Abstract
The strychnine-sensitive glycine receptor (GlyR) is a ligand-gated chloride channel and a member of the superfamily of cysteine loop (Cys-loop) neurotransmitter receptors, which also comprises the nicotinic acetylcholine receptor (nAChR). Within the extracellular domain (ECD), the eponymous Cys-loop harbors two conserved cysteines, assumed to be linked by a superfamily-specific disulfide bond. The GlyR ECD carries three additional cysteine residues, two are predicted to form a second, GlyR-specific bond. The configuration of none of the cysteines of GlyR, however, had been determined directly. Based on a crystal structure of the nAChRα1 ECD, we generated a model of the human GlyRα1 where close proximity of the respective cysteines was consistent with the formation of both the Cys-loop and the GlyR-specific disulfide bonds. To identify native disulfide bonds, the GlyRα1 ECD was heterologously expressed and refolded under oxidative conditions. By matrix-assisted laser desorption ionization time-of-flight mass spectrometry, we detected tryptic fragments of the ECD indicative of disulfide bond formation for both pairs of cysteines, as proposed by modeling. The identity of tryptic fragments was confirmed using chemical modification of cysteine and lysine residues. As evident from circular dichroism spectroscopy, mutagenesis of single cysteines did not impair refolding of the ECD in vitro, whereas it led to partial or complete intracellular retention and consequently to a loss of function of full-length GlyR subunits in human embryonic kidney 293 cells. Our results indicate that the GlyR ECD forms both a Cys-loop and a GlyR-specific disulfide bond. In addition, cysteine residues appear to be important for protein maturation in vivo.
Introduction
The glycine receptor (GlyR)2 is a ligand-gated ion channel that mediates fast neuronal inhibition predominantly in the spinal cord and brain stem (1). In the mammalian central nervous system, five gene variants of homologous GlyR subunits have been identified (2), four belong to the α-type (α1–α4) and one to the β-type. The GlyR complex presents as a rosette-like assembly of five subunit polypeptides surrounding a central anion pore. During development of the spinal cord, embryonic α2-homopentamers are largely replaced by adult heteromeric isoforms (3), composed of 3 β- and 2 α-subunits (4).
The GlyR belongs to the superfamily of cysteine loop (Cys-loop) receptors, which also comprises GABAA/C (γ-aminobutyric acid) receptors, nicotinic acetylcholine receptors (nAChR), and 5-hydroxytryptamine type 3 receptors. Structural predictions for Cys-loop receptors rely on sequence-based predictions and mutational studies. Crystal structures of the Lymnaea stagnalis acetylcholine-binding protein (AChBP), the Torpedo marmorata nAChR, the extracellular domain (ECD) of the murine nAChRα1, and prokaryotic proton-gated channels indicate a common, conserved protein fold (5–8). Accordingly, each subunit of a Cys-loop receptor comprises a large amino-terminal ECD adopting a twisted β-sandwich structure that precedes four α-helical transmembrane segments (TM1–TM4), each connected to its neighbors via intra- and extracellular loops. TM2 is an amphipathic helix that forms the inner lining of the channel pore. The ligand-binding pocket is located at the interface between adjacent ECDs and is covered by the flexible loop C (9). Consequently, channel gating is mediated through an interaction of the ECD with the TMs.
The Cys-loop, a loop within the ECD harboring two conserved cysteines, is eponymous for the superfamily of these ligand-gated ion channels. The Cys-loop extends into the interface between the ECD and TM domains, suggesting an important role in channel gating (10). It is generally assumed that these cysteine residues form a family-specific disulfide bond stabilizing the Cys-loop. Their functional role for the nAChR was explored using reducing agents and site-directed mutagenesis, which demonstrated influences on receptor expression and physiology (11, 12). Direct experimental evidence for the formation of the disulfide bond in the Cys-loop comes from chemical modification of informative peptides of purified Torpedo nAChR and from crystallography of the ECD of the murine nAChRα1 subunit (7, 13).
In the GlyR ECD, in addition to the Cys-loop, loop C is also thought to be stabilized by a second, unique disulfide bond. This assumption is based on recombinant expression studies with mutant receptor constructs (14). In the human GlyRα1 ECD, putative formation of the Cys-loop disulfide bond (Cys138–Cys152) and the GlyR-specific disulfide bond (Cys198–Cys209) would leave one cysteine residue (Cys41) unpaired. To our knowledge, there is no direct experimental evidence for disulfide bond formation in the GlyR ECD.
Starting from a multiple sequence alignment, we generated a model for the GlyRα1 ECD, consistent with specific formation of both the Cys-loop and the GlyR-specific disulfide bond. Making use of isolated GlyRα1 ECD obtained from Escherichia coli expression and functional refolding (15), we identified tryptic fragments corresponding to the predicted disulfide bonds by matrix-assisted laser-desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry. To elucidate their functional relevance, we generated serine mutants for each cysteine residue, preventing the formation of the respective disulfide bonds in isolated ECD and full-length receptor constructs. Our results suggest that the cysteine residues play important roles in receptor maturation.
EXPERIMENTAL PROCEDURES
Sequence Alignment and Homology Modeling
For sequence alignment, amino acid sequences of mature human nAChRα2, α3, α5, α7, and β2 subunits, human GlyRα1, α2, and α3 subunits, the murine nAChRα1 subunit (mm-ECD), and the L. stagnalis AChBP were obtained from the Swiss Prot data base (16). For transmembrane proteins, ECD sequences were used as annotated. A multiple sequence alignment was performed with ClustalW (17) followed by minimal manual corrections. Based on the crystal structure of the murine nAChRα1 ECD (9), the homology model of the human GlyRα1 ECD was generated using Swiss Model (18). Appropriate cysteine bonds were manually added followed by 100 steps of conjugate gradient energy minimization in Sybyl 7.3 (Tripos Inc., Munich, Germany).
