Abstract
Coactivator-associated arginine methyltransferase 1 (CARM1) is a dual functional coregulator that facilitates transcription initiation by methylation of Arg17 and Arg26 of histone H3 and also dictates the subsequent coactivator complex disassembly by methylation of the steroid receptor coactivator family coactivators and p300/cAMP-response element-binding protein-binding protein. However, the regulation of CARM1 enzymatic activity and substrate specificity remains largely unknown. In this study, we report that CARM1 function is regulated by phosphorylation at Ser217, a residue completely conserved in the type I protein arginine methyltransferase (PRMT) family of enzymes. Comparative analysis of the published CARM1 crystal structures reveals that the hydroxyl group of Ser217 forms a strong hydrogen bond with the carbonyl oxygen atom of Tyr154 to lock the cofactor S-adenosylmethionine inside the binding cavity. Phosphorylation of Ser217 disrupts this hydrogen bond and subsequently abolishes S-adenosylmethionine binding and its methyltransferase activity. Importantly, Tyr154 is also conserved in the type I PRMT family of enzymes, suggesting a general role of this hydrogen bond in maintaining the holo structure of the type I PRMT catalytic domain. Moreover, we found that phosphorylation at Ser217 also promoted CARM1 cytoplasmic localization and that this translocation occurred mainly during mitosis. We propose that phosphorylation at Ser217 serves as a molecular switch for controlling CARM1 enzymatic activity during the cell cycle.
Introduction
Nuclear hormone receptors are ligand-activated transcription factors that regulate gene expression through a series of events triggered by high affinity ligand binding (1, 2). Most of the nuclear receptors contain three functional domains: the NH2-terminal activation domain (AF1), a DNA binding domain that is highly conserved and located in the middle of the receptor, and a flexible hinge region that links the DNA binding domain to the carboxyl-terminal ligand binding domain that is responsible for hormone binding and initial recruitment of coactivators. Over the past 15 years, many transcriptional coactivators have been identified, including the steroid receptor coactivator (SRC)2 family of coactivators (3, 4). It has been well established that agonist-bound steroid receptors directly recruit the SRC/p160 family coactivators, which subsequently recruit secondary coactivators, including the E1A binding protein p300 and its homolog, the cAMP-response element-binding protein-binding protein (CBP), as well as the coactivator-associated arginine methyltransferase 1 (CARM1) (5–7). p300/CBP contains potent histone acetyltransferase activity (8), whereas CARM1 has more potent histone methyltransferase activity.
CARM1 was initially identified as a SRC-2/GRIP1 binding protein in a yeast two-hybrid screening, and it belongs to the protein arginine methyltransferase (PRMT) family (6). As a type I PRMT enzyme, CARM1 catalyzes the transfer of methyl groups from S-adenosyl-l-methionine to the guanidine nitrogens of arginine, producing asymmetric dimethylated arginine as its final product. This is in contrast to the type II PRMT enzyme, which produces symmetric dimethylated arginine as its final product (9, 10). CARM1 can function as a secondary coactivator for nuclear receptor-mediated transcription by methylating arginine residues 17 and 26 of histone H3 (6, 11–13). The indispensable role of CARM1 in the estrogen signaling pathway has been established by loss of estrogen receptor (ER) activity in CARM1-deficient mouse embryonic fibroblast cells (14).
In addition to modifying the core histones at promoters, CARM1 can regulate the functions of transcriptional coactivators by methylation of their arginine residues. For instance, CARM1 can methylate various arginines in p300/CBP and thereby modulate the interactions between p300/CBP, steroid receptors, and cAMP-response element-binding protein (15). CARM1 also can methylate arginines close to the KIX domain of CBP and consequently modulate its coactivator function (16). Moreover, methylation of the GRIP1 binding region of p300 by CARM1 leads to an attenuated interaction between GRIP1 and p300 (17). Studies from our laboratory provided in vitro and in vivo evidence to show that SRC-3, as well as SRC-1 and SRC-2, is a natural substrate for CARM1 and that SRC-3 methylation is induced by estrogen signaling (18). Methylation of SRC-3 by CARM1 induces dissociation between CARM1 and SRC-3. The combined results indicate that CARM1 is a dual functional coactivator that facilitates transcription initiation by methylation of histone H3 and also dictates the subsequent coactivator complex disassembly by methylation of SRC-3 and p300/CBP (18).
Besides histones and transcriptional coregulators, CARM1 also can methylate RNA binding proteins such as PABP1, HuR, HuD, and thymocyte cyclic AMP-regulated phosphoprotein (19–22), as well as splicing factors such as CA150, SAP49, and SmB (23). The broad range of CARM1 substrates correlates with its functional diversity. It has been reported that CARM1 plays an important role in alternative splicing through methylation of the splicing factors (23). It also has been shown that CARM1 plays a critical role in promoting differentiation of early thymocyte progenitors, possibly through methylating thymocyte cyclic AMP-regulated phosphoprotein (22). In addition, CARM1 has been shown to directly regulate expression of cell growth genes such as E2F1 and cyclin E1 (24, 25). Consistently, CARM1 has been implicated in tumorigenesis in several studies. For instance, overexpression of CARM1 was involved in the development of prostate carcinoma as well as androgen-independent prostate carcinoma, and the mRNA level of CARM1 was found to be elevated in grade 3 breast tumors (25, 26). It has been proposed that CARM1 may represent a novel therapeutic target in cancers (26).
