylosylphosphotransferase activity in any system.
Introduction
Fungi generate a wide variety of glycans, many of which differ from those of higher eukaryotes. In pathogenic fungal species, these unique structures and their synthetic pathways represent possible drug targets. One example of these distinctive cellular glycans is the extensive polysaccharide capsule of the basidiomycete, Cryptococcus neoformans. An environmental yeast, C. neoformans can be isolated from soil, avian excreta, and certain trees, but it is also capable of causing disease in mammals following the inhalation of spores or small yeast cells (1). C. neoformans is typically neutralized within the lung by the immune system without any symptomatic evidence of infection. Under conditions of compromised host immunity, however, the organism may disseminate from its primary site of infection in the lungs to more distal sites in the body, demonstrating a particular tropism for the central nervous system. If C. neoformans reaches the brain, it can form lesions called cryptococcomas and cause a meningoencephalitis that is fatal if not treated.
Although other yeasts bear polysaccharide capsules, members of the C. neoformans species complex are the only known pathogenic fungi with this feature. The capsule, which is required for virulence (2), is primarily composed of two polysaccharides: glucuronoxylomannan (3) and glucuronoxylomannogalactan (4, 5). In addition to this distinguishing structure, C. neoformans synthesizes a diverse array of glycans, which differ from those of its mammalian host as well as the “model” yeast Saccharomyces cerevisiae. For example, the core oligosaccharide of N-glycans is truncated in C. neoformans such that it lacks the three terminal Glc residues present on the same structure in most eukaryotes, including S. cerevisiae and mammals (6). Cryptococcal O-glycans also differ from the oligomannose structures found in S. cerevisiae (7); studies in the environmental yeast Cryptococcus laurentii have identified O-glycans that are branched and contain Gal, xylose (Xyl),3 and mannose (Man) residues (8). These oligosaccharides are thus more like mammalian O-glycans in complexity, although the array of components is quite distinct (9). In addition, some glycan structures found in fungi do not have an equivalent in mammalian cells, such as the lipid-linked glycosylinositol phosphorylceramides (GIPCs). In S. cerevisiae, GIPCs contain only a single Man residue, whereas cyptococcal GIPCs contain residues of Man, Xyl, Gal, and glucosamine (10). Despite the obvious significance of glycan biosynthetic pathways as potential drug targets among pathogenic fungi, however, few have been studied in detail.
In order to investigate glycan biosynthesis, our laboratory studies the central enzymes in these pathways, known as glycosyltransferases. These enzymes generally catalyze the transfer of a monosaccharide moiety from an activated sugar donor to a specific acceptor molecule, forming a particular linkage. Donors of sugar molecules can include nucleotide mono- and diphosphosugars, sugar phosphates, and dolichol-linked sugars; proteins, lipids, and other saccharides can all serve as acceptors in transferase reactions. Glycosyltransferases are divided into families based on their tertiary structure and catalytic mechanism, including their requirements for metal ion cofactors (11). Conservation in sequence at the amino acid level occurs only between some closely related glycosyltransferases.
We are particularly interested in the addition of Xyl residues to the cellular glycans of C. neoformans. This stems from a series of studies on the synthesis of UDP-Xyl, the Xyl donor in eukaryotic cells, which is formed upon the decarboxylation of UDP-glucuronic acid (UDP-GlcA) by Uxs1p. The cryptococcal gene encoding Uxs1p was identified in our laboratory (12). Deletion of UXS1 in C. neoformans yields a strain that generates no UDP-Xyl (13); as a consequence, isolated glucuronoxylomannan contains no detectable Xyl (14), capsule fibers appear short and thickened when viewed by electron microscopy (13), and cellular GIPCs lacks Xyl and are truncated (15, 16). Importantly, uxs1Δ strains are avirulent in a murine model of cryptococcal infection (14), indicating that the incorporation of Xyl residues into cellular glycans is required for C. neoformans to cause disease. This compelling result led us to investigate xylosyltransferases in C. neoformans.
In this report, we document the discovery of a novel xylosyltransferase activity in C. neoformans that generates a highly unusual product, xylosylphosphomannose. Below, we describe the initial identification of this activity, our determination of the corresponding gene, and our characterization of this unique and intriguing enzyme.
EXPERIMENTAL PROCEDURES
Materials
DNA polymerases (Taq and AccuPrimeTM Pfx) and oligonucleotides used for PCR (supplemental Table 1) were from Invitrogen; DNA purification kits were from Qiagen. GDP-[2-3H]mannose (22.2 Ci/mmol), UDP-[1-3H]glucuronic acid (10.2 Ci/mmol), and UDP-[14C(U)]xylose (151 mCi/mmol) were from PerkinElmer; UDP-[6-3H]galactose (60 Ci/mmol), UDP-[1-3H]glucose (15 Ci/mmol), and UDP-[6-3H]N-acetylglucosamine (34.8 Ci/mmol) were from American Radiolabeled Chemicals. α-1,2-d-Mannobiose and α-1,6-d-mannobiose were from Sigma; α-1,3-d-mannobiose was from Carbohydrate Synthesis (Oxford, UK); α-1,4-d-mannobiose was from V-Labs. Restriction enzymes were from New England BioLabs. Unless specified, all other chemicals or reagents were obtained from Sigma.
Strains and Cell Growth
C. neoformans strains (Table 1) were grown in liquid culture at 30 °C in YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose) with shaking (230 rpm) or at 30 °C on YPD agar plates (YPD medium with 2% (w/v) agar). As appropriate, media included 100 μg/ml nourseothricin (NAT; Werner BioAgents) or Geneticin® (G418; Invitrogen). For RNA interference (RNAi) studies, strains were cultured at 30 °C in Gal-5FOA minimal medium (1.34% (w/v) yeast nitrogen base without amino acids, 2% (w/v) galactose, 0.087% (w/v) complete dropout mix without uracil, 1.2% (w/v) uracil, 1 mg/ml 5-fluoroorotic acid, 1 mm NaOH) or at 30 °C on Gal-5FOA agar plates (Gal-5FOA minimal medium with 2.5% (w/v) agar). Genetic crosses were performed at room temperature on V8 agar plates (5% (v/v) V8 juice, 0.05% KH2PO4, pH 7.0, 4% (w/v) agar).
TABLE 1.
C. neoformans strains used in these studies
| Namea,b | Origin |
|---|---|
| CAP67 | Ref. 26 |
| CAP67 cxt1Δ | Ref. 17 |
| CAP67 cxt2Δ | J. S. Klutts and T. L. Doering, manuscript in preparation |
| CAP67 cxt1Δ cxt2Δ | J. S. Klutts and T. L. Doering, manuscript in preparation |
| CAP67 cxt1Δ pIB103 | This study |
| CAP67 cxt1Δ pXPT1i | This study |
| JEC21 | Ref. 43 |
| KN99α | Ref. 44 |
| KN99a | Ref. 44 |
| KN99α cxt1Δ cxt2Δ | J. S. Klutts and T. L. Doering, manuscript in preparation |
| KN99α xpt1Δ | This study |
| KN99α xpt1Δ pXPT1 | This study |
| KN99α xpt1Δ pXPT1-HA | This study |
a All strains are MATα, except KN99a is MATa.
b All KN99 strains are serotype A; all other strains are serotype D.
Escherichia coli strains were grown in liquid culture at 37 °C in LB medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl) with shaking (250 rpm) or at 37 °C on LB agar plates (LB medium with 2% (w/v) agar). As appropriate, media included 100 μg/ml ampicillin or 60 μg/ml kanamycin.