Constructs and Mutagenesis
For bacterial expression, the pET30a vector (EMD, San Diego, CA) containing the isolated human GlyRα1 ECD was used. The full-length human GlyRα1 receptor subunit in pRK5 was used for expression in HEK293 cells. Single amino acid substitutions were introduced using PCR-based site-directed mutagenesis. All plasmids were sequence verified.
Bacterial Protein Expression and Refolding
Recombinant expression of the isolated GlyRα1 ECD constructs and refolding under oxidative conditions (using CuCl2) was carried out essentially as described (15). Deviating from this protocol, RosettaGami2 cells (EMD) were used for expression and affinity purification was omitted.
Tryptic Proteolysis and Chemical Modification
For tryptic proteolysis of the refolded ECD, the proteins were dialyzed against cleavage buffer (25 mm NH4HCO3). The protease-to-protein ratio was adjusted to 1:100 and cleavage was carried out at 37 °C overnight to obtain complete digestion. For limited proteolysis, protease-to-protein ratios of 1:500 or less were used. Where needed, dithiothreitol (5 mm final concentration) was added and reduction of the disulfide bonds was performed at 56 °C for 30 min. Modification of cysteine residues was performed for 1 h with a 40-fold molar excess of N-maleoyl-β-alanine (Sigma) over the concentration of dithiothreitol used. Lysine modification was performed with O-methylisourea using the proteomass guanidination kit (Sigma). All peptide samples were cleaned using Zip-Tip reversed phase microcolumns (Millipore, Billerica) prior to MALDI-TOF mass spectrometry.
MALDI-TOF Mass Spectrometry of Digested Protein Samples
Tryptically digested protein samples were dried using a vacuum concentrator (Eppendorf, Hamburg). The pellet was dissolved in 0.1% (v/v) trifluoroacetic acid. 2 μl of the sample were mixed with 2 μl of 2% (v/v) trifluoroacetic acid and 2 μl of dihydroxyacetophenone matrix (15 mg/ml of 2,5-dihydroxyacetophenone, 5 mg/ml of diammonium hydrogen citrate in 75% ethanol). An aliquot of 0.5 μl was spotted on a stainless steel target and allowed to dry at ambient temperature. MALDI-TOF mass spectrometry was performed on a Bruker Autoflex (Bruker Daltonik, Bremen, Germany), equipped with a nitrogen laser (λ = 337 nm). Positive ions were analyzed in the reflector mode after acceleration by 20 kV. External calibration was performed using the peptide calibration standard (Bruker Daltonik, Bremen, Germany). For each displayed mass spectrum, at least 250 individual spectra obtained from several positions on a spot were averaged. Spectrum analysis was performed using Flex Analysis software (Bruker Daltonik).
CD Spectroscopy
Circular dichroism spectra were recorded on a Jasco J810 spectropolarimeter (Jasco, Groß-Umstadt) equipped with a Peltier element. Data were recorded from wild-type as well as cysteine-mutant GlyRα1 ECDs in 10 mm sodium phosphate buffer, pH 7.4, at 20 °C from 260 to 190 nm with a step size of 0.2 nm and an integration time of 2 s in a quartz cell with 0.1-cm path length. Concentrations were determined by measuring the absorbance at 280 nm and ranged from 2.0 to 5.6 μm. A blank spectrum with buffer only was recorded under identical conditions and subtracted from the spectra. Spectra were smoothed using a Savitzky-Golay filter with a window size of 11 data points. Secondary structure contents were estimated using the DICHROWEB service (19, 20) applying the CDSSTR algorithm with the SP175(190) reference data base (21).
[3H]Ligand Binding Assays
Binding of [3H]strychnine and [3H]glycine (PerkinElmer Life Sciences) to refolded proteins was performed as described (15). To achieve retention of the refolded protein on GF/C filters, it was precipitated with an equal volume of 15% PEG400 in binding buffer (25 mm potassium Pi, pH 7.4, 200 mm KCl). Filters were washed with binding buffer after application of the protein; short washing times were essential. Unlabeled strychnine (200 μm) and glycine (200 mm) were used for competition and unspecific binding was subtracted. The affinity of [3H]strychnine binding was determined by Scatchard analysis (15).
Expression in HEK293 Cells, Membrane Preparation, and Western Blot
HEK293 cells were transfected according to a modified calcium phosphate protocol. Where necessary, cells were cotransfected with enhanced green fluorescent protein. For Western blot analysis, cells were harvested and homogenized in PBS at 4 °C. After centrifugation for 30 min at 100,000 × g, the pellet was resuspended in 20 mm potassium phosphate buffer containing protease inhibitors (Complete EDTA-free; Roche Applied Science). The protein concentration was determined and equal amounts were subjected to SDS-PAGE and Western blot. For detection of GlyRα1, the mouse monoclonal antibody mAb4a (supernatant of hybridoma cells) was used. Goat anti-mouse antibody coupled to Cy5 (1:250) or horseradish peroxidase (1:10,000) was used for visualization (Dianova, Hamburg).
Immunocytochemistry and Electrophysiology of HEK293 Cells
For immunocytochemistry, transfected HEK293 cells were washed 3 times in PBS and fixed for 30 min with paraformaldehyde (3% w/v in PBS) followed by quenching with 50 mm ammonium chloride in PBS. Blocking was performed with 5% (v/v) sheep serum in PBS for 30 min at room temperature. To perform staining of intracellular GlyRs, 0.1% (v/v) Triton X-100 was added to the blocking solution to permeabilize the plasma membrane. For staining of the ECD of GlyRα1, purified mouse monoclonal antibody mAb2b was used in blocking solution (1:200). Transfected cells were incubated with the primary antibody for 1 h at room temperature and washed 3 times with PBS. This procedure was repeated with goat anti-mouse antibody coupled to Alexa 488 (1:400 in blocking solution; Invitrogen). Finally, cells were embedded in Mowiol and subjected to confocal microscopy on a DMIRE2 confocal microscope (Leica, Wetzlar). Electrophysiology was carried out essentially as described (22). Under the conditions used, recombinant GlyRs showed little desensitization, consistent with earlier reports (23).
