Abstract
Dos from Escherichia coli is a bacterial gas sensor protein comprising a heme-containing gas sensor domain and a phosphodiesterase catalytic domain. Using a combination of static light scattering and gel filtration experiments, we established that, as are many other sensor proteins, the full-length protein is dimeric. The full-length dimer (association constant <10 nm) is more stable than the dimeric heme domain (association constant ∼1 μm), and the dimer interface presumably includes both sensor and catalytic domains. Ultrafast spectroscopic studies showed little influence of the catalytic domain on kinetic processes in the direct vicinity of the heme. By contrast, the properties of ligand (CO and O2) binding to the heme in the sensor domain, occurring on a microsecond to second time scale, were found to be influenced by (i) the presence of the catalytic domain, (ii) the dimerization state, and in dimers, (iii) the ligation state of the other subunit. These results imply allosteric interactions within dimers. Steady-state titrations demonstrated marked cooperativity in oxygen binding to both the full-length protein and the isolated heme domain, a feature not reported to date for any dimeric sensor protein. Analysis of a variety of time-resolved experiments showed that Met-95 plays a major role in the intradimer interactions. The intrinsic binding and dissociation rates of Met-95 to the heme were modulated ∼10-fold by intradimer and sensor-catalytic domain interactions. Dimerization effects were also observed for cyanide binding to the ferric heme domains, suggesting a similar role for Met-95 in ferric proteins.
Introduction
Dos from Escherichia coli (EcDos)4 is a modular gas sensor protein in which phosphodiesterase activity is coupled with the binding and release of external ligands in an associated heme-binding sensor domain (1–3). Cyclic AMP (4) and cyclic diGMP act as substrates of this enzyme, the latter with much higher activity (5). EcDos has been found to display 6–7-fold enhanced catalytic activity toward cyclic diGMP after binding of gaseous molecules such as O2, CO, and NO to the heme.
Although the heme-containing PAS sensor domains of EcDos and the rhizobial sensor protein FixL (6) share a conserved structural fold, the ligand-induced regulation of both proteins differs considerably. In contrast to EcDos, FixL is an oxygen-specific sensor that couples the status of its sensor domain to the activity of a histidine kinase. This enzymatic activity is decreased to a great extent upon O2 binding (2). Furthermore, whereas in the ferrous deoxy form of FixL the heme is pentacoordinate, in EcDos the heme iron is coordinated to a proximal histidine (His-77) and a distal methionine (Met-95). Met-95 can be replaced by small gaseous molecules and plays a crucial role in the regulation of the catalytic activity.
The three-dimensional structures of the Fe(III), Fe(II), and Fe(II)-O2 complexes of the isolated heme sensor domain of EcDosH (EcDosH) have been determined (7, 8). These structures have revealed the importance of several key amino acids near the distal heme-binding site involved in changes in the heme environment upon O2 binding. In particular, upon its replacement by O2 as a heme ligand, the internal ligand, Met-95, undergoes a major structural rearrangement toward a position where it points out of the heme pocket. Rearrangements occur equally for Arg-97 and Phe-113, which are both involved in stabilizing the polar Fe–O2 bond by hydrogen bond interactions. The crucial roles of these residues in regulating the catalytic domain have been studied (9–11). In the ferric form of crystallized EcDosH, a water molecule rather than Met-95 is bound in the distal heme site (7).
The heme sensor domain EcDosH crystallizes as a dimer (7, 8) and is also dimeric in solution (1). Surprisingly, based on gel filtration chromatography, full-length EcDos has been characterized as a tetramer (12, 13). This contrasts with many other heme-based sensor enzymes, including FixL or the CO sensor CooA, which have been described as stable dimers (14–17).
The reactivity of EcDos to various gaseous signaling ligands (O2, NO, CO) is determined by their binding and release kinetics to the heme domain and by intermediates formed during the kinetic process. For the full-length protein, O2, CO, and cyanide binding studies on the time scale of seconds have been reported that indicate an influence of the presence of the enzymatic domain (10, 18). However, kinetic spectroscopic studies of the relevant processes on the femtosecond to millisecond time scales have been performed to date essentially only on the isolated heme domain (1, 11, 17, 19–21).
We have now heterologously expressed the full-length protein EcDos in high quality and quantity suited for detailed static and time-resolved optical spectroscopic measurements. Here, we present a study of the combined oligomerization and ligand binding properties of EcDos and its sensor domain EcDosH, as well as a comparison with the isolated heme domain of FixL (FixLH). We determined the molar masses and hydrodynamic diameters of the above proteins by size exclusion fast protein liquid chromatography coupled with multi-angle laser light scattering (MALLS). We concluded that full-length EcDos is a dimeric protein, as are the sensor domains EcDosH and FixLH. The monomer-monomer interfaces for the truncated proteins were found to be considerably less stable than for the full-length protein. The microscopic binding constants of O2, CO, and Met-95 were measured for EcDos, EcDosH, and two EcDosH mutant proteins with the substitutions M95I and M95H. We present evidence for substantial allosteric interactions between the two proteins constituting the dimer, leading to modulation of ligand binding parameters and in particular to a cooperative mechanism for O2 binding.
MATERIALS AND METHODS
Expression and Purification of EcDos and EcDosH
The gene coding for the full-length protein EcDos (corresponding to codons 1–799) was amplified by PCR using the primers 5′-atg aag cta acc gat gcg gat-3′ (forward) and 5′-tca gat ttt cag cgg taa cac-3′ (reverse) and subsequently cloned into a pBAD TOPO TA cloning vector (Invitrogen). The construct obtained under control of an arabinose-inducible promoter was verified by DNA sequencing and eventually transformed into E. coli HB101 or BL21DE3 for protein expression in autoinducible medium (22) or with 0.2% arabinose induction. 1 mm δ-aminolevulinic acid was added to the culture medium. Cultures were grown for 48 h at 32 °C and 250 rpm agitation. The protein was purified with an Akta purifier system (GE Healthcare) using a 5-ml HisTrap affinity column (GE Healthcare) equilibrated with PBS, pH 7.5, and was eluted at a concentration of 250 mm imidazole. The samples obtained were loaded on a desalting Sephadex G-25 column (GE Healthcare) to eliminate imidazole and subsequently suspended in PBS, pH 7.5, or 50 mm Tris/HCl, pH 7.6.
The DNA fragment corresponding to codons 1–142, coding for the PAS heme domain (EcDosH), was amplified using the primers 5′-tta ctc cat atg aag cta acc gat gcg gat aa-3′ (forward) and 5′-cga gtg gga tcc cta aac ggc aat aat caa ttg tc-3′ (reverse) and cloned in pET-3a (Novagen). The final constructs were transformed into E. coli BL21DE3 for expression in autoinducible medium (Overnight Express Instant TB medium, Novagen), and 1 mm δ-aminolevulinic acid was added to the culture medium. Cultures were grown for 24 h at 32 °C and 250 rpm agitation. The EcDosH proteins were purified with an Akta purifier system on a Hitrap DEAE-Sepharose column (Amersham Biosciences). Because of the low pI of EcDosH, the samples were loaded on the column equilibrated with 50 mm Tris/HCl buffer, pH 8.5, and the protein was eluted at 50 mm NaCl. The concentrated material was loaded on a SuperoseTM 12 HR 16/50 (GE Healthcare) column equilibrated with PBS, pH 7.5. Expression and purification of the mutant proteins EcDosH M95I and M95H were carried out as described previously (20).
The gene fragment encoding FixLH was amplified and cloned as described previously (23). The protein was expressed in autoinducible medium (Overnight Express Instant TB medium), and 1 mm δ-aminolevulinic acid was added to the culture medium. Cultures were grown for 24 h at 32 °C and 250 rpm agitation. The protein purification protocol was the same as for EcDos.
Pure ferric EcDos, EcDosH, and FixLH were found to have absorption ratios (A280 nm/AmaxSoret) of 1, 0.36, and 0.24, respectively, very similar to those published previously (1, 13, 24). These comparisons, as well as the comparison with predicted spectra based on protein composition (not shown), indicate that the heme to monomer ratio was close to 1 for all of our preparations.