Total Membrane Preparation and Detergent Extraction
For membrane preparation and detergent extraction, all steps were performed at 4 °C. C. neoformans was cultured overnight in 50 ml of YPD to late log phase (∼1 × 108 cells/ml), harvested by centrifugation (3,000 × g; 10 min), and washed in 40 ml of Tris-EDTA buffer (100 mm Tris-HCl, pH 8.0, 0.1 mm EDTA). The washed cell pellet was resuspended in an equal volume of Tris-EDTA buffer, and 800-μl aliquots were transferred to 2-ml screw-cap microcentrifuge tubes. Samples were bead-beaten with 800 μl of 0.5-mm glass beads (BioSpec Products) in 1-min bursts on a Mini-Bead Beater (BioSpec Products), alternating with 2 min on ice. Once ∼75% of the cells were disrupted (as assessed by microscopy), the lysate was transferred to a 15-ml conical tube, and the glass beads were rinsed with 800 μl of Tris-EDTA buffer; this buffer and three more rinses were pooled with the lysate. The pooled material was subjected to a clearing centrifugation step (1,000 × g; 20 min), and the resulting supernatant fraction was transferred to an ultracentrifuge tube. Total membranes were isolated by ultracentrifugation (60,000 × g; 45 min), resuspended in 50–200 μl of Tris buffer (100 mm Tris-HCl pH 8.0), and stored at 4 °C. Protein concentrations of the total membrane samples were determined using the Bio-Rad protein assay.
For detergent extraction, Triton X-100 was added to the membranes to a final concentration of 1%, and the sample was incubated on ice for 30 min with vortex mixing every 5 min. Particulate material was removed by ultracentrifugation (75,000 × g; 30 min), and the supernatant was stored at 4 °C. Protein concentrations of the detergent-extracted cellular membrane samples were determined using the Bio-Rad detergent-compatible protein assay.
For substrate titration studies, Triton X-100 extracts were prepared in bulk to minimize any variability in manganese-dependent xylosyltransferase activity levels between membrane preparations. Detergent extracts were prepared as above from a 1-liter culture of KN99α cxt1Δ cxt2Δ; glycerol was added to a final concentration of 15%, and the material was stored in aliquots at −80 °C. For xylosyltransferase reactions, an aliquot of this extract was thawed at 37 °C and immediately placed on ice. Glycerol was removed by subjecting the sample to two rounds of 10-fold dilution with Tris Buffer and concentration using an Amicon® Ultra-15 centrifugal filter device (30,000 molecular weight cut-off; Millipore), and protein concentration was determined using the Bio-Rad detergent-compatible protein assay.
Xylosyltransferase Activity Assays
Enzyme activity was assayed by monitoring the transfer of [14C]Xyl from a UDP-[14C]Xyl donor to an α-1,3-d-mannobiose acceptor (α-1,3-Man2). Standard reactions included 625 μg of protein (from total membranes or Triton X-100 extracts), 1 μm UDP-[14C]Xyl, 8.8 mm α-1,3-Man2, and 7.5 mm MnCl2 in 100 mm Tris-HCl, pH 6.5, and were incubated at 20 °C for either 4 h (for substrate titration) or overnight (for other studies). The reaction products were isolated and visualized as described in Ref. 17. Briefly, unincorporated UDP-[14C]Xyl was removed by passing the sample over an 800-μl column of AG® 2-X8 resin (Bio-Rad) with water elution. The product was resolved by TLC using Silica Gel 60 plates (EM Sciences) and developed in a solvent system of 5:4:1 1-propanol/acetone/water; plates were sprayed with En3Hance® spray (PerkinElmer Life Sciences), and radiolabeled products were visualized by autoradiography.
Isolation of the Manganese-dependent Reaction Product
Xylosyltransferase reactions using Triton X-100 extracts of CAP67 cxt1Δ membranes were performed with 0.5 μm UDP-[14C]Xyl plus 14.7 mm non-radioactive UDP-Xyl and incubated at 20 °C for 36 h. Samples were processed and resolved by TLC as above, and the product of interest was localized using a System 200A imaging scanner (Bioscan Inc.). The appropriate region of silica was scraped from the plate, and the product eluted from the silica with water and purified using an Envi-Carb solid phase extraction column (Supelco) as in Ref. 17.
Additional Product Analysis
For some studies, the products of standard xylosyltransferase reactions were treated with or without acetic acid (final concentration of 1%) at 100 °C for 1 h. For others, samples were incubated at 25 °C overnight with or without jack bean mannosidase (final concentration of 21 μg/ml in 150 mm citric acid, pH 5.0). Treated samples were resolved by TLC and visualized by autoradiography as above.
One-dimensional 1H and Two-dimensional 1H-1H Nuclear Magnetic Resonance Spectroscopy
The product of the manganese-dependent xylosyltransferase reaction (∼0.5 mg) was deuterium-exchanged by repeated lyophilization from D2O and then dissolved in 0.5 ml of D2O for NMR analysis. One-dimensional 1H NMR, two-dimensional 1H-1H gCOSY, and two-dimensional 1H-1H TOCSY spectra were acquired at 25 °C on a Varian Unity Inova 500-MHz spectrometer (Varian Inc.), using standard acquisition software available in the Varian VNMR software package. Proton chemical shifts were referenced to internal acetone (δ = 2.225 ppm).
Electrospray Ionization Mass Spectrometry
Mass spectrometry was performed in both positive and negative ion modes on a linear ion trap (LTQ; Thermo Fisher Scientific), with sample introduction via direct infusion in 50% methanol in water (for the native manganese-dependent xylosyltransferase reaction product) or 100% methanol (for the permethylated manganese-dependent xylosyltransferase reaction product) and sample concentration of ∼100 ng/μl.
RNAi Targeting
The construct for RNAi in C. neoformans, pIB103, is diagrammed in supplemental Fig. 1; its construction is described elsewhere.4 The plasmid contains two opposing pairs of promoters and terminators cloned from the GAL7 locus (NCBI accession number U16994) flanking a ∼250-bp fragment of the URA5 gene (NCBI accession number AF140188). Bidirectional transcription across this segment produces double-stranded RNA that effectively silences URA5, allowing cell growth in the presence of 5-fluoroorotic acid. I-SceI digestion linearizes the plasmid while exposing telomeric sequences (18).
To isolate total RNA, an overnight culture of the C. neoformans strain JEC21 was harvested by centrifugation (3,000 × g; 10 min) and washed with 50 ml of diethylpyrocarbonate-treated water; the pellet was frozen in a dry ice/methanol bath and lyophilized overnight. Four milliliters of 0.5-mm glass beads were added to the dried pellet, and the sample was beaten to a fine powder using a vortex mixer, mixed with 4 ml of TRIzol® (Invitrogen), and incubated at room temperature for 5 min. Next, the sample was mixed with 800 μl of chloroform, incubated at room temperature for 3 min, and centrifuged at 3,000 × g for 30 min. The upper aqueous phase was mixed with an equal volume of 70% ethanol and applied to an RNeasy® Maxi column (RNeasy® Maxi Kit, Qiagen). Total RNA was isolated according to the manufacturer's protocol and used to generate cDNA using the SuperscriptTM first strand synthesis system for reverse transcription-PCR (Invitrogen).