RESULTS
Modeling of the GlyRα1 ECD
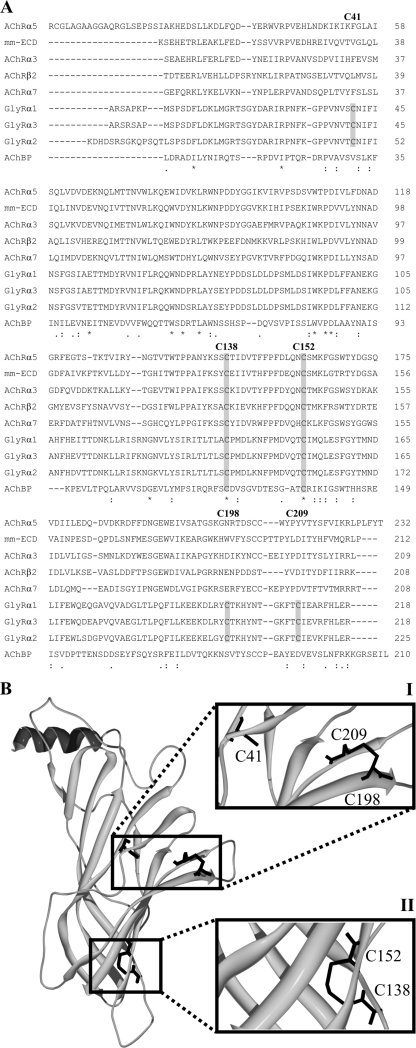
The human GlyRα1 ECD contains five cysteine residues (Cys41, Cys138, Cys152, Cys198, and Cys209). We generated a multiple sequence alignment for ECDs of several nAChR and GlyR subunit variants as well as the AChBP (Fig. 1A). This confirmed that only the cysteines within the Cys-loop (C138, C152) were conserved among the proteins analyzed, which is in agreement with previous alignments (4, 6).
FIGURE 1.
Alignment and modeling of the GlyRα1 ECD. A, human nAChRα2, α3, α5, α7, and β2 subunits, human GlyRα1, α2, and α3 subunits, the murine nAChRα1 subunit (mm-ECD), and the L. stagnalis AChBP are used for multiple sequence alignments with ClustalW (17). Cysteine residues are boxed in gray. Numbers correspond to the respective position in the processed human GlyRα1. Asterisks indicate identical, colons conserved, and dots semi-conserved substitutions. B, model of the human GlyRα1 ECD based on the crystal structure of the murine nAChRα1 ECD (9) after manual bonding of adjacent cysteines and energy minimization. Details covering the positions of the cysteine residues are enlarged (I and II). The model is consistent with the conjunction of four cysteines to form two distinct disulfide bonds. Cysteines 138 and 152 form the Cys-loop disulfide bond (I), whereas Cys198 and Cys209 form the GlyR-specific disulfide bond (II). One cysteine (C41) remains unpaired (I).
Based on the crystal structure of the murine nAChRα1 (7), a model of the human GlyRα1 ECD was generated. This model indicated the opposition of both conserved cysteine residues in the Cys-loop as well as two GlyR-specific cysteines in the loop C (Cys198 and Cys209), allowing disulfide bond formation. The model also predicted that cysteine residue Cys41 is buried, suggesting that it is not involved in disulfide bond formation (Fig. 1B).
Probing for Disulfide Bonds in the GlyRα1 ECD with MALDI-TOF Mass Spectrometry
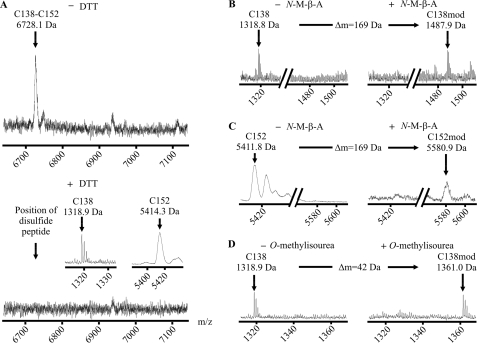
To analyze the status of the GlyR extracellular cysteines under in vitro conditions, the isolated GlyRα1 ECD was expressed in E. coli and refolded from inclusion bodies under oxidative conditions as described (15), obtaining native-like ligand-binding capability. Then, the ECD was digested with trypsin and analyzed by MALDI-TOF mass spectrometry, yielding a signal of 6728.1 Da matching the calculated mass of the tryptic fragments holding Cys138 and Cys152 linked by a disulfide bond (Fig. 2A). When the peptide mixture was chemically reduced to break disulfide bonds, this molecular mass disappeared, leaving signals (1318.9 and 5414.3 Da), which matched the calculated masses of the respective reduced fragments (Fig. 2A). Apparently, the pseudo-ion observed in the non-reduced sample contained a bond cleavable under mild reducing conditions, as expected for a disulfide bond. Respective single fragments, however, were still detectable in the non-reduced samples, suggesting that cysteine bonding was not fully completed during in vitro refolding.
FIGURE 2.
The Cys-loop disulfide bond and its associated peptide fragments. A, MALDI-TOF mass spectra of unmodified tryptic fragments of the GlyRα1 ECD refolded under oxidative conditions. The signal matching the theoretical mass of an ion composed of two disulfide-linked peptides carrying Cys138 and Cys152 is shown (calculated average isotopic mass 6728.9 Da, upper spectrum). Upon reduction, this signal is abolished, confirming its origin from a disulfide-linked pair of peptides (lower spectrum). The corresponding single peptides are shown (calculated masses 1318.7 Da (monoisotopic) and 5412.2 Da (average), inset). B and C, spectra of tryptic fragments carrying Cys138 (B) and Cys152 (C) before (left) and after (right) cysteine modification with N-maleoyl-β-alanine (N-M-β-A) (expected mass shift +169 Da). D, spectra of the fragment carrying Cys138 before (left) and after (right) lysine modification with O-methylisourea (expected mass shift +42 Da). As predicted from their sequence, both peptides are shifted with N-maleoyl-β-alanine and the peptide holding Cys138 is shifted with O-methylisourea, confirming the signal identities.