Although it has been established that CARM1 plays critical roles in diverse biological processes, very little is known about how CARM1 enzymatic functions are regulated by different physiological signaling pathways. Based on the general importance of CARM1 and on the hypothesis that nuclear receptor coregulators are usually regulated by a “post-translational modification (PTM) code,” we characterized the phosphorylation sites of CARM1 by mass spectrometry. Here, we show that CARM1 was indeed phosphorylated at Ser217 both in vivo and in vitro and that this phosphorylation inactivated CARM1 methyltransferase activity by disrupting a hydrogen bond with Tyr154 and caused cytoplasmic localization of CARM1 protein. Interestingly, phosphorylation of CARM1 at Ser217 appears to occur mainly during cell mitosis, suggesting that phosphorylation at Ser217 serves as a molecular switch for controlling CARM1 enzymatic activity during the cell cycle.
EXPERIMENTAL PROCEDURES
Plasmids
A mammalian expression vector for mouse CARM1 (pSG5-HA-CARM1) was provided by Michael R. Stallcup (University of Southern California). CARM1 mutants S217E, S217A, Y154A, Y154F, and Y154R were created by PCR-based site-directed mutagenesis (Stratagene, La Jolla, CA) and confirmed by sequencing analysis. The pSG5-FLAG-SRC-3 and ERE-Luc reporter vectors have been described previously (18).
Nano-HPLC-coupled Tandem Mass Spectrometry Analysis
In-gel digestion and nano-HPLC-coupled tandem mass spectrometry for peptide identification were carried out as described before (27). The GelCode blue-stained bands of one-dimensional SDS-polyacrylamide gels were excised and destained with 50 mm ammonium bicarbonate solution in 50% methanol. Gel pieces were then washed in HPLC water overnight. After the wash procedure, gel pieces were digested with 100 ng of trypsin in 50 mm NH4HCO3 (pH 8.5) for 4 h in a volume of 15 μl. After digestion, peptides were extracted by the addition of 200 μl of acetonitrile. The supernatants were dried in a SpeedVac dryer (Thermo Savant). Each dried sample was dissolved in 20 μl of 5% methanol, 95% water, 0.01% formic acid solution and loaded onto an Zorbax 300SB-C18 trap column (0.3 × 5 mm; Agilent) equilibrated in 0.01% trifluoroacetic acid in water at 10 μl/min and washed for 5 min at the same flow rate, and then the trap column was switched in-line with a 50 mm × 75 μm C18 column (BioBasic C18, 5 μm, 300-Å pore diameter, PicoFritTM; New Objective) equilibrated in 0.01% formic acid and water. The peptides were separated with a 35-min discontinuous gradient of methanol and 0.01% formic acid (5–90% methanol for 20 min) at a flow rate of 200 nl/min. Separated peptides were directly electrosprayed into a mass spectrometer (Finnigan LTQTM; ThermoFinnigan) using a nano-spray source with a voltage of 2.0 kV applied to the liquid junction. The mass spectrometer was operated in the data-dependant mode acquiring fragmentation spectra of the top 20 strongest ions. The resulted tandem mass spectra were analyzed against the modified NCBI Reference Protein Sequence Database using the BioWorks data base search engine (BioWorksBrowser version 3.2; Thermo Electron). All peptide identification with stringent BioWorksBrowser filtering criteria (peptide probability > 1 × 10−6 and Xcorr score > 2.0) was manually examined, and all peptides had to be identified by consecutive b- or y- ions so that false identifications were eliminated.
Immunoprecipitation and Western Blot Analysis
Preparation of cell lysates, immunoprecipitation, and Western blot analysis were performed as previously described (18). Anti-CARM1 antibody was obtained form Bethyl Laboratories. Anti-hemagglutinin (HA) rat monoclonal antibody was purchased from Roche Applied Science. Anti-Myc mouse monoclonal antibody was purchased from NeoMarkers (Fremont, CA). Anti-histone H3 phospho-Ser10 antibody was purchased from Cell Signaling Technology (Beverly, MA). Anti-histone H3 antibody was obtained from Abcam (Cambridge, MA). Anti-histone H3 methylated Arg17 antibody was purchased from Upstate Biotechnology (Lake Placid, NY). HA peptide, anti-FLAG, and anti-β-actin antibodies were purchased from Sigma. For generation of Ser217 phosphorylation-specific antibody, peptide N-“CIYAVEAS(P)TMAQHAE”-C with the phosphorylated serine was synthesized and used to immunize the rabbits. The antibody was affinity-purified from the sera with the immobilized phosphopeptide. λ-Phosphatase was purchased from New England Biolabs (Beverly, MA). The precipitated samples were treated by λ-PPase according to the manufacturer's instructions.
In Vitro Methylation Assay and S-Adenosylmethionine (AdoMet) Binding Assay
The in vitro methylation assays were performed as described previously (28). Briefly, ∼100 ng of CARM1 proteins were incubated with substrates in the presence of 20 mm Tris-HCl (pH 8.0), 4 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, and 1 μl of [3H]AdoMet (13.3 Ci/mm; PerkinElmer Life Sciences). The reactions were incubated at 30 °C for 1 h before the proteins were separated in a 4–15% SDS-polyacrylamide gel. After fixation in a mixture containing 50% methanol and 10% acetic acid for 30 min, the gels were treated with autoradiography Amplify reagent (Amersham Biosciences) for 20 min, dried, and exposed to x-ray films. The substrate proteins used included core histones, SRC-3, and p300 recombinant proteins. Core histones were a gift from Yi Zhang (University of North Carolina at Chapel Hill), whereas SRC-3 and p300 proteins were generated from the baculoviral system as previously described (18).
AdoMet binding assay was performed using CARM1 proteins purified from HEK293T cells. Specifically, HA-tagged wild-type CARM1 and phospho-mutants were transfected into HEK293T cells using FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. Three days later, cells were harvested and lysed, and CARM1 proteins were immunoprecipitated with anti-HA antibody followed by elution with HA peptide. The eluted CARM1 proteins were quantified by Western blot analysis. In the AdoMet binding assay, equal amounts of CARM1 proteins were incubated with 1 μl of [3H]AdoMet (13.3 Ci/mm) in binding buffer (20 mm Tris-HCl (pH 8.0), 4 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 0.5 mm dithiothreitol) for 30 min on ice. Then the samples were cross-linked on ice using GS Gene Linker (Bio-Rad) at 200 mJ for 15 min. AdoMet-bound CARM1 was separated by SDS-PAGE, fixed, dried, and exposed to x-ray films for 2–4 weeks.
