Abstract
Williams-Beuren syndrome (WBS), an autosomal dominant genetic disorder, is characterized by a unique cognitive profile and craniofacial defects. WBS results from a microdeletion at the chromosomal location 7q11.23 that encompasses the genes encoding the members of TFII-I family of transcription factors. Given that the haploinsufficiency for TFII-I is causative to the craniofacial phenotype in humans, we set out to analyze the effect of post-transcriptional silencing of TFII-I during BMP-2-driven osteoblast differentiation in the C2C12 cell line. Our results show that TFII-I plays an inhibitory role in regulating genes that are essential in osteogenesis and intersects with the bone-specific transcription factor Runx2 and the retinoblastoma protein, pRb. Identification of pathways regulated by TFII-I family transcription factors may begin to shed light on the molecular determinants of WBS.
Introduction
The last decade has brought major progress in our understanding of the molecular basis of osteogenic differentiation. A large number of signaling pathways and transcriptional regulators have been shown to play a role in bone development. However, a precise understanding of the molecular mechanisms for osteogenic differentiation is still lacking, elucidation of which would help us to comprehend the pathogenesis of bone and skeletal diseases and could lead to the development of targeted therapies for them.
Runx2 (also known as Cbfa1, AML-3, PEBP2αA, and Osf2) is a transcription factor belonging to the runt family, which in mammals contains three members: Runx1, Runx2, and Runx3 (1). Runx2 is a key regulator of chondroblast and osteoblast differentiation and of bone development in vivo (2, 3). Targeted disruption of Runx2 in mice resulted in a complete lack of ossification due to the failure of maturation in osteoblasts and resulted in death shortly after birth (4–6). Runx2 regulates the expression of major extracellular matrix genes including alkaline phosphatase, osteopontin, osteocalcin, type I collagen, and bone sialoprotein expressed by chondroblasts and osteoblasts (2, 7).
Findings from Thomas et al. (8) indicate a novel function for Runx2. Runx2 physically interacts with the retinoblastoma protein (pRb),4 which is a tumor suppressor that frequently undergoes loss-of-function mutations in many human cancers including osteosarcoma, retinoblastoma, and small cell lung and bladder carcinomas (9). Runx2 and pRb associate with the promoters of the osteoblast-specific genes osteocalcin and osteopontin in vivo. In reporter assays with osteocalcin (OCN), pRb augmented Runx2-dependent transcription from the OCN promoter (8). More support for the role of pRb in osteogenesis is provided by the finding that recombinant bone morphogenetic protein (BMP-2)-induced osteogenic differentiation is blocked in RB−/− mouse embryonic fibroblasts in comparison with wild type mouse embryonic fibroblasts, as indicated by lower alkaline phosphatase activity, lower osteocalcin mRNA expression, and less intense staining for Alazarin Red, which detects the mineralization of the extracellular matrix of osteoblasts. Taken together these data suggest that pRb acts as a necessary co-activator for Runx2-induced osteogenic differentiation (8).
WBS is a neurodevelopmental disorder with multisystem manifestations, including supravalvular aortic stenosis, hypercalcemia in infancy, mild to moderate mental retardation, and characteristic craniofacial features. Despite the fact that the complex features of WBS are ultimately due to the decreased copy number of several of these genes, the critical downstream biological pathways that alter development and adult function are unknown. Two of these genes, GTF2I and GTF3/GTF2IRD1/WBSCR11, encode vertebrate-specific transcription factors TFII-I and BEN (10–12), which appear to contribute to the craniofacial and cognitive defects observed in WBS (13–16). For instance, in mice, homozygous loss of Gtf2ird1 (BEN) results in craniofacial and cognitive abnormalities reminiscent of human WBS (16, 17). Three genotype-phenotype studies with patients that had an atypical smaller deletion of the telomeric region, containing the cluster of TFII-I family genes, provided a strong correlation between cognitive and craniofacial defects and the heterozygous deletion of GTF2I (TFII-I) and GTF2IRD1 (BEN) (13–15). Interestingly, a patient with only a small deletion of the GTF2IRD1 locus exhibited mild facial dysmorphology of WBS (16), suggesting that the haploinsufficiency for both BEN and TFII-I might be necessary for more severe WBS pathology to occur. However, the molecular basis for the involvement of TFII-I family members in craniofacial defects is currently unknown.
