Abstract
Post-translational modification of protein cysteine residues is emerging as an important regulatory and signaling mechanism. We have identified numerous putative targets of redox regulation in the unicellular green alga Chlamydomonas reinhardtii. One enzyme, isocitrate lyase (ICL), was identified both as a putative thioredoxin target and as an S-thiolated protein in vivo. ICL is a key enzyme of the glyoxylate cycle that allows growth on acetate as a sole source of carbon. The aim of the present study was to clarify the molecular mechanism of the redox regulation of Chlamydomonas ICL using a combination of biochemical and biophysical methods. The results clearly show that purified C. reinhardtii ICL can be inactivated by glutathionylation and reactivated by glutaredoxin, whereas thioredoxin does not appear to regulate ICL activity, and no inter- or intramolecular disulfide bond could be formed under any of the conditions tested. Glutathionylation of the protein was investigated by mass spectrometry analysis, Western blotting, and site-directed mutagenesis. The enzyme was found to be protected from irreversible oxidative inactivation by glutathionylation of its catalytic Cys178, whereas a second residue, Cys247, becomes artifactually glutathionylated after prolonged incubation with GSSG. The possible functional significance of this post-translational modification of ICL in Chlamydomonas and other organisms is discussed.
Introduction
The importance of redox regulation and redox signaling linked to post-translational modifications of cysteine residues of proteins starts to be widely recognized. Indeed, cysteine residues can undergo different states of oxidation, such as sulfenic, sulfinic, and sulfonic acid, but also protein disulfide bridges (intra- or intermolecular), S-thiolation (mainly glutathionylation), or nitrosylation. These post-translational redox modifications are mainly under the control of two types of ubiquitous disulfide oxidoreductases: thioredoxins (TRXs)2 and glutaredoxins (GRXs).
Oxido-reduction of intra- or interprotein disulfide bridges is probably the most extensively studied redox modification. It is mainly controlled by TRXs, which play a major role in redox signaling and oxidative stress responses. In photosynthetic organisms, different approaches led to the identification of ∼400 putative TRX targets implicated in nearly all cell processes (1, 2).
Besides TRX-dependent regulation of the redox state of protein disulfide bonds, glutathionylation has recently emerged, among other thiol-based post-translational modifications, as an important redox-based signaling mechanism (3–5). This modification consists in the formation of a mixed disulfide between an accessible free thiol on a protein and a molecule of glutathione. In mammals, glutathionylation occurs under oxidative stress conditions and can protect specific cysteine residues from irreversible oxidation but can also modulate, either positively or negatively, the activity of numerous proteins. Although the precise mechanisms leading to glutathionylation in vivo is still unknown, the reverse reaction, named deglutathionylation, is catalyzed by GRX, proteins belonging to the thioredoxin superfamily (6–8).
We have identified numerous putative targets of redox regulation in the unicellular green alga Chlamydomonas reinhardtii, a convenient model organism for the study of basic biological processes, including photosynthesis and eukaryotic flagella and basal body functions (9). A proteomic approach, based on the use of a monocysteinic TRX affinity columns, allowed the identification of 55 putative TRX targets in Chlamydomonas total extracts (10). More recently, we employed a strategy based on radiolabeling of the glutathione pool by [35S]cysteine to identify 25 proteins undergoing S-thiolation in vivo in Chlamydomonas cells (11). One enzyme, isocitrate lyase (ICL), was identified both as a putative TRX target and as an S-thiolated protein in vivo.
ICL (EC 4.1.3.1) catalyzes the first committed step of the glyoxylate cycle. The function of the glyoxylate cycle was first proposed by Kornberg and Krebs (12) to account for microbial growth on two-carbon compounds as a sole carbon source. In oil-rich seed plants, the glyoxylate cycle is used during seedling development to convert lipids to carbohydrates in the growing seedling (13). In this cycle, ICL cleaves a C–C bond of isocitrate to form glyoxylate and succinate, and malate synthase uses glyoxylate and acetyl-CoA to form malate. These two enzymes, specific of the glyoxylate cycle, are widely distributed among microorganisms, including bacteria, protozoa, fungi, and algae (14). In most eukaryotic organisms, ICL is localized in glyoxysomes, a specialized form of peroxisome.