Gel Filtration
Gel filtration experiments were carried out using a fast protein liquid chromatograph (Gilson) equipped with a SuperoseTM 12 HR 10/300 GL column (GE Healthcare) at 25 °C in buffer containing 30 mm PBS, pH 7.5, and 100 mm NaCl. The elution time was determined at the peak half-height; a void volume of 7.8 ml and an internal volume for the gel bed of 25 ml were used to calculate the molecular sieve coefficients (Kav). High reproducibility of the loaded sample volumes was obtained using a Gilson autoinjector device; for an injection volume of 70 μl, a dilution factor of 10 after elution was calculated. The absorbance of the eluent was registered at 415 and 280 nm.
We analyzed the elution profiles for each sample in different ligation states. The oxygenated forms were studied directly after purification to avoid heme oxidation, and 2 mm dithiothreitol was added to ensure full reduction of the heme proteins. The oxidized forms were obtained by heme oxidation with an excess of potassium ferricyanide after deoxygenation of the sample, to favor oxidation over O2 binding and to limit the time of reaction with the oxidant finally removed after loading the samples onto a desalting Sephadex G-25 column (GE Healthcare). Completion of cyanide binding to the ferric protein was checked spectrophotometrically.
Dynamic Light Scattering
The particle size was measured with a Zetasizer Nano-ZS (Malvern Instruments), based on dynamic light scattering. Size distribution by volume was used to interpret the results. Measurements were performed at 20 °C in 30 mm PBS, pH 7.5, 0.10 m NaCl, and the diameter was determined from the average of triplicate measurements. Hydrodynamic diameters of the protein were estimated relative to those of standard proteins, specifically recombinant dehaloperoxidase (15.5 kDa), myoglobin (16.1 kDa) (Sigma-Aldrich), recombinant neuroglobin (16.5 kDa), hemoglobin (64.7 kDa), diaspirin-cross-linked hemoglobin (Baxter Healthcare Corp., Deerfield, IL), albumin monomer standard (67 kDa) (Sigma-Aldrich), and a recombinant octameric hemoglobin (131.7 kDa).
Size Exclusion by Fast Protein Liquid Chromatography and Multi-angle Laser Light Scattering
The molar masses in solution were determined by MALLS coupled with size exclusion chromatography (SEC-MALLS). Gel filtration separation reactions were carried out using an EttanTM LC liquid chromatography system (GE Healthcare) equipped with a SuperoseTM 12 HR 10/300 GL column (GE Healthcare). Isocratic elution was performed at a flow rate of 0.39 ml/min using a mobile phase of 30 mm PBS, pH 7.5, 100 mm NaCl, and 0.03% sodium azide at 25 °C. Light scattering analysis was performed using an EttanTM LC HPLC system with an automatic degasser and thermostatted autosampler, connected in-line to a DAWN® HELEOSTM II 18-angle static light scattering detector; this was equipped with a quasi-elastic light scattering instrument (QELS; Wyatt Technology, Santa Barbara, CA) and an Optilab® rEX differential refractometer with a Peltier temperature-regulated flow cell maintained at 25 °C (Wyatt Technology, Santa Barbara, CA). Calibration of the light scattering detector was verified using an albumin monomer standard, recombinant neuroglobin, myoglobin, and diaspirin-cross-linked hemoglobin. The molar mass was calculated from the light scattering data using a specific refractive index increment (dn/dc) value of 0.183 ml/g. The light scattering detector was controlled and analyzed using ASTRA V software (version 5.3.4.13) (Wyatt Technology).
Spectra and Ligand Binding/Dissociation Kinetics
Steady-state spectral measurements were performed with a Varian Cary 400 or a HP 8453 diode array spectrophotometer. All ligand binding experiments were performed in 150 mm Tris acetate, 50 mm NaCl, pH 7.5, or in PBS, pH 7.5. Five mm dithiothreitol was added to all EcDos samples to avoid misfolded proteins and folding intermediates due to the formation of disulfide bonds.
Association rates of cyanide to heme were obtained by monitoring changes of absorption in PBS, pH 7.5, at 25 °C with a HP 8453 diode array spectrophotometer. The same apparatus was used for measuring the CO dissociation from EcDos after replacement by either 1 atm of NO or O2.
The bimolecular binding kinetics after heme ligand photolysis were measured using a Nd:YAG Big Sky laser CFR-300 (Quantel) generating 8 ns/30 mJ pulses at 532 nm. The laser beam and the detection monochromatic light were directed to the sample-containing optical cuvette by long length glass optical fibers. We also used a repeat sequence of pulses at 10 Hz frequency during ∼10 s to photoincubate the EcDos protein in the hexacoordinated state by flashing off the CO.
Stopped-flow rapid mixing experiments were performed with SFM-3 Bio-Logic equipment. The methods used to assess hexacoordination and bimolecular CO and O2 rate constants have been described previously (25). Samples from 1 to 10 μm, as determined on the basis of heme absorption, were measured in 4-mm optical path length quartz cells, whereas more highly concentrated samples (>10 μm) were measured in 1-mm cells.
Multicolor femtosecond absorption spectroscopy was performed as described (21) with a 30-fs pump pulse centered at 565 nm and a <30-fs white light continuum probe pulse at a repetition rate of 30 Hz and a temperature of 20 °C. The proteins were prepared to a sample concentration of 50 to 70 μm.
Equilibrium Oxygen Binding Curves
O2 binding curves at equilibrium were measured using a HEMOX analyzer (TCS Scientific Corp.), allowing the simultaneous measurement of absorption and oxygen tension by a Clark-type electrode upon deoxygenation with nitrogen. A dual wavelength measurement is monitored at 560 and 576 nm, which closely matches the maximum and minimum of the absorption difference between the O2 and deoxy hexacoordinated spectra. Nevertheless, the calculation of the fractional saturation can be more complex if another spectral component is involved during the deoxygenation process, especially for the partially liganded species. Therefore control measurements of the whole absorption spectra in the visible region at different O2 fractional saturation were performed. Similar O2 binding curve shapes were measured for EcDos and EcDosH, and several isosbestic points were found. A further complication arises from the rate of auto-oxidation of EcDos, which increases at low partial O2 pressure. For this reason we used an enzymatic system with ferredoxin as the terminal electron donor (26), achieving full reduction of the oxidized proteins within 1 min.
RESULTS
Size Exclusion Chromatography and Light Scattering
The theoretical molar mass of monomeric full-length EcDos and the heme domain EcDosH is 93,000 (including the His6 tag at a molar mass ∼2,000 g/mol) and 16,200 g/mol, respectively. EcDos and EcDosH were characterized previously by gel filtration as tetrameric and dimeric, respectively (1, 12). Although gel filtration is useful for estimating equilibrium constants between protein subunits, it is considerably less accurate for determining the absolute Mr and aggregation state of a protein, unless the Mr markers belong to the same protein family as the protein of interest (see “Discussion”). For this reason we used in parallel the static light scattering technique (MALLS), which allows the measuring of absolute molar masses and sizes of molecules without having to rely on the calibration of standards and assumptions of their conformation.
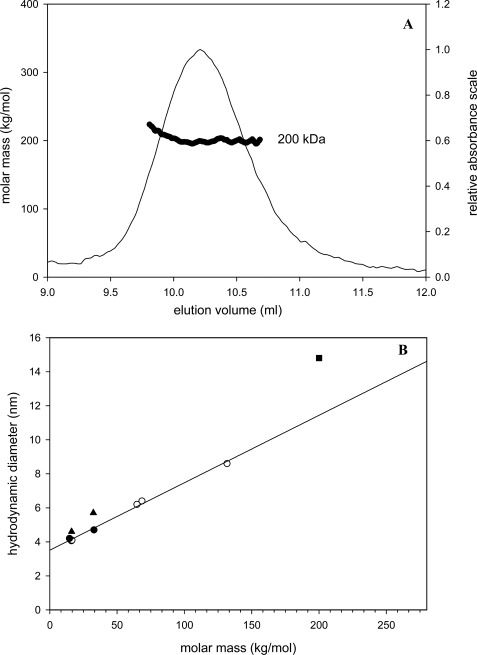
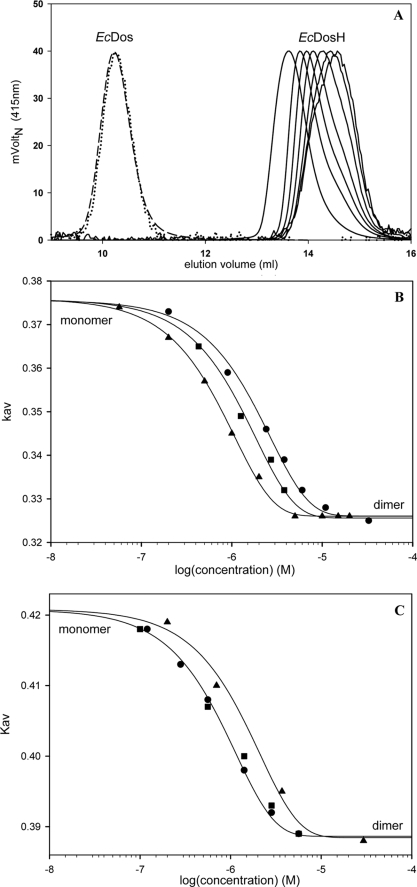
Molar masses determined by MALLS analysis coupled with size exclusion chromatography yielded a Mr of 199,000 ± 4,000 g/mol for the full-length protein EcDos (Fig. 1A) and 32,000 ± 100 g/mol for both the EcDosH and FixLH heme domains (supplemental Fig. 1) for heme concentrations of several μm (EcDos) and several tens of μm (EcDosH and FixLH). No evidence of a tetrameric structure was found for any sample, even at high concentrations (∼40 μm; instead, our data indicated dimeric forms for all three proteins. The heme domains EcDosH and FixLH show a marked dependence on the protein size as measured by gel filtration in the heme concentration range of 40 to 0.05 μm, indicating a change in their oligomerization state (Fig. 2). The elution profile for EcDos is concentration-independent in the range of 1 to 0.01 μm (Fig. 2A).