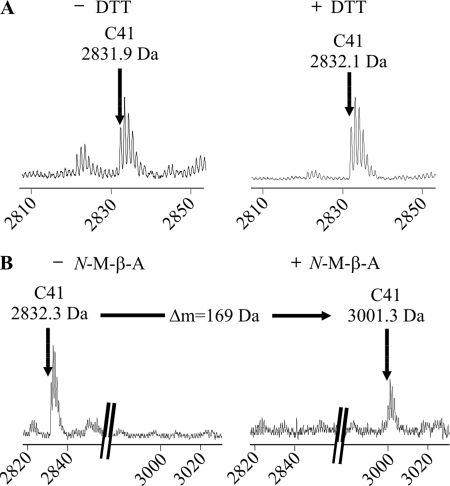
In some experiments, the tryptic peptides were chemically modified using side chain reactive agents. When the single fragments were modified with N-maleoyl-β-alanine, a mass shift of +169 Da occurred, consistent with the presence of a single sulfhydryl group in each fragment (Fig. 2, B and C). Finally, the reduced fragment holding Cys138 was modified once with O-methylisourea (+42 Da), which is in agreement with the presence of a single lysine residue, further corroborating the identity of this fragment (Fig. 2D).
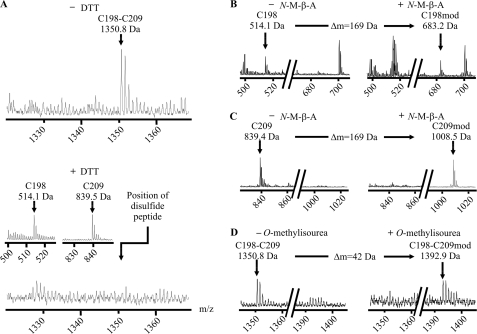
Furthermore, we found a second informative proton adduct of 1350.8 Da, matching the theoretical mass of the tryptic fragments harboring Cys198 and Cys209 linked by a disulfide bond. This bonded fragment, again, disappeared after chemical reduction, leaving the corresponding single fragments (514.1 and 839.5 Da) (Fig. 3A). Under oxidative conditions, signals representing single fragments were low in MALDI-TOF mass spectrometry.
FIGURE 3.
The GlyR-specific disulfide bond and its associated peptide fragments. A, MALDI-TOF mass spectra of unmodified tryptic fragments of the GlyRα1 ECD refolded under oxidative conditions. The signal matching the theoretical mass of an ion composed of two disulfide-linked peptides carrying Cys198 and Cys209 is shown (calculated monoisotopic mass 1350.6 Da, upper spectrum). Upon reduction, this signal is abolished, confirming its origin from a disulfide-linked pair of peptides (lower spectrum). The corresponding single peptides are shown (calculated monoisotopic masses 514.2 and 839.4 Da, inset). B and C, spectra of tryptic fragments carrying Cys198 (B) and Cys209 (C) before (left) and after (right) cysteine modification with N-maleoyl-β-alanine (N-M-β-A) (expected mass shift +169 Da). D, spectra of the disulfide-linked fragment before (left) and after (right) lysine modification with O-methylisourea (expected mass shift +42 Da). As predicted from their sequence, both peptides are shifted with N-maleoyl-β-alanine and the linked fragment is shifted with O-methylisourea, confirming the signal identities.
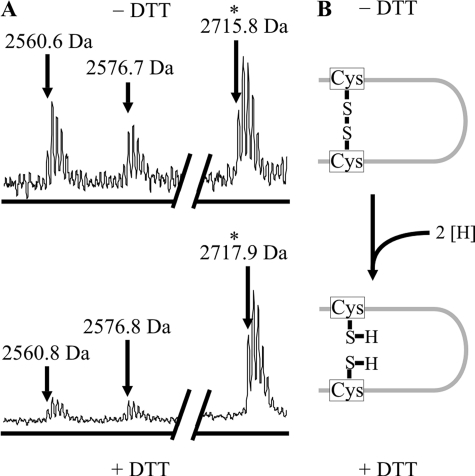
Mass shifts upon modification with N-maleoyl-β-alanine and O-methylisourea indicated the presence of cysteine residues in both single fragments as well as the presence of a lysine residue in the bonded fragment (Fig. 3, B–D). After limited tryptic proteolysis, we also found a proton adduct corresponding to the mass of a single fragment with 3 missed cleavages harboring both Cys198 and Cys209 (Fig. 4). The mass of this fragment was increased by 2 Da upon reduction, indicating cleavage of an intrapeptide disulfide bond. This observation confirmed our previous results.
FIGURE 4.
The GlyR-specific disulfide bond in limited proteolysis. A, MALDI-TOF mass spectra of the unmodified tryptic fragments of the GlyRα1 ECD refolded under oxidative conditions. The signal matching the theoretical mass of a fragment with 3 missed cleavages that carries both Cys198 and Cys209 (asterisk) before (upper spectrum, calculated monoisotopic mass: 2715.3 Da) and after (lower spectrum, calculated monoisotopic mass: 2717.3 Da) reduction with dithiothreitol (DTT) is shown. The signal is shifted by +2 Da, as expected from reduction of an intrapeptide disulfide bond. This confirms the formation of the GlyR-specific disulfide bond. Note that neighboring mass signals remain unchanged upon reduction, indicating the accuracy of the measurement. B, illustration of the reduction for an intrapeptide disulfide bond, showing that addition of two hydrogens results in a +2 Da mass shift as observed in A.
In all of our spectra, we exclusively found signals indicating bonds formed between Cys138 and Cys152, as well as Cys198 and Cys209, respectively. In particular, no alternative bond between these cysteines or to Cys41 was observed in any of the experiments. A fragment harboring Cys41 could be readily detected and modified (Fig. 5). The identities of all detected lysine and cysteine harboring fragments were confirmed by chemical modification. Taken together, identifiable fragments covered more than 95% of the protein sequence (Fig. 6). These data indicate selective formation of the predicted Cys-loop and GlyR-specific disulfide bond in the GlyRα1 ECD.