Cell Culture and Transfection
MCF-7, CV-1, and HEK293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. CARM1+/+ and CARM1−/− mouse embryonic fibroblast cell lines were a gift from Mark T. Bedford (University of Texas M. D. Anderson Cancer Center). FuGENE 6 transfection reagent was used for all transient transfections. For luciferase gene reporter assays, cells were maintained in phenol red-free medium containing 5% charcoal/dextran-stripped fetal calf serum until hormone addition. Transfected cells were treated with 10 nm estradiol and harvested after a 24-h incubation. Luciferase activity was determined using the Promega luciferase assay kit according to the manufacturer's instructions (Promega Corp., Madison, WI).
Cell Synchronization and Immunofluorescent Staining
To enrich HeLa cells during mitosis, cells were incubated with 2 mm thymidine (Sigma) for 18 h, released for 3 h, and then incubated with 100 ng/ml nocodazole for 12 h. To obtain cells synchronized at the G1/S boundary, HeLa cells were treated with 2 mm thymidine for 18 h, followed by a 9-h release in thymidine-free medium, and then treated again with 2 mm thymidine for 17 h to arrest cells at the G1/S boundary. After an extensive washing, the synchronized cells were released in fresh medium and harvested every 2 h. Immunofluorescent staining was performed as previously described (29).
RNA Isolation and Quantitative Reverse Transcription-PCR Analysis
Total RNA was extracted from MCF-7 cells with TriReagent (Molecular Research Center). To measure the relative mRNA levels, real-time reverse transcription-PCR was performed in an ABI 7700 real-time PCR system (Applied Biosystems, Foster City, CA). 18 S rRNA was used as an internal control. The primers and probe for pS2 mRNA were as follows: forward, 5′-GGTCGCCTTGGAGCAGA; reverse, 5′-GGGCGAAGATCACCTTGTT; and probe, 5′-(FAM)-TCCATGGTGGCCATTGCCTCCT-6-(TAMRA). The primer and probe set for 18 S rRNA was purchased from Applied Biosystems.
RESULTS
Identification of a Novel CARM1 Phosphorylation Site
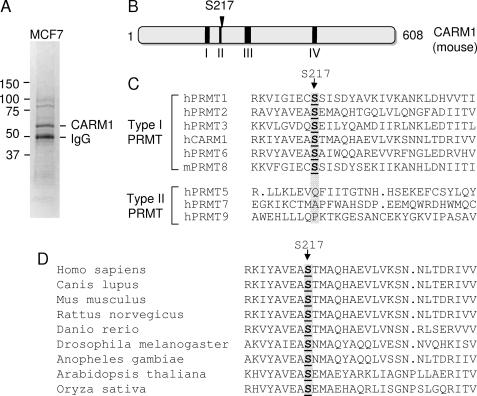
To identify in vivo functional CARM1 phosphorylation sites, human breast carcinoma MCF-7 cells were used to purify CARM1 protein for mass spectrometry analysis. Endogenous CARM1 protein was immunoprecipitated from whole cell lysate, separated by SDS-PAGE, and analyzed by mass spectrometry to identify potential phosphorylation sites on CARM1 (Fig. 1A). Surprisingly, only one phosphorylation site, Ser216 (Ser217 in mouse primary sequence; this designation will be used thereafter) was identified in one out of the seven peptides recovered. The specific peptide contained amino acid residues 201–227. In the CARM1 primary sequence, Ser217 next to the conserved region II, which is conserved in all PRMT family proteins and is important for their methyltransferase activity (Fig. 1B). Sequence alignment of PRMT family members reveals that Ser217 is completely preserved in all type I PRMT enzymes, including PRMT1, PRMT2, PRMT3, PRMT6, and PRMT8, but not in type II enzymes, including PRMT5, PRMT7, and PRMT9 (Fig. 1C). In addition, Ser217 in CARM1 is evolutionarily conserved from plant to human (Fig. 1D), indicating that Ser217 is likely important for CARM1 function.
FIGURE 1.
Human CARM1 is phosphorylated at Ser217 in MCF-7 cells. A, purification of CARM1 protein from MCF-7 cells. Endogenous CARM1 protein was immunoprecipitated from MCF-7 whole cell lysate using anti-CARM1 antibody. Following SDS-PAGE, the gel was stained with GelCode blue stain reagent to illustrate the precipitated bands. B, schematic representation of mouse CARM1 functional domains conserved in the PRMT family of enzymes. C, primary sequence alignment of the PRMT family of proteins. The accession numbers for the PRMTs are as follows (where h is human and m is mouse): hPRMT1 (AAF62893), hPRMT2 (AAH00727), hPRMT3 (AAC39837), hCARM1 (NP_954592), hPRMT5 (AAF04502), hPRMT6 (Q96LA8), hPRMT7 (NP_061896), mPRMT8 (DAA01382), and hPRMT9 (AAH64403). D, primary sequence alignment of CARM1 proteins from different species. The accession numbers for CARM1 sequences are as follows: Homo sapiens CARM1 (NP_954592), Canis lupus CARM1 (XP_853774), Mus musculus CARM1 (NP_067506), Rattus norvegicus CARM1 (NP_001029258), Danio rerio CARM1 (NP_001003645), Drosophila melanogaster CARM1 (NP_649963), Anopheles gambiae CARM1 (XP_318375), Arabidopsis thaliana CARM1 (NP_974913), and Oryza sativa CARM1 (NP_001060600).