FIGURE 1.
A, molar mass measured by multi-angle light scattering after gel filtration for EcDos (5 μm) using SEC-MALL. B, correlation between hydrodynamic diameters (measured by dynamic light scattering, Malvern Instrument and Wyatt Technology) and the molar mass of six globular proteins (○). FixLH hydrodynamic diameters (monomeric, 0.5 μm; dimeric, 25 μm) (●) fit well with the hydrodynamic diameters of globular proteins, whereas monomeric (0.5 μm) and dimeric (30 μm) EcDosH PAS (▴) and full-length EcDos (■) hydrodynamic diameters deviate from the correlation line.
FIGURE 2.
A, normalized gel filtration elution profiles of EcDos (1 μm (– – –) and 0.01 μm (····)) and the heme domain, EcDosH, in a concentration range between 15 and 0.05 μm. B and C, protein size dependence on concentration, as measured by gel filtration. Molecular sieve coefficients, Kav, are plotted versus protein concentration for EcDosH (B) and FixLH (C). Three different forms of each protein were plotted: oxy-ferrous (▴), cyanide-ferric (■), and ferric (●).
We estimated the hydrodynamic radius (Rh) using two dynamic light scattering instruments (Wyatt Technology and Malvern Instruments). The Rh values for monomeric EcDosH (2.3 nm), dimeric EcDosH (2.8 nm), and full-length EcDos (6.5 nm) were plotted against their Mr along with several globular proteins (Fig. 1B). Different from FixLH, the values for EcDosH and EcDos do not appear to correlate well with the linear relation found between Rh and Mr for the other proteins, suggesting a less compact and globular-like geometry of EcDosH compared with FixLH and the globins used for comparison. The interpretation of gel filtration experiments, assuming a direct relation between hydrodynamic volumes and Mr, explains the previous assessment of a tetrameric structure for EcDos (12, 13). For EcDos, the Rh distribution (at a few μm) was identical in the ferric and ferrous hexacoordinated form, as well as in the CO ferrous form. It is therefore reasonable to assume that EcDos was fully dimeric in our functional studies whatever the ligation state of the heme domain. Because three-dimensional structures are only available for the heme domain of EcDos, we cannot assess to date whether the difference in the stability of the dimeric structure between EcDosH and EcDos stems from a change at the heme domain interface or rather from the additional interface between the two enzymatic domains.
Fig. 2, B and C, shows the partition coefficients measured by gel filtration chromatography for EcDosH and FixLH in the ferric (Fe(III)), ferric cyanide (Fe(III)-CN), and ferrous oxy forms. The equilibrium binding constants for association of monomers into dimers were 1 and 0.4 μm for EcDosH and 0.8 and 1.4 μm for FixLH in their oxidized and oxygenated forms, respectively. This difference represents a stabilization of the EcDosH heme domain interface of 0.5 kcal/mol upon oxygen binding (subunit association free energy: −8.7 kcal/mol), whereas for cyanide binding, the energy change is very small compared with the hexacoordinated oxidized form (Fig. 2B). This energy change obviously results from a structural change at the subunit interface. FixLH presents a weak destabilization of the heme domain interface of 0.35 kcal/mol for oxygen binding (−8 kcal/mol) with no difference in free energy at the interface upon cyanide binding compared with the pentacoordinated oxidized form.
Cyanide Binding Kinetics
To further investigate the nature of the distal ligand in the ferric state in EcDos proteins in solution (a water molecule in the EcDosH crystal structures (7, 8)), we compared the cyanide binding kinetics of EcDos, EcDosH, the M95I mutant, and myoglobin. The binding kinetics of cyanide were pseudo-first order. We did not observe any biphasic pattern indicative of a heme in equilibrium between significant amounts of pentacoordinated (or hexacoordinated to a low affinity ferric ligand either a water molecule or a hydroxide) and a strong hexacoordinated state with the methionine. The bimolecular cyanide binding rates were, respectively, 117 m−1·s−1 for myoglobin; 113 m−1·s−1 for M95I EcDosH; 30.5 m−1·s−1 for EcDosH at 0.3 μm and 9 m−1·s−1 for EcDosH at 17 μm; and 0.9 m−1·s−1 for full-length EcDos (supplemental Fig. 2). Overall, these rates are in the same range as those measured previously (19, 27), although quantitative differences were observed (see “Discussion”). We note that the rate for the M95I EcDosH mutant protein is very similar to that for myoglobin, consistent with a similar ligation of the ferric heme. The binding rate for EcDosH is markedly lower. Our results show that for EcDosH the association rates depend on the protein quaternary state (monomer/dimer). Under the hypothesis that in ferric EcDosH in solution the heme would be coordinating Met-95, this difference may be explained by the difference in the microscopic association and dissociation rates for the methionine residue between the EcDosH monomer and dimer (see below), with the EcDosH dimer exhibiting ligand binding properties closer to those of the full-length protein EcDos (see also “Discussion”).
Ultrafast External and Internal Ligand Rebinding Kinetics
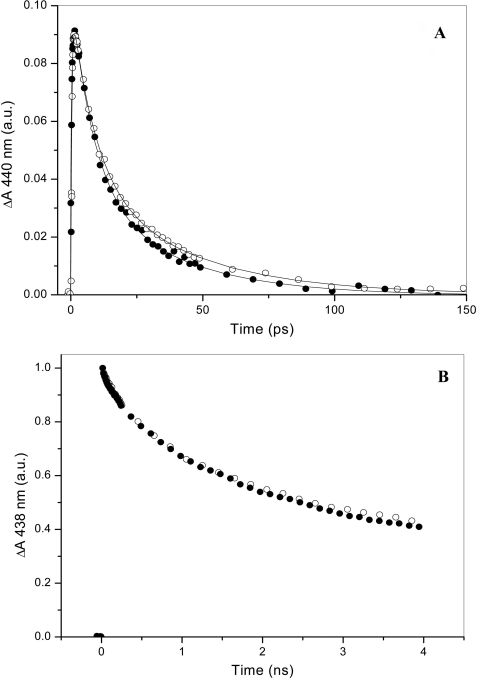
The dynamics of internal and external ligand binding in the heme domain were investigated using femtosecond spectroscopy under conditions in which all investigated proteins were predominantly dimeric. Fig. 3 compares the kinetics of rebinding of the internal ligand, Met-95, and CO after photolysis from the heme in EcDosH and in the full-length protein EcDos. The Met-95 rebinding kinetics are biexponential with time constants of ∼7 and ∼35 ps (20), presumably reflecting two configurations of Met-95 that do not strongly interchange on the time scale of the experiment (21). In the full-length protein the overall rebinding occurs moderately, but significantly, faster than in the isolated heme domain (Fig. 3A). In particular, the relative amplitude of the faster component is higher. This effect is qualitatively similar to the effect of glycerol on the heme domain (21) and suggests that the presence of the enzymatic domain influences the relative population of the two Met-95 configurations in the heme domain.
