Abstract
Previous work from our laboratory has focused on mitochondrial DNA (mtDNA) repair and cellular viability. However, other events occur prior to the initiation of apoptosis in cells. Because of the importance of mtDNA in ATP production and of ATP in fuel cell cycle progression, we asked whether mtDNA damage was an upstream signal leading to cell cycle arrest. Using quantitative alkaline Southern blot technology, we found that exposure to menadione produced detectable mtDNA damage in HeLa cells that correlated with an S phase cell cycle arrest. To determine whether mtDNA damage was causatively linked to the observed cell cycle arrest, experiments were performed utilizing a MTS-hOGG1-Tat fusion protein to target the hOGG1 repair enzyme to mitochondria and enhance mtDNA repair. The results revealed that the transduction of MTS-hOGG1-Tat into HeLa cells alleviated the cell cycle block following an oxidative insult. Furthermore, mechanistic studies showed that Chk2 phosphorylation was enhanced following menadione exposure. Treatment of the HeLa cells with the hOGG1 fusion protein prior to menadione exposure resulted in an increase in the rate of Chk2 dephosphorylation. These results strongly support a direct link between mtDNA damage and cell cycle arrest.
Introduction
Mitochondria are often described as the powerhouses of the cell, producing ATP through oxidative phosphorylation for cellular utilization. However, this process can produce reactive oxygen species (ROS)2 as a result of electron leakage from the electron transport chain. It is estimated that 1–5% of the electrons from electron transport join with molecular oxygen to produce superoxide (1, 2). The superoxide molecule can then undergo dismutation, either spontaneously or aided by manganese superoxide dismutase, to form hydrogen peroxide. Although superoxide has a very short half-life, hydrogen peroxide is much more stable and can diffuse freely across membranes. Once hydrogen peroxide reacts with iron, it can undergo Fenton chemistry and produce the hydroxyl radical. Because the hydroxyl radical is so highly reactive, it reacts in close proximity to its production site. Because of the location of ROS production from electron transport in the mitochondrial inner membrane, mtDNA can become oxidatively damaged. Indeed, research has shown that mitochondrial DNA is a sensitive target for mitochondria-derived ROS production (3–6).
The mammalian mitochondrion contains 2–10 copies of mtDNA, a circular double-stranded DNA molecule that encodes 13 proteins, 22 tRNAs, and 2 rRNAs (7). The integrity of the mtDNA must be maintained to ensure proper electron transport chain function. The repair of lesions in mtDNA is especially important because it has no introns and almost all of the mtDNA must be transcribed. Therefore, efficient repair of lesions in this DNA is essential to ensure that the mitochondria-encoded proteins for the respiratory chain are produced and that efficient electron transport is maintained.
To ensure genomic stability and remove oxidative base lesions, mtDNA is repaired through base excision repair (BER), which requires a stepwise removal of the damaged base and its replacement with the correct base (1, 8, 9). Failure to repair mtDNA damage has been shown previously to initiate cell death by apoptosis (6, 10–12). However, there are changes in cellular function that can occur prior to the initiation of cell death to allow the cell to repair damage that it has sustained. One such example is the initiation of a cell cycle arrest. Because of the importance of mtDNA in ATP production and the production of ROS that can occur from aberrant electron transport, it is likely that mitochondria exert some level of control over cellular proliferation. Recent evidence supports the concept of a mitochondrial checkpoint (13). Therefore, our experiments explored whether a link exists between mtDNA integrity and the cell cycle. Experiments utilized menadione to produce ROS in HeLa cells to determine whether there is a link between mtDNA integrity and cell cycle arrest. mtDNA BER was modulated using a fusion protein containing human 8-oxoguanine DNA glycosylase-1 (hOGG1) targeted to mitochondria to enhance mtDNA repair. The results show that mtDNA damage is linked to cell cycle arrest.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Exposure
HeLa cells and RCSN-3 cells were incubated at 37 °C in 5.0% CO2 in a humidified chamber. HeLa cells, a human cervical cancer cell line, were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 2 mm glutamine (Sigma), and 100 mg/ml penicillin/streptomycin (Sigma). RCSN-3 cells, a neuronal cell line derived from the substantia nigra of 4-month-old rat brains, were cultured in 1:1 F12/Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 50 mg/ml gentamicin sulfate (Sigma), and 6.0 g/liter glucose (14–16). In all experiments, cells were plated 24 h prior to the start of the experiment. For menadione exposure, menadione sodium bisulfite (Sigma) was dissolved in Hanks' balanced salt solution (HBSS; Sigma) and diluted to the desired concentrations. The medium in the dishes was removed, cells were washed once in HBSS, and the HBSS containing menadione (or HBSS alone for control dishes) was added to the dishes for 1 h. Following the 1-h exposure, the menadione was removed, the dishes were washed once with HBSS, and the cells were either harvested for analysis immediately (e.g. mtDNA or nuclear DNA damage, etc.), or fresh medium was placed into the cell dishes for 24 h (e.g. cell cycle analysis, trypan blue dye exclusion viability, etc.). Carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) treatment followed the same protocol as for menadione, except that CCCP was dissolved in 100% dimethyl sulfoxide prior to diluting into HBSS; control CCCP samples contained only dimethyl sulfoxide in HBSS. N-Methyl-N-nitrosourea (MNU) treatment followed the same protocol as menadione, except that MNU was diluted into citrate buffer, pH 4.2, prior to dilution into HBSS. Control MNU samples contained only citrate buffer in HBSS, with a final citrate buffer concentration of 1%.
Cell Viability and Proliferation
Cells were plated into tissue culture vessels at a subconfluent density. Cells were allowed to grow for 24 h in normal medium prior to experimentation. Twenty-four hours after plating, the cells were exposed to varying concentrations of menadione following the exposure regimen described above. At the appropriate time, the cells were trypsinized, pelleted, and resuspended. An aliquot was taken from the cell suspension and mixed 1:1 with 0.4% trypan blue. Cells then were counted in a hemacytometer to determine the number of cells present. Viability was calculated, and proliferation was determined based on the sample collected at T = 0 h, the time that the chemical exposure began.