ICL has been isolated and characterized from a variety of sources, and biochemical studies have shown that the enzymes from different species have similar catalytic properties and kinetic mechanisms (15). In C. reinhardtii, ICL allows growth on acetate as a sole carbon source, and its expression was shown to be regulated by light at the mRNA level (16). The activity of C. reinhardtii ICL (CrICL) could be increased in total soluble extracts by TRX-mediated reduction, suggesting that the enzyme might contain cysteine residues involved in the formation of a regulatory disulfide (10). Moreover, purified recombinant ICL was shown to undergo glutathionylation in vitro in the presence of GSSG, which promoted formation of two glutathione adducts on the protein with a concomitant loss of enzyme activity (11). These results strongly suggest that CrICL, and therefore the assimilation of acetate in Chlamydomonas, are redox-regulated. The existence of two redox active cysteines has also been suggested for Phycomyces blakasleeanus ICL (17). In the case of CrICL, it is clear that the enzyme contains one or more redox-sensitive cysteines whose oxidation state could affect protein activity. However, the precise molecular mechanisms of this redox regulation of ICL as well as the role of thiol/disulfide interchange and/or glutathionylation in this regulation remain unclear.
In this work, we analyzed the effects of different types of oxidizing molecules, such as GSSG, oxidized DTT, or H2O2, on Chlamydomonas ICL and determined the regulatory role of TRXs and GRXs. Moreover, we investigated the glutathionylation using activity assays, mass spectrometry analyses, site-directed mutagenesis, and Western blotting. The results presented detail the molecular mechanism of redox regulation of Chlamydomonas ICL and clarify the type of redox post-translational modifications involved and their effect on the enzyme activity.
EXPERIMENTAL PROCEDURES
Materials/Chemicals
NAP-5 columns and 5,5′-dithiobis-2-nitrobenzoic acid were purchased from GE Healthcare and Pierce, respectively. GSH, GSSG, trypsin, chymotrypsin, and Glu-C endoproteases were from Roche Applied Science. All of the other reagents were from Sigma.
Biotinylation of GSSG
A water-soluble biotinylation reagent, EZ link sulfosuccinimidobiotin (Sulfo-NHS-Biotin, Perbio Science (Cramlington, UK)), was used to generate biotinylated oxidized glutathione (BioGSSG) by coupling biotin to the primary amino groups of glutathione disulfide (Sigma) under mild alkaline conditions. Specifically, the biotinylation reagent (50 μl, 48 mm) was added to GSSG (50 μl, 32 mm) in 50 mm potassium phosphate buffer (pH 7.2), and the mixture was left to derivatize for 1 h at room temperature. After incubation, any remaining biotinylation reagent was quenched by adding 35 μl of 0.6 m ammonium carbonate buffer (NH4HCO3).
Production and Purification of Recombinant Proteins
Production and purification of recombinant cytosolic GRX1 and TRXh1 from C. reinhardtii and NADPH-thioredoxin reductase b from Arabidopsis thaliana were previously reported (6, 18, 19). Recombinant Chlamydomonas ICL was produced and purified as previously described (11). Briefly, the pET-3c-HIS/Escherichia coli BL21 expression system was used. Protein expression was induced at 27 °C for 16 h by the addition of 200 μm isopropyl-β-d-thiogalactopyranoside to exponentially growing cells. The recombinant protein was purified by affinity chromatography using an Ni2+ HiTrap chelating resin (HIS-Select® nickel affinity gel, Sigma) according to the manufacturer's instructions. The molecular mass and purity of the protein were analyzed by SDS-PAGE and Coomassie Blue staining after dialysis against 50 mm HEPES-NaOH buffer, pH 7.2. The protein concentration was determined spectrophotometrically using a molar extinction coefficient at 280 nm of 64,540 m−1·cm−1. The resulting homogeneous protein was stored at −20 °C. The specific activity of purified ICL was 9.5 ± 0.2 μmol·min−1·mg−1, a value within the upper range reported for ICL from diverse sources (20).