FIGURE 3.
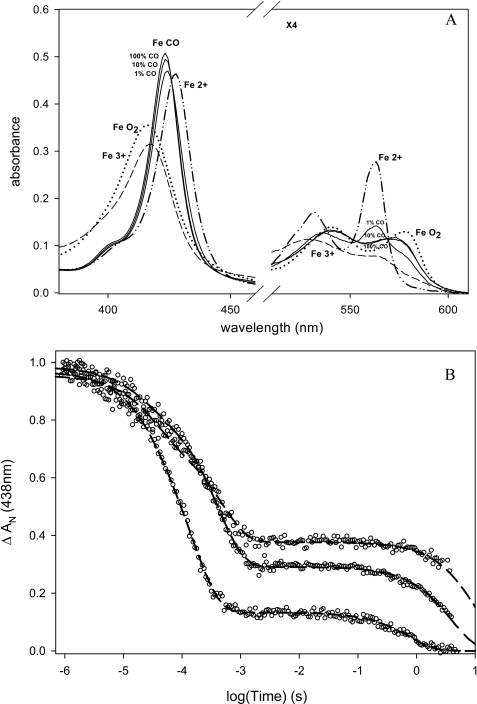
Transient absorption experiments showing geminate rebinding of Met-95 in fully reduced (A) and of CO (decay, 64%; time constant, 1.6 ns) in reduced CO-bound (B) EcDos (●) and EcDosH (○). The lines in A indicate fits with biexponential decays with time constants of 6 and 30 ps (58 and 42%) and 7 and 35 ps (53 and 47%) for EcDos and EcDosH, respectively.
In contrast, the partial geminate rebinding of CO to the heme occurring on the picosecond-to-nanosecond time scale is very similar in EcDosH and EcDos (Fig. 3B). These kinetics reflect competition between low energy barrier rebinding to the heme and thermally activated escape from the heme pocket (21), suggesting that the initial escape route for CO is not influenced by the enzymatic domain.
Microsecond to Second CO Rebinding Kinetics for EcDosH
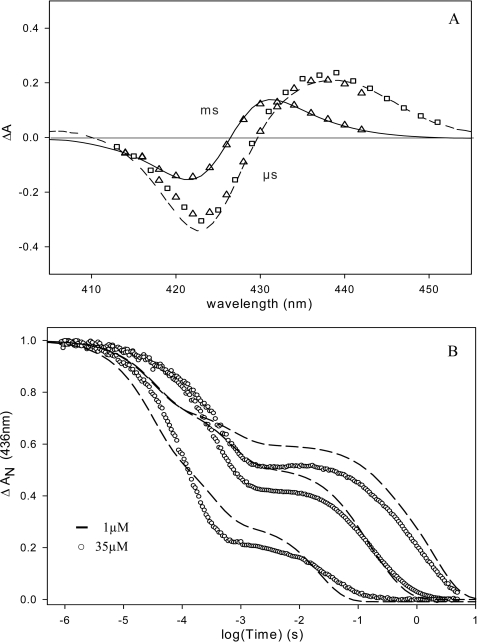
Fig. 4A shows that the shape of the transient absorption spectrum measured at 0.1 μs for EcDos and EcDosH is close to that measured at 2 orders of magnitude faster time. After CO migration out of the protein, Met-95 can rebind on the 100 μs time scale, in competition with bimolecular CO rebinding (20). The shape of the transient spectrum changes and is well simulated by the difference of the steady-state spectra: deoxy hexacoordinated His-heme-Met minus His-heme-CO (Fig. 4A). Subsequent replacement of Met-95 by CO from solution takes place in milliseconds to seconds (20). To investigate the influence of dimerization on the intrinsic rate constants for Met-95 and CO binding and dissociation in EcDosH, we monitored Met-95 and CO binding kinetics after flash photolysis as a function of the CO concentration for two different heme concentrations. At 35 μm EcDosH is almost exclusively dimeric, whereas at 1 μm it is significantly displaced toward the monomeric form (see above).
FIGURE 4.
A, transient absorption spectra after flash photolysis of the heme-CO complex of EcDos (▵) and EcDosH (□) at 0. 1 μs and ms time scales. – – –, transient spectrum at 4 ns; —, difference between steady-state spectra: deoxy hexacoordinated His-Fe2+-Met minus His-Fe2+-CO. B, flash photolysis kinetics for EcDosH at 436 nm and 25 °C. Recombination kinetics at different CO concentrations, from top to bottom: 0.01, 0.1, and 1 atm of CO. After flash photolysis of CO, the first phase represents a competitive binding between CO and Met to the heme. The second phase is a slow replacement reaction of Met by CO to return to the preflash steady state. Heme concentration dependence on CO kinetics is observed, due to the presence of two protein structures in equilibrium: monomeric and dimeric with different Met on- and off-rates.
Any proteins with hemes coordinating not CO but Met-95 prior to the flash will not contribute to the signal, because after flash photolysis Met-95 rebinds (Fig. 3A) on a subnanosecond time scale. Equilibration with 0.01 atm of CO leads to complete CO saturation of the protein. The rebinding occurs in two main phases reflecting the above described binding and replacement processes under all conditions. However, the dependence of the kinetics on both protein and CO concentration (Fig. 4B) is complex. For the fully dimeric CO-saturated samples, the kinetics of each phase can be satisfactorily described by a single exponential. For the mixed dimeric/monomeric samples, we observed two rates in the binding phase and replacement phase each, at all CO concentrations. Thus, the microscopic reaction rates are different in the monomeric and dimeric heme domains. Analysis of the CO dependence on the binding reaction allows us to extract the konMet and konCO values, which then permits the estimation of the koffMet value with analytical and numerical approaches as described previously for other hexacoordinated heme proteins (25). This analysis indicates that konCO is 5.106 m−1·s−1 for both monomers and dimers, a value very similar to that determined previously (20). On the other hand, konMet and koffMet are both 1 order of magnitude higher for monomers (30,000 and 400 s−1) than for CO-saturated (see below) dimers (2000 and 50 s−1) (Table 1). Thus, whereas the affinities for methionine are close for monomers and dimers, the monomer binding rates of Met-95 are 10 times faster, indicating a more flexible heme pocket.
TABLE 1.
O2 and CO binding parameters for full-length EcDos and the EcDosH sensor domain
The experimental conditions are: 150 mm Tris acetate, 50 mm NaCl, pH 7.5, at 25 °C (20 °C for BjFixL).
| Protein | konCO | koffCO | KCOa | konO2 | koffO2 | KO2a | konMet | koffMet | KMet |
|---|---|---|---|---|---|---|---|---|---|
| /m/s | /s | mmHg | /m/s | /s | mmHg | /s | /s | ||
| EcDos | 4 × 106 | 0.007 | 1.3/3.0b | 2.0 × 107 | 1.1 | 30/(11/69)c | 2,000 | 2 | 1,000 |
| 30,000 | 9d | 3,300 | |||||||
| EcDosH monomer | 5 × 106 | 2.0 × 107 | 30,000 | 400 | 75 | ||||
| EcDosH dimer | 5 × 106 | 2.0 × 107 | 1.5 | 1.6/(1.6/11)c | 2,000 | 50 | 40 | ||
| 30,000 | 70d | 430d | |||||||
| EcDosH M95I | 3.4 × 106 | 2.0 × 107 | 1.2 | 0.03 | |||||
| EcDosH M95H | 8 × 106 | 3.0 × 107 | 2.1 | 0.2 | 150(His) | 40(His) | 4(His) | ||
| BjFixLH | 1.6 × 104 | 1.9 × 105 | 6 | 18 |
a Because of the competition for heme binding between CO or O2 and Met, the overall ligand affinities differ from the intrinsic affinities (kon/koff) and are calculated from the following formula: Kligand = (kon ligand/koff ligand)/(1 + KMet). Solubilities are 1.82 10−6 μm/mmHg and 1.36 10−6 μm/mmHg for O2 and CO, respectively.
b This equilibrium constant corresponds to the CO partial pressure of the binding sites at half-saturation.
c In parentheses are shown O2 affinities measured at equilibrium for the two binding steps. Note that the oxygen binding enthalpy was measured at equilibrium equal to −9.0 ± 0.5 kcal/mol.
d Estimated from CO binding kinetics by a combination of stopped-flow measurements to probe the unliganded state (see “Results”) and flash photolysis measurements for the binding rates of CO and Met. For this latter value, we used the fast Met binding rate measured for the partially liganded species (see “Results”). The other values present in this table were measured by flash photolysis from the fully CO-liganded species.