DNA Isolation and Quantitative Southern Blot Analysis
Cells were plated and exposed to menadione as described above. At the appropriate time, cells were lysed in DNA lysis buffer (0.3 mg/ml proteinase K (Roche Applied Science), 0.5% SDS, 10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0) and incubated overnight at 37 °C. To each sample, 5 m NaCl was added to a final NaCl concentration of 1 m, and DNA was extracted twice with an equal volume of SEVAG (24:1, chloroform:iso-amyl alcohol). To the resulting aqueous layer, 10.5 m ammonium acetate was added to a final ammonium acetate concentration of 2.1 m. The samples were mixed by inversion; then twice the sample volume of 100% ice-cold ethanol was added to each sample and mixed by inversion, and the DNA was allowed to precipitate at −20 °C overnight. The precipitated samples then were pelleted and resuspended in distilled H2O, and the sample was treated with RNase (final concentration, 1.25 μg/ml; Roche Applied Science) and BamHI restriction enzyme (Roche Applied Science). The restriction was allowed to proceed for at least 6 h at 37 °C. Following restriction, samples were precipitated using 10.5 m ammonium acetate and 100% ice-cold ethanol at −20 °C overnight. The precipitated DNA samples were pelleted, resuspended in 1× TE (10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0), and quantitated using calf thymus DNA as the standard in a Hoefer DyNA Quant 200 fluorometer and a Hoechst 33258 fluorescent solution. The quantitated total cellular DNA was heated to 70 °C for 15 min, cooled to room temperature for 10 min, and treated for 30 min with 5 units/μg endonuclease III and 5 units/μg formamidopyrimidine glycosylase (New England Biolabs). Then, NaOH was added to a final concentration of 0.1 n, and the samples were incubated at 37 °C for 20 min. Alkaline loading dye (10 mm EDTA, 25% Ficoll, 0.25% bromcresol purple, 500 mm NaOH in H2O) was added to each sample, and the samples were loaded on a 0.6% alkaline-agarose gel (0.6% agarose, 1 mm EDTA, pH 8.0, 30 mm NaOH) in alkaline running buffer (1 mm EDTA, pH 8.0, 45 mm NaOH). The gels ran for 16 h at 30 V (1.5 V/cm gel length). The resulting gel was stained with ethidium bromide and imaged using UV light to obtain a gel picture. The gel picture was saved, and the gel was then washed twice with 0.25 m HCl, twice with an alkaline wash (0.5 m NaOH, 1.5 m NaCl), and twice with neutral buffer (0.5 m Tris, 1.5 m NaCl, pH 7.5) for 10 min each. The gel was transferred to a Zeta-Probe GT nylon membrane (Bio-Rad) using 10× SSC (1.5 m NaCl, 0.15 m sodium citrate) and cross-linked using a GS Gene Linker UV chamber (Bio-Rad). The membrane was hybridized to a species-specific mitochondrial radioactive probe in 7% SDS, 0.25 m Na2HPO4 at 55 °C overnight.
Following hybridization, the membrane was washed twice in 5% SDS, 20 mm Na2HPO4, pH 7.2, and twice in 1% SDS, 20 mm Na2HPO4 for 15 min each. The membrane was dried and placed on film for exposure and analysis. Film density was determined either by Molecular Analyst software (Bio-Rad) or using Fujifilm Image Gauge 4.0 software. Break frequencies were calculated using the Poisson equation, Bf = −ln(Po), where Bf is the number of breaks per fragment and Po is the fraction of fragments free of breaks. All results are shown as lesions per mitochondrial genome. Samples of the resulting Southern blot images are included in supplemental Fig. 1.
Quantitative Alkaline Gel Electrophoresis (QAGE)
The gel picture obtained during the quantitative alkaline Southern blotting procedure was scanned into a computer. The film was then analyzed using Fujifilm Image Gauge 4.0 software to determine the peak fluorescent intensity size, which represents the area of greatest intensity along the DNA sample lane. The location then was compared with the lambda band standard (Promega) to determine the size of the peak fluorescent intensity. Nuclear DNA damage causes a decrease in the fragment size of the peak fluorescent intensity compared with the control. Note that the DNA visualized by QAGE was treated enzymatically with formamidopyrimidine glycosylase and endonuclease III as described under “Quantitative Southern Blot Analysis” (“Experimental Procedures”). Samples of the resulting QAGE images are included in supplemental Fig. 1.
Mitochondrial Membrane Potential Analysis
HeLa cells exposed to menadione or CCCP were collected by trypsinization, pelleted, and resuspended at a concentration of 1 million cells/ml in serum-free medium. At this point, positive controls (previously untreated cells) were treated with 1 μl of 50 mm CCCP for 5 min at 37 °C (final concentration of 50 μm). To each sample, 6 μl of 200 μm JC-1 dye was added, and the samples were incubated for 20 min at 37 °C. Following incubation, cells were washed once with serum-free medium, pelleted, and resuspended in fresh serum-free medium. Samples were immediately analyzed by flow cytometry, with 10,000 events collected for each sample.
ADP/ATP Ratio Determination
ADP/ATP ratios were determined using the ApoSENSOR ADP/ATP ratio assay kit (Alexis Biochemicals). Cells were plated in 96-well dishes with 2 × 103 cells/well and allowed to grow for 24 h. Following the growth period, cells were treated with menadione or CCCP at varying concentrations and exposure times. Following chemical exposure, cells were lysed and analyzed using the protocol provided. ATP and ADP levels were determined using a reporter luminometer (Turner Designs), with ATP analysis preceding ADP analysis.
MTS-hOGG-Tat Construction
The sequence for hOGG1-α was obtained and codon-optimized for efficient expression in Escherichia coli (GenScript Corp.). The MTS sequence is from human manganese superoxide dismutase and is 24 amino acids in length (17). The C-terminal extension, HA-Tat-His10, was generated from synthetic oligonucleotides using an overlapping PCR technique. All components then were constructed together using overlapping PCR. The final construct (MTS-hOGG1-HA-Tat-10His) was assembled in the pBluescriptII (Stratagene) derivative vector, with the expression driven by the Plac promoter. This vector was introduced into E. coli BL21(DE3) cells for protein expression. The bacteria were grown in liquid culture at 15 °C for 42 h in the presence of 0.1 mm isopropyl β-d-1-thiogalactopyranoside. The bacteria were pelleted by centrifugation at 1800 × g for 15 min, and the pellet was resuspended in purification buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm phenylmethylsulfonyl fluoride (no EDTA), 1× EDTA-free protease inhibitor mixture, 5 mm imidazole). The bacterial suspension was sonicated for 15 s with 45 s in between using a Branson Sonifier 250. Sonication was repeated five times. The bacterial lysates were then centrifuged at 22,500 × g for 30 min at 4 °C. The clear lysates were mixed with 1 ml of nickel-nitrilotriacetic acid-agarose suspension (Qiagen) and incubated for 1 h at 4 °C on a slow orbital shaker. A polystyrene column was prepared for the lysate suspension, and the nickel-nitrilotriacetic acid-agarose column that was formed was washed with 2–3 volumes of wash buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 30 mm imidazole, 10% glycerol). A sample of the flow-through was taken for the absorbance measurement using a spectrophotometer, with an A280 of <0.05 obtained before the elution buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10% glycerol, 300 mm imidazole) was applied. Once the A value was obtained, the elution buffer was applied, and timed fractions of eluting protein were collected from the column in 1.5-ml Eppendorf tubes. The protein obtained then was desalted to remove imidazole using a desalting buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 10% glycerol), and the protein was concentrated. The amount of protein was measured using a spectrophotometer (Bio-Rad SmartSpec 3000), the purity of the eluted protein was assessed using SDS-PAGE and Coomassie staining, and a Western blot was performed to confirm the correct size of the protein (45.9 kDa).