FIGURE 5.
Identification of the tryptic peptide containing the unpaired cysteine 41. A, MALDI-TOF mass spectra of unmodified tryptic fragments of the GlyRα1 ECD refolded under oxidative conditions. The signal matching the theoretical mass of the tryptic peptide harboring Cys41 is shown (calculated monoisotopic mass 2832.3 Da). Upon reduction with dithiothreitol (DTT) (right), this signal is not shifted, as expected for a fragment holding a reduced cysteine residue. B, spectra of tryptic fragments carrying Cys41 before (left) and after (right) cysteine modification with N-maleoyl-β-alanine (N-M-β-A) (expected mass shift +169 Da). The peptide signal is shifted with N-maleoyl-β-alanine, confirming the presence of a single cysteine residue.
FIGURE 6.
Summary of all detected GlyRα1 ECD peptides and their modifications. The complete amino acid sequence of the bacterial expression construct is shown. Bars indicate observed tryptic fragments as detected by MALDI-TOF mass spectrometry. Theoretical monoisotopic molecular masses (Da) are given for each fragment. Where essential, corresponding average molecular masses are given in brackets. Redundant fragments resulting from limited proteolysis are omitted. The number of asterisks (*) indicates the number of observed lysine modifications for each peptide. Cysteine residues are confirmed by mass shifts of the respective fragments upon N-maleoyl-β-alanine (N-M-β-A) modification and serine substitution (#). Brackets connect cysteine residues that are found to form disulfide bonds. The total sequence coverage is greater than 95%.
Mutagenesis of the GlyRα1 ECD Construct
To analyze the process of disulfide formation in vitro, we generated single cysteine-to-serine substitutions of the ECD constructs. The mutant ECDs were expressed in E. coli, forming inclusion bodies, and subjected to refolding under oxidative conditions. Surprisingly, all mutants appeared to readily refold, yielding considerable concentrations of soluble protein. None of the mutants displayed increased precipitation, essentially resembling the wild-type situation (data not shown). This suggested that the overall fold of the ECD may be independent of disulfide bond formation. Subsequently, the mutant ECDs were subjected to tryptic digestion and analysis by MALDI-TOF mass spectrometry.
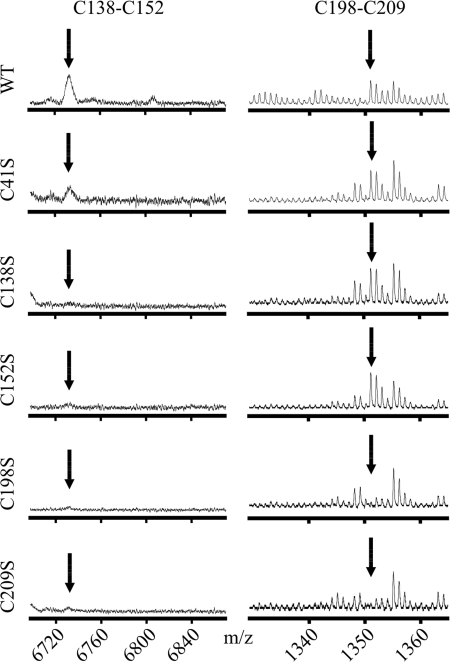
The single fragments holding mutant residues showed the expected mass shifts resulting from mutagenesis. The pseudo-ion corresponding to the bonding of Cys198 and Cys209 was observed for wild-type and mutant constructs C41S, C138S, and C152S, but not for C198S and C209S (Fig. 7). The bonding of Cys138 and Cys152, however, was only observed for wild-type and C41S, indicating that, in vitro, formation of the Cys-loop disulfide bond may require the presence of the GlyR-specific bond.
FIGURE 7.
Disulfide bond formation in mutant GlyRα1 ECDs. MALDI-TOF mass spectra of unmodified tryptic fragments of different GlyRα1 ECD variants refolded under oxidative conditions. The signals corresponding to the Cys-loop (left, Cys138–Cys152) and GlyR-specific disulfide bond (right, Cys198–Cys209) are shown (see Figs. 2 and 3 for reference). Mutant C41S shows unaltered cysteine bond formation as compared with the wild-type domain (WT). As expected, for mutants C138S and C152S the signal for Cys198–Cys209 but not Cys138–Cys152 is observed. In contrast, mutants C198S and C209S display no signal for disulfide bond formation, indicating the requirement of the GlyR-specific for the effective formation of the Cys-loop disulfide bond in vitro.
Secondary Structure Analysis of Wild-type and Mutant GlyRα1 ECD
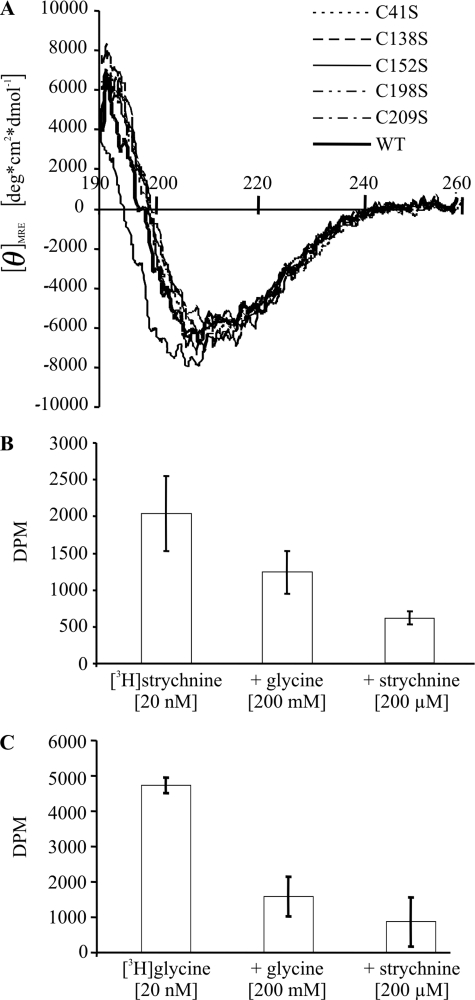
To analyze secondary structure elements of GlyRα1 ECDs, the refolded proteins were subjected to circular dichroism spectroscopy. The resulting spectrum of wild-type ECD was consistent with previous observations (15) and very well superimposed with spectra obtained from each of the mutants (Fig. 8A), indicating an overall comparable secondary structure content (Table 1). This result is in line with an equally efficient in vitro refolding of all ECD variants. Furthermore, the composition of secondary structure elements is in good agreement with our proposed model of the GlyRα1 ECD.