TFII-I is a signal-induced multifunctional transcription factor that activates a number of genes (18–22). However, TFII-I also acts as a repressor (23–25). Given the phenotypic role of TFII-I in WBS pathology, we investigated its potential involvement in osteogenic differentiation using an in vitro differentiation model. Through posttranscriptional silencing of TFII-I, in this study, we uncovered a potential function of TFII-I in regulating genes important for the osteogenic pathway. We show that TFII-I regulates expression of marker genes of osteoblast differentiation: alkaline phosphatase (ALP), which is an early gene, and OCN, which is a late gene. Interestingly, we show that TFII-I functions as a repressor of OCN and ALP gene expression, possibly by preventing the recruitment of the Runx2-pRb complex to the nearby OSE2 site of the OCN promoter. This mechanistic study of TFII-I-mediated regulation of osteocalcin gene via interference of Runx2-pRb function might provide an insight into the molecular mechanism of how the hemizygous deletion of TFII-I can lead to the craniofacial pathology observed in WBS.
MATERIALS AND METHODS
Luciferase Assay
Cos7 cells were transiently transfected with a combination of 600 ng of luciferase reporter construct mOG1.3 containing 1.3 kb of osteocalcin promoter; 0.35 ng of Renilla pRL-TK; and increasing amounts (250, 500, and 1000 ng) of wild type TFII-I (pEBG-TFII-I), FLAG-Runx2, HA-pRb, or pEBG vector to normalize the amount of DNA per condition, using PolyFect transcription reagent (Qiagen) according to the manufacturer's instructions. Transfections and measurements were performed in triplicate. Cells were lysed 48 h after transfection, and relative luciferase and Renilla activities were measured using the Dual-Luciferase assay kit (Promega) according to the manufacturer's instructions. Western blot analysis was performed to confirm the expression of all the proteins.
Generation of TFII-I Knockdown
TFII-I knockdown clones of the C2C12 cell line were established using the same shRNA methodology that was employed to generate TFII-I knockdown in NIH 3T3 cells (18). Several TFII-I knockdown and control (non-silencing shRNA or GFP-shRNA, supplemental Table S2) clonal cell lines were established. The level of knockdown of each clone was assessed using anti-TFII-I rabbit polyclonal Ab (Sigma). Although several TFII-I knockdown clones were tested for ALP activity, TFII-I knockdown clone 15 (KD) was chosen for the present studies.
Quantitative RT-PCR
To test for the levels of gene expression in TFII-KD (KD) versus control (KDC) and wild type parental C2C12 cell line (WT) without BMP-2, the growth medium was replaced with a fresh growth medium containing 10% FBS the day after plating (day 0). Subsequently, the cells were harvested at 24, 72, and 144 h. To test for levels of gene expression in differentiated cells, the KD, KDC, and WT cells were treated with 300 ng/ml recombinant human BMP-2 (rhBMP-2) (R&D Systems) in DMEM with 0.2% FBS and harvested at 24, 48, 72, and 144 h.
A QIAShredder and an RNeasy mini kit (Qiagen) were used to extract RNA from cells. To remove any DNA contamination, the columns were treated with an RNase-free DNase set (Qiagen). 1 μg of total RNA of each sample was used for cDNA synthesis. Relative gene expression of ALP and OCN was measured by real-time PCR using the Power SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's protocol. As an internal control, 18 S gene expression was measured. All reactions were carried out in triplicate. All primers sequences are found in supplemental Table S1. The data were analyzed using GeneAmp® 7300 SDS software. The values for each of the genes were normalized to 18 S, and the -fold change relative to WT value at 24 h was calculated using 2−ΔΔCt as described in Ref. 26.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer's protocol (Upstate Biotechnology) and as described (8, 18, 27). To assess the recruitment of TFII-I, Runx2, or pRb to osteocalcin promoter, the lysates were incubated overnight at 4 °C with no antibody, anti-mouse TFII-I (BD Biosciences), anti-mouse pRb (1 μg each of Ab-4 and Ab-5, Oncogene Science), anti-CBFA1 (2 μg, Abcam), or control antibody IgG (2 μg of anti-rabbit IgG). DNA samples from ChIP were analyzed by quantitative PCR in triplicate using the Taqman gene expression master mix (Applied Biosystems). The sequence of the primers and Taqman probe are provided in supplemental Table S3.