Site-directed Mutagenesis and Construction of Single, Double, and Triple Mutants of ICL
A total of four cysteine residues in ICL, at positions 165, 178, 247, and 301, were altered by PCR site-directed mutagenesis using as a template the pET-3c-HIS/ICL vector previously obtained (11). The couples of primers listed in supplemental Table 1 were used according to the procedures of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The C165S/C247S double mutant of ICL was obtained by PCR using the primers for Cys247 mutagenesis and the plasmid containing the C165S mutation as template. The ICL triple mutant vector C165S/C247S/C301S was obtained by subcloning the NcoI/BamHI region of the C301S vector into the C165S/C247S plasmid. All mutations were confirmed by sequencing. The recombinant plasmids were introduced into E. coli BL21 (DE3) for overexpression, and production and purification procedures were performed as described above for the wild type (WT) protein.
Enzymatic Assay
ICL activity was assayed according to Michelet et al. (11). Briefly, the reaction mixture contained 50 mm HEPES-NaOH, pH 7.2, 10 mm MgCl2, 4 mm threo-dl-isocitrate, 4 mm phenylhydrazine, and enzyme in a final volume of 1 ml. Isocitrate cleavage was measured by the change in absorbance at 324 nm associated with the formation of glyoxylate phenylhydrazone (molar extinction coefficient at 324 nm = 17,000 m−1·cm−1).
Inactivation of ICL by Oxidant Treatments
Before each treatment, recombinant WT ICL and its variants were treated with 20 mm reduced dithiothreitol (DTTred) for 1 h at room temperature and desalted on a NAP-5 Sephadex G-25 column equilibrated with 50 mm HEPES-NaOH, pH 7.2. Inactivation treatments were performed at room temperature by treating 10 μm recombinant protein with oxidized DTT (10 mm alone or in the presence of 10 μm TRXh1), GSSG (5 mm), H2O2 (1 or 0.1 mm), or 0.1 mm H2O2 in the presence of 0.5 mm GSH. At the indicated times, aliquots were withdrawn in order to assay enzyme activity monitored as described above. The reversibility of the different treatments was assessed by incubating for 20 min the treated enzymes in the presence 20 mm DTTred at room temperature.
Reactivation of ICL in the Presence of TRXh1 or GRX1
To prepare inactive proteins, WT ICL and its variants were treated with 5 mm GSSG for 16 h at room temperature in 50 mm HEPES-NaOH (pH 7.2) and extensively dialyzed at 4 °C against the same buffer. All reactivation treatments were performed in 50 mm HEPES-NaOH, pH 7.2, at room temperature. Reaction mixtures contained ICL (10 μm) and DTTred (20 or 0.2 mm) or 5 mm GSH. ICL reactivation was also tested in the presence of TRXh1 (20 μm) supplemented with either 0.2 mm DTTred or 0.3 mm NADPH and 0.22 μm A. thaliana NADPH-thioredoxin reductase b. The reactivation of ICL was also determined in the presence of GRX1 (1 μm) supplemented with either 0.2 mm DTTred or 5 mm GSH with or without the addition of 0.2 mm NADPH and 6 μg/ml yeast glutathione reductase. At the indicated times, aliquots were withdrawn in order to assay enzyme activity monitored as described above.
In Vitro Glutathionylation of WT ICL and Its Variants Using BioGSSG
The prereduced proteins were incubated in 100 mm Tris-HCl (pH 7.9) in the presence of 2 mm BioGSSG. After a 1-h incubation, BioGSSG-treated samples were alkylated in the presence of 100 mm iodoacetamide (IAM) or treated with 20 mm DTTred for 30 min to assess the reversibility of the reaction. Control samples were incubated with 100 mm IAM for 30 min in the dark prior to incubation in the presence of 2 mm BioGSSG. All treatments were performed at room temperature. Proteins were then loaded on non-reducing SDS-PAGE and analyzed by Western blot using monoclonal anti-biotin antibodies (1:5000 dilution; Sigma). Signals were visualized by enhanced chemiluminescence (21).