Subcellular Fractionation
HeLa cells were treated with 50 μg/ml MTS-hOGG1-Tat or the desalting buffer for 3 h at 37 °C in an incubator. Following a 3-h protein transduction, HeLa cells were collected by trypsinization and centrifuged at 200 × g for 5 min. Cells were washed twice with 1× PBS and centrifuged at 200 × g for 5 min after each wash. The pelleted cells were resuspended in ice-cold 0.25 m sucrose in 1× TKM (10 mm Tris, 3 mm MgCl2, 15 mm KCl, pH 7.4). Cells were allowed to sit for 40 min on ice to allow for cell swelling. Cells were homogenized using a Dounce homogenizer, and the homogenates were centrifuged at 14,000 × g for 20 min at 4 °C to pellet nuclear and mitochondrial organelles. The supernatant was discarded, and the pellet was resuspended in 1 ml of 0.25 m sucrose in 1× TKM. The cell suspension then was mixed with 2 ml of 2.3 m sucrose in 1× TKM, giving a final sucrose concentration of ∼1.62 m sucrose. The cells then were loaded onto a cushion of 1 ml of 2.3 m sucrose in 1× TKM in 5-ml Ultracentrifuge tubes, and 1 ml of 0.25 m sucrose in 1× TKM was placed on the very top of the column. Samples were centrifuged at 124,000 × g for 30 min at 4 °C using a swinging bucket rotor. The tubes were removed, and the mitochondrial fraction (at the 0.25 and 1.62 m sucrose interface) and the nuclear fraction (at the 1.62 and 2.3 m sucrose interface) were collected into separate tubes and centrifuged at 14,000 × g for 20 min at 4 °C. Mitochondrial and nuclear pellets were lysed in lysing buffer (20 mm HEPES, pH 7.6, 1 mm EDTA, 5% glycerol, 0.2% Triton X-100, 5 mm dithiothreitol, 300 mm KCl) with 5 μl/ml protease inhibitors (Sigma) added to each sample. Lysates were centrifuged at 14,000 × g for 20 min at 4 °C to pellet the unlysed organelles and membranes. The supernatant from the spin was collected and saved as protein material to be analyzed.
OGG Activity Assay
A 5′-end radiolabeled 8-oxoguanine oligonucleotide was created by adding the following components together: 14.5 μl of H2O, 2.0 μl of 10× T4 kinase buffer (Promega), 0.5 μl (5 pmol total) of 8-oxoguanine oligonucleotide (Bio-Synthesis), 1.0 μl of T4 kinase (Promega), and 1.5 μl of [33γ]ATP (PerkinElmer Life Sciences). The 8-oxoguanine oligonucleotide has an 8-oxoguanine lesion at the 11th position. The mixture was incubated at 37 °C for 30 min and then heated to 90 °C for 2 min to inactivate the T4 kinase. At that point, 0.5 μl (5 pmol total) of the complementary oligonucleotide (Bio-Synthesis) was added, and the samples were cooled to 4 °C until used.
The protein samples from subcellular fractionations were quantitated using the Bradford assay (Bio-Rad). Activity samples were mixed together using the following components: 20 μg of protein and 0.2 pmol of 8-oxoguanine oligonucleotide in 1× REC buffer (10 mm HEPES, pH 7.4, 100 mm KCl, 10 mm EDTA, 100 μg/ml bovine serum albumin). Samples were incubated at 37 °C for 6 h and then stored at 4 °C until gel analysis. For controls, formamidopyrimidine glycosylase (6.4 units; New England Biolabs) and purified MTS-hOGG1-Tat (5 μg) were utilized.
For gel analysis, 4 μl of the activity samples was mixed with 2 μl of 3× alkaline dye (300 mm NaOH, 97% formamide, 0.2% bromphenol blue) and heated to 95 °C for 2 min, and 4 μl was loaded on a 7 m urea, 20% acrylamide gel and run at 200 V in 1× Tris borate-EDTA buffer. The gel then was placed on film and put at −70 °C for exposure.
Protein Isolation
Cells were exposed to menadione as described, and following exposure, the cells were trypsinized and centrifuged at 200 × g for 5 min. Cells were washed twice with 1× PBS and pelleted again by centrifugation. Cells then were lysed for 80 s using ice-cold digitonin (0.4 mg/ml digitonin, 2.5 mm EDTA, 250 mm mannitol, 17 mm MOPS, pH 7.4). Lysates were added to 2.5× mannitol-sucrose buffer (525 mm mannitol, 175 mm sucrose, 12.5 mm Tris, 112.5 mm EDTA) to a final concentration of 1×. Lysates were centrifuged at 14,000 × g for 20 min to pellet organelles. The supernatant was collected and used for protein analysis of cytosolic proteins relating to the cell cycle block. These samples were stored at −70 °C until analyzed by SDS-PAGE.
Western Blot Analysis
Western blots were performed as described previously (4). Briefly, protein samples were quantitated using the Bradford assay, aliquoted, and brought to volume with 1× TE and 5× SDS loading dye (0.225 m Tris-HCl, pH 6.8, 50% glycerol, 5% SDS, 0.05% bromphenol blue, 0.25 m dithiothreitol). Samples were boiled for 5 min, cooled, and resolved by 12 or 7.5% SDS-PAGE. Resolved proteins were transferred to a polyvinylidene difluoride membrane (Millipore) by standard procedures, and membranes were blocked in 5% nonfat dried milk in 1× TBS-T (20 mm Tris, 137 mm NaCl, 0.1% Tween 20, pH 7.6) for 1 h. Membranes then were exposed to primary antibodies overnight at 4 °C. Membranes were rinsed with 5% blocking solution, incubated with the appropriate secondary antibody for 1 h at room temperature in 5% blocking solution, and then rinsed with 1× TBS-T. Membranes then were exposed to chemiluminescent substrates (West Dura or West Pico-Pierce) for 5 min and placed on film. The following antibodies were used: HA (Sigma), succinate dehydrogenase (Invitrogen), lamin A (Santa Cruz Biotechnology), actin (Sigma), phosphorylated Thr-68 and total Chk2 (R&D Systems), and secondary antibodies of anti-mouse, anti-goat, and anti-rabbit (Santa Cruz Biotechnology).
Phospho-proteome Profiler
To determine the signaling pathways activated following menadione and CCCP exposure, the human phospho-kinase array kit was utilized (R&D Systems). HeLa cells were plated as described previously and exposed to either menadione (200 μm) or CCCP (50 μm) for 1 h. Following exposure, the cells were lysed according to the supplied protocol and reagents. Protein concentrations were determined using the Bradford assay, and 150 μg of protein was placed on each membrane. The membranes were prepared and exposed to film following the supplied protocol, and the densities of the resulting spots on the array were determined using Image Gauge software (Fujifilm).
Cell Cycle Analysis
Cells were plated and exposed to menadione as described above. The cells were trypsinized at the appropriate time point and then pelleted by centrifugation. The pelleted cells were resuspended in 70% ice-cold ethanol at a concentration of 1–3 million cells/ml and stored at 4 °C until the day of analysis. Analysis was performed within less than 3 weeks. On the day of analysis, the cells were centrifuged at 200 × g for 5 min, washed twice in 1× PBS, and pelleted after each wash by centrifugation. After the second 1× PBS wash, the cells were resuspended in serum-free medium (i.e. medium without fetal bovine serum added) at a concentration of ∼1 million cells/ml. At this point, 5 μg of RNase (10 μl of 500 μg/ml RNase) was added to the cell suspension, and the cells were incubated at 37 °C for 30 min. Following the incubation, propidium iodide was added to each cell suspension to a final concentration of 50 μg/ml, and the cells were stored on ice in the dark until analysis by flow cytometry. Approximately 10,000 cells were analyzed by flow cytometry, and the resulting distribution of cells was analyzed by the ModFit program to determine the percentage of cells in G0-G1, S, or G2-M phases of the cell cycle.