Alkaline Phosphatase Staining
WT, KD, and KDC were plated in 12-well dishes at 2 × 104 cells/well. Cells were left untreated or treated with 300 ng/ml BMP-2 in DMEM containing 0.2% FBS for 6 days. On day 6, cells were fixed in methanol for 2 min at −20 °C and rehydrated in wash buffer (100 mm Tris-HCl (pH 9.5), 100 mm NaCl, 10 mm MgCl2) for 10 min. Then the cells were stained with Fast BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium) (B5655, Sigma) for 15 min and rinsed in double-distilled H2O for 5 min.
Alkaline Phosphatase Activity
Alkaline phosphatase activity was determined as described previously (28). C2C12 cells were grown in 6-well dishes and were either treated with 300 ng/ml BMP-2 in DMEM containing 0.2% serum or left untreated. Alkaline phosphatase activity was expressed as nanomoles of p-nitrophenol generated per microgram of total protein as determined by Bradford assay.
Western Blot
Whole cell lysates were loaded onto 10% SDS-PAGE and subjected to Western blot analysis. The anti-TFII-I polyclonal Ab at 1:2500, anti-GST Ab (Sigma) at 1:3300, anti-green fluorescent protein antibody (JL-8, BD Biosciences) at 1:2000, and anti-FLAG Ab (Sigma) and anti-HA Ab (Sigma) at 1:2500 dilution were used. The anti-β-Actin Ab (Sigma) was used at a 1:5000 dilution. In each case, the secondary anti-mouse or anti-rabbit horseradish peroxidase-linked antibodies were used at a 1:10,000 dilution in Tris-buffered saline with Tween.
RESULTS
TFII-I Knockdown Augments Expression of Osteoblastic Marker Genes
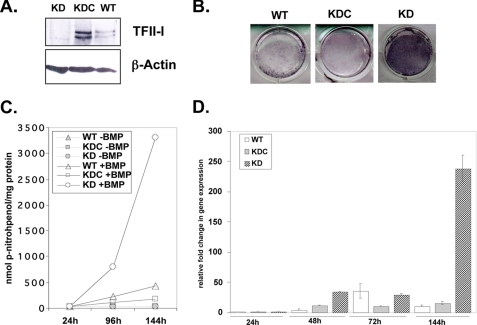
To assess the potential role of TFII-I in osteoblastic differentiation, we employed the C2C12 mesenchymal cell line, which has been widely used as an in vitro differentiation model (30). We stably silenced TFII-I by shRNA in these cells (KD) and obtained significantly lower protein expression as compared with control cells, cells infected with the non-silencing shRNA (KDC), or the uninfected parental wild type cells (WT) (Fig. 1A). We used BMP-2 in C2C12 cells to induce osteoblast differentiation in vitro (29, 30). Stimulation of C2C12 cells with BMP-2 leads to a rise in alkaline phosphatase activity by day 7 (30). We ascertained ALP activity by performing the histochemical staining of the cells with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium. Upon treatment with BMP-2, we found that on day 6 (144 h), TFII-I KD cells showed more intense staining than both WT and KDC lines (Fig. 1B). Six TFII-I KD clones were assessed for this phenotype with similar results (data not shown).
FIGURE 1.