Titration of Free Sulfhydryl (-SH) Groups
ICL (50 μm) was reduced with 20 mm DTTred for 1 h at room temperature and desalted on NAP-5 columns equilibrated with 50 mm HEPES-NaOH, pH 7.2. Reduced ICL was subsequently treated with 5 mm GSSG for 5 h at room temperature, followed by a dialysis against 50 mm HEPES-NaOH buffer, pH 7.2. The number of free thiols in prereduced or GSSG-treated samples was determined spectrophotometrically under non-denaturing conditions with 5,5′-dithiobis-2-nitrobenzoic acid (22). Briefly, 5–10 μm protein was added to a solution containing 200 μm 5,5′-dithiobis-2-nitrobenzoic acid in 30 mm Tris-HCl, pH 7.9. After 10 min at room temperature, the absorbance at 412 nm was determined. A molar extinction coefficient of 14,150 m−1·cm−1 was used to calculate the number of titrated sulfhydryl groups.
Analysis of Reduced or Glutathionylated ICL by CD
The effect of ICL glutathionylation on enzyme secondary structure was examined by CD. Measurements were carried out on a 202 AVIV Associates spectrometer (Lakewood, NJ) using a semimicro quartz rectangular 1 × 10 × 40-mm cuvette. ICL samples (10 μm in 5 mm HEPES-NaOH, pH 7.2) were maintained at 22 °C. Spectra were recorded while scanning in the far-UV region (190–260 nm), with bandwidth of 1.0 nm, step size of 1.0 nm, integration time of 30 s, and three repeats. The output of the CD spectrometer was converted into molar ellipticity units (degrees·cm2·dmol−1) based on the protein concentration, amino acid content, and cuvette thickness.
MALDI-TOF Mass Spectrometry
For molecular mass determinations, WT ICL and its variants were reduced as described previously. Prereduced proteins were treated for 1, 5, and 24 h at room temperature with 5 mm GSSG. At the indicated times, MALDI-TOF mass spectrometry analyses were performed before and after treatment in the presence of 20 mm DTTred for 30 min at room temperature. For that purpose, 1 μl of protein solution was mixed with 1.5 μl of a saturated solution of sinapinic acid in 30% acetonitrile containing 0.3% trifluoroacetic acid, and 1.5 μl of this premix was deposited onto the sample plate and allowed to dry under a gentle air stream at room temperature. Mass determination of ICL and its variants were carried out on whole proteins as described (23).
Determination of glutathionylated cysteines was performed after overnight proteolytic cleavage of ICL treated with 5 mm GSSG for 24 h and dialyzed. 10 μl of glutathionylated ICL were mixed for 20 min at 20 °C in the dark with 2 μl of 80 mm IAM and 2 μl of 500 mm ammonium bicarbonate. 13 mg of urea were also added, and incubation was prolonged for 20 min under the same conditions. Urea was partially removed by precipitation at 0 °C for 10 min. For acidic digestion, 3 μl of supernatant were mixed with 4.5 μl of 250 mm ammonium acetate (pH 4.0) and 2 μl of Glu-C endoproteinase at a concentration of 100 ng/μl in double-distilled H2O. All volumes were adjusted to 12 μl with double-distilled H2O. For peptide mass fingerprints, 1 μl of digests, before and after 10 mm DTTred treatment at alkaline pH, was mixed with 9 μl of a half-saturated solution of α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile containing 0.3% trifluoroacetic acid. 1.5 μl of this final solution was deposited onto the sample plate and allowed to dry under a gentle air stream at room temperature. Mass spectra were acquired as described previously (24).
Replicates
All of the results reported are representative of at least three independent experiments and expressed as mean ± S.D.