Statistics
All experiments were analyzed using a one-way analysis of variance and the Tukey post hoc test. However, when only two data points were compared, a two-tail Student's t test was used. Data and statistics were analyzed using GraphPad Prism version 4 software. Graphical results represent mean ± S.E. Results were deemed statistically significant at the level of p < 0.05 (as labeled with an asterisk in Figs. 1–4 and 6–8), and experiments were done with n ≥ 3.
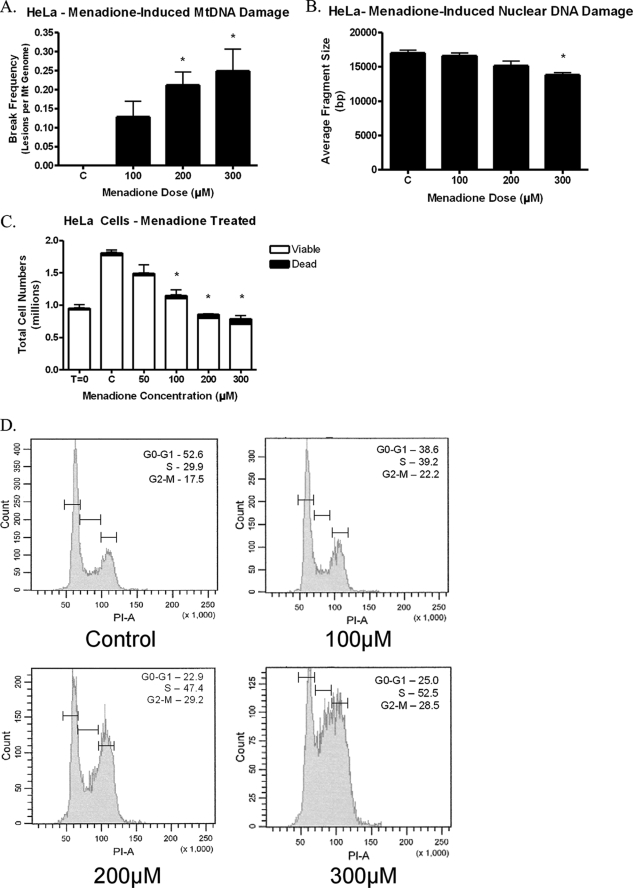
FIGURE 1.
Menadione treatment of HeLa cells. HeLa cells were treated with menadione for 1 h. A, menadione induced mtDNA damage in HeLa cells following 1 h of treatment (n = 6). B, menadione did not produce detectable nuclear DNA damage as determined by QAGE (n = 4). C, HeLa cells were exposed to menadione and then allowed to proliferate for 24 h in normal culture media. Cell counts in the presence of trypan blue dye showed that menadione caused a decrease in the total number of cells with no significant changes in the percentage of viable cells, suggesting a cell cycle arrest (n = 3). D, HeLa cells were treated with menadione and allowed to proliferate for 24 h. Cells were collected and analyzed using propidium iodide and flow cytometry. The percentage of cells in each phase is shown for each sample. The results reveal that menadione induces a cell cycle arrest with the primary arrest in the S phase and a smaller yet significant block in the G2-M phase (n = 9).
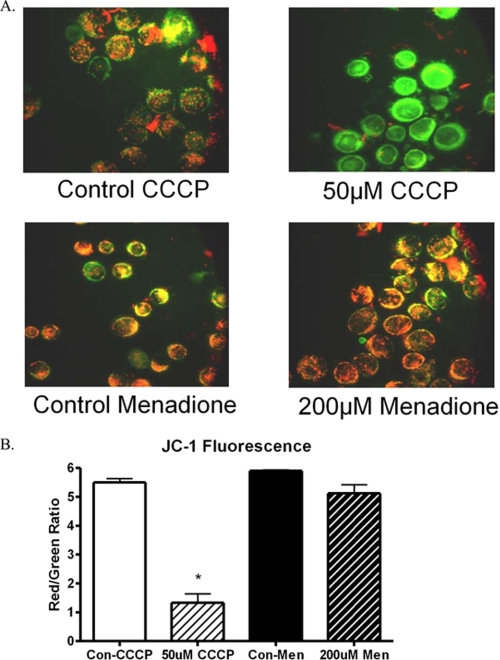
FIGURE 2.
Mitochondrial membrane potential following CCCP and menadione exposure in HeLa cells. HeLa cells were treated for 1 h with either menadione (200 μm) or CCCP (50 μm). The cells were collected and stained with JC-1 dye, which fluoresces green in the monomeric form and red as aggregates form as a result of membrane potential. CCCP-treated cells show a reduction in red fluorescence as seen by fluorescent microscopy. Menadione-treated cells did not undergo any change in mitochondrial membrane potential. A, visual representation of mitochondrial membrane potential. B, quantitative results of JC-1 flow cytometry analysis.
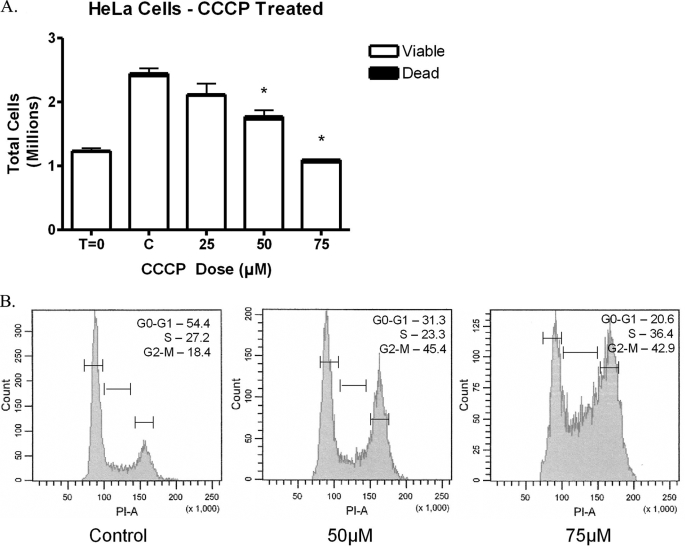
FIGURE 3.
CCCP treatment in HeLa cells. HeLa cells were exposed to CCCP for 1 h and then allowed to proliferate for 24 h in normal media. A, cells were collected and analyzed by trypan blue dye exclusion. The results show that CCCP caused a decrease in the total number of cells with no significant changes in the percentage of viable cells, thus suggesting a cell cycle arrest (n = 4). B, HeLa cells were analyzed using propidium iodide and flow cytometry, and the percentage of cells in each phase is shown for each sample. The results reveal that CCCP induced a cell cycle arrest with the primary arrest in the G2-M phase and a secondary block in the S phase (n = 8).
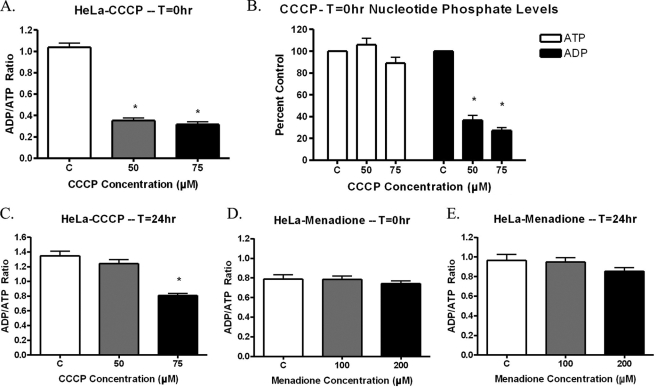
FIGURE 4.