Ser217 Phosphorylation Abrogates CARM1 Methyltransferase Activity
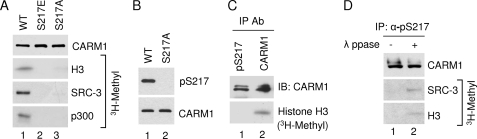
Because CARM1 is an arginine methyltransferase, and Ser217 localizes next to the conserved region that is essential for its methyltransferase activity, we asked whether Ser217 phosphorylation could affect CARM1 enzymatic activity. Mutants mimicking constitutively phosphorylated (S217E) and non-phosphorylated (S217A) proteins were analyzed in the in vitro methyltransferase assay. Three known substrates of CARM1, including histone H3, SRC-3, and p300, were tested in the methylation assay. Interestingly, neither mutant showed any obvious activity against any of the three substrates (Fig. 2A). Mutation of Ser217 to Thr or Cys also abolished CARM1 methyltransferase activity (data not shown). Our results indicate that Ser217 is essential for CARM1 methyltransferase activity. Modification or substitution of this residue leads to broad and significant loss of its enzymatic activity.
FIGURE 2.
Phosphorylation of Ser217 abrogates CARM1 methyltransferase activity. A, Ser217 mutants of CARM1 lost methyltransferase activity. Wild-type (WT), S217E, and S217A CARM1 were exogenously expressed in 293T cells. CARM1 proteins were purified by immunoprecipitation followed by elution with HA peptide. Their methyltransferase activities were analyzed by in vitro methylation assay. Purified core histones, recombinant SRC-3, and p300 were used as substrates. Western blot analysis was used to determine the amount of CARM1 proteins used for the methylation assay. B, characterization of CARM1 Ser217 phosphospecific antibody. Exogenously expressed wild-type and S217A mutant CARM1 were purified from HEK293T cells as described in A. The pSer217 and CARM1 protein levels were determined by Western blot analysis. C, Ser217 phosphorylation abrogates CARM1 methyltransferase activity. Total CARM1 protein or CARM1 specifically phosphorylated at Ser217 was immunoprecipitated (IP) from HeLa cell lysate with the indicated antibodies (Ab). Their activities were analyzed by in vitro methylation assay using core histones as substrate. Western blot analysis showed the amount of CARM1 protein used in the assay. IB, immunoblot. D, Ser217 phosphorylation inhibits CARM1 methyltransferase activity. Endogenous Ser217-phosphorylated CARM1 was immunoprecipitated from HeLa cell lysate and treated with or without λ-phosphatase (λ ppase). Following extensive washing, precipitated CARM1 was used for in vitro methylation assay. Total CARM1 protein was determined by Western blot analysis.
We next determined whether Ser217 phosphorylation would affect the methyltransferase activity of endogenous CARM1 protein. For this purpose, we generated a Ser217 phosphorylation-specific rabbit polyclonal antibody. After affinity purification of the sera with phosphorylated CARM1 peptide, the quality of antibody was tested by immunoprecipitation-Western blot analysis. As shown in Fig. 2B, our antibody only recognized wild-type CARM1 but not the S217A mutant protein immunoprecipitated from cell lysate, indicating that the antibody specifically recognizes Ser217-phosphorylated CARM1. This antibody allowed us to immunoprecipitate Ser217-phosphorylated CARM1 from HeLa cell lysate. When compared with a similar amount of immunoprecipitated total CARM1 protein in an in vitro methylation assay, we found that Ser217-phosphorylated CARM1 displayed negligible methyltransferase activity (Fig. 2C). However, after we treated the Ser217-phosphorylated CARM1 protein with λ-phosphatase, the CARM1 protein regained a modest level of methyltransferase activity toward SRC-3 and histone H3 in the in vitro methylation assay (Fig. 2D). These results established that Ser217 phosphorylation negatively regulates CARM1 enzymatic activity toward histone H3, p300, and SRC-3. It is interesting to point out that endogenous CARM1 immunoprecipitated from HeLa cells exhibits double bands on gels, but CARM1 proteins isolated from MCF-7 cells or exogenously expressed CARM1 from HEK293T cells migrate as a single band, suggesting that CARM1 may contain additional PTM sites in certain type of cells (e.g. HeLa cells).
Ser217 Is Critical for AdoMet Binding Activity of CARM1
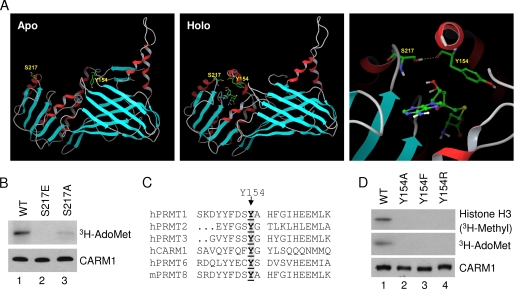
The crystal structures of the CARM1 catalytic core in the apo and holo states (30, 31) suggest that Ser217 may directly participate in binding of the methyl group donor, AdoMet (Fig. 3A). In the apo structure, Ser217 is at its open position, and its hydroxyl group is exposed in solution (Fig. 3A, left panel). In contrast, in its holo structure, the hydroxyl group of Ser217 is packed in, forms a strong hydrogen bond with the carbonyl oxygen atom of Tyr154, and becomes an integral part of the cofactor binding cavity (Fig. 3A, middle and right panels). Therefore, we tested whether the Ser217 mutation would affect the AdoMet binding ability. The UV cross-linking experiment revealed that indeed the AdoMet binding ability of the phosphorylation mutants was significantly compromised (Fig. 3B). These results indicate that the loss of CARM1 methyltransferase activity by Ser217 phosphorylation is likely due to the loss of cofactor AdoMet binding.
FIGURE 3.