RESULTS
Sequence and Phylogenetic Analysis of Chlamydomonas ICL
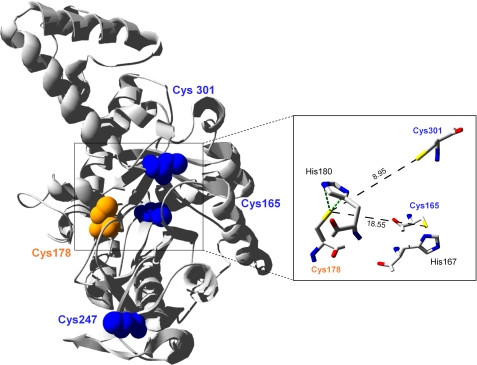
Multiple sequence alignments and phylogenetic analyses indicate that CrICL is more closely related to bacterial ICL than to higher plant ICL (supplemental Fig. 1). The sequence of Chlamydomonas ICL shares higher sequence identity with the E. coli enzyme (61%) than with its A. thaliana counterpart (31%). Two major groups can be distinguished in the ICL family, as previously observed (25, 26). One corresponds to bacterial-type ICLs and includes the Chlamydomonas enzyme, whereas the second group corresponds to eukaryotic-type ICLs and contains sequences from organisms such as higher plants or yeast. Moreover, ICL sequences from this second group differ from those of Chlamydomonas and bacteria by an insertion of ∼100 amino acids. Higher plants ICLs are localized to peroxisomes and harbor a peroxisomal targeting signal (PTS). CrICL does not present any obvious PTS1 or PTS2 sequence and is therefore considered to be localized in the cytosol (16). Similarly, in the yeast Saccharomyces cerevisiae, ICL does not contain any PTS, and the protein was demonstrated, by fractionation and GFP fusion, to be only located in the cytoplasm and not associated with organelles, even under growth conditions that induce peroxisome proliferation (27, 28). CrICL contains four cysteines, two of which (Cys165 and Cys247) are unique to Chlamydomonas (supplemental Fig. 1A). Cys301 is only conserved in bacterial-type ICLs, whereas Cys178 is conserved in all organisms, consistent with its role in catalysis (29, 30) Because two cysteine residues of ICL have been previously suggested to be sensitive to redox modifications (10, 11, 17), based on sequence conservation, Cys178 and Cys301 appear as the most likely candidates. Three-dimensional modeling of CrICL suggests that these two residues are the two closest cysteines in the enzyme but are too distant to allow formation of a disulfide bridge (Fig. 1). The analysis of available ICL structures also suggests that the distance between cysteines of different monomers is not compatible with the formation of intermolecular disulfide bonds. In addition, two strictly conserved histidine residues, His167 and His180, which have been shown to be important for the activity of the E. coli enzyme (31), are located in proximity of Cys165 and Cys178 and could enhance the reactivity of these cysteines.
FIGURE 1.
Three-dimensional modeling of C. reinhardtii ICL. Modeling was made using Swiss-Model workspace (available on the World Wide Web) based on the known structure of ICL from Mycobacterium tuberculosis (Protein Data Bank code 1F61). The structure was generated with the Swiss-PDB viewer software and rendered with POV-Ray (available on the World Wide Web). The catalytic cysteine 178 appears in orange, and the three other cysteines are shown in blue. Inset, black broken lines represent the distance between sulfur atoms (in Å). Two histidine residues potentially important for ICL reactivity (His167 and His180) and located near Cys165 and Cys178 are also represented.
Effect of Oxidized and Reduced DTT on ICL Activity
We have previously identified CrICL among putative TRX targets retained on a monocysteinic TRX affinity column, and reduced TRX was found to increase ICL activity in total extracts (10). This suggested the existence of a TRX-reducible disulfide bond in CrICL whose reduction would increase ICL activity. In order to investigate the formation of a disulfide bond in recombinant Chlamydomonas ICL, the enzyme was prereduced with 20 mm DTTred in 50 mm Hepes-NaOH (pH 7.2), desalted in the same buffer using an NAP-5 column, and subsequently treated with different oxidizing molecules. Non-reducing SDS-PAGE revealed that no intermolecular disulfide bond could be formed after oxidant treatment (data not shown). When the reduced protein was incubated for 30 min in the presence of 20 mm oxidized DTT alone or in the presence of cytosolic TRXh1, no inhibition of ICL activity was observed compared with untreated samples (data not shown). These results suggest that no disulfide bond can be formed in the presence of oxidized DTT alone or supplemented with TRXh1.