FIGURE 8.
A, circular dichroism spectra of refolded wild-type (WT) and cysteine mutant GlyRα1 ECDs. Superimposition of CD spectra of wild-type and cysteine mutant ECDs indicates an overall comparable content of secondary structure. B and C, binding (±S.D.) of 20 nm [3H]strychnine (B) and 20 nm [3H]glycine (C) to refolded wild-type GlyRα1 ECD. Binding of both radioligands was inhibited by an excess of either unlabeled glycine or strychnine. For the GlyRα1 ECD preparation shown, the KD for [3H]strychnine was 25 nm.
TABLE 1.
Calculated secondary structure from CD spectroscopy
Deconvolution of the CD spectra reveals a high β-sheet content and a small fraction of α-helix for each protein variant analyzed. These results are consistent with the structural model proposed. Overall, only very small deviations can be observed indicating essential identical folds of wild-type and mutant proteins.
| Sample | α-Helix | β-Sheet | Turn | Unordered |
|---|---|---|---|---|
| % | ||||
| Wild-type | 8 | 36 | 12 | 43 |
| C41S | 9 | 37 | 12 | 42 |
| C138S | 11 | 36 | 12 | 41 |
| C152S | 8 | 35 | 12 | 43 |
| C198S | 7 | 38 | 11 | 42 |
| C209S | 9 | 38 | 12 | 41 |
Ligand Binding of Refolded GlyRα1 ECD
In a filtration assay, the refolded wild-type GlyRα1 ECD showed high affinity [3H]strychnine binding (Fig. 8B) with KD values ranging from 25 to 70 nm, similar to our earlier report (15). In addition, [3H]strychnine binding was partially displaceable by an excess of unlabeled glycine. Conversely, we observed strychnine-displaceable [3H]glycine binding (Fig. 8C). With the filtration assay used (15), saturation of [3H]glycine binding was not achieved, consistent with the low affinity of the glycine site (2, 4). This precluded a determination of the corresponding KD value. Taken together, this indicates that a mutually exclusive binding site for glycine and strychnine had formed in the refolded ECD. Apparently, the ECD folded into its native-like, functional conformation.
Cysteine Mutagenesis in Full-length GlyR Constructs
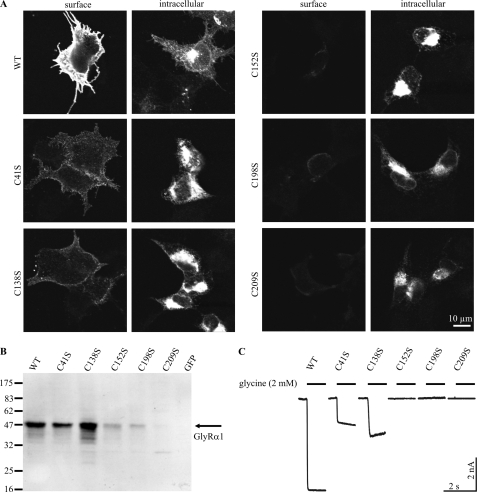
To address the function of the disulfide bonds, we performed single cysteine-to-serine substitutions in the ECD of full-length GlyRα1 constructs. The constructs were expressed in HEK293 cells and immunostained with mAb2b recognizing the extracellular domain of GlyRα1. Discrimination between surface expression and intracellular retention of the GlyR protein was achieved through facultative permeabilization of the cells prior to immunostaining. In confocal immunocytochemistry of transfected, non-permeabilized HEK293 cells, mutant GlyRs C41S and C138S showed reduced surface expression. In contrast, mutants C152S, C198S, and C209S showed no detectable surface expression (Fig. 9A). Accordingly, we found pronounced perinuclear staining for all mutants, indicating an increased intracellular retention. This finding contrasts the situation in vitro, where refolding appeared to be independent of the cysteine residues. The number of cells positive for intracellular GlyR stains was markedly reduced for C152S, C198S, and C209S (data not shown). Consequently, we found low GlyR immunoreactivity for these mutants in membrane preparations analyzed by Western blots of transfected HEK293 cells (Fig. 9B).
FIGURE 9.
Characterization of full-length GlyRα1 variants. A, immunocytochemistry of transfected HEK293 cells, surface-stained or permeabilized for intracellular staining of GlyRα1 with mAb2b. The wild-type receptor (WT) shows intensive surface and intracellular staining. Note that surface expression is significantly reduced for mutants C41S and C138S, and abolished for C152S, C198S, and C209S. This reduction coincides with an intracellular accumulation of the GlyRα1 antigen in perinuclear regions, implying defects in maturation. B, Western blot analysis of membrane preparations of HEK293 cells using mAb4a reflects reduced total expression of C152S, C198S, and C209S, which may correlate with disturbed maturation. Note the faint band at about 30 kDa for C209S indicating a reduced stability of this receptor variant. C, examples of whole cell patch clamp recordings of transfected HEK293 cells with saturating glycine application (2 mm) are shown (n ≥ 6). In contrast to WT, C41S, and C138S, no activity is observed for receptor mutants C152S, C198S, and C209S, which correlates well with abolished surface expression.