Phosphorylation of CARM1 at Ser217 abolishes its AdoMet binding activity. A, the crystal structures of the CARM1 catalytic core in the apo (left panel) and holo (middle and right panels) states. The right panel shows a close look at the hydrogen bond between Ser217 and Tyr154. B, Ser217 mutants lost AdoMet binding ability. CARM1 proteins were prepared as described for Fig. 2A. Their AdoMet binding activities were visualized by autoradiography. C, conservation of Tyr154 shown by the primary sequence alignment of the type I PRMT proteins. D, Tyr154 mutants lost methyltransferase activity and AdoMet binding activity. CARM1 proteins were prepared as described for Fig. 2A. Core histones were used as substrates for the methyltransferase assay. Purified CARM1 proteins were determined by Western blot analysis. WT, wild-type CARM1; h, human; m, mouse.
Sequence alignment of PRMT family members reveals that Tyr154 also is preserved in all type I PRMT enzymes (Fig. 3C). We tested whether the Tyr154 mutation would affect CARM1 AdoMet binding and/or catalytic activity. The in vitro methyltransferase assay and AdoMet binding assay results indicate that mutations of Tyr154 to Ala, Phe, and Arg all lost AdoMet binding and methyltransferase activity (Fig. 3D). These results demonstrate that side chain of Tyr154 is critical for CARM1 cofactor binding and its enzymatic activity, in agreement with the observation that the phenyl ring of Tyr154 engages in hydrophobic interactions with the ribose and sulfur atom of S-adenosylhomocysteine (30, 31). In addition, loss of activity by mutation of Y154F demonstrates an essential role of the hydroxyl group of Tyr154. Further examination of the published structure reveals that the hydroxyl group of Tyr154 forms a hydrogen bond with the carboxyl of another conserved residue, Glu267 (30, 31), which is to neutralize the positive charge of the substrate guanidino group.
Ser217 Phosphorylation Mutations Do Not Affect CARM1 Dimerization and Coactivator Complex Formation
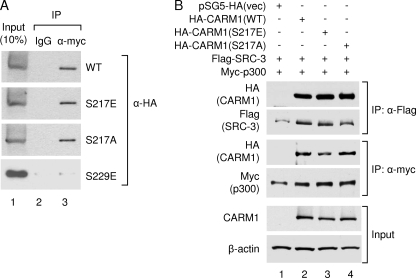
Previous studies reported that phosphorylation of Ser229 disrupts CARM1 dimerization (32). Because Ser217 is located in the proximity of the CARM1 dimerization surface, we investigated whether Ser217 plays any role in CARM1 dimerization. We performed a co-immunoprecipitation assay to determine whether the S217E or S217A mutation affects interaction between wild-type and mutant CARM1. As shown in Fig. 4A, neither mutation had any effect on CARM1 dimerization. As a control, the S229E mutant lost dimerization in agreement with a previous report (32). Our results indicate that mutation of the Ser217 phosphorylation site does not affect CARM1 dimerization.
FIGURE 4.
Ser217 phosphorylation does not affect CARM1 dimerization or coactivator association. A, CARM1 dimerization is not affected by Ser217 phosphorylation. Myc-tagged wild-type CARM1 was coexpressed with HA-tagged wild-type (WT) CARM1 or the S217E, S217A, or S229E mutant in HEK293T cells. Following immunoprecipitation (IP) with antibody against the Myc tag, the precipitated proteins were separated on SDS-polyacrylamide gel and analyzed by Western blot analysis with antibody against the HA tag. B, CARM1 coactivator association is not affected by Ser217 phosphorylation. HEK293T cells were transfected with the indicated plasmids. Following immunoprecipitation with anti-FLAG or anti-Myc antibodies, the precipitated proteins were separated on SDS-polyacrylamide gel, and associated coactivators were determined by Western blot analysis. vec, vector.
Because CARM1 forms a coactivator complex with other transcriptional coactivators in cells to facilitate transcription, we determined whether Ser217 phosphorylation mutation would affect its interaction with coactivators p300 and SRC-3. Shown in Fig. 4B, neither S217E nor S217A had a significant effect on interactions with p300 or SRC-3, suggesting that Ser217 phosphorylation does not regulate CARM1 coactivator complex formation.
Ser217 Phosphorylation Mutations Abolish CARM1 Transcriptional Coactivator Activity
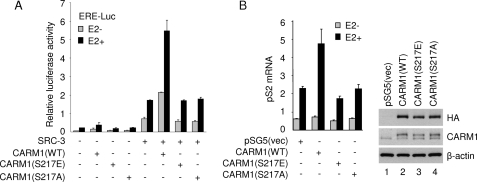
Because CARM1 enzymatic activity is known to be required for its coactivator function (33), we then asked whether Ser217 phosphorylation would affect CARM1 coactivator function in a cultured cell system. As might be expected, we observed that both CARM1 S217A and S217E mutants completely lost their coactivator activity in an ER-dependent transcriptional assay, either in the absence or presence of SRC-3 (Fig. 5A). Moreover, overexpression of wild-type but not S217E or S217A mutant CARM1 in MCF-7 cells increased the expression of an authentic endogenous ER target gene pS2 (Fig. 5B, left panel). Equal expression of wild-type and mutant CARM1 was confirmed by Western blot analysis (Fig. 5B, right panel). These results further substantiate that methyltransferase activity is critical for in vivo CARM1 coactivator function and that phosphorylation of Ser217 negatively regulates its activity.
FIGURE 5.