ADP/ATP Ratios in menadione- or CCCP-treated HeLa cells. A–C, HeLa cells were treated with varying concentrations of CCCP for 1 h. Following CCCP exposure, the cells were lysed and analyzed for ATP and ADP relative light units. CCCP reduced the ADP/ATP ratio at T = 0 h (A), which was the result of decreased ADP levels, without any change in ATP levels (B) (n = 6). The CCCP-induced decrease in ADP/ATP ratios is reversible, and HeLa cells can recover following 24 h in normal media (C). d and E, HeLa cells were treated with varying concentrations of menadione for 1 h. Following menadione treatment, the cells were lysed and analyzed for ATP and ADP relative light units. The results show that menadione does not affect the ADP/ATP ratio at either T = 0 h (D) or T = 24 h (E) (n = 6).
FIGURE 6.
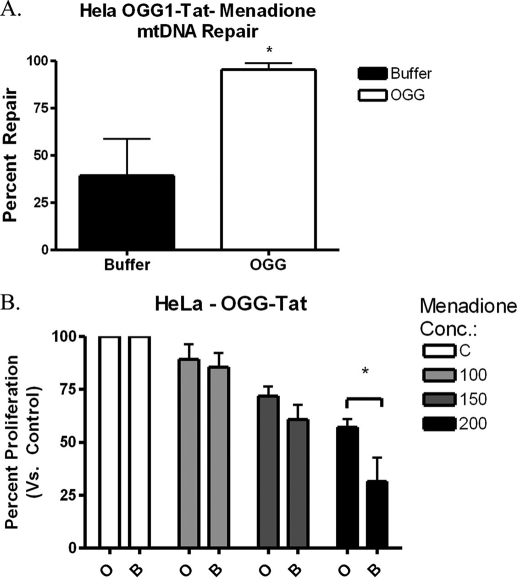
MTS-hOGG1-Tat protein activity in HeLa cells. A, HeLa cells were pretreated for 3 h with 50 μg/ml hOGG1 fusion protein and then treated with 200 μm menadione for 1 h. HeLa cells were collected 6 h post-menadione treatment and analyzed for mtDNA repair. The hOGG1 fusion protein enhanced mtDNA repair of menadione-induced damage (n = 3). B, HeLa cells pretreated with the hOGG1 fusion protein displayed enhanced cellular proliferation compared with the buffer-only controls 24 h post-menadione exposure (n = 4).
FIGURE 7.
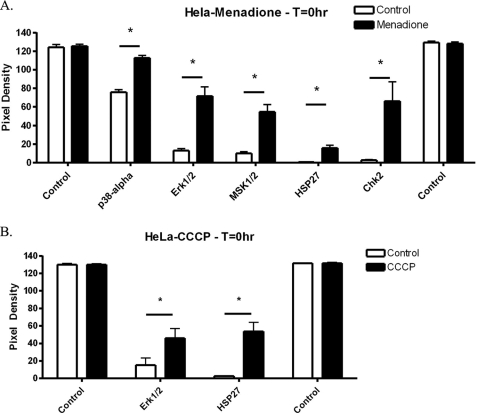
Phospho-proteome analysis of menadione- or CCCP-treated HeLa cells. A, HeLa cells were treated for 1 h with menadione. Phospho-proteome analysis was performed as described under “Experimental Procedures.” Quantitation of the resulting films revealed five proteins that underwent increased phosphorylation following menadione treatment (n = 3). B, HeLa cells were treated for 1 h with CCCP, and then phospho-proteome analysis was performed. Quantitation of the resulting films revealed two proteins that underwent increased phosphorylation following CCCP treatment (n = 3).
FIGURE 8.
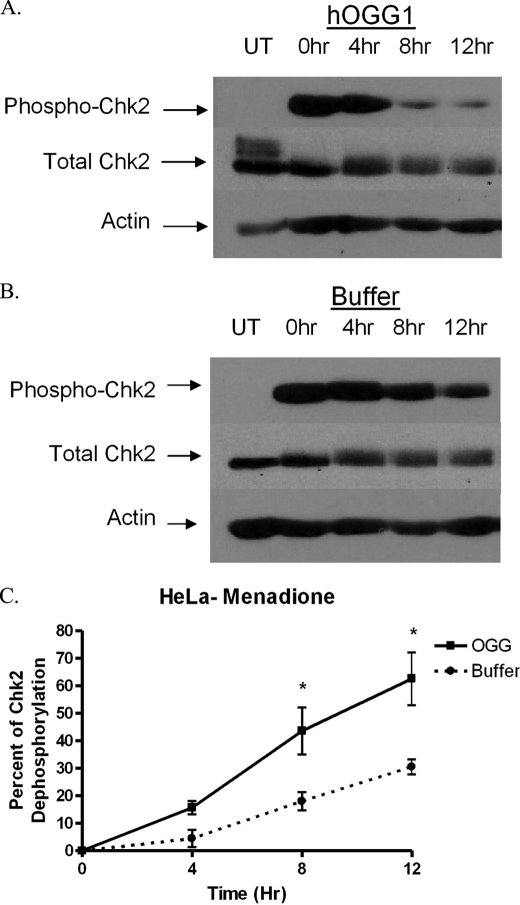
MTS-hOGG1-Tat mediates inactivation of Chk2. HeLa cells were pretreated with the hOGG1 fusion protein and then exposed to menadione (200 μm). The results show that hOGG1 (A) can decrease phosphorylation of Chk2 (Thr-68) more rapidly than buffer-treated controls (B), thereby demonstrating an effect resulting from enhanced mitochondrial BER. UT, samples untreated by menadione. Times shown are the number of hours following menadione treatment that the cells were harvested. C, quantitation of the density of the bands from the resulting films showing a statistical change in the phosphorylation state in the hOGG1-treated samples compared with buffer-treated controls (n = 3).
RESULTS
mtDNA Damage and Cell Cycle Arrest
To determine the effects of menadione on mtDNA damage, HeLa cells were treated with menadione, lysed, and analyzed using the quantitative alkaline Southern blot technique. Menadione caused mtDNA damage in a dose-dependent manner (Fig. 1A). Also, mtDNA damage was observed at menadione concentrations that were lower than the concentrations at which cell death had been observed previously (4). Furthermore, when the same samples were analyzed for nuclear DNA damage using QAGE, there was detectable nuclear DNA damage only at menadione doses greater than 300 μm (Fig. 1B). As a result of this finding, further studies were performed at dosage levels at which no nuclear DNA damage was detected (i.e. 200 μm or less). HeLa cells then were exposed to menadione to determine the effects menadione had on cell proliferation and viability. Menadione-treated HeLa cells were collected and analyzed for cell numbers in the presence of trypan blue dye, with viable cells excluding the trypan blue dye. Menadione exposure produced a decrease in the total number of cells present without a significant increase in the percentage of dead cells, with all doses analyzed having more than 95% viability (Fig. 1C). To verify that the decrease in total cell numbers was a cell cycle arrest, HeLa cells were treated with menadione and analyzed using propidium iodide and flow cytometry (Fig. 1D). The results verified that menadione does cause a cell cycle arrest in HeLa cells by arresting cells in the S phase and in a minor, yet significant way in the G2-M phase. Therefore, menadione produced a cell cycle arrest at doses at which mtDNA damage was observed, and no nuclear DNA damage was detected.