Two regions of XPT1 (nucleotides 724–1267 and 2551–2966) were targeted for RNAi. Each was PCR-amplified from JEC21 cDNA using primer pairs that introduced SpeI restriction sites at both ends (CNJ001/CNJ002 and MCR035/MCR036). When the CNJ001/CNJ002 amplicon was cloned into the pCR®2.1-TOPO® vector (TOPO TA Cloning® Kit, Invitrogen) it formed the plasmid TOPO XPT1 RNAi, which was then transformed into TOP10 E. coli cells. The pTOPO XPT1 RNAi and pIB103 plasmids were isolated and digested with SpeI; the latter was also treated with calf intestinal alkaline phosphatase (Fisher). The RNAi target insert and the linearized vector were purified, and the fragments were ligated with T4 DNA ligase (Roche Applied Science) to form the XPT1 RNAi construct pXTP1i, which was then transformed into DH5α E. coli cells (Invitrogen).
Purified pXPT1i was linearized with I-SceI and transformed into the C. neoformans strain CAP67 cxt1Δ by electroporation (19). Colonies were selected on YPD G418 agar and then screened for growth on Gal 5-fluoroorotic acid agar.
Confirmation of the XPT1 Transcript
The coding sequence of XPT1 was PCR-amplified in overlapping fragments using primers directed against predicted exon sequences (C. neoformans var. grubii H99 data base maintained by the Broad Institute), and cDNA were generated as above but from strain KN99α. The resulting DNA segments were cloned into the pCR®2.1-TOPO® vector and sequenced. The 5′- and 3′-UTRs of the XPT1 transcript were verified using the GeneRacerTM kit (Invitrogen). The XPT1 transcript sequence has been submitted to the NCBI data base (accession number GQ403790).
XPT1 Deletion Strain and Complementation
Regions of ∼1 kb immediately 5′ to the start codon of XPT1 (5′-UTR) and immediately 3′ to the stop codon of XPT1 (3′-UTR) were PCR-amplified from KN99α genomic DNA prepared as in Ref. 20 using the primer pairs MCR063/MCR101 and MCR104/MCR068, respectively. The NAT resistance cassette (NATR) was amplified from the plasmid pGMC200-MCS (13) by PCR using primers MCR102 and MCR103. To form the XPT1 deletion (xpt1Δ) construct, the purified 5′-UTR, NATR, and 3′-UTR amplicons were assembled into a single sequence by overlap PCR (21) using primers MCR069 and MCR070.
The xpt1Δ construct was biolistically transformed into KN99α cells (22). Genomic DNA was prepared from NAT-resistant transformants and screened by PCR for loss of the native XPT1 locus and presence of the xpt1Δ construct. Finally, candidate deletion strains were confirmed by Southern blot analysis (23). Verified KN99α xpt1Δ strains were crossed to KN99a on V8 agar as described in Ref. 24, and the resulting spores were plated on YPD NAT plates; a NATR MATa progeny strain was then back-crossed to KN99α. This crossing procedure (switching mating type) was repeated three times with spore selection on YPD NAT plates. The progeny of two independent KN99α xpt1Δ strains were confirmed to lack Xpt1p activity and used for further study.
For plasmid-based complementation of the xpt1Δ strain, XPT1 with ∼1 kb of flanking sequence on either side was PCR-amplified using primers MCR063 and MCR068 from KN99α genomic DNA prepared as above. The amplicon was cloned into the PCR®-XL-TOPO® vector (TOPO® XL PCR Cloning Kit; Invitrogen) to form the plasmid pXL-TOPO XPT1, which was then transformed into TOP10 E. coli cells, purified, and sequenced to ensure that no errors were introduced during PCR. Confirmed XPT1 was released from pXL-TOPO XPT1 by digestion with SpeI and XbaI, blunted with T4 DNA polymerase (from New England Biolabs), and cloned into KpnI-digested and calf intestinal alkaline phosphatase-treated pIB103; the resulting pXPT1 plasmid was then transformed into DH5α E. coli cells. pXPT1 was isolated, linearized with I-SceI, and transformed into the C. neoformans strain KN99α xpt1Δ by electroporation (25). Genomic DNA prepared from G418-resistant transformants was screened by PCR and candidate KN99α xpt1Δ pXPT1 clones were assayed for recovery of xylosyltransferase activity as above.
Generation of Epitope-tagged XPT1 Strains
Xpt1p was epitope-tagged by replacing a region extending from the middle of the gene through the 3′-UTR with the same sequences modified to incorporate HA at the C terminus. The replacement cassette was created by overlap PCR (21), which combined two PCR products: a ∼1.5-kb region immediately 5′ to the stop codon of XPT1 (PCR-amplified from the plasmid pXL-TOPO XPT1 using the primer pair MRC127 and MCR120) and a ∼650-bp region including and immediately 3′ to the stop codon of XPT1 (PCR-amplified from pXL-TOPO XPT1 using the primer pair MCR121 and MCR149). Primers MCR120 and MCR121 each introduced sequence encoding the HA epitope, enabling fragment joining by overlap PCR using primers MCR127 and MCR149. The resulting product and pXL-TOPO XPT1 were both digested with SacII and NheI; the insert and the linearized vector were purified, and the fragments were ligated with T4 DNA ligase to form pXL-TOPO XPT1-HA, containing the complete and tagged sequence. This was transformed into DH5α E. coli cells and confirmed by sequencing.
To express XPT1-HA in C. neoformans, the complete tagged sequence was released from pXL-TOPO XPT1-HA by digestion with SpeI and XbaI, blunted with T4 DNA polymerase, and cloned into KpnI-digested and calf intestinal alkaline phosphatase-treated pIB103. The resulting pXPT1-HA plasmid was linearized with I-SceI and transformed into the C. neoformans strain KN99α xpt1Δ by electroporation (25). G418-resistant transformants were screened by PCR, and candidate KN99α xpt1Δ pXPT1-HA clones were assayed for recovery of xylosyltransferase activity as above.
Immunoprecipitation Studies
Detergent extracts of total membranes from KN99α xpt1Δ pXPT1 and KN99α xpt1Δ pXPT1-HA were prepared as above. Total protein (5 mg) was rotated at 4 °C for 1 h with 50 μl of anti-HA MicroBeads from the μMACSTM HA epitope tag protein isolation kit (Miltenyi Biotec) in a total of 500 μl of Tris-EDTA buffer. The samples were then applied to μColumns placed in the magnetic field of a μMACS Separator, and the columns were washed with 200 μl of Tris buffer five times. The μColumns were removed from the magnetic field of the μMACS separator, and 100 μl of Tris buffer was applied to each column to elute the anti-HA MicroBeads and associated material. The eluates were assayed for xylosyltransferase activity as above.
RESULTS
Xylosyltransferase Activities in C. neoformans
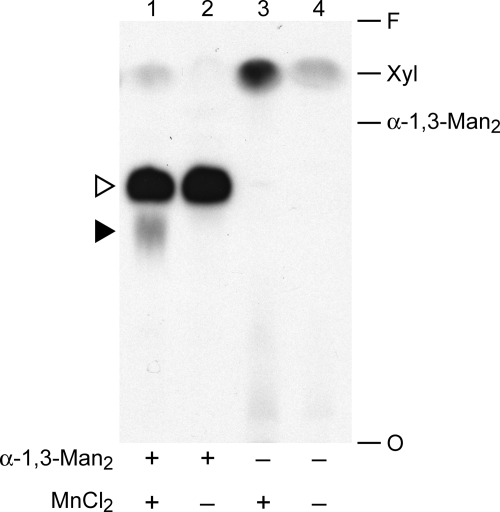
We are interested in identifying and characterizing glycosyltransferases that synthesize the unique glycan structures of C. neoformans in order to better understand these fungus-specific processes. We have focused in particular on the transfer of Xyl residues because of the requirement for this moiety in virulence (14). In one assay of Xyl modification, we monitored the transfer of [14C]Xyl from UDP-[14C]Xyl to an α-1,3-Man2 acceptor. As shown in lane 1 of Fig. 1, this assay yielded two distinct products (indicated by the arrowheads) as well as a small amount of free Xyl near the solvent front, presumably formed by degradation of the radiolabeled precursor. The generation of both products depended on the presence of α-1,3-Man2 (Fig. 1, compare lanes 1 and 3); the slower migrating product (filled arrowhead) additionally required MnCl2 for its formation (Fig. 1, compare lanes 1 and 2). CAP67, an acapsular strain mutated in the CAP59 gene (26), was used here because these cells are more readily lysed than wild-type. Deletion of CAP59 or several other genes (CAP10, -60, and -64) (27–29) that are required for capsule synthesis and suggested to be glycosyltransferases (30) did not affect the formation of either product (data not shown).