TFII-I knockdown accelerates early osteoblast differentiation in C2C12 cells. A, whole-cell lysates of TFII-I KD, non-silencing control knockdown (KDC), and uninfected C2C12 (WT) cells were probed in a Western blot with anti-TFII-I (upper panel) and with anti-β-Actin antibody as a loading control (lower panel). B, 6 days following the treatment with BMP-2, cells were stained for the presence of ALP activity. One representative experiment out of three experiments is shown. C, C2C12 cell lines were plated in 6-well dishes in triplicate for each condition. Following the treatment with BMP-2 or no treatment, WT, KDC, and KD lysates were collected at 24, 96, and 144 h, and ALP activity was measured. Each data point represents a mean ± S.D. (error bars) of one representative experiment performed in triplicate wells. Three experiments were performed with similar results. D, KD cells show a higher level of ALP gene expression at 144 h than both WT and KDC cell lines by quantitative RT-PCR assay. Data shown are the mean ± S.D. (error bars) of duplicate wells of one representative experiment, performed in duplicate four times with similar results.
To quantify the differences in ALP, we performed an enzymatic assay using p-nitrophenyl phosphate as a substrate of ALP. Consistent with the staining results, we found that TFII-I KD exhibited much higher activity starting at 96 h, and this difference was even more striking at 144 h as compared with the KDC and WT cells (Fig. 1C). These data suggest that TFII-I KD cells undergo early osteoblast differentiation faster and more efficiently in vitro than either KDC or WT cells, indicating that TFII-I may inhibit this differentiation. Furthermore, to assay for the effect of TFII-I KD on gene expression of ALP, we performed quantitative RT-PCR. We found that at 144 h, there was a large increase in the ALP mRNA in the TFII-I KD as compared with the WT and KDC (Fig. 1D), which was consistent with our observation that the TFII-I KD exhibited higher ALP activity (Fig. 1, A and C).
TFII-I Represses Runx2-pRb-induced Transcription of Osteocalcin
To further investigate the effect of TFII-I knockdown during osteogenic differentiation in vitro, we performed quantitative RT-PCR to assay for the expression of the OCN gene, which is a late marker of osteoblast differentiation (29). The OCN gene is exclusively expressed in osteoblasts, and it is responsible for the formation of mineralization, i.e. calcium deposits in the extracellular matrix (2). Strikingly, even without BMP-2 treatment, we found that at 144 h, the TFII-I KD cells showed an increase in OCN gene expression as compared with the KDC and WT samples (Fig. 2A). This indicates that even in the absence of BMP-2 stimulus, silencing of TFII-I alone is sufficient to induce the expression of the late osteoblastic genes.
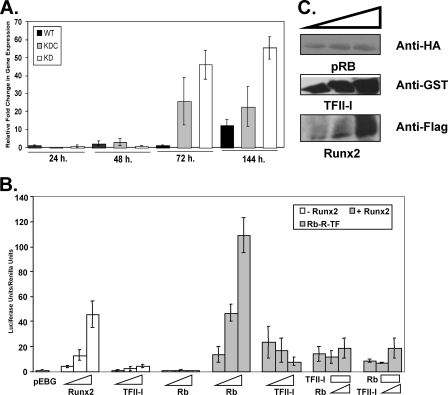
FIGURE 2.
TFII-I inhibits Runx2-Rb-induced transcription of osteocalcin gene. A, KD, KDC, and WT C2C12 cells were seeded in 6-well dishes. The RNA lysates were harvested at the following time points: 24, 48, 72, and 144 h. Quantitative RT-PCR was performed using with the following primers: OCN and 18 S. The data are the mean ± S.D. (error bars) of triplicate wells of one experiment, performed three times with similar results. B, Cos7 cells were transiently transfected in triplicate wells with 1.3 kbp of osteocalcin promoter-luciferase construct (mOG1.3) and Renilla (pRL-TK). The following amounts of expression plasmids were transfected: for TFII-I, 250, 500, and 1000 ng; for Runx2, 250, 500, and 1000 ng; and for Rb, 250, 500, and 1000 ng. pEBG empty vector was used to normalize the amount of DNA. The data are the mean ± S.D. (error bars) of triplicate wells of a representative experiment of four total experiments. C, Western blot analysis was performed to confirm the expression of GST-tagged TFII-I, HA-tagged pRb, and FLAG-tagged Runx2 proteins.