Interestingly, the contribution of a faster reactive conformation does not increase only with a decrease in the protein concentration but also at a low CO saturation level for high protein concentration (data not shown). This indicates that the monoliganded species (one CO per EcDosH dimer) behaves differently than the diliganded species (konMet 30,000 s−1 versus 2,000 s−1; koffMet is less affected) and displays Met-95 binding similar to the monomer of EcDosH. This implies the presence of allosteric interactions between the two hemes of the dimer. For instance at 10% CO-heme saturation, the faster rebinding rate represents only 30% of the binding phase. Given that the dominant liganded species is the singly liganded form (90% for a binomial distribution, less for a cooperative system), this result reflects the allosteric equilibrium of the singly liganded dimer with at least one-third in the rapid state for Met rebinding.
Microsecond to Second CO Rebinding Kinetics for EcDos
The absorption spectra of Fe(III), Fe(II), and Fe(II)-CO (0.01, 0.1, and 1 atm) and of Fe(II)-O2 complexes of EcDos are shown in Fig. 5A. Equilibration with 0.01 and 0.1 atm of CO leads to incomplete CO saturation of the protein with, respectively, a 25 and 5% presence of the hexacoordinated Met-95-bound form.
FIGURE 5.
A, absorbance spectra of EcDos in 150 mm acetate buffer, 50 mm NaCl, pH 7.5. – – –, ferric hexacoordinated form; (····, deoxy ferrous hexacoordinated form; ··—··, CO form at 0.01, 0.1, and 1 atm and ferrous oxy form. B, flash photolysis kinetics measurement for EcDos at 438 nm and 25 °C. Recombination kinetics at different CO concentrations is shown (from top to bottom: 0.01, 0.1, and 1 atm of CO). After flash photolysis of CO, the first phase represents competitive binding between CO and Met to the heme sites. The second phase is a slow replacement reaction of Met by CO to return to the preflash steady state.
The characteristics of the CO rebinding kinetics for the full-length protein EcDos (Fig. 5B) resemble those measured for EcDosH at a high rather than a low protein concentration. This observation is consistent with the finding that EcDos is fully dimeric (see above). At 1 atm of CO (full CO saturation), the kinetics of both phases are close to single exponential. At high CO concentration, the binding phase occurs predominantly at a [CO]-dependent rate (∼6000 s−1 at 1 atm of CO; Fig. 6A). Despite the full CO saturation, the binding of a few percentages of the heme occurs much faster at an almost [CO]-independent rate of 30,000 s−1 (see below). The Met-95 replacement phase is 1 order of magnitude slower than in EcDosH (Fig. 4B) and limited by the koffMet (∼2 s−1). At lower CO concentrations the kinetics become increasingly multi-exponential. We attribute this to the fact that the samples are not fully CO-saturated and contain mixtures of diliganded and monoliganded dimers with different kinetic parameters, in particular konMet (any dimers not binding CO do not contribute to the signal at this time scale). Analysis of the ensemble of data indicates that for both configurations the konCO is 5.106 m−1·s−1 (close to the value found for EcDosH) and the konMet is 2,000 s−1 (di-CO dimers) (Fig. 6A and Table 1) and 30,000 s−1 (mono-CO dimers) as in EcDosH. The fact that the allosteric effect of CO binding in the two proteins of the dimer is very similar for EcDosH and full-length EcDos indicates that the dimer interface between the heme domains is similar for both proteins. Our results clearly indicate a conformational difference at the dimer interface between di- and mono-CO-containing dimers. Such a difference can be expected also for unliganded ferrous dimers. Therefore, a change in conformation and in the associated konMet is expected to occur after CO dissociation. The finding that strong differences in kinetics between di- and mono-CO-containing dimers are observed at the submillisecond time range indicates that these changes occur in a range longer than milliseconds. Indeed, in our previous work on CO binding to EcDosH (20), we had already noted a difference in the replacement phase between flash photolysis and stopped-flow mixing (starting from fully unliganded protein) that we attributed to a slow conformational change. Such a difference is also observed in the full-length protein (Fig. 6B). To further investigate the time scale of the conformational changes, we performed photoincubation experiments in which the sample was brought to an unliganded state (hexacoordinate Met-95 bound) by a series of laser flashes at 10 Hz, and the replacement reaction was subsequently monitored. In single-flash experiments this replacement takes place in ∼0.5 s at 1 atm of CO to ∼10 s at 0.01 atm of CO (Figs. 5B and 6B), limiting the time of the unliganded proteins for possible conformational changes. Note that at 0.01 atm of CO, after a single-flash photodissociation, the Met replacement reaction by CO is biphasic, because in the unliganded form the transition between the fast and the slow reactive conformation occurs on the seconds time scale (only 30% of the fast component was measured). Note that identical results were obtained in stopped-flow and photoincubation experiments (Fig. 6B); the rates were unchanged for 1 atm of CO and decreased 6-fold for 0.01 atm of CO compared with flash photolysis experiments. The conformational change at a low CO concentration is most significant, as at 1 atm of CO the rate is limited by the koffMet. We conclude that the conformational change takes place on the time scale of a few seconds or faster. In fact the curves could be fitted well with the kon and koff values for methionine keeping the same CO kon and koff values measured for the liganded EcDos form. konCO was measured by flash photolysis after subtracting the contribution of konMet determined in Fig. 6A, whereas koffCO was measured by replacing it with a large excess of NO or O2 (data not shown). For the slow reactive conformation with a konMet value of 30000 s−1, measured for the partially liganded species, the replacement rate data set was simulated with a koffMet of 9 s−1.
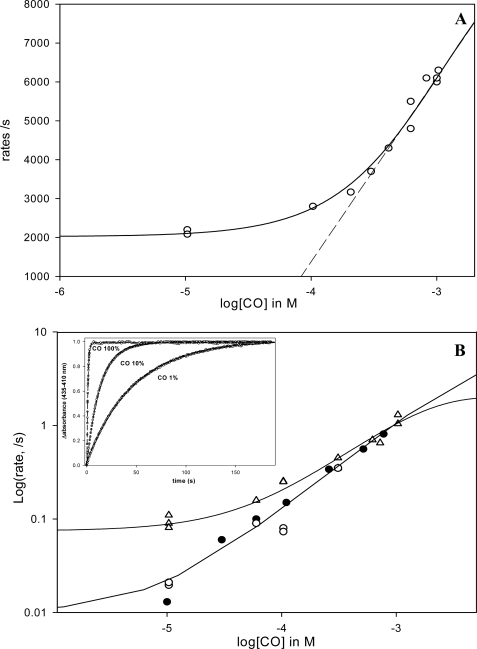
FIGURE 6.
A, fast phase rate for the CO binding kinetics versus [CO] for EcDos. As this phase represents the sum of konMet + konCO at low [CO], one expects the curve to reach a plateau if the Met association becomes the rate-limiting step (konMet ≫ konCO). By contrast, at high [CO] konMet becomes negligible; the asymptote of the hyperbola gives the bimolecular rate. Note that the faster rate for the Met (30,000 s−1), which increases in amplitude at low [CO] as much as the unbound fraction increased at equilibrium, is not shown. Indeed, in this case konMet is much higher than konCO in the range of [CO] investigated, and no [CO] dependence is measurable. B, rate of the slow phase corresponding to methionine replacement by CO by flash photolysis (▵, upper curve). The lower curve shows the rates of methionine replacement by CO measured by stopped-flow (deoxygenated samples are mixed with a CO-equilibrated buffer) (●) and by photoincubation of the deoxy form with repeat flash cycle (10-Hz laser frequency during several seconds) (○). The difference between these curves suggests the presence of two conformations for Met binding to heme with regard to the initial ligation state of the deoxy heme versus liganded heme CO. Inset, replacement of methionine by CO for EcDos after photoinduction (10 s, 10-Hz laser frequency) at different CO concentrations.
In EcDosH, in similar photoinduction experiments, or after mixing with CO by stopped-flow, the binding kinetics are also slower than those measured after single-pulse flash photolysis (supplemental Fig. 3) (20). This implies that EcDosH also rapidly reaches another conformational state after external ligand removal.