Mutation of CARM1 Ser217 phosphorylation site diminishes its coactivator function. A, CARM1 Ser217 mutants showed little coactivator activity for ERα-mediated transcription. CV-1 cells were transiently transfected with 200 ng of ERE-Luc reporter, 6 ng of pCR3.1-ERα, 50 ng of pSG5-FLAG-SRC-3, and 100 ng of pSG5-HA-CARM1 wild type (WT) or indicated mutant in each well of a 12-well plate. 10 nm estradiol (E2) was added 24 h after transfection, and luciferase activity was measured at 48 h post-transfection. Luciferase activity was normalized against total cell lysate protein. B, CARM1 Ser217 phosphorylation mutants lost coactivator activity on expression of an endogenous ER targeting gene. MCF-7 cells in 6-well plates were transiently transfected with 2 μg of pSG5-HA-CARM1 wild type or indicated mutant. 48 h after transfection, cells were treated with 10 nm estradiol for 18 h to induce endogenous ER target gene expression. The mRNA level of pS2 was determined by real-time reverse transcription-PCR, and the relative amount is shown. The 18 S rRNA was used as an internal control. +, present; −, absent. The exogenously expressed CARM1 proteins were examined by Western blot analysis, as shown in the right panel. vec, vector.
Ser217 Phosphorylation Promotes Cytoplasmic Localization and Occurs Mainly during Mitosis
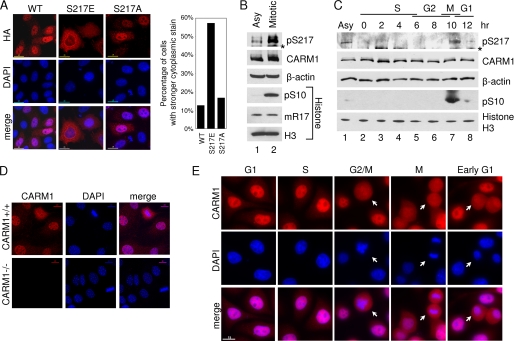
As a transcriptional coactivator, CARM1 is mainly a nuclear protein. We next examined whether Ser217 phosphorylation could regulate CARM1 subcellular localization. HA-tagged wild-type CARM1 and S217E and S217A mutants were transiently expressed in HeLa cells, and immunofluorescence staining was performed with anti-HA antibody. As shown in Fig. 6A, the S217E mutant was predominantly located in the cytoplasm, whereas wild-type CARM1 and the S217A mutant localized mainly in the nucleus. Our results suggest that Ser217 phosphorylation may promote CARM1 cytoplasmic localization.
FIGURE 6.
Ser217 phosphorylation promotes CARM1 cytoplasmic localization and occurs at cell mitosis. A, Ser217 phosphorylation of CARM1 changes its subcellular localization. HA-tagged CARM1 Ser217 mutants and wild-type (WT) protein were exogenously expressed in HeLa cells. HA staining shows the CARM1 protein, and 4′,6-diamidino-2-phenylindole (DAPI) shows the DNA. 200 positively stained cells were counted for each sample, and the percentage of cells showing stronger cytoplasmic signal is shown. B, CARM1 Ser217 phosphorylation level greatly increases during cell mitosis. HeLa cells were cycle-synchronized at M phase using thymidine/nocodazole treatment. Ser217 phosphorylation level was determined by direct immunoprecipitation with pSer217 antibody followed by Western blot analysis against CARM1. CARM1 and β-actin levels shown by Western blot analysis. Histones were prepared by acid extraction method, and the amount of Ser10 phosphorylation and Arg17 methylation of histone H3 in different samples was determined by Western blot analysis. IgG heavy chain is labeled with an asterisk. C, Ser217 phosphorylation of CARM1 is a mitotic event. HeLa cells were synchronized at the G1/S boundary by double thymidine treatment. After being released into fresh medium, cells were collected every 2 h, and the level of Ser217 phosphorylation (pSer217) of CARM1 was determined by immunoprecipitation-Western blot analysis, as described for B. Direct Western blot analysis also shows the amount of CARM1, β-actin, pSer10, and total histone H3 in different samples. IgG heavy chain is labeled with an asterisk. D, characterization of CARM1 antibody in immunofluorescent staining. CARM1+/+ and CARM1−/− mouse embryonic fibroblast cells were used for immunofluorescent staining by CARM1 antibody. CARM1 protein is shown in red, whereas DNA is shown in blue (DAPI). E, CARM1 localizes in the cytoplasm during M and early G1 phases. HeLa cells were synchronized at the G1/S boundary by double thymidine block. After being released into regular medium, immunofluorescent staining was performed at 0, 3, 7, 10, and 12 h to show CARM1 cellular localization at late G1, S, G2, M, and early G1 phases, respectively. CARM1 is labeled in red, and DNA is shown in blue (DAPI). Typical cells at the designated cell cycle stage are indicated out by white arrows.
Interestingly, a previous study of DART4 (Drosophila homolog of CARM1) shows that DART4 accumulates strongly in the cytoplasm during mitosis, when transcription is quiescent for young embryos (34). Because the CARM1 phosphorylation mimic mutant S217E also showed predominant cytoplasmic localization, we asked whether Ser217 phosphorylation of human CARM1 also occurs during mitosis. Indeed, we found that the Ser217 phosphorylation level was dramatically increased during mitosis when HeLa cells were arrested in mitosis by thymidine/nocodazole treatment (Fig. 6B). Moreover, when HeLa cells were synchronized at the G1/S boundary by double thymidine treatment and then released to fresh medium to continue the cell cycle, Ser217 phosphorylation only peaked during mitosis and quickly disappeared after cells entered G1 phase (Fig. 6C). Next, we asked where endogenous Ser217-phosphorylated CARM1 localizes in cells. Unfortunately, the pSer217 antibody was not useful for immunofluorescent staining. Consequently, we chose a CARM1 antibody that specifically recognizes CARM1 in immunofluorescent staining (Fig. 6D). Consistent with the previous observation, immunofluorescent staining of endogenous CARM1 protein revealed that it is mainly a nuclear protein during G1, S, and G2 phases. With the condensation of chromatin and breakup of nuclear membrane during mitosis, CARM1 accumulated in the cytoplasm and remained there until two daughter cells separated completely at early G1 phase (Fig. 6E). These results indicated that phosphorylation of CARM1 at Ser217 is cell cycle-dependent and mainly occurs during mitosis. Consequently, they suggest that the CARM1 methyltransferase activity is turned off during mitosis when gene transcription is silent and turned on in G1 phase when gene transcription again becomes active.