Finally, the physiological activity of the mutant receptors was measured in whole cell voltage-clamp recordings, applying saturating glycine concentrations (Fig. 9C). As expected from total and surface expression, mutant constructs C41S and C138S, but not C152S, C198S, and C209S were active in electrophysiology. Partially conserved maturation of mutant C41S is in agreement with the in vitro data, which shows that Cys41 is neither involved in disulfide bond formation nor in folding stability of the ECD. Surprisingly, the differences observed between Cys-loop mutants C138S and C152S indicate a functional asymmetry of the single cysteines during maturation of the protein, as substitution of Cys152 exerts a stronger impact on surface accumulation and channel function.
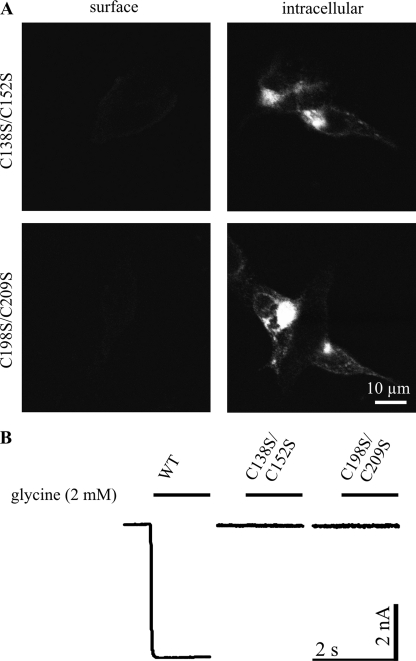
To address whether the observed maturation defects resulted from exposure of unpaired cysteines, we generated double mutants C138S/C152S and C198S/C209S. The double mutant C198S/C209S neither showed surface expression nor functionality in electrophysiology, essentially resembling the respective single mutants (Fig. 10). This indicates that intracellular retention did not result from exposed, unpaired cysteines 198 or 209. Likewise, the double mutant C138S/C152S was also retained, exhibiting neither surface expression nor ion channel activity, as observed for C152S but not for C138S. Hence, for surface expression, the mutation C152S is dominant over C138S. These data confirm that intracellular retention did not result from exposure of single unpaired cysteine residues.
FIGURE 10.
Characterization of full-length GlyRα1 double mutants. A, immunocytochemistry of transfected HEK293 cells, surface-stained or permeabilized for intracellular staining of GlyRα1 with mAb2b. No surface expression is detected for double mutants C138S/C152S and C198S/C209S, whereas intracellular accumulation in perinuclear regions is observed, implying defects in maturation. B, examples of whole cell patch clamp recordings of transfected HEK293 cells with saturating glycine application (2 mm) are shown (n = 6). Both double mutants are inactive, as expected from abolished surface expression.
DISCUSSION
Based on primary sequence analysis, GlyRs have been predicted to carry both a Cys-loop and a GlyR-specific disulfide bond (24). Here, we present experimental evidence for the formation of these disulfide bonds in the recombinant ECD of GlyRα1 and for their role in GlyR biogenesis.
Homology modeling of the human GlyRα1 ECD strongly suggested formation of both bonds, as the respective cysteines were directly opposed to each other. To test this hypothesis, we used the refolded ECD as a model for the receptor. Upon refolding under oxidative conditions, the ECD construct gains a native-like fold and attains high affinity strychnine binding (15). As evident from glycine displaceable [3H]strychnine binding and strychnine-displaceable [3H]glycine binding, the GlyRα1 ECD carries determinants for both agonist and antagonist recognition (25).
The purity of the ECD constructs allowed for reproducible detection of tryptic fragments covering nearly the entire sequence of the protein. Upon random disulfide bond formation between the five cysteine residues present, 15 different bonds (10 inter- or intrasubunit and 5 intersubunit bonds) are possible. Of these, only the predicted two disulfide bonds were found. This strongly indicates specificity in the formation of the disulfide bonds.
Several lines of evidence confirm the identity of the detected proton adducts. (i) The deviation of the informative signals from the calculated m/z values was below 1 Da. (ii) The signals were detectable under oxidative conditions only, as expected for ions holding a disulfide bond. (iii) The signals were absent from tryptic digests of the respective cysteine mutants. For the GlyR-specific bond, further evidence comes (iv) from the chemical modification of a lysine residue and (v) from the detection of a signal corresponding to a fragment with 3 missed cleavages. This fragment carried an intrapeptide disulfide bond that was readily reduced to obtain the predicted mass shift of +2 Da. We conclude that the GlyR holds both the conserved bond in the Cys-loop between Cys138 and Cys152, as well as the GlyR-specific bond in loop C between Cys198 and Cys209.
Single-cysteine mutagenesis did not disturb refolding in vitro, indicating that the overall native fold is not strongly dependent on disulfide bond formation. Circular dichroism spectroscopy revealed a comparable secondary structure content of both wild-type (15) and mutant ECDs, indicating that disulfide bond disruption remained without major structural changes. A similar secondary structure distribution was also observed for a chimeric glycine-binding protein, derived from human GlyRα1 and AChBP (26). Thus, achieving the overall native-like fold of the isolated GlyRα1 ECD during in vitro refolding appeared to be independent of disulfide bond formation. Curiously, however, the formation of its Cys-loop disulfide bond was dependent on the presence of Cys198 and Cys209 forming the GlyR-specific bond. This suggests that the formation of the GlyR-specific bond leads to a minor conformational shift allowing for the eponymous Cys-loop bond to form.
The effect of disulfide bond formation on the function of the GlyR has been studied using reducing and oxidizing agents. The oxidative status of the receptor has no effect on the affinity to strychnine and glycine, but modifies the Hill coefficient of binding (27). Furthermore, oxidation and reduction have opposing effects on whole cell currents in rat retinal ganglion cells (28). This finding is contrasted by a recent study detecting no effect of redox agents on glycinergic currents in mouse hippocampal neurons (29). Taken together, no gross reproducible functional defect of surface-expressed receptors was observed with reducing agents. None of these studies, however, determined the oxidative status of the single cysteines after manipulation.