The microscopic rate values are summarized in Table 1. On the basis of the kinetic experiments, EcDos shows evidence of different reactive states for methionine depending on the ligation states with CO. At least two different states are involved during the overall ligand binding reaction with CO for which methionine affinity differs by a factor of 3 to 4.
Microsecond to Second O2 Rebinding Kinetics for EcDos and EcDosH
The O2 binding and dissociation kinetics were investigated using CO-liganded samples in the presence of O2. By analogy with the above described photoincubation experiments, we chose experimental conditions that allow O2 to compete with CO for heme rebinding after flash photolysis. Because the yield of O2 escape to the solvent after heme-O2 dissociation is very low, the bimolecular reaction with O2 can best be studied upon CO photodissociation in the presence of O2; O2 will then bind, and eventually be replaced by CO (supplemental Fig. 4). The microscopic binding constants kon and koff for the external ligand O2 (and CO) are almost identical for EcDosH and EcDos (Table 1). Thus the intrinsic O2 binding affinities are also similar. The measured overall binding affinities (P50) depend both on the competition with the Met-95 residue and on the intrinsic O2 affinity and are therefore different for EcDosH and EcDos (Table 1). The intrinsic O2 affinities for EcDosH and EcDos are ∼3 orders of magnitude higher than FixLH and full-length FixL (24). Although it is obvious that the gas-sensing PAS domains in FixL and EcDos tune their (low) O2 affinities by different mechanisms, namely, large steric hindrance for ligand binding in FixL and competition with a constitutive internal residue for heme binding in EcDos, they nevertheless exhibit very similar overall affinities for oxygen.
Microsecond to Second Ligand Rebinding Kinetics for EcDosH Mutants M95I and M95H
Substitution of Met-95 by isoleucine gives rise to a pentacoordinated heme in the deoxy state (19, 21). This allows the rate of ligand binding to heme to be measured directly without competition with an internal ligand. Indeed, in contrast to wild type EcDosH (Fig. 4B), in the M95I mutant protein, the kinetics of bimolecular CO rebinding after photolysis are monophasic. The rates we determined for CO and O2 bimolecular binding and dissociation (in the same order as those determined by Gonzalez et al. (19)) are very similar to those of wild type EcDosH (Table 1), as is CO geminate rebinding (21). Thus the presence of Met-95 does not influence the intrinsic ligand affinities. In particular, the high intrinsic oxygen affinity is confirmed by these direct measurements, indicating that the heme pocket is designed for a stable O2 binding. It has been shown that the distal arginine 97 acts as a key determinant in the heme pocket binding by forming a hydrogen bound with the bound O2 molecule. Mutation of this residue leads to a large increase in the O2 dissociation rate and thus to auto-oxidation (28) similar to that observed for the analogous arginine 220 in the PAS domain of FixL (29).
In the EcDosH mutant M95H, the histidine residue is able to form a reversible bond with the heme (17, 21) but is much less flexible than the native methionine residue, as witnessed by the differences in ultrafast binding kinetics (21). To investigate the influence of this property on ligand replacement reactions, we determined the bimolecular binding rates for CO and O2, as well as the His-95 binding and dissociation rates, in a manner similar to wild type EcDosH (Table 1). Intrinsic CO and O2 binding was found to be similar to the wild type sensor domain. By contrast, the histidine binding affinity for the ferrous heme is lower than for methionine because of a large decrease of the residue association rate from the microsecond to the millisecond range. This finding indicates that the greater flexibility of Met-95 over His-95 allows more rapid switching in the sensor domain. It should be noted that in globin proteins the fastest His binding rate is also about 1 ms (25).
Oxygen Binding Curves at Equilibrium
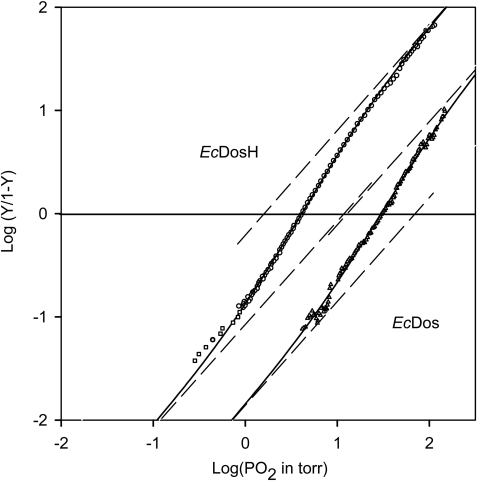
Fig. 7 shows the oxygen binding curve for EcDos. Remarkably, it does not follow a simple nHill = 1 binding curve but requires a two-binding site model. A good fit was obtained with two intrinsic oxygen binding affinities differing by a factor of about 6 (11 and 69 torr). This compares reasonably well with the two Met affinities from the CO rebinding kinetics, which differ by a factor of 3 to 4. Consequently, the O2 binding of a first ligand to EcDos gives rise to a cooperative binding for the second ligand. The cooperativity index (nHill) reaches a maximum value of 1.5 at half-saturation (compared with the maximum allowed value of 2). This also implies that at equilibrium the monoliganded species is always weakly populated with respect to the sum of the diliganded and unliganded hexacoordinated species. The same behavior was observed for EcDosH dimers (Fig. 7). Two intrinsic oxygen binding affinities (1.6 and 11 torr) were observed that were lower than those for the full-length protein, in agreement with the kinetic data. Indeed EcDosH and the full-length protein EcDos exhibit the same intrinsic binding constants except for the methionine dissociation rate, which is 1 order of magnitude faster for EcDosH, leading to a decrease in the internal ligand affinity and a less efficient competition with O2 for heme binding.
FIGURE 7.
Oxygen binding curve for EcDos and EcDosH at [heme] ≥ 10 μm in PBS buffer (with 2 mm dithiothreitol for EcDos) and the enzymatic reducing system from Hayashi et al. (26). The data are simulated with a two-binding site model (dark line). The dashed straight lines with a slope of unity indicate the two oxygen affinities at the intercept with the x axis.
A summary of the data obtained on bimolecular binding is presented in Table 1 and in Fig. 8. Flash photolysis probes the ligand affinity of the liganded state, whereas the oxygen affinity measured at equilibrium reveals cooperative behavior. Although ligand binding data have been measured for EcDosH and EcDos (18, 19), a direct comparison is difficult because these studies did not take into account the complexities we have revealed here: (i) the functional differences between EcDosH monomers and dimers; (ii) the slow protein relaxation upon ligand release, which is the basis of ligand binding cooperativity; and finally, (iii) the functional differences between the mono- and diliganded species, both of which have properties that support ligand binding cooperativity.
FIGURE 8.
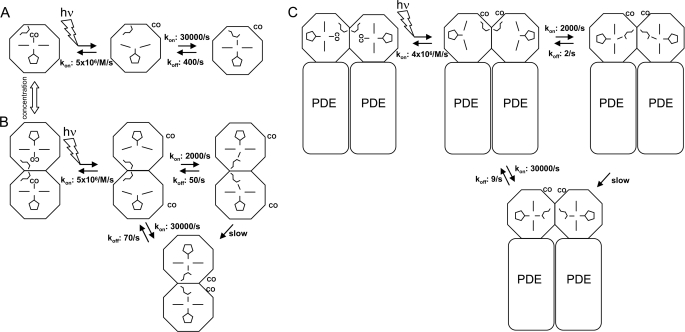
Schemes for CO and Met-95 binding with monomeric EcDosH (A), dimeric EcDosH (B), and full-length EcDos (C) containing the heme domain in tandem with the phosphodiesterase domain (PDE). The rate constants were deduced from CO flash photolysis experiments from the microsecond-to-second time scales. The different Met-95 positions depict two allosteric states. For simplicity, the geminate CO rebinding phases and the picosecond Met-95 binding phases are not included in the scheme nor are the intradimer interactions between mixed CO-liganded and nonliganded heme domains.
DISCUSSION
Different from the heme domain, we expressed the full-length EcDos sensor protein from a construct that puts the gene under the control of a bacterial pBAD promoter that can be regulated by arabinose. The thusly obtained stable EcDos in high quality and quantity in solution allowed an extensive comparison with the isolated heme domain of both its static properties and its fast kinetic processes. Most importantly, we show that the full-length protein is dimeric in solution, as is the heme domain at high concentrations, and that allosteric interactions exist between the two units, allowing cooperative binding of sensed ligands. These points are discussed in more detail below.