DISCUSSION
The PTM Code, a Key Determinant of Coregulator Function
Nuclear receptor coregulators play diverse functions in regulation of eukaryotic gene expression (35, 36). Recent studies have demonstrated that post-translational modifications such as phosphorylation, methylation, acetylation, sumoylation, and ubiquitination are key determinants of coregulator activity and stability (37). For instance, SRC-3 has been shown to be phosphorylated on multiple residues, and different combinations of these phosphorylation sites constitute a PTM code, which is able to regulate both activity and stability of SRC-3 protein (35, 38). Different signaling pathways such as those of steroid or growth factors initiate separate kinase cascades, which result in different patterns of phosphorylation of SRC-3 and subsequently lead to formation of specific coactivator complexes on different target genes. As a result, signaling from specific environmental stimuli leads to distinct subsets of gene expression and diverse cellular responses.
Our current study on regulation of CARM1 function further broadens the concept of the PTM code by identifying Ser217 phosphorylation as a regulatory switch for CARM1 enzymatic function during the cell cycle. We found that phosphorylation of Ser217 does not affect CARM1 dimerization or its interaction with other coactivators such as SRC-3 and p300/CBP. However, phosphorylation of Ser217 specifically disrupts cofactor AdoMet binding and subsequently abolishes its methyltransferase activity. Importantly, mutation of Ser217 to Ala also dramatically reduces its AdoMet binding and enzymatic activity, indicating that the hydroxyl group of Ser217 is essential for maintaining the intact structure and function of CARM1. Indeed, based on the crystal structures of the CARM1 catalytic core in the apo and holo states, binding of the methylation intermediate S-adenosylhomocysteine causes reorientation of the hydroxyl group of Ser217 and forms a strong hydrogen bond with a carbonyl oxygen atom of Tyr154 (31). Our results demonstrate that this hydrogen bond is critical for the cofactor AdoMet binding in CARM1. Moreover, both Ser217 and Tyr154 are completely conserved in the type I but not type II PRMT family of enzymes, suggesting that this hydrogen bond plays a general role in maintaining the functions of type I PRMT enzymes. By examining the previously published structure of the conserved core of PRMT3, another member of the type I PRMT enzyme family (39), we found that a hydrogen bond is formed between the corresponding Ser284 and Tyr221. It is likely that similar hydrogen bonds may form in some or all of the type I PRMT enzymes, and phosphorylation of a corresponding Ser residue may be a general mechanism in functional regulation of type I PRMT family members.
Another interesting observation is that CARM1 S217E mutant protein is predominantly localized in the cytosol, raising a question as to whether CARM1 plays an unknown function in the cytoplasm. However, because Ser217 phosphorylation inactivates CARM1 methyltransferase activity, any potential cytoplasmic function should be independent of its enzymatic activity.
In addition to our current study, it was reported that CARM1 can be inactivated by phosphorylation at Ser229 (32). However, Ser229 is not conserved in other members of the PRMT family, and phosphorylation of Ser229 disrupts dimerization and thereby abolishes methyltransferase activity by a separate mechanism (32), indicating that Ser217 and Ser229 play different roles in regulation of CARM1 function. Thus, two distinct mechanisms could exist for phospho-inhibition of CARM1 function.
Regulation of CARM1 Activity by Phosphorylation
Partially due to the highly condensed state of chromatin, gene transcription is silenced globally during mitosis. As a transcriptional coactivator that contains arginine methyltransferase activity, it may be advantageous to attenuate the nuclear function of CARM1 during mitosis to avoid off-target methylation. Importantly, in the current study, we found that phosphorylation at Ser217 mainly occurred at mitosis and that this phosphorylation inactivated CARM1 enzymatic activity and coactivator function. Our results indicate that phosphorylation at Ser217 could be an important adjunct means for controlling CARM1 function during cell cycle. However, it remains unknown if CARM1 Ser217 is quantitatively phosphorylated due to technical limitations.
One immediate question raised by our finding is which kinase is responsible for CARM1 Ser217 phosphorylation. Because Ser217 is mainly phosphorylated during mitosis, we examined several kinases that are known to be active during mitosis. However, by using relevant kinase inhibitors, we were not able to specifically inhibit Ser217 phosphorylation. Thus, the specific kinase(s) that catalyzes the Ser217 phosphorylation remains to be determined. Additionally, because protein phosphorylation is a reversible process, once the cells exit mitosis, CARM1 function could be restored by dephosphorylation at Ser217 and either reused as a transcriptional coactivator or degraded. Another interesting question raised is where Ser217 phosphorylation occurs. A small percentage of wild-type CARM1 is found in the cytoplasm, whereas the majority of CARM1 is localized in the nucleus, suggesting that CARM1 could be a protein that shuttles between the nucleus and cytoplasm. It is possible that CARM1 is phosphorylated at Ser217 in the nucleus and subsequently exported to the cytoplasm. It is also possible that the nuclear localized CARM1 redistributes throughout the whole cell at the time of mitotic nuclear envelope breakdown and that exposure of CARM1 to cytoplasmic kinase(s) leads to Ser217 phosphorylation.
This work was supported, in whole or in part, by National Institutes of Health NIDDK National Research Service Award Individual Postdoctoral Fellowship 5F32DK078473 (to Q. F.), NIDDK Mentored Research Scientist Development Award 1K01DK081446 (to B. H.), NIDDK Grant 5P01 DK059820 and NCI Baylor College of Medicine Cancer Center Cores Grant P30CA125123 (to B. W. O.), and NIDDK-NURSA (Nuclear Receptor Signaling Atlas) Grant U19 DK062434 (to B. W. O., M.-J. T., and S. Y. T.). This work was also supported by a Baylor College of Medicine start-up fund (to Y. S.) and Welch Foundation Grant Q-1521 (to B. W. O.).