Glutathionylation of ICL in the Presence of GSSG or GSH Plus Hydrogen Peroxide
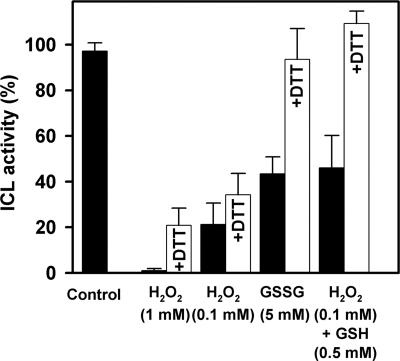
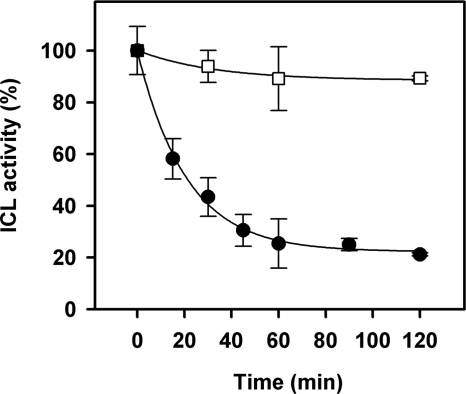
We have previously observed that a prolonged incubation of ICL for 24 h in the presence of 5 mm GSSG resulted in a complete inactivation of the enzyme, which could be reverted by DTTred (11). Here, we have explored the kinetics of GSSG inactivation of ICL. CrICL was incubated in the presence of 5 mm GSSG for 30 min, and protein activity was assayed before and after DTTred treatment as described above for other oxidizing molecules. As shown in Fig. 2, ICL activity was inhibited to 50% and fully reverted by DTTred. In order to better understand the kinetics of inactivation, CrICL was incubated with 5 mm GSSG, and the activity was followed for 2 h (Fig. 3). In these conditions, a loss of catalytic activity was observed, and after 1 h of incubation, CrICL retained 20% of its initial activity, decreasing further to 10% after 2 h.
FIGURE 2.
Effect of oxidative treatments on CrICL activity. Reduced CrICL (control) was incubated for 30 min in the presence of different oxidants as indicated. The reversibility of CrICL inactivation was assessed by incubation in the presence of 20 mm DTT (+DTT). Data are represented as mean percentage of maximal activity ± S.D. (n = 3–5). 100% corresponds to the initial activity of reduced ICL.
FIGURE 3.
Kinetics of inactivation of CrICL by GSSG. Prereduced CrICL (10 μm) was incubated in the presence (closed circles) or absence (open squares) of 5 mm GSSG. Aliquots were withdrawn at the indicated times to assay enzyme activity. Activities are represented as mean percentage ± S.D. (n = 3–5) of the initial activity measured before treatments.
Besides thiol disulfide exchange mediated by GSSG, it has been reported that protein glutathionylation can also be achieved in the presence of GSH and oxidants, such as H2O2, conditions promoting the conversion of protein thiols into sulfenic acids, which then react with GSH to give rise to mixed disulfides (32–34). Incubation of CrICL with 0.1 mm H2O2 and 0.5 mm GSH for 30 min resulted in a decrease of protein activity comparable with that obtained in the presence of GSSG (Fig. 2). The addition of DTTred to H2O2/GSH-treated CrICL provided full reactivation of enzyme activity as observed for the GSSG-treated enzyme. By contrast, the exposure of CrICL to 1 mm H2O2 for 30 min resulted in an almost complete inactivation of the enzyme, whereas in the presence of a 10-fold lower concentration of H2O2 (0.1 mm), CrICL retained 20% of total activity after 30 min of incubation (Fig. 2). This inactivation could not be significantly reverted by incubation with 20 mm DTTred for 20 min. This irreversible inactivation is most likely to primary oxidation of one or several cysteine residues to sulfenic acid (-SOH), which can be further oxidized to irreversible states (i.e. sulfinic (-SO2H) and sulfonic acids (-SO3H)). The full recovery observed for H2O2/GSH-treated CrICL suggests that GSH, reacting with sulfenic acid, can protect the protein from H2O2-mediated irreversible inactivation by glutathionylation. However, we cannot exclude the possibility that glutathionylation of CrICL in the presence of GSSG or GSH plus H2O2 might constitute an intermediary step leading to disulfide formation, as observed for a soybean tyrosine-specific protein phosphatase (35).
Reactivation of GSSG-treated