Previous studies have shown that pRb physically interacts with Runx2 and acts as its co-activator by augmenting the Runx2-induced transcription of the OCN promoter (8). The osteocalcin promoter contains three cis-regulatory elements termed A, B, and C that bind osteoblast-specific complexes (31). The well conserved element A, known as osteoblast-specific cis-acting element 1 (OSE1), is located between −74 and −47 in the OCN promoter. It was also shown that OSE1 is present in the promoter of several osteoblast-specific genes including Cbfa1/Runx2. Interestingly, we observed that the OSE1 element overlaps with the classical Inr-like element (YYANWYY) that can bind TFII-I. The presence of this Inr-like sequence in OSE1 suggests that TFII-I might interfere with Runx2-pRb binding to the OSE1 and thus provide a functional basis for the observed enhancement in OCN expression upon silencing of TFII-I. Hence, we tested whether the TFII-I repressive effects are due to inhibition of pRb-Runx2 transcription functions.
For transient transfection reporter assays, we used Cos7 cells because they do not express any endogenous Runx2 protein (32, 33) and have low levels of the endogenous TFII-I protein (34). The ectopic expression of either GST-tagged TFII-I or HA-tagged pRb had no effect on the osteocalcin reporter gene expression (Fig. 2B). Consistent with the published data (38), transfection of increasing concentrations of FLAG-Runx2 construct alone resulted in the dose-dependent activation of the reporter (Fig. 2B). When increasing concentrations of pRb were co-expressed with a fixed amount of Runx2, a dose-dependent increase in the activation of OCN promoter was observed (Fig. 2B). Most importantly, when increasing amounts of TFII-I were co-expressed with a fixed amount of Runx2, TFII-I inhibited the activation of the OCN reporter in a dose-dependent manner (Fig. 2B), indicating that TFII-I interferes with the Runx2-induced activation of the OCN promoter. We co-transfected an increasing amount of TFII-I, together with fixed amounts of pRb and Runx2, and found that even under these conditions, TFII-I inhibited the Runx2-induced activation of the reporter (Fig. 2B). To determine whether pRb counteracts this inhibition, we co-transfected the cells with increasing concentrations of pRb together with a fixed amount of TFII-I and Runx2. We found that TFII-I-mediated repression of the reporter was independent of pRb as TFII-I still repressed Runx2 even in the presence of pRb.
Inverse Recruitment of TFII-I and Runx2-pRb to the Osteocalcin Promoter
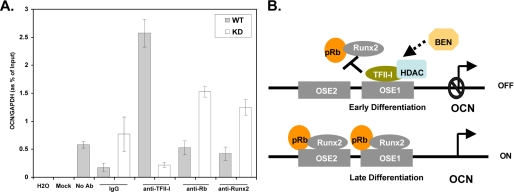
To further establish whether TFII-I competes with the Runx2-pRb complex at the osteocalcin promoter, we performed a ChIP assay. The primers and probe were selected to amplify the proximal regions of the OSE1 that harbors the Inr-like element. We harvested the KD and WT C2C12 cells that were not treated with BMP-2 and immunoprecipitated the DNA-protein complexes with the indicated antibodies. We found that TFII-I was specifically recruited to the proximal promoter of OCN and that in the C2C12 cells, which were not treated with BMP-2, there was a slight recruitment (less than ∼2-fold) of pRb and no recruitment of Runx2 to the promoter (Fig. 3A). Strikingly, upon knockdown of TFII-I, there was a significantly enhanced recruitment of pRb and Runx2 to the promoter (Fig. 3A). Together, these data suggest that TFII-I in the absence of BMP-2 constitutively binds to the proximal region of the osteocalcin promoter, thus hindering the association of Runx2 and pRb with the promoter.
FIGURE 3.