Quaternary Characterization of EcDos, EcDosH, and FixLH
Many heme sensor proteins such as FixLH, CooA, and soluble guanylate cyclases are known to be homo- or heterodimeric (14–17). The heme-containing EcDosH sensor domain was characterized initially as a dimer with a molar mass of native EcDosH of ∼36 kDa based upon gel filtration assays (1). We have now determined that the association constants for various forms of EcDosH are ∼1 μm as is also the case for FixLH (Fig. 2).
Characterization of the quaternary structure of the full-length protein EcDos and the heme sensor PAS domains EcDosH and FixLH by a combination of analytical gel chromatography and static and dynamic light scattering allowed us to show that the full-length protein EcDos is a dimeric protein in solution (Fig. 2A). This result contrasts with the previous assessment by Sasakura et al. (12) of a tetrameric structure based on gel filtration chromatography, which assumes a linear correlation between molar mass and elution volumes of protein markers. No correlation between the hydrodynamic radius and the molar mass of EcDos and EcDosH could be determined using different proteins from the globin family as comparison (Fig. 2B). This might be because of the less compact overall structure of the EcDos proteins, which would explain the poor accuracy in estimating the Mr of these proteins based on gel filtration chromatography. Our static light scattering experiments unambiguously characterized the complex in solution as dimeric.
The association constant for full-length EcDos is less than 10 nm, implying that the dimer-dimer interface is more stable than in isolated PAS domains (∼1 μm). The similarity of the observed allosteric ligand binding interactions between isolated heme domains and full-length EcDos (see below) suggests that the dimer interface between the heme domains in both complexes is similar. A plausible explanation for the observation of a more stable full-length dimer is an additional intersubunit interface arising from additional protein parts, notably the catalytic domain (no three-dimensional structure is available for the full-length protein EcDos). In the available crystal structures of dimeric EcDosH (7, 8), the C-terminal β-strands are oriented in the same direction, so that the two catalytic domains are likely to share an interaction surface.
In EcDosH and FixLH the monomer-dimer equilibrium is also influenced by the nature of the ligand in the heme pocket. Binding of O2 represents a stabilization of the heme domain interface in EcDosH, whereas it destabilizes the FixLH heme domain interface (Fig. 2, B and C). There is a weaker transition upon cyanide binding to the hexacoordinated form of EcDosH and no change is observed for FixLH. The modulation of the monomer-monomer interface due to the ligation state can probably be explained by the rearrangement of key amino acids that occurs in the heme pocket of EcDosH and FixLH (8, 29–31). Cusanovich and Meyer (32) suggested that a signaling event initiated within a PAS domain is transmitted from the ligand-binding site to the dimer interface via distortion of the central β-sheet to the interacting N and C termini (involved in monomer-monomer contacts) and their associated hydrophobic residues. For FixLH it has been suggested that oxygen binding to the heme may result in steric hindrance, which may cause distortion of the central β-sheet and alter its interaction with the N- and C-terminal helices (32), although recently Ayers and Moffat (33) showed minimal crystal lattice effects on the integrity of the CO-bound FixLH structure. Binding of different ligands to the heme causes modulation of the monomer-monomer interface of the PAS sensor domain, leading to stabilization or destabilization. As the interaction of PAS domains is known to influence the activity of the catalytic domain (13), this finding may play a role in signaling to the catalytic domain in the stable dimeric full-length proteins.
Cyanide Binding and Heme Coordination State in the Absence of External Ligand
Our data show that dimerization effects on ligand binding properties occur not only in ferrous proteins, but also in ferric EcDosH, for cyanide binding (supplemental Fig. 2). This finding may explain moderate differences in cyanide binding constants reported by Gonzalez et al. (19) and Watanabe et al. (27), where the monomer-dimer equilibrium was not taken into account.
In view of the above discussed role of Met-95 in the dimerization effects of ligand binding in the ferrous species, such a role might also be inferred for the ferric species. In the ferrous species, the allosteric effect is most likely brought about by the large rearrangement imposed by the replacement of Met-95 by an external ligand. However, in crystals, the heme in ferric EcDosH ligates a water molecule rather than Met-95 (7), and a large rearrangement of Met-95 upon binding external ligands is not expected to occur. Spectroscopic investigations indicate that in solution Met-95 may possibly bind to the ferric heme (17, 19). The literature concerning the steady-state spectroscopic resolution of this issue, dating from before the x-ray structure determinations, is conflicting (17, 19, 34, 35). The increase in the rate of cyanide binding to ferric EcDosH upon replacement of Met-95 by isoleucine that we measured, also observed upon replacement of Met-95 by alanine and leucine (27), is consistent with the suggestion that Met-95 binds to the heme and is more difficult to replace by cyanide than a water molecule or a hydroxide, ligating to the heme in the mutant proteins. Altogether, our observations have the potential to reopen the question of the heme ligation state of ferric EcDos proteins.
Conformational Changes for EcDos upon Ligand Dissociation and Association
Picosecond and early nanosecond kinetic events initiated by ligand dissociation from the heme reflect processes in the heme environment prior to ligand exchange with the protein environment (36). Our ultrafast kinetics did not show strong differences between the internal dynamics of EcDosH and EcDos protein (Fig. 3), indicating that the enzymatic domain does not play a significant role in the early molecular signaling events which occur after dissociation of methionine or an external ligand from the heme. For both, EcDosH and EcDos, we observed two decay constants (7 and 35 ps) for methionine rebinding, arising from the possible presence of two configurations of this residue in the heme domain, one of which has been proposed to be active in signal transmission (21). The finding that the relative population of these steady-state configurations depends (moderately) on the presence of the catalytic domain is fully consistent with this proposal, as it suggests a mechanistic link between the Met-95 configuration and the catalytic domain.
When the heme binds an external ligand, Met-95 is swapped out of the heme pocket (7, 8). Dissociation of CO then leads to binding of Met-95 (konMet rate) involving an important reorganization event and hence taking place on a much slower time scale (microseconds) than after dissociation of Met-95 (20). For both dimeric proteins, two Met binding rates also were measured after CO photodissociation (33 and 500 μs), although these were not necessarily related to the two phases observed in the ultrafast Met-95 dissociation experiments. However, for the EcDosH monomer only the faster rebinding rate was observed, leading us to propose that the dimeric structure is necessary for at least one conformation. Interestingly, we found that the mono-CO-liganded species and the bi-CO-liganded species behave differently with regard to methionine rebinding; the liganded subunit of the asymmetric (CO unliganded) dimer in both EcDosH and EcDos rebinds methionine with a rate close to that measured for the EcDosH monomer. We also found that the methionine dissociation rate was affected by the dimerization and the ligation state. Probing of the hexacoordinated species reactivity for ligand rebinding after prolonged CO dissociation from EcDosH and EcDos (by photoincubation or simply after mixing the deoxy species with a CO buffered solution by stopped-flow) showed a decrease in methionine replacement by the external ligand. If we assume that the binding rates for the external ligand do not change, this involves a concomitant change for both the association and dissociation rates of the methionine residue. Indeed, this assumption is supported by the finding that the other binding parameters we measured for CO and O2 for the EcDosH monomer and dimer, as well as for full-length EcDos after photodissociation of the fully liganded protein-CO complexes, were identical. Altogether our experiments thus strongly indicate that the intrinsic parameters governing the binding of external ligands to the heme do not strongly “sense” the environment of the heme domain but that, by contrast, the methionine dynamic parameters are influenced by (i) the presence of the monomer-monomer interface, (ii) the ligation state of the dimer (one or two ligands), and (iii) the presence of the enzymatic domain because the methionine affinity increases for EcDos by 1 order of magnitude compared with EcDosH. This marked role of methionine in allosteric interactions is in line with the above discussed link with the catalytic domain.
The binding properties mediated by the dynamics of methionine indicate a heme-heme interaction mechanism upon ligand binding or release. Indeed the O2 binding measurements at equilibrium demonstrate unambiguously the presence of positive cooperativity. The second oxygen molecule will bind to EcDos with a 6-fold higher intrinsic affinity than the first one, and the fraction of the asymmetric monoliganded species will be much lower than that with two ligands or no ligand at all, whatever the oxygen fractional saturation. On a molecular level, this indicates that the replacement of Met-95 by O2 and its swapping out of the heme pocket in one unit lead to a modification at the dimer interface, presumably leading to decreased Met-95 affinity for heme (and hence increased net O2 affinity) in the other subunit. In view of the predominant role of Met-95 in the allosteric effects, we suggest that binding of other ligands (CO and NO) is also cooperative, although they are more difficult to measure experimentally.