By site-directed mutagenesis, we tested the influence of the cysteine residues on receptor expression and function. For mutant C41S, we observed reduced surface expression and activity, indicating that beyond disulfide bond formation, cysteines play important roles in the process of receptor maturation. In contrast to C152S, C138S is to some extent surface-expressed and functional. This discrepancy shows that two cysteines involved in the formation of a single disulfide bond can have different significance for receptor maturation and function. As observed for C152S, mutants C198S and C209S were not only inactive, but also lacked surface expression. This is contrasted by a previous report of a surface-expressed, but inactive, mutant C209S (14). Nevertheless, both studies sustain the hypothesis of functional asymmetry of cysteine residues. In our hands, none of the inactive mutants were surface expressed. Additionally, cysteine mutagenesis might interfere with glycine-gated chloride channel function, i.e. ligand binding or channel open probability, which cannot readily be separated from ion channel biogenesis in heterologous cell expression. Intracellular retention and dysfunctionality of double mutants C138S/C152S and C198S/C209S suggests that exposure of an unpaired cysteine was not the cause for the intracellular retention of the single mutants. Moreover, refolding experiments with mutated constructs showed that the formation of disulfide bonds may be interdependent, even though the overall stability of the fold is not dramatically influenced by bond formation as shown by circular dichroism spectroscopy. Hence, the cysteines seem to have important functions in the maturation of the receptor in vivo that go beyond the formation of disulfide bonds. Eventually, it is tempting to speculate that disulfide bond formation occurs as a late step in folding the ECD. Taken together, this study does not only provide experimental evidence for the formation of both the Cys-loop and the GlyR-specific disulfide bond, but also offers a paradigm for the analysis of the folding mechanism of the ECD.
Acknowledgments
We thank Drs. Andreas Humeny, Martin Eberhardt, and Stephan Hupfer for helpful discussions and critically reading this manuscript. We thank Dr. Kristina Becker for help with site-directed mutagenesis, and Drs. Heike Meiselbach and Heinrich Sticht for help with modeling. Technical assistance of Claudia Sass and Rosa Weber is gratefully acknowledged.
This work was supported by European Union Grant Neurocypres, HEALTH-F4-2008-202088, Elitenetzwerk Bayern, Lead Structures of Cell Function, Grant K-BM-2003-85, and the Deutsche Forschungsgemeinschaft.
- GlyR
- glycine receptor
- ECD
- extracellular domain
- TM
- transmembrane domain
- GABA
- γ-aminobutyric acid
- nAChR
- nicotinic acetylcholine receptor
- AChBP
- acetylcholine binding protein
- MALDI-TOF
- matrix-assisted laser-desorption-ionization time-of-flight
- PBS
- phosphate-buffered saline
- mAb
- monoclonal antibody.
REFERENCES
- 1.Betz H., Laube B. (2006) J. Neurochem. 97, 1600–1610 [DOI] [PubMed] [Google Scholar]
- 2.Breitinger H. G., Becker C. M. (2002) ChemBioChem 3, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 3.Becker C. M., Hoch W., Betz H. (1988) EMBO J. 7, 3717–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grudzinska J., Schemm R., Haeger S., Nicke A., Schmalzing G., Betz H., Laube B. (2005) Neuron 45, 727–739 [DOI] [PubMed] [Google Scholar]
- 5.Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J. P., Corringer P. J. (2007) Nature 445, 116–119 [DOI] [PubMed] [Google Scholar]
- 6.Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 7.Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 8.Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 9.Gay E. A., Yakel J. L. (2007) J. Physiol. 584, 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 11.Rojas L., Zuazaga C., Steinacker A. (1991) Brain Res. 551, 10–15 [DOI] [PubMed] [Google Scholar]
- 12.Sumikawa K., Gehle V. M. (1992) J. Biol. Chem. 267, 6286–6290 [PubMed] [Google Scholar]
- 13.Kao P. N., Karlin A. (1986) J. Biol. Chem. 261, 8085–8088 [PubMed] [Google Scholar]
- 14.Rajendra S., Vandenberg R. J., Pierce K. D., Cunningham A. M., French P. W., Barry P. H., Schofield P. R. (1995) EMBO J. 14, 2987–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitinger U., Breitinger H. G., Bauer F., Fahmy K., Glockenhammer D., Becker C. M. (2004) J. Biol. Chem. 279, 1627–1636 [DOI] [PubMed] [Google Scholar]
- 16.The UniProt Consortium (2008) Nucleic Acids Res. 36, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 18.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 19.Whitmore L., Wallace B. A. (2008) Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 20.Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lees J. G., Miles A. J., Wien F., Wallace B. A. (2006) Bioinformatics 22, 1955–1962 [DOI] [PubMed] [Google Scholar]
- 22.Oertel J., Villmann C., Kettenmann H., Kirchhoff F., Becker C. M. (2007) J. Biol. Chem. 282, 2798–2807 [DOI] [PubMed] [Google Scholar]
- 23.Legendre P., Muller E., Badiu C. I., Meier J., Vannier C., Triller A. (2002) Mol. Pharmacol. 62, 817–827 [DOI] [PubMed] [Google Scholar]
- 24.Grenningloh G., Rienitz A., Schmitt B., Methfessel C., Zensen M., Beyreuther K., Gundelfinger E. D., Betz H. (1987) Nature 328, 215–220 [DOI] [PubMed] [Google Scholar]
- 25.Becker C. M. (1992) in Handbook of Experimental Pharmacology (Herken H., Hucho F. eds) pp. 539–575, Springer, Heidelberg [Google Scholar]
- 26.Liu Z., Ramanoudjame G., Liu D., Fox R. O., Jayaraman V., Kurnikova M., Cascio M. (2008) Biochemistry 47, 9803–9810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz-Gómez A., Vaello M. L., Valdivieso F., Mayor F., Jr. (1991) J. Biol. Chem. 266, 559–566 [PubMed] [Google Scholar]
- 28.Pan Z. H., Bähring R., Grantyn R., Lipton S. A. (1995) J. Neurosci. 15, 1384–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thio L. L., Zhang H. X. (2006) Neuroscience 139, 1315–1327 [DOI] [PubMed] [Google Scholar]