Enhanced recruitment of TFII-I to the OCN promoter results in reduced Runx2-pRb recruitment. A, quantitative ChIP assay of KD and WT cells, where TFII-I, pRb, and Runx2 were immunoprecipitated with anti-TFII-I, anti-HA, and anti-FLAG antibodies, respectively, or rabbit IgG or no antibody as negative controls. The Mock lane is without either lysate or antibody and serves as a negative control for cross-contamination. The quantitative PCR was performed in triplicate with primers and probe for OCN promoter and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The data are the mean ± S.D. (error bars) of triplicate wells of one representative experiment out of three. B, proposed model. Constitutive recruitment TFII-I to the OSE1 site, containing the Inr-like element, interferes with the recruitment of Runx2 and pRb to the OSE1 site, and given the resolution of the ChIP assay, potentially to the nearby OSE2, which also contains the Runx2 binding site, resulting in the inhibition of pRb-Runx2-induced transcriptional activation of the OCN gene during early differentiation. TFII-I may also recruit HDACs to the promoter, thereby further contributing to the transcriptional inhibition. At the later stage of osteoblast differentiation, TFII-I is displaced from the promoter (by an as yet unknown mechanism), resulting in relief of inhibition. BEN may also potentially contribute to the inhibitory effect.
DISCUSSION
Our study has revealed an unexpected role of TFII-I as a potential negative regulator of osteoblast differentiation and of the Runx2-induced transcription of the osteocalcin gene, crucial in bone development. We investigated the effect of silencing of TFII-I function by creating a stable knockdown of TFII-I with shRNA in the mesenchymal precursor C2C12 cell line that differentiates into osteoblasts in response to BMP-2 signaling in vitro (30). In this in vitro system, the knockdown of TFII-I results in high levels of ALP activity relative to the wild type (uninfected) and non-silencing shRNA controls in response to BMP-2 treatment (Fig. 1, A and B). The amount of ALP activity is a hallmark for the extent of early osteoblast differentiation. Thus, this phenotype suggests an inhibitory function of TFII-I in early osteoblast differentiation. To eliminate clonal variability, these results were also confirmed in multiple clones of TFII-I knockdowns (data not shown).
The potential importance of TFII-I in the osteogenic pathway is further underscored by our findings that TFII-I down-regulates both early (ALP) and late (OCN) genetic markers of osteoblast differentiation (Figs. 1 and 2). Using quantitative RT-PCR, we showed that remarkably, even without BMP-2 treatment, the gene expression of OCN was significantly up-regulated upon the knockdown of TFII-I relative to the wild type controls (Fig. 3A). Further evidence in support of our proposed model (Fig. 3B) is provided by real-time ChIP results. Thus, knockdown of TFII-I in C2C12 cells results in more recruitment of Runx2 and pRb to the promoter relative to the wild type control, whereas in the wild type control, there was no recruitment of Runx2 and little recruitment of pRb (Fig. 3A). Importantly, we showed that TFII-I was recruited to the promoter in the wild type cells but not in the knockdown cells.
Although the transcriptional effects of TFII-I could be solely due to its function to prevent Runx2-pRb from binding to the OSE sites, considering that TFII-I was shown to physically interact with HDAC3 (35), it is also possible that TFII-I may be recruiting HDAC3 to the promoter, thereby contributing to the observed inhibitory effect. Future experiments could assess whether HDAC3 is also recruited to the same region of the OCN promoter. If TFII-I prevents the recruitment of Runx2-pRb to the osteocalcin promoter, thus inhibiting the transcription of OCN, then how is it possible that wild type osteoblasts still express OCN and are still able to mineralize by forming calcium deposits in the extracellular matrix? Although the precise answer to this question remains untested, it is possible that the interplay between various signaling pathways in late osteoblast development relieves the inhibition of TFII-I, allowing more Runx2-Rb to be recruited to the promoter to activate OCN.
Because haploinsufficiency in TFII-I is associated with the characteristic craniofacial and neurodevelopmental pathologies of WBS, it is of considerable interest to elucidate the molecular pathways that TFII-I control. Moreover, heterozygous deletion in the Gtf2i gene in mice exhibits craniofacial and general osteogenic defects (36). Although the precise mechanistic role for TFII-I in osteogenic differentiation is currently unknown, the repression of osteogenic markers OCN and ALP by TFII-I reported here and the inhibitory role of TFII-I in blocking Runx2 and pRb recruitment to the OCN promoter suggest evidence for a link between TFII-I and Runx2, which is essential for bone development because the loss-of-function of Runx2 leads to cleidocranial dysplasia (5, 37). Therefore, a thorough analysis of the function of Runx2-pRb and TFII-I in cells derived from bone-specific conditional knock-out mice of these proteins might help to further elucidate the direct role of TFII-I in craniofacial development and potentially in the pathogenesis of WBS.