FIGURE 1.
Xylosyltransferase activities of C. neoformans. Total membranes prepared from CAP67 cells were assayed as described under “Experimental Procedures” with UDP-[14C]Xyl in the presence (+) or absence (−) of the α-1,3-Man2 acceptor and MnCl2 cofactor as indicated. An autoradiograph of the products resolved by TLC is shown, with the migration positions of free Xyl and α-1,3-Man2 standards indicated at the right. O, origin; F, solvent front. In this and subsequent figures, the filled arrowhead indicates the product of the manganese-dependent xylosyltransferase activity, and the open arrowhead indicates the products of unrelated xylosyltransferase activities (see “Results”).
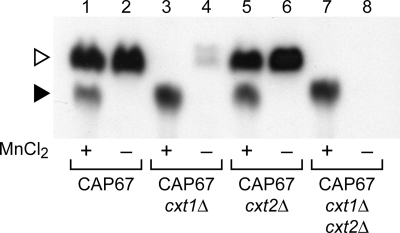
In previous work, we identified a protein responsible for the formation of the majority of the manganese-independent product (Fig. 1, open arrowhead) as Cxt1p (17). Deletion of the corresponding gene (CXT1) significantly reduced this signal (Fig. 2, compare lanes 2 and 4). The small amount of similarly migrating product seen in a cxt1Δ strain is generated by a related enzyme, Cxt2p,5 and disappeared in a cxt2Δ strain (Fig. 2, compare lanes 4 and 8). In this investigation, we focused on the activity responsible for forming the manganese-dependent assay product (Fig. 2, filled arrowhead). The migration of this product in our TLC system differed significantly from that of the Cxt1p/Cxt2p product (open arrowhead), indicating that the carbohydrate structure is not the same. Further, the manganese requirement for its formation suggested that it is made by a distinct enzymatic activity. To confirm this hypothesis, we assessed formation of the lower mobility material in strains deleted for CXT1 and CXT2. As shown in Fig. 2 (compare lanes 1 and 7), these deletions did not compromise formation of the manganese-dependent product but rather enhanced it, presumably by eliminating competition for the reaction donor or substrate. Targeting other members of the Cxt1p enzyme family (31) by RNAi also failed to alter production of the Xpt1p product (data not shown). These studies confirmed the novelty of the activity under investigation. The increased formation of the manganese-dependent product by cxt1Δ membranes also suggested that these cells would be useful in additional studies; this strain is used in several experiments below.
FIGURE 2.
Xylosyltransferase activities in CXT1 and CXT2 mutants. Xylosyltransferase activity assays of membranes from the strains indicated were performed with UDP-[14C]Xyl and α-1,3-Man2 in the presence (+) or absence (−) of MnCl2. Only the relevant region of the TLC plate is shown; no signal was detected in other regions beyond minor amounts of free Xyl (see Fig. 1). Symbols are as in Fig. 1.
The manganese-dependent xylosyltransferase product was formed over a broad pH range (4–7) and at temperatures of 4–30 °C (data not shown). For our assays, we generally used pH 6.5, since that also allowed for efficient formation of the Cxt1p/Cxt2p product as a convenient internal control for membrane activity. Standard assays were performed overnight at 20 °C to maximize product yield.
Manganese-dependent Xylosyltransferase Product Analysis
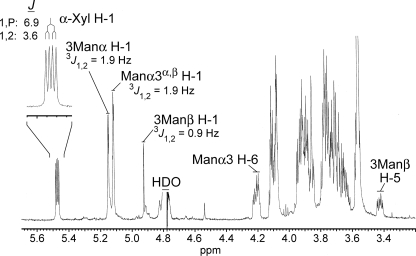
In our standard assay, the Cxt1p and Cxt2p activities generated a trisaccharide product of Xyl-linked β-1,2 to the reducing Man of the α-1,3-Man2 (17).5 The manganese-dependent xylosyltransferase product migrated nearer to the TLC plate origin than the Cxt1p/Cxt2p trisaccharide in our solvent system (Fig. 1, lane 1). This behavior suggested that the manganese-dependent product was more polar but was also potentially consistent with products that differ only with respect to the linkage of the Xyl residue to α-1,3-Man2. To determine the structure of the manganese-dependent product, the reaction was scaled up, and the product was recovered from the TLC plate as described under “Experimental Procedures.” Glycosyl composition analysis by GC/MS was consistent with material containing Man and Xyl in a 2:1 ratio (data not shown), as would be expected for the addition of a single Xyl residue to the α-1,3-Man2 acceptor substrate. Surprisingly, the 1H NMR spectrum of the product (Fig. 3) did not exhibit a simple doublet resonance corresponding to H-1 of an added β-Xyl residue anywhere near the chemical shift (4.3–4.6 ppm) expected for this scenario. Instead, in addition to the expected resonances for the dimannosyl substrate, a doublet of doublets resonance was detected far downfield at 5.471 ppm (J = 3.6, 6.9 Hz). A possible explanation was suggested by comparison with our published spectrum of UDP-Xyl (12), which showed a similar doublet of doublets signal for H-1 of the α-Xyl-1-O-phosphate residue at 5.517 ppm (3J1,2 = 3.5 Hz and 3J1,P = 7.0 Hz). The nearly identical splitting pattern in the manganese-dependent xylosyltransferase product indicated that it included an α-Xyl-1-O-phosphate, presumably derived from UDP-Xyl, linked to the dimannose substrate.
FIGURE 3.
Xpt1p product composition. A 500-MHz 1H NMR spectrum of the manganese-dependent xylosyltransferase reaction product (D2O, 25 °C) with selected resonance assignments is shown. Inset, expansion of the α-Xyl H-1 resonance with a summary of the J-coupling analysis.