mtDNA Damage and Cell Cycle Arrest Correlation Are Not Restricted to Chemical or Cell Type
To determine whether the correlation between mtDNA damage and cell cycle arrest was dependent upon the cell type or chemical used, experiments were performed in an unrelated cell line. RCSN-3 cells are a dopaminergic neuronal cell line derived from rat substantia nigra (14–16). Experiments using these cells revealed a similar link between mtDNA damage and cell cycle arrest, with no detectable nuclear DNA damage (supplemental Fig. 2). In addition, experiments were performed with MNU in the HeLa cell line (supplemental Fig. 3). The results showed that MNU, an alkylating agent that has previously been shown to cause mtDNA damage, produced a decrease in cellular proliferation and no change in cell viability at doses at which mtDNA damage, but not nuclear DNA damage, had previously been detected (18). These results demonstrated that the link between mtDNA damage and cell cycle arrest is not cell type-dependent, nor is it a characteristic of the chemical utilized.
ADP/ATP Analysis
Because of the importance of mitochondria for cellular ATP production, it was possible that changes in available ATP could be the cause of the cell cycle arrest. A cell lacking in sufficient nutrients could have arrested proliferation until nutrients become more available. To determine the effect of mitochondrial electron transport on cellular proliferation, CCCP was utilized. CCCP is a potent mitochondrial uncoupler that is lipid-soluble and readily dissipates the mitochondrial membrane potential. CCCP action was verified initially by the reduction of mitochondrial membrane potential as determined by alterations in JC-1 dye aggregation. CCCP was found to reduce mitochondrial membrane potential as seen using JC-1 dye; but interestingly, when menadione was used, mitochondrial membrane potential was not altered (Fig. 2). To determine whether CCCP exposure causes mtDNA damage, quantitative alkaline Southern blots were employed over a range of doses of CCCP. CCCP did not produce detectable mtDNA damage in HeLa cells over the range of doses analyzed; in addition, no nuclear DNA damage was observed by QAGE analysis (data not shown). Next, HeLa cells were treated with CCCP to determine the effect of CCCP on HeLa cell proliferation. Cell counts performed in the presence of trypan blue dye revealed that CCCP produced a decrease in total cell number without a significant increase in cell death (Fig. 3A). This suggested that CCCP inhibits cellular proliferation. To verify cell cycle arrest, CCCP-treated HeLa cells were analyzed using propidium iodide and flow cytometry (Fig. 3B). CCCP did produce a cell cycle arrest, but the profile did not correspond to that of menadione. CCCP produced a primary G2-M block, with only a secondary S phase block at higher doses. These results suggest that there is a difference in the mechanism of action of menadione and CCCP, but they do not exclude the possibility that both compounds may alter ATP levels similarly.
To determine the effects that menadione and CCCP have on ATP and ADP levels, a luminescent assay was utilized. HeLa cells treated with CCCP underwent a substantial decrease in their ADP/ATP ratios (Fig. 4A). This decrease was caused by a sharp decline in available ADP, with no detectable alterations in ATP levels (Fig. 4B). In addition, the effects of CCCP treatment were reversible, with ADP levels recovering following 24 h of maintenance in normal media (Fig. 4C). This indicated that the ADP/ATP ratio was affected during CCCP treatment. In contrast, menadione treatment did not alter ADP or ATP levels following initial chemical exposure (T = 0 h) or 24 h following menadione treatment (T = 24 h) (Fig. 4, D and E, respectively). These results provide additional evidence that menadione and CCCP act through different mechanisms. However, when taken together these data reveal that the available ATP levels in menadione-treated HeLa cells are not altered and, thus, are not the cause of the observed cell cycle arrests.
MTS-hOGG1-Tat Fusion Protein Localization and Activity
Previous work utilized stable transfection of mitochondria-targeted hOGG1 into HeLa cells to enhance viability following menadione treatment (4). Because hOGG1 previously had been shown to work by enhancing BER in the mitochondria of HeLa cells, it was used again in the following experiments. However, the present experiments used the protein transduction domain of the HIV-Tat protein to enable a mitochondrially targeted hOGG1 protein to localize to and repair mtDNA. The targeting of proteins to mitochondria by this method has been described previously (19). For these studies, the MTS-hOGG1-Tat fusion protein was column-purified to a sole band of protein at the expected molecular mass of 45.9 kDa (Fig. 5A).
FIGURE 5.
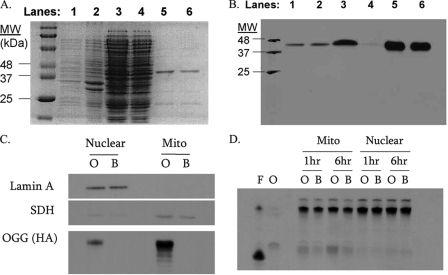
MTS-hOGG1-Tat fusion protein. MTS-hOGG1-Tat fusion proteins were extracted from E. coli and purified as described. Protein extracts were analyzed by Coomassie stain (A) and Western blot (B) against the HA tag in the fusion protein. Lane 1 is a whole cell lysate. Lanes 2 and 3 are the pellet and supernatant, respectively, following sonication. Lane 4 shows column flow-through prior to wash. Lane 5 is the column wash using imidazole, and lane 6 is the final hOGG1 fusion protein following imidazole desalt step. C, HeLa cells were treated with 50 μg/ml fusion protein for 3 h and then fractionated by digitonin and sucrose gradients. Lamin A was used as a marker for nuclear proteins, and succinate dehydrogenase (SDH) was used as a marker for mitochondrial proteins. The HA tag in the fusion protein was used to localize the fusion protein without cross-reactivity with endogenous OGG. O and B correspond to hOGG1-treated and buffer-treated HeLa cells, respectively. D, HeLa cells were pretreated with fusion protein and fractionated on sucrose gradients. In this activity assay, the 8-oxoguanine oligonucleotide is cleaved by the active OGG enzyme, with darker bands corresponding to higher activity. The oligonucleotide was incubated at 37 °C for 1 or 6 h in the presence of the subcellular protein fractions. F and O, formamidopyrimidine glycosylase and OGG enzyme controls, respectively.
To verify that the enzymatically active fusion protein was directed to mitochondria, HeLa cells were plated as described and treated with 50 μg/ml hOGG1 fusion protein for 3 h prior to any experimental manipulation. For protein localization, HeLa cells treated with hOGG1 were collected, lysed, and centrifuged through a sucrose gradient to separate mitochondrial and nuclear fractions. The enriched mitochondrial and nuclear protein fractions were analyzed by PAGE and Western blot (Fig. 5B). The Western blot revealed that the MTS-hOGG1-Tat fusion protein was present in mitochondria of HeLa cells following 3 h of exposure. A minimal amount of fusion protein and the mitochondrial protein succinate dehydrogenase were found in the nuclear fraction. This likely was caused by contamination of mitochondrial proteins in the nuclear fraction.