This work was supported, in whole or in part, by National Institutes of Health Grants AG020208 (to P. H.) and HD04603 (to A. L. R.)

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
- pRb
- retinoblastoma protein
- WBS
- Williams-Beuren syndrome
- ALP
- alkaline phosphatase
- OCN
- osteocalcin
- OSE
- osteoblast-specific cis-acting element 1
- HDAC
- histone deacetylase
- shRNA
- short hairpin RNA
- FBS
- fetal bovine serum
- HA
- hemagglutinin
- ChIP
- chromatin immunoprecipitation
- DMEM
- Dulbecco's modified Eagle's medium
- Ab
- antibody
- GST
- glutathione S-transferase
- RT-PCR
- real-time PCR
- KD
- knockdown
- KDC
- knockdown control
- WT
- wild type
- Inr
- Initiator.
REFERENCES
- 1.van Wijnen A. J., Stein G. S., Gergen J. P., Groner Y., Hiebert S. W., Ito Y., Liu P., Neil J. C., Ohki M., Speck N. (2004) Oncogene 23, 4209–4210 [DOI] [PubMed] [Google Scholar]
- 2.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 3.Fujita T., Azuma Y., Fukuyama R., Hattori Y., Yoshida C., Koida M., Ogita K., Komori T. (2004) J. Cell Biol. 166, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 5.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 6.Choi J. Y., Pratap J., Javed A., Zaidi S. K., Xing L., Balint E., Dalamangas S., Boyce B., van Wijnen A. J., Lian J. B., Stein J. L., Jones S. N., Stein G. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee C., McCabe L. R., Choi J. Y., Hiebert S. W., Stein J. L., Stein G. S., Lian J. B. (1997) J. Cell. Biochem. 66, 1–8 [DOI] [PubMed] [Google Scholar]
- 8.Thomas D. M., Carty S. A., Piscopo D. M., Lee J. S., Wang W. F., Forrester W. C., Hinds P. W. (2001) Mol. Cell 8, 303–316 [DOI] [PubMed] [Google Scholar]
- 9.Tiemann F., Hinds P. W. (1998) EMBO J. 17, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinsley T. A., Cunliffe P., Tipney H. J., Brass A., Tassabehji M. (2004) Protein Sci. 13, 2588–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy A. L. (2001) Gene 274, 1–13 [DOI] [PubMed] [Google Scholar]
- 12.Pérez Jurado L. A., Wang Y. K., Peoples R., Coloma A., Cruces J., Francke U. (1998) Hum. Mol. Genet 7, 325–334 [DOI] [PubMed] [Google Scholar]
- 13.Morris C. A., Mervis C. B., Hobart H. H., Gregg R. G., Bertrand J., Ensing G. J., Sommer A., Moore C. A., Hopkin R. J., Spallone P. A., Keating M. T., Osborne L., Kimberley K. W., Stock A. D. (2003) Am. J. Med. Genet. A 123A, 45–59 [DOI] [PubMed] [Google Scholar]
- 14.Hirota H., Matsuoka R., Chen X. N., Salandanan L. S., Lincoln A., Rose F. E., Sunahara M., Osawa M., Bellugi U., Korenberg J. R. (2003) Genet Med. 5, 311–321 [DOI] [PubMed] [Google Scholar]
- 15.Gagliardi C., Bonaglia M. C., Selicorni A., Borgatti R., Giorda R. (2003) J. Med. Genet 40, 526–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tassabehji M., Hammond P., Karmiloff-Smith A., Thompson P., Thorgeirsson S. S., Durkin M. E., Popescu N. C., Hutton T., Metcalfe K., Rucka A., Stewart H., Read A. P., Maconochie M., Donnai D. (2005) Science 310, 1184–1187 [DOI] [PubMed] [Google Scholar]