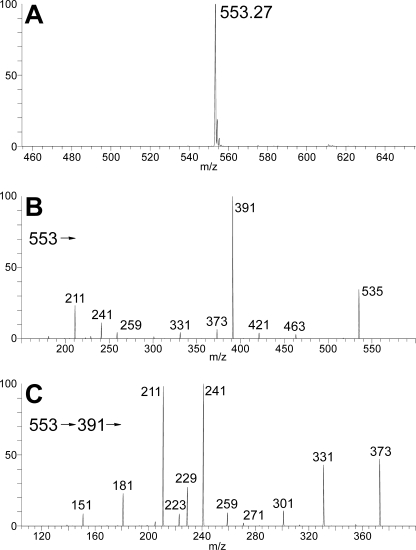
The presence of phosphate in the manganese-dependent xylosyltransferase product was confirmed by positive mode electrospray ionization mass spectrometry in a linear ion trap; an abundant ion was observed in MS1 at m/z 599, consistent with a molecular salt/adduct ion (M(Na) + Na)+ corresponding to Man2Xyl + P (where P represents phosphate ester; data not shown). Furthermore, in the negative mode analysis, a deprotonated molecular ion (M − H)− was observed abundantly at m/z 553 (Fig. 4A), consistent with a molecular mass of 554 units for the product; this is 98 units more than that expected for the trisaccharide alone, again consistent with the presence of phosphate. In the MS2 spectrum of m/z 553 (Fig. 4B), the major product ion was observed at m/z 391, consistent with a loss of one Man residue, leaving Xyl-P-Man. Structurally significant product ions were observed in pairs at m/z 259/241 and m/z 229/211; the same products were observed in an MS3 spectrum acquired by isolation and activation of the m/z 391 fragment generated in MS2 (Fig. 4C). These product ions correspond to (Man-P)−/(Man-P − H2O)− and (Xyl-P)−/(Xyl − H2O)− product pairs, respectively, which can only be generated if the Man and Xyl residues share the same phosphate residue via a phosphodiester linkage. Consistent with this assignment, treatment of the manganese-dependent xylosyltransferase product with mild acid resulted in the release of free [14C]Xyl due to hydrolysis of the phosphate bond (data not shown).
FIGURE 4.
Xpt1p product structure. Negative ion mode electrospray ionization quadrupole ion trap mass spectroscopy analysis of the manganese-dependent xylosyltransferase reaction product is shown. A, MS1 molecular ion profile showing (M-H)− at m/z 553; B, MS2 product spectrum of precursor ion m/z 553; C, MS3 product spectrum of intermediate precursor ion m/z 391 (m/z 553→391→).
Neither positive nor negative mode ion mass spectra yielded information regarding the specific hydroxyl of either Man residue to which the Xyl-P is linked. However, analysis of two-dimensional 1H-1H NMR spectra (gCOSY and TOCSY; data not shown) supported linkage of the phosphate to the 6-hydroxyl of the nonreducing Man residue. First, long range TOCSY cross-peaks showed the complex resonance observed at 4.211 ppm (Fig. 3) to be in the same spin system as the H-1 centered at 5.116 ppm; this was observed as a pair of closely overlapping doublets, which clearly belong to the non-reducing terminal Man, slightly perturbed by the equilibrium exchange between the α- and β-anomers of the reducing terminal Man (β-anomer H-1 at 4.925 ppm, 3J1,2 = 0.9 Hz; α-anomer H-1 at 5.149 ppm, 3J1,2 = 1.9 Hz). Second, connectivity analysis of this resonance showed it to be one of the exocyclic H-6 resonances of the Man residue in question. Third, analysis of the splitting pattern of this resonance showed one more coupling than expected for a Man H-6 resonance; in other words, like the α-Xyl H-1, it exhibited an additional coupling with the phosphate atom (3J6,P = 5.8 Hz, whereas 3J5,6 = 2.0 Hz and 2J6,6 = 11.3 Hz). Finally, along with the additional coupling, the far downfield shift of this resonance compared with its position in the spectrum of the unmodified dimannose substrate (e.g. Fig. 3A of Ref. 31) was consistent with O-6 of the corresponding Man residue as the linkage point for the Xyl-P moiety.
We also treated the manganese-dependent xylosyltransferase product with a mannosidase isolated from jack bean known to release non-reducing terminal mannoses from α-1,2, α-1,3, or α-1,6 linkages (32). This treatment did not affect the manganese-dependent xylosyltransferase product, although it did cleave the α-1,3-Man2 substrate when it was modified on the reducing mannose (as in the Cxt1p activity product; data not shown). This further supported the linkage of the Xyl-P to the non-reducing mannose of the α-1,3-Man2 substrate. In sum, our results indicated that the product of the manganese-dependent xylosyltransferase activity is Xyl-α-1-phosphate-6-Man-α-1,3-Man, and the enzyme responsible was a xylosylphosphotransferase (Xpt1p).
Identification of XPT1
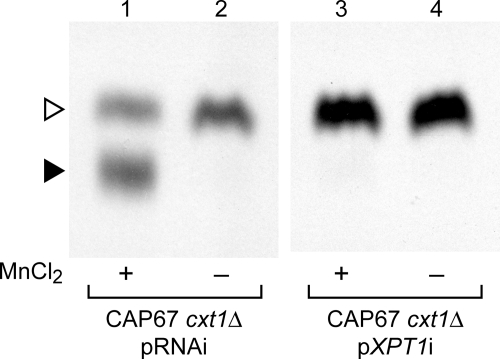
To determine the enzyme responsible for the manganese-dependent xylosylphosphotransferase activity, we looked to potentially related enzyme sequences for clues. We first reviewed the literature for reports of glycosylphosphotransferases from other organisms and then compared the sequences of these enzymes with the genome of C. neoformans (C. neoformans var. neoformans JEC21 data base, maintained by The Institute for Genomic Research). As shown in Table 2, the four probe sequences identified five candidate loci with widely varying degrees of homology. Each of the candidate sequences was targeted by RNAi in CAP67 cxt1Δ, a strain with both the manganese-dependent Xpt1p activity and the Cxt2p activity. In the presence of an empty RNAi vector containing no target gene sequence (pRNAi), products of both enzymes were detected in standard assays (Fig. 5, lanes 1 and 2). Excitingly, RNAi targeting sequence from one candidate locus (XP_567569), a weak homolog of the α/β subunit of the human GlcNAc-1-phosphate transferase, eliminated all detectable Xpt1p product (Fig. 5, lane 3), whereas the Cxt2p product was unaffected. A second RNAi construct directed against an independent region of the same target locus caused a similarly striking reduction in the manganese-dependent xylosyltransferase activity (data not shown); this demonstrated that the decrease in Xpt1p activity was due to reduced transcription of the targeted gene and was not the result of off-target interference. Targeting the other four candidate loci, including one encoding a predicted protein with 32% homology to Xpt1p (XP_567514), did not alter the generation of either the Xpt1p or Cxt2p products (data not shown). We concluded that the XP_567569 candidate locus probably encodes the Xpt1p activity and termed it XPT1.
TABLE 2.
Known glycosylphosphotransferases from other organisms
| Glycosylphosphotransferases |
Nearest cryptococcal homologs |
||
|---|---|---|---|
| Protein | NCBI locus identifier | NCBI locus identifier | e valuea |
| S. cerevisiae mannosylphosphate transferase (Ktr1p/Mnn6p) | NP_015272 | XP_568891 | 4e-58 |
| Homo sapiens UDP-GlcNAc-1-phosphotransferase α/β subunits | NP_077288 | XP_567514 | 0.04b |
| XP_567569 | 0.21b | ||
| H. sapiens UDP-GlcNAc-1-phosphotransferase γ subunit | NP_115909 | XP_566800 | 2e-09 |
| H. sapiens UDP-GlcNAc:dolichyl-phosphate N-acetylglucosaminephosphotransferase | NP_001373 | XP_567597 | 2e-81 |
a Results indicated are from searches run with NCBI Blastp version 2.2.20 on July 17, 2009, using the entire protein sequence of the indicated known enzymes against all cryptococcal sequences in the NCBI database.
b Not significant; see “Discussion.”
FIGURE 5.
RNA interference targeting of the XPT1 gene. Xylosyltransferase activity assays were performed as in Fig. 2 using total membranes prepared from strains containing either a control RNAi plasmid (pRNAi) or a plasmid targeting nucleotides 724–1267 of the XPT1 coding sequence (pXPT1i).