To determine whether the hOGG1 protein was enzymatically active in the mitochondria, the enriched fractions were incubated for either 1 or 6 h in the presence of an end-labeled 8-oxoguanine-containing duplex oligonucleotide. These samples were loaded onto a denaturing gel, and hOGG1 enzyme activity was evaluated as the amount of cleaved substrate (Fig. 5C). The OGG activity assay revealed that the MTS-hOGG1-Tat fusion protein was active and that it enhanced OGG activity in the mitochondrially enriched fractions compared with buffer-only samples; there was no detectable change in nuclear OGG activity in this assay. These experiments revealed that the hOGG1 fusion protein localizes to mitochondria in HeLa cells and augments OGG activity in mitochondrial fractions.
Finally, to determine the effects of the hOGG-1 fusion protein on mtDNA repair, the quantitative alkaline Southern blot technique was utilized. The results showed that there was enhanced mtDNA repair (∼95% in hOGG1 versus ∼45% in buffer only) 6 h after exposure to menadione (Fig. 6A). Therefore, these results show enhanced mtDNA repair by the mitochondrial hOGG1 fusion protein.
hOGG1-Tat Promotes Proliferation
The fusion protein was utilized to determine the effects of augmenting mitochondrial BER on cellular proliferation. HeLa cells were pretreated with the hOGG1 fusion protein and then were treated with menadione for 1 h. Cells were allowed to proliferate for 24 h and then collected and counted (Fig. 6B). Cell counts in the presence of trypan blue dye revealed that the addition of the hOGG1 fusion protein enhanced cellular proliferation by up to 20% in cultures treated with 200 μm menadione, thus establishing a link between mtDNA damage and cellular proliferation.
Mechanism of Action
Although a causal link had been demonstrated between mtDNA damage and cell cycle arrest, the mechanism of action still needed to be elucidated. Therefore, an investigation into the signaling pathway responsible was initiated. For these studies, a phospho-proteome assay was utilized. This assay allowed for the rapid detection of over 40 different signaling molecules to determine which pathways might be involved and to rule out other possible phosphorylation events unrelated to menadione or CCCP exposure. (For full results, see supplemental Tables 1 and 2.) HeLa cells were treated with either 200 μm menadione or 50 μm CCCP for 1 h and then lysed. The lysed cell samples next were analyzed for changes in phosphorylated signaling responses. When menadione was used, multiple increases in protein phosphorylation were detected, including Chk2, p38α, ERK1/2, MSK1/2, and HSP27 (Fig. 7A). Because the lysates were collected immediately following chemical exposure, the results represent immediate responses by the HeLa cells to menadione treatment. Of the five proteins that were found to have increased phosphorylation, Chk2 had the largest increase. In addition, CCCP treatment also induced a couple of significant phosphorylation events, including ERK1/2 and HSP27 (Fig. 7B). Interestingly, CCCP treatment did not promote phosphorylation of Chk2, p38α, or MSK1/2. The resulting differences between the two responses reflect a unique mechanism of action for menadione, which only partially overlaps that of CCCP, because similar phosphorylation events were detected in both sets.
hOGG1-Tat Promotes Chk2 Dephosphorylation
Of the proteins detected in the proteome arrays, Chk2 provides a potential functional link between DNA damage and the cell cycle. It can cause the same S phase cell cycle block that was seen in the menadione studies. Chk2 is a mediator kinase that acts in concert with other proteins to mediate the response of the ATM/ATR pathway. In addition to the experiments showing that menadione could increase Chk2 phosphorylation, experiments in HeLa cells also revealed an increase in Chk2 phosphorylation following exposure to MNU (supplemental Table 2). Therefore, because previous experiments with the MTS-hOGG1-Tat fusion protein showed an increase in HeLa cell proliferation compared with buffer controls, the fusion protein was again used to determine whether enhancing mitochondrial BER could alter signaling through Chk2 phosphorylation at threonine 68 (Thr-68). Western blots revealed no changes in the initial Chk2 phosphorylation levels; however, pretreatment with hOGG1 fusion protein resulted in a more rapid dephosphorylation of Chk2 when compared with buffer-treated HeLa cells alone (Fig. 8). These experiments demonstrated that menadione, but not CCCP, could increase phosphorylation of Chk2 and that enhancement of mitochondrial BER could modulate Chk2 dephosphorylation following menadione-induced mtDNA damage.
DISCUSSION
The results of this research show for the first time that mtDNA damage is an upstream signal of cell cycle arrest and that modulating mtDNA integrity can be used to alter cell cycle progression. In this cellular model, a single 1-h exposure to menadione caused detectable mtDNA damage. The use of menadione to produce mtDNA damage has been well characterized, and the mtDNA damage it produces can be repaired by BER (4–6). In these experiments, HeLa cells underwent an S phase cell cycle arrest following menadione exposure.
Further experimentation revealed that menadione exposure produced a marked increased in Chk2 phosphorylation in HeLa cells. Chk2 phosphorylation is a marker for ATM pathway activation following DNA damage (20). When ATM is activated, it phosphorylates Chk2 to ensure further activation of downstream effectors. When phosphorylated, Chk2 initiates a cell cycle arrest at an intra-S phase or G2-M transition, similar to the block found in the menadione cell cycle arrest profile. Interestingly, there have been no reports showing Chk2 in mitochondria from any cell. This suggests that signals must come from the mitochondria to the cytoplasm or nucleus to cause phosphorylation of Chk2. Clearly, more experimentation is warranted to fully elucidate this point.
The changes in Chk2 phosphorylation may be the result of two possible events. The first is that Chk2 becomes dephosphorylated due to the inhibition of a kinase; this is possibly, but not necessarily, limited to ATM. The phosphorylation of Chk2 at threonine 68 is an ATM kinase-characterized event, and no other kinases have been shown to phosphorylate this site (20, 21). However, this does not discount the possibility of a previously uncharacterized function of another kinase. Experimentation into a role for ATM in the response to mtDNA damage in HeLa cells did not reveal any changes in ATM phosphorylation at the Ser-1981 site following menadione exposure (data not shown). The second possible explanation for Chk2 dephosphorylation could be the activation of a phosphatase. Phosphatases have a well documented role in cell cycle control, and the possible link between phosphatase activity and mtDNA damage is currently an active area of investigation (22).
Interestingly, the secondary response to menadione exposure was a G2-M cell cycle block and ERK1/2 activation. The nature of the ERK1/2 activation in these experiments is not known. ERK has been characterized as being a proproliferative factor, the phosphorylation and activation of which constitute a necessary factor in cellular proliferation (23). However, it also has been shown that overphosphorylation of ERK can have a negative effect on cell cycle progression, and therefore ERK may mediate the G2-M cell cycle block (24, 25). Further research will be necessary to determine the role of ERK1/2 in menadione and CCCP cell cycle blocks.
The results of the proteome profiler assay with menadione- and CCCP-treated HeLa cells provided sufficient signaling information to develop a possible model for mtDNA damage-initiated cell cycle arrest. One possible explanation for these results is that menadione exposure produces ROS that damage mtDNA and thereby activate the Chk2 response, leading to a substantial increase in the phosphorylation and activation of Chk2. This initiates a cell cycle block in the S phase. Following induction of the S phase block, menadione initiates a second cell cycle block at the G2-M transition to prevent cells from undergoing mitosis. This G2-M block involves the phosphorylation of ERK1/2 and the phosphorylation of HSP27, although these two proteins may not be involved directly with the block and may be a result of the block. Because CCCP does not produce detectable mtDNA damage, its effect on the cell cycle will be an ERK1/2-mediated G2-M arrest. Therefore, in this model, mtDNA damage causes an S phase cell cycle block.