- SRC
- steroid receptor coactivator
- CBP
- cAMP-response element-binding protein-binding protein
- CARM1
- coactivator-associated arginine methyltransferase 1
- PRMT
- protein arginine methyltransferase
- ER
- estrogen receptor
- PTM
- post-translational modification
- HPLC
- high pressure liquid chromatography
- HA
- hemagglutinin.
REFERENCES
- 1.Evans R. M. (1988) Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng Z., Allan G. F., Thaller C., Cooney A. J., Tsai S. Y., O'Malley B. W., Tsai M. J. (1994) Endocrinology 135, 248–252 [DOI] [PubMed] [Google Scholar]
- 3.Li X., Lonard D. M., Jung S. Y., Malovannaya A., Feng Q., Qin J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2006) Cell 124, 381–392 [DOI] [PubMed] [Google Scholar]
- 4.Oñate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (1995) Science 270, 1354–1357 [DOI] [PubMed] [Google Scholar]
- 5.Xu J., O'Malley B. W. (2002) Rev. Endocr. Metab. Disord. 3, 185–192 [DOI] [PubMed] [Google Scholar]
- 6.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 7.Chen D., Huang S. M., Stallcup M. R. (2000) J. Biol. Chem. 275, 40810–40816 [DOI] [PubMed] [Google Scholar]
- 8.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 9.Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedford M. T. (2007) J. Cell Sci. 120, 4243–4246 [DOI] [PubMed] [Google Scholar]
- 11.Stallcup M. R., Chen D., Koh S. S., Ma H., Lee Y. H., Li H., Schurter B. T., Aswad D. W. (2000) Biochem. Soc. Trans. 28, 415–418 [PubMed] [Google Scholar]
- 12.Ma H., Baumann C. T., Li H., Strahl B. D., Rice R., Jelinek M. A., Aswad D. W., Allis C. D., Hager G. L., Stallcup M. R. (2001) Curr. Biol. 11, 1981–1985 [DOI] [PubMed] [Google Scholar]
- 13.Bauer U. M., Daujat S., Nielsen S. J., Nightingale K., Kouzarides T. (2002) EMBO Rep. 3, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav N., Lee J., Kim J., Shen J., Hu M. C., Aldaz C. M., Bedford M. T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W., Chen H., Du K., Asahara H., Tini M., Emerson B. M., Montminy M., Evans R. M. (2001) Science 294, 2507–2511 [DOI] [PubMed] [Google Scholar]
- 16.Chevillard-Briet M., Trouche D., Vandel L. (2002) EMBO J. 21, 5457–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y. H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3611–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q., Yi P., Wong J., O'Malley B. W. (2006) Mol. Cell. Biol. 26, 7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J., Bedford M. T. (2002) EMBO Rep. 3, 268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Park S., Kilburn B., Jelinek M. A., Henschen-Edman A., Aswad D. W., Stallcup M. R., Laird-Offringa I. A. (2002) J. Biol. Chem. 277, 44623–44630 [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara T., Mori Y., Chu D. L., Koyama Y., Miyata S., Tanaka H., Yachi K., Kubo T., Yoshikawa H., Tohyama M. (2006) Mol. Cell. Biol. 26, 2273–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J., Lee J., Yadav N., Wu Q., Carter C., Richard S., Richie E., Bedford M. T. (2004) J. Biol. Chem. 279, 25339–25344 [DOI] [PubMed] [Google Scholar]
- 23.Cheng D., Côté J., Shaaban S., Bedford M. T. (2007) Mol. Cell 25, 71–83 [DOI] [PubMed] [Google Scholar]
- 24.Frietze S., Lupien M., Silver P. A., Brown M. (2008) Cancer Res. 68, 301–306 [DOI] [PubMed] [Google Scholar]
- 25.El Messaoudi S., Fabbrizio E., Rodriguez C., Chuchana P., Fauquier L., Cheng D., Theillet C., Vandel L., Bedford M. T., Sardet C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13351–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumder S., Liu Y., Ford O. H., 3rd, Mohler J. L., Whang Y. E. (2006) Prostate 66, 1292–1301 [DOI] [PubMed] [Google Scholar]
- 27.Jung S. Y., Li Y., Wang Y., Chen Y., Zhao Y., Qin J. (2008) Anal. Chem. 80, 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 29.Feng Q., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Zhang Y. (2002) Mol. Cell. Biol. 22, 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yue W. W., Hassler M., Roe S. M., Thompson-Vale V., Pearl L. H. (2007) EMBO J. 26, 4402–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troffer-Charlier N., Cura V., Hassenboehler P., Moras D., Cavarelli J. (2007) EMBO J. 26, 4391–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higashimoto K., Kuhn P., Desai D., Cheng X., Xu W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12318–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teyssier C., Chen D., Stallcup M. R. (2002) J. Biol. Chem. 277, 46066–46072 [DOI] [PubMed] [Google Scholar]
- 34.Urwyler O., Zhang L., Li X., Imboden H., Suter B. (2007) Differentiation 75, 757–765 [DOI] [PubMed] [Google Scholar]
- 35.Oh A. S., Lahusen J. T., Chien C. D., Fereshteh M. P., Zhang X., Dakshanamurthy S., Xu J., Kagan B. L., Wellstein A., Riegel A. T. (2008) Mol. Cell. Biol. 28, 6580–6593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 37.Han S. J., Lonard D. M., O'Malley B. W. (2009) Trends Endocrinol. Metab. 20, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu R. C., Smith C. L., O'Malley B. W. (2005) Endocr. Rev. 26, 393–399 [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Zhou L., Cheng X. (2000) EMBO J. 19, 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]