There is no doubt that the heme ligand binding properties will greatly influence enzymatic catalysis in vivo, because a decrease of c-diGMP phosphodiesterase activities upon oxygen release has been demonstrated (9). A cooperative mechanism will obviously enhance the sensitivity to perceive and react to changes of the ambient tension of O2 (and other sensed ligands) in the natural environment of E. coli (). To our knowledge, our assessment of cooperative O2 binding in EcDos constitutes the first direct evidence for cooperativity in a heme-based sensor protein. More generally, only a few examples of cooperativity in a dimeric heme protein have been reported thus far, for a globin homodimer from the mollusc Scapharca inaequivalis (37) and a truncated globin homodimer from Mycobacterium tuberculosis (38).
This work was supported by INSERM, Délégation Générale pour l'Armement (DGA) Contract 07.34.004, and Universités Paris VI et XI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- EcDos
- E. coli direct oxygen sensor
- EcDosH
- heme sensor domain of EcDos
- FixLH
- heme sensor domain of FixL
- PBS
- phosphate-buffered saline
- MALLS
- multi-angle laser light scattering.
REFERENCES
- 1.Delgado-Nixon V. M., Gonzalez G., Gilles-Gonzalez M. A. (2000) Biochemistry 39, 2685–2691 [DOI] [PubMed] [Google Scholar]
- 2.Gilles-Gonzalez M. A., Gonzalez G. (2005) J. Inorg. Biochem. 99, 1–22 [DOI] [PubMed] [Google Scholar]
- 3.Sasakura Y., Yoshimura-Suzuki T., Kurokawa H., Shimizu T. (2006) Acc. Chem. Res. 39, 37–43 [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura-Suzuki T., Sagami I., Yokota N., Kurokawa H., Shimizu T. (2005) J. Bacteriol. 187, 6678–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi H., Shimizu T. (2006) Chem. Lett. 35, 970–971 [Google Scholar]
- 6.Gilles-Gonzalez M. A., Ditta G. S., Helinski D. R. (1991) Nature 350, 170–172 [DOI] [PubMed] [Google Scholar]
- 7.Kurokawa H., Lee D. S., Watanabe M., Sagami I., Mikami B., Raman C. S., Shimizu T. (2004) J. Biol. Chem. 279, 20186–20193 [DOI] [PubMed] [Google Scholar]
- 8.Park H., Suquet C., Satterlee J. D., Kang C. (2004) Biochemistry 43, 2738–2746 [DOI] [PubMed] [Google Scholar]
- 9.Tanaka A., Takahashi H., Shimizu T. (2007) J. Biol. Chem. 282, 21301–21307 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka A., Shimizu T. (2008) Biochemistry 47, 13438–13446 [DOI] [PubMed] [Google Scholar]
- 11.El-Mashtoly S. F., Nakashima S., Tanaka A., Shimizu T., Kitagawa T. (2008) J. Biol. Chem. 283, 19000–19010 [DOI] [PubMed] [Google Scholar]
- 12.Sasakura Y., Hirata S., Sugiyama S., Suzuki S., Taguchi S., Watanabe M., Matsui T., Sagami I., Shimizu T. (2002) J. Biol. Chem. 277, 23821–23827 [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura T., Sagami I., Sasakura Y., Shimizu T. (2003) J. Biol. Chem. 278, 53105–53111 [DOI] [PubMed] [Google Scholar]
- 14.Jain R., Chan M. K. (2003) J. Biol. Inorg. Chem. 8, 1–11 [DOI] [PubMed] [Google Scholar]
- 15.Hao B., Isaza C., Arndt J., Soltis M., Chan M. K. (2002) Biochemistry 41, 12952–12958 [DOI] [PubMed] [Google Scholar]
- 16.Chan M. K. (2001) Curr. Opin. Chem. Biol. 5, 216–222 [DOI] [PubMed] [Google Scholar]
- 17.Sato A., Sasakura Y., Sugiyama S., Sagami I., Shimizu T., Mizutani Y., Kitagawa T. (2002) J. Biol. Chem. 277, 32650–32658 [DOI] [PubMed] [Google Scholar]
- 18.Taguchi S., Matsui T., Igarashi J., Sasakura Y., Araki Y., Ito O., Sugiyama S., Sagami I., Shimizu T. (2004) J. Biol. Chem. 279, 3340–3347 [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez G., Dioum E. M., Bertolucci C. M., Tomita T., Ikeda-Saito M., Cheesman M. R., Watmough N. J., Gilles-Gonzalez M. A. (2002) Biochemistry 41, 8414–8421 [DOI] [PubMed] [Google Scholar]
- 20.Liebl U., Bouzhir-Sima L., Kiger L., Marden M. C., Lambry J. C., Négrerie M., Vos M. H. (2003) Biochemistry 42, 6527–6535 [DOI] [PubMed] [Google Scholar]
- 21.Yamashita T., Bouzhir-Sima L., Lambry J. C., Liebl U., Vos M. H. (2008) J. Biol. Chem. 283, 2344–2352 [DOI] [PubMed] [Google Scholar]
- 22.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 23.Liebl U., Bouzhir-Sima L., Negrerie M., Martin J. L., Vos M. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilles-Gonzalez M. A., Gonzalez G., Perutz M. F., Kiger L., Marden M. C., Poyart C. (1994) Biochemistry 33, 8067–8073 [DOI] [PubMed] [Google Scholar]
- 25.Uzan J., Dewilde S., Burmester T., Hankeln T., Moens L., Hamdane D., Marden M. C., Kiger L. (2004) Biophys. J. 87, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi A., Suzuki T., Shin M. (1973) Biochim. Biophys. Acta 310, 309–316 [DOI] [PubMed] [Google Scholar]
- 27.Watanabe M., Matsui T., Sasakura Y., Sagami I., Shimizu T. (2002) Biochem. Biophys. Res. Commun. 299, 169–172 [DOI] [PubMed] [Google Scholar]
- 28.Ishitsuka Y., Araki Y., Tanaka A., Igarashi J., Ito O., Shimizu T. (2008) Biochemistry 47, 8874–8884 [DOI] [PubMed] [Google Scholar]
- 29.Balland V., Bouzhir-Sima L., Kiger L., Marden M. C., Vos M. H., Liebl U., Mattioli T. A. (2005) J. Biol. Chem. 280, 15279–15288 [DOI] [PubMed] [Google Scholar]
- 30.El-Mashtoly S. F., Takahashi H., Shimizu T., Kitagawa T. (2007) J. Am. Chem. Soc. 129, 3556–3563 [DOI] [PubMed] [Google Scholar]
- 31.Ito S., Araki Y., Tanaka A., Igarashi J., Wada T., Shimizu T. (2009) J. Inorg. Biochem. 103, 989–996 [DOI] [PubMed] [Google Scholar]
- 32.Cusanovich M. A., Meyer T. E. (2003) Biochemistry 42, 4759–4770 [DOI] [PubMed] [Google Scholar]
- 33.Ayers R. A., Moffat K. (2008) Biochemistry 47, 12078–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomita T., Gonzalez G., Chang A. L., Ikeda-Saito M., Gilles- Gonzalez M. A. (2002) Biochemistry 41, 4819–4826 [DOI] [PubMed] [Google Scholar]
- 35.Hirata S., Matsui T., Sasakura Y., Sugiyama S., Yoshimura T., Sagami I., Shimizu T. (2003) Eur. J. Biochem. 270, 4771–4779 [DOI] [PubMed] [Google Scholar]
- 36.Vos M. H., Battistoni A., Lechauve C., Marden M. C., Kiger L., Desbois A., Pilet E., de Rosny E., Liebl U. (2008) Biochemistry 47, 5718–5723 [DOI] [PubMed] [Google Scholar]
- 37.Royer W. E., Jr., Fox R. A., Smith F. R. (1997) J. Biol. Chem. 28, 5689–5694 [DOI] [PubMed] [Google Scholar]
- 38.Couture M., Yeh S. R., Wittenberg B. A., Wittenberg J. B., Ouellet Y., Rousseau D. L., Guertin M. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]