The findings of this research, along with previous research, support a model in which an increase in ROS can initiate, at different thresholds, a cell cycle arrest that is followed by the induction of apoptosis. As the oxidative stress increases, the mtDNA will be subjected to increasing amounts of ROS. These ROS will increasingly deplete endogenous antioxidants to the point that the ROS will begin to damage other biomolecules such as mtDNA. When mtDNA becomes damaged, the mitochondria will rely on BER to repair the resulting damage. At the initiation of mtDNA damage, the cell will begin to initiate a cell cycle arrest that will give it time to repair the damage. After the cell is arrested and the damage to mtDNA continues to intensify, the cell will initiate apoptosis through the intrinsic pathway. This model reveals a therapeutic window of opportunity to prevent apoptosis once the cell has under gone cell cycle arrest. The cell cycle arrest may be useful as an early marker in proliferating cells that are under oxidative stress but have not yet committed to apoptosis.
In summary, the results of the present study indicate that mtDNA damage plays a role in cell cycle progression. This research has immediate implications in the field of cancer, where there is a growing interest in the relationship between mitochondria and the development of cancer. The association of this response with Chk2 also links cell cycle control by mtDNA integrity to an already known pathway with ties to cancer development. In addition, the modulation of mtDNA BER can alter the cellular proliferative response to mtDNA damage by either promoting or inhibiting cellular proliferation depending on the application. Future work will be directed toward exploring both of these possibilities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
- ROS
- reactive oxygen species
- BER
- base excision repair
- HBSS
- Hanks' balanced salt solution
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- MNU
- N-methyl-N-nitrosourea
- QAGE
- quantitative alkaline gel electrophoresis
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- MOPS
- 4-morpholinepropanesulfonic acid
- ATM
- ataxia telangiectasia-mutated
- ERK
- extracellular signal-regulated kinase
- OGG
- 8-oxoguanine DNA glycosylase
- hOGG
- human OGG
- MTS
- mitochondrial targeting sequence.
REFERENCES
- 1.Mandavilli B. S., Santos J. H., Van Houten B. (2002) Mutat. Res. 509, 127–151 [DOI] [PubMed] [Google Scholar]
- 2.Spiteller G. (2002) Ann. N.Y. Acad. Sci. 959, 30–44 [DOI] [PubMed] [Google Scholar]
- 3.Hollensworth S. B., Shen C., Sim J. E., Spitz D. R., Wilson G. L., LeDoux S. P. (2000) Free Radic. Biol. Med. 28, 1161–1174 [DOI] [PubMed] [Google Scholar]
- 4.Dobson A. W., Xu Y., Kelley M. R., LeDoux S. P., Wilson G. L. (2000) J. Biol. Chem. 275, 37518–37523 [DOI] [PubMed] [Google Scholar]
- 5.Rachek L. I., Grishko V. I., Musiyenko S. I., Kelley M. R., LeDoux S. P., Wilson G. L. (2002) J. Biol. Chem. 277, 44932–44937 [DOI] [PubMed] [Google Scholar]
- 6.Druzhyna N. M., Hollensworth S. B., Kelley M. R., Wilson G. L., LeDoux S. P. (2003) Glia 42, 370–378 [DOI] [PubMed] [Google Scholar]
- 7.Wallace D. C. (2005) Gene 354, 169–180 [DOI] [PubMed] [Google Scholar]
- 8.Fortini P., Pascucci B., Parlanti E., D'Errico M., Simonelli V., Dogliotti E. (2003) Biochimie 85, 1053–1071 [DOI] [PubMed] [Google Scholar]
- 9.de Souza-Pinto N. C., Wilson D. M., 3rd, Stevnsner T. V., Bohr V. A. (2008) DNA Repair 7, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druzhyna N. M., Musiyenko S. I., Wilson G. L., LeDoux S. P. (2005) J. Biol. Chem. 280, 21673–21679 [DOI] [PubMed] [Google Scholar]
- 11.Grishko V., Rachek L., Musiyenko S., LeDoux S. P., Wilson G. L. (2005) Free Radic. Biol. Med. 38, 755–762 [DOI] [PubMed] [Google Scholar]
- 12.Harrison J. F., Hollensworth S. B., Spitz D. R., Copeland W. C., Wilson G. L., LeDoux S. P. (2005) Nucleic Acids Res. 33, 4660–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh K. K. (2006) Ann. N.Y. Acad. Sci. 1067, 182–190 [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Alvarado P., Dagnino-Subiabre A., Paris I., Metodiewa D., Welch C. J., Olea-Azar C., Caviedes P., Caviedes R., Segura-Aguilar J. (2001) Biochem. Biophys. Res. Commun. 283, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 15.Paris I., Martinez-Alvarado P., Perez-Pastene C., Vieira M. N., Olea-Azar C., Raisman-Vozari R., Cardenas S., Graumann R., Caviedes P., Segura-Aguilar J. (2005) J. Neurochem. 92, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 16.Paris I., Lozano J., Cardenas S., Perez-Pastene C., Saud K., Fuentes P., Caviedes P., Dagnino-Subiabre A., Raisman-Vozari R., Shimahara T., Kostrzewa J. P., Chi D., Kostrzewa R. M., Caviedes R., Segura-Aguilar J. (2008) Neurotox. Res. 13, 221–230 [DOI] [PubMed] [Google Scholar]
- 17.Shimoda-Matsubayashi S., Matsumine H., Kobayashi T., Nakagawa-Hattori Y., Shimizu Y., Mizuno Y. (1996) Biochem. Biophys. Res. Commun. 226, 561–565 [DOI] [PubMed] [Google Scholar]
- 18.LeDoux S. P., Shen C. C., Grishko V. I., Fields P. A., Gard A. L., Wilson G. L. (1998) Glia 24, 304–312 [PubMed] [Google Scholar]
- 19.Shokolenko I. N., Alexeyev M. F., LeDoux S. P., Wilson G. L. (2005) DNA Repair 4, 511–518 [DOI] [PubMed] [Google Scholar]
- 20.Chaturvedi P., Eng W. K., Zhu Y., Mattern M. R., Mishra R., Hurle M. R., Zhang X., Annan R. S., Lu Q., Faucette L. F., Scott G. F., Li X., Carr S. A., Johnson R. K., Winkler J. D., Zhou B. B. (1999) Oncogene 18, 4047–4054 [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka S., Huang M., Elledge S. J. (1998) Science 282, 1893–1897 [DOI] [PubMed] [Google Scholar]
- 22.Heideker J., Lis E. T., Romesberg F. E. (2007) Cell Cycle 6, 3058–3064 [DOI] [PubMed] [Google Scholar]
- 23.Meloche S., Pouysségur J. (2007) Oncogene 26, 3227–3239 [DOI] [PubMed] [Google Scholar]
- 24.Tombes R. M., Auer K. L., Mikkelsen R., Valerie K., Wymann M. P., Marshall C. J., McMahon M., Dent P. (1998) Biochem. J. 330, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbott D. W., Holt J. T. (1999) J. Biol. Chem. 274, 2732–2742 [DOI] [PubMed] [Google Scholar]