The XPT1 locus was annotated as a hypothetical predicted protein in both The Institute for Genomic Research database of the JEC21 genome and the Broad Institute data base of the H99 genome. Several expressed sequence tag sequences corresponding to this locus had been reported, indicating that it was transcribed, but these did not represent the entire predicted transcript. To confirm the XPT1 transcript and the predicted Xpt1p sequence, we generated cDNA for sequencing and 5′-rapid amplification of cDNA ends and 3′-rapid amplification of cDNA ends of the XPT1 transcript. The encoded 864-amino acid Xpt1 protein sequence contains a single predicted transmembrane domain and demonstrated no significant homology to any known proteins or recognized protein domains.
Deletion of XPT1 and Episomal Complementation of the xpt1Δ Strain
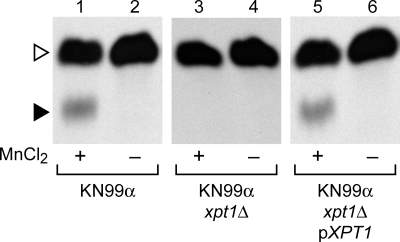
To confirm the association between XPT1 and the manganese-dependent xylosyltransferase activity, we replaced the XPT1 locus in the wild-type strain KN99α with a drug resistance cassette (see “Experimental Procedures”). Consistent with our RNAi studies, deletion of XPT1 yielded a corresponding loss of the manganese-dependent xylosyltransferase product (Fig. 6, compare lanes 1 and 3). This loss of activity was observed in two independent deletion strains and was maintained through a series of back-crosses (data not shown), indicating that the change correlated with the deletion of XPT1 and was not due to unrelated alterations in the genome potentially generated during the transformation process (33). Complementation of the KN99α xpt1Δ strain by episomal expression of XPT1 under the control of its native promoter (pXPT1) restored the manganese-dependent xylosyltransferase activity to wild-type levels (Fig. 6, lane 5).
FIGURE 6.
Deletion of XPT1. Xylosyltransferase activity assays were performed as in Fig. 2 using total membranes from the wild-type (KN99α), mutant (KN99α xpt1Δ), or complemented mutant (KN99α xpt1Δ pXPT1) strains indicated.
Xpt1p Reaction Components
Several glycosylphosphotransferases have been described (34, 35), but we are aware of no other reports of a xylosylphosphotransferase activity. For this reason, we investigated the specificity of the novel Xpt1p activity. Our standard Xpt1p reaction utilizes a UDP-[14C]Xyl donor, α-1,3-Man2 acceptor, and MnCl2 cofactor. The preference of the Xpt1p activity for each of these reaction components was assessed.
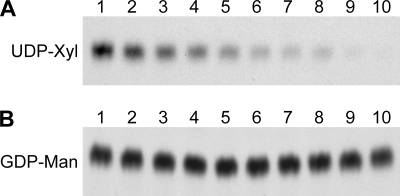
To explore the specificity of the Xpt1p activity for UDP-Xyl as the donor, we first performed standard xylosyltransferase reactions in the presence of non-radiolabeled nucleotide sugars or their components; membrane proteins were prepared from KN99α cxt1Δ cxt2Δ to prevent any competition from Cxt1p or Cxt2p activities. We were particularly interested in whether Xpt1p could use GDP-Man as a donor molecule given that transfer of sugar phosphate from this nucleotide sugar has been reported in S. cerevisiae (34). As expected, the addition of increasing concentrations of unlabeled UDP-Xyl reduced formation of the radiolabeled Xpt1p product (Fig. 7A). In contrast, no inhibition of the [14C]Xyl product was seen in the presence of GDP-Man (Fig. 7B), suggesting that Xpt1p does not utilize that donor. Inhibition of radiolabeled product formation was observed in the presence of UDP-Gal, UDP-Glc, UDP-GlcA, and UDP-GlcNAc, but the pattern of inhibition in each case was similar to that induced by UDP alone (data not shown). GDP alone did not inhibit Xpt1p activity; nor did monosaccharide Xyl (data not shown).
FIGURE 7.
Competition of Xpt1p activity. Xylosyltransferase activity assays were performed using detergent extracts of KN99α cxt1Δ cxt2Δ cells (see “Experimental Procedures”). All reactions contained standard amounts of UDP-[14C]Xyl, α-1,3-Man2, and MnCl2 with varying amounts of UDP-Xyl (A) or GDP-Man (B). Lane 1, 0 μm; lane 2, 1 μm; lane 3, 5 μm; lane 4, 10 μm; lane 5, 25 μm; lane 6, 50 μm; lane 7, 75 μm; lane 8, 100 μm; lane 9, 250 μm; lane 10, 500 μm. Only the region of the TLC plate with the Xpt1p product is shown; no signal was detected in other regions. F, solvent front.
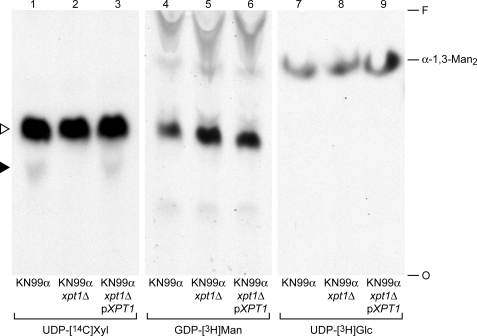
Although our competition studies indicated that Xpt1p does not utilize GDP-Man, we were unable to distinguish between the various UDP-sugars as potential donors because all were inhibitory (probably due to the shared UDP moiety). To further examine Xpt1p donor substrate specificity, we analyzed the products of transferase reactions in which UDP-[14C]Xyl was replaced with other radiolabeled nucleotide sugars. To assess Xpt1p-dependent reaction products, we compared products from assays of membranes from XPT1 wild-type, mutant or complemented mutant strains (KN99α, KN99α xpt1Δ, or KN99α xpt1Δ pXPT1, respectively). Only those reactions in which UDP-[14C]Xyl was supplied as a donor demonstrated any Xpt1p-dependent variation in the pattern of radiolabeled products (Fig. 8, lanes 1–3, filled arrowhead). In contrast, no radiolabeled products generated by reactions containing GDP-[3H]Man, UDP-[3H]Glc, UDP-[3H]Gal, UDP-[3H]GlcA, or UDP-[3H]GlcNAc (Fig. 8 and data not shown) were dependent on the presence of Xpt1p. Together, these studies suggested specificity for UDP-Xyl as the reaction donor for Xpt1p.
FIGURE 8.
Donor substrates of Xpt1p. Xpt1p activity assays were performed as in Fig. 2, using total membranes from wild-type, mutant, or complemented mutant strains as indicated. All reactions contained α-1,3-Man2, MnCl2, and the radiolabeled nucleotide sugar donor indicated. Labels are as in Fig. 1, although the arrowheads refer only to lanes 1–3; the identity of the radiolabeled products in lanes 4–9 has not been determined.
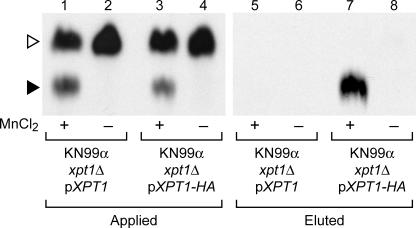
Having identified UDP-Xyl as the preferred xylose donor in Xpt1p-mediated reactions, we considered the source of the phosphate moiety in the Xpt1p product. With other known glycosylphosphotransferases, the phosphate is derived from the nucleotide sugar donor (36). It is possible, however, that the phosphate in the Xpt1p product is independently derived, in a step preceding xylose transfer, from some other compound present in the assayed membranes or extracts. To generate more pure material to test in our transferase assays, we added sequence encoding a C-terminal HA epitope tag to pXPT1 and isolated the tagged protein by magnetic immunoprecipitation methods (see “Experimental Procedures”). The addition of the HA epitope did not alter the overall activity of Xpt1p (Fig. 9, compare lanes 1 and 3 and data not shown) and did mediate specific isolation of the manganese-dependent activity (Fig. 9, compare lanes 5 and 7). This demonstration of [14C]Xyl-P transfer mediated by affinity-purified protein strongly suggested that the phosphate moiety found in the Xpt1p product is derived from the UDP-[14C]Xyl donor.