- 17.Young E. J., Lipina T., Tam E., Mandel A., Clapcote S. J., Bechard A. R., Chambers J., Mount H. T., Fletcher P. J., Roder J. C., Osborne L. R. (2008) Genes Brain Behav. 7, 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desgranges Z. P., Ahn J., Lazebnik M. B., Ashworth T., Lee C., Pestell R. C., Rosenberg N., Prives C., Roy A. L. (2005) Mol. Cell. Biol. 25, 10940–10952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakre S., Tussie-Luna M. I., Ashworth T., Novina C. D., Settleman J., Sharp P. A., Roy A. L. (2006) Mol. Cell 24, 301–308 [DOI] [PubMed] [Google Scholar]
- 20.Hong M., Lin M. Y., Huang J. M., Baumeister P., Hakre S., Roy A. L., Lee A. S. (2005) J. Biol. Chem. 280, 16821–16828 [DOI] [PubMed] [Google Scholar]
- 21.Cheriyath V., Desgranges Z. P., Roy A. L. (2002) J. Biol. Chem. 277, 22798–22805 [DOI] [PubMed] [Google Scholar]
- 22.Grueneberg D. A., Henry R. W., Brauer A., Novina C. D., Cheriyath V., Roy A. L., Gilman M. (1997) Genes Dev. 11, 2482–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogura Y., Azuma M., Tsuboi Y., Kabe Y., Yamaguchi Y., Wada T., Watanabe H., Handa H. (2006) Genes Cells 11, 373–381 [DOI] [PubMed] [Google Scholar]
- 24.Crusselle-Davis V. J., Vieira K. F., Zhou Z., Anantharaman A., Bungert J. (2006) Mol. Cell. Biol. 26, 6832–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crusselle-Davis V. J., Zhou Z., Anantharaman A., Moghimi B., Dodev T., Huang S., Bungert J. (2007) FEBS J. 274, 6065–6073 [DOI] [PubMed] [Google Scholar]
- 26.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27.Lazebnik M. B., Tussie-Luna M. I., Roy A. L. (2008) J. Biol. Chem. 283, 11078–11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asahina I., Sampath T. K., Hauschka P. V. (1996) Exp. Cell Res. 222, 38–47 [DOI] [PubMed] [Google Scholar]
- 29.Peng Y., Kang Q., Luo Q., Jiang W., Si W., Liu B. A., Luu H. H., Park J. K., Li X., Luo J., Montag A. G., Haydon R. C., He T. C. (2004) J. Biol. Chem. 279, 32941–32949 [DOI] [PubMed] [Google Scholar]
- 30.Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., Suda T. (1994) J. Cell Biol. 127, 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducy P., Karsenty G. (1995) Mol. Cell. Biol. 15, 1858–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou G., Zheng Q., Engin F., Munivez E., Chen Y., Sebald E., Krakow D., Lee B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19004–19009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ducy P., Starbuck M., Priemel M., Shen J., Pinero G., Geoffroy V., Amling M., Karsenty G. (1999) Genes Dev. 13, 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheriyath V., Novina C. D., Roy A. L. (1998) Mol. Cell. Biol. 18, 4444–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tussié-Luna M. I., Bayarsaihan D., Seto E., Ruddle F. H., Roy A. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enkhmandakh B., Makeyev A. V., Erdenechimeg L., Ruddle F. H., Chimge N. O., Tussie-Luna M. I., Roy A. L., Bayarsaihan D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou G., Chen Y., Zhou L., Thirunavukkarasu K., Hecht J., Chitayat D., Gelb B. D., Pirinen S., Berry S. A., Greenberg C. R., Karsenty G., Lee B. (1999) Hum. Mol. Genet 8, 2311–2316 [DOI] [PubMed] [Google Scholar]
- 38.Lee J. S., Thomas D. M., Gutierrez G., Carty S. A., Yanagawa S., Hinds P. W. (2006) J. Bone Miner. Res. 21, 921–933 [DOI] [PubMed] [Google Scholar]