FIGURE 9.
Xylosyltransferase activities of Xpt1p-HA. Xylosyltransferase activity assays were performed as in Fig. 2. Samples were either detergent extracts from membranes of KN99α xpt1Δ pXPT1 or KN99α xpt1Δ pXPT1-HA prior to anti-HA affinity isolation or eluted material, as indicated.
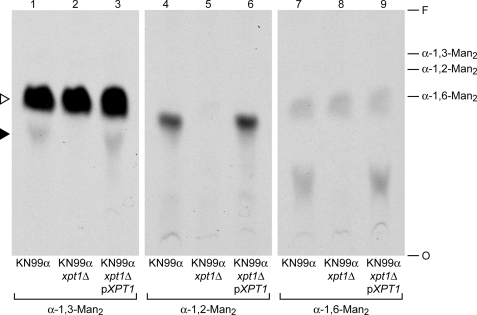
We next used a series of dimannose molecules to examine the acceptor preferences of Xpt1p. As with the donor specificity experiments above, xylosyltransferase reactions were prepared using membranes from KN99α, KN99α xpt1Δ, or KN99α xpt1Δ pXPT1; the standard α-1,3-Man2 reaction substrate was replaced with various potential acceptors. We observed Xpt1p-dependent modification of the dimannose compounds α-1,2-Man2, α-1,4-Man2, and α-1,6-Man2 in addition to the α-1,3-Man2 acceptor used in our earlier experiments (Fig. 10 and data not shown). Similar studies demonstrated Xpt1p utilization of d-Man as an acceptor molecule but not of d-Gal, d-Glc, d-GlcNAc, or d-Xyl (data not shown). It appeared that Xpt1p was specific for Man as an acceptor but was flexible as to the structural context of the Man residue.
FIGURE 10.
Acceptor substrates of Xpt1p. Xpt1p activity assays were performed as in Fig. 2 using total membranes from wild-type, mutant, or complemented mutant strains, as indicated. All reactions contained UDP-[14C]Xyl, MnCl2 and the indicated mannose dimer acceptor. The migration positions of the unmodified acceptors are indicated at the right; all other labels are as in Fig. 8. Assays with no acceptor show no radiolabeled products (see Fig. 1, lane 3). F, solvent front.
Finally, we tested product formation in xylosyltransferase assays containing a panel of chloride salts in place of the MnCl2 cofactor using membranes prepared from KN99α cxt1Δ cxt2Δ. Although the most robust Xpt1p activity was seen in the presence of MnCl2, some similarly migrating product was detected in the presence of both CoCl2 and MgCl2 (∼5 and ∼15% of the MnCl2 product, respectively; data not shown). Assays using other cations (CaCl2, CuCl2, Fe(II)Cl2, NiCl2, and ZnCl2) did not yield any detectable product, and including EDTA in the reactions resulted in the absence of any detectable product regardless of which cation was present (data not shown). MnCl2 was therefore the preferred metal ion cofactor of Xpt1p.
DISCUSSION
We have discovered a unique xylosylphosphotransferase activity in C. neoformans and the gene encoding it, which we named XPT1 (for xylosylphosphotransferase 1). This activity appears to be specific for UDP-Xyl, making it distinct from previously reported mannosylphosphotransferases and N- acetylglucosaminylphosphotransferases (34, 35). The modification of Man with Xyl-P is novel; to our knowledge, this structure has not been reported in any biological system, nor have activities similar to those of Xpt1p been previously observed.
Our studies indicate that the Xpt1p is specific for UDP-Xyl. However, we cannot exclude the possibility that this enzyme can also utilize some uncommon UDP-sugar donor that we did not include in our test panel. Although there are no data to indicate that C. neoformans synthesizes any such donor, comprehensive studies have not been performed in this system. Additionally, our studies with affinity-purified protein suggest that the only reaction components needed for Xpt1p-dependent Xyl-P transfer are UDP-Xyl, a Man-containing substrate molecule, and MnCl2. This suggests that UDP-Xyl serves as the source for both the phosphate and the sugar moiety, as with the GlcNAc-1-phosphate transferase (36).
The predicted 100-kDa protein encoded by XPT1 does not resemble other known xylosyltransferases and has no conserved domains as identified by either NCBI or other publicly available search engines. A cluster of hydrophobic amino acids at residues 83–103 of Xpt1p may comprise a transmembrane domain, consistent with the observed enrichment of the manganese-dependent xylosyltransferase activity in detergent extracts of cryptococcal membranes (data not shown). The Xpt1p sequence contains several possible DXD motifs, sites of metal ion binding commonly found in glycosyltransferases (37). There are also potential N- and O-linked glycosylation sites present within the protein sequence (38). Interestingly, Xpt1p demonstrates strong homology to hypothetical proteins found in the basidiomycetous fungi Postia placenta (brown wood rot; 28% identity overall) and Ustilago maydis (corn smut; 27% identity overall). Although no biological role or biochemical activity has been attributed to these two proteins, it is possible that they perform similar catalytic functions to Xpt1p.
Overall, Xpt1p has extremely limited homology to proteins of known function. We identified its sequence based on a 43-amino acid stretch (residues 407–449 of Xpt1p) that is 38% identical to a region in the α/β subunit of UDP-GlcNAc phosphotransferase. The UDP-GlcNAc phosphotransferase, which has an α2β2γ2 subunit structure, catalyzes the initial step in the mannose-6-phosphate modification of lysosomal hydrolases that is necessary for targeting these proteins to the lysosome. The α and β subunits are encoded by a single gene and are thought to contain the catalytic portion of the protein (39), whereas the γ subunit is encoded by a separate gene and is thought be a regulatory element (40, 41). Based on our studies, Xpt1p cannot utilize UDP-GlcNAc as a donor. We propose that the region of homology between the α/β subunit of UDP-GlcNAc phosphotransferase and Xpt1p may be involved in sugar-phosphate transfer from a nucleotide sugar donor.
Kudo et al. (39) first speculated that a region of the α/β subunit of UDP-GlcNAc phosphotransferase (residues 321–432) “might be involved in the binding of nucleotide sugar or transfer of sugar phosphate” because of its similarity to sequence elements found in bacterial capsule polymerases. This possibility was revisited in a broad in silico analysis of predicted glycosyltransferases in pathogen genomes (42). Several proteins identified by Sperisen et al. (42) in that study were previously implicated in the biosynthesis of exopolysaccharide phosphoglycans, although the functions of these proteins were indicated only by indirect association with a given phenotype rather than by functional assays. Among the identified hypothetical loci were two from C. neoformans, including XPT1. The region of homology between the α/β subunit of UDP-GlcNAc phosphotransferase and Xpt1p was included both within the potential domain described by Kudo et al. (39) and the conserved regions identified by Sperisen et al. (42).
A key question for the future is the biological role of Xpt1p. As mentioned earlier, the cryptococcal cell contains a variety of glycan structures, several of which are unique to this pathogen. The best studied of these is the cryptococcal capsule, but analyses of the polysaccharide components of this structure have not indicated the presence of xylophosphomannose linkages (3–5). Preliminary studies suggest that Xpt1p acts in the modification of cryptococcal proteins (data not shown), but further work will be required to elucidate the cellular function of this novel and intriguing enzyme.
Acknowledgments
Use of the 500-MHz NMR facility of the Department of Chemistry, University of New Hampshire, as well as the mass spectrometry facilities of the University of New Hampshire Glycomics Center (Vernon N. Reinhold, Primary Investigator) is gratefully acknowledged. We thank David Haslam, Igor Almeida, and Andrew Lovering for helpful insights on this project; Matthew Williams and Elizabeth Held for technical assistance; and Aki Yoneda for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM071007 (in support of the studies of cryptococcal glycan synthesis in the Doering laboratory).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) GQ403790.
I. Bose and T. L. Doering, manuscript in preparation.
J. S. Klutts and T. L. Doering, manuscript in preparation.
- Xyl
- xylose
- gCOSY
- gradient-selected correlation spectroscopy
- GIPC
- glycosylinositol phosphorylceramide
- GlcA
- glucuronic acid
- G418
- geneticin
- Man
- mannose
- NAT
- nourseothricin
- NATR
- NAT resistance cassette
- TOCSY
- total correlation spectroscopy
- RNAi
- RNA interference
- UTR
- untranslated region
- HA
- hemagglutinin.
REFERENCES
- 1.Casadevall A., Perfect J. (1998) Cryptococcus neoformans, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Fromtling R. A., Shadomy H. J., Jacobson E. S. (1982) Mycopathologia 79, 23–29 [DOI] [PubMed] [Google Scholar]
- 3.Cherniak R., O'Neill E. B., Sheng S. (1998) Infect. Immun. 66, 2996–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaishnav V. V., Bacon B. E., O'Neill M., Cherniak R. (1998) Carbohydr. Res. 306, 315–330 [DOI] [PubMed] [Google Scholar]
- 5.Heiss C., Klutts J. S., Wang Z., Doering T. L., Azadi P. (2009) Carbohydr. Res. 344, 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto M. (2007) Biosci. Biotechnol. Biochem. 71, 1415–1427 [DOI] [PubMed] [Google Scholar]
- 8.Schutzbach J., Ankel H., Brockhausen I. (2007) Carbohydr. Res. 342, 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wopereis S., Lefeber D. J., Morava E., Wevers R. A. (2006) Clin. Chem. 52, 574–600 [DOI] [PubMed] [Google Scholar]
- 10.Heise N., Gutierrez A. L., Mattos K. A., Jones C., Wait R., Previato J. O., Mendonça-Previato L. (2002) Glycobiology 12, 409–420 [DOI] [PubMed] [Google Scholar]
- 11.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 12.Bar-Peled M., Griffith C. L., Doering T. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12003–12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith C. L., Klutts J. S., Zhang L., Levery S. B., Doering T. L. (2004) J. Biol. Chem. 279, 51669–51676 [DOI] [PubMed] [Google Scholar]
- 14.Moyrand F., Klaproth B., Himmelreich U., Dromer F., Janbon G. (2002) Mol. Microbiol. 45, 837–849 [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez A. L., Farage L., Melo M. N., Mohana-Borges R. S., Guerardel Y., Coddeville B., Wieruszeski J. M., Mendonca-Previato L., Previato J. O. (2007) Glycobiology 17, 1C–11C [DOI] [PubMed] [Google Scholar]
- 16.Castle S. A., Owuor E. A., Thompson S. H., Garnsey M. R., Klutts J. S., Doering T. L., Levery S. B. (2008) 7, 1611–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klutts J. S., Levery S. B., Doering T. L. (2007) J. Biol. Chem. 282, 17890–17899 [DOI] [PubMed] [Google Scholar]
- 18.Liu X., Hu G., Panepinto J., Williamson P. R. (2006) Mol. Microbiol. 61, 1132–1146 [DOI] [PubMed] [Google Scholar]
- 19.Wickes B. L., Moore T. D., Kwon-Chung K. J. (1994) Microbiology 140, 543–550 [DOI] [PubMed] [Google Scholar]
- 20.Nelson R. T., Hua J., Pryor B., Lodge J. K. (2001) Genetics 157, 935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., D'Souza C., Wang P., Heitman J. (2002) Microbiology 148, 2607–2615 [DOI] [PubMed] [Google Scholar]
- 22.Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., Perfect J. R. (1993) J. Bacteriol. 175, 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown T. (2004) in Current Protocols in Molecular Biology, pp. 2.9.1–2.9.20, John Wiley & Sons, Inc., New York [Google Scholar]
- 24.Yan Z., Li X., Xu J. (2002) J. Clin. Microbiol. 40, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edman J. C., Kwon-Chung K. J. (1990) Mol. Cell. Biol. 10, 4538–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson E. S., Ayers D. J., Harrell A. C., Nicholas C. C. (1982) J. Bacteriol. 150, 1292–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y. C., Kwon-Chung K. J. (1999) J. Bacteriol. 181, 5636–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Y. C., Kwon-Chung K. J. (1998) Infect. Immun. 66, 2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Y. C., Penoyer L. A., Kwon-Chung K. J. (1996) Infect. Immun. 64, 1977–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doering T. L. (2009) Annu. Rev. Microbiol. 63, 223–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klutts J. S., Doering T. L. (2008) J. Biol. Chem. 283, 14327–14334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y. T. (1967) J. Biol. Chem. 242, 5474–5480 [PubMed] [Google Scholar]
- 33.Kwon-Chung K. J., Goldman W. E., Klein B., Szaniszlo P. J. (1998) Med. Mycol. 36, Suppl. 1, 38–44 [PubMed] [Google Scholar]
- 34.Jigami Y., Odani T. (1999) Biochim. Biophys. Acta 1426, 335–345 [DOI] [PubMed] [Google Scholar]
- 35.Braulke T., Pohl S., Storch S. (2008) J. Inherit. Metab. Dis., Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 36.Bao M., Elmendorf B. J., Booth J. L., Drake R. R., Canfield W. M. (1996) J. Biol. Chem. 271, 31446–31451 [DOI] [PubMed] [Google Scholar]
- 37.Wiggins C. A., Munro S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gemmill T. R., Trimble R. B. (1999) Biochim. Biophys. Acta 1426, 227–237 [DOI] [PubMed] [Google Scholar]
- 39.Kudo M., Bao M., D'Souza A., Ying F., Pan H., Roe B. A., Canfield W. M. (2005) J. Biol. Chem. 280, 36141–36149 [DOI] [PubMed] [Google Scholar]
- 40.Raas-Rothschild A., Cormier-Daire V., Bao M., Genin E., Salomon R., Brewer K., Zeigler M., Mandel H., Toth S., Roe B., Munnich A., Canfield W. M. (2000) J. Clin. Invest. 105, 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohl S., Tiede S., Castrichini M., Cantz M., Gieselmann V., Braulke T. (2009) Biochim. Biophys. Acta 1792, 221–225 [DOI] [PubMed] [Google Scholar]
- 42.Sperisen P., Schmid C. D., Bucher P., Zilian O. (2005) PLoS Comput. Biol. 1, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon-Chung K. J., Wickes B. L., Stockman L., Roberts G. D., Ellis D., Howard D. H. (1992) Infect. Immun. 60, 1869–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen K., Cox G. M., Wang P., Toffaletti D. L., Perfect J. R., Heitman J. (2003) Infect. Immun. 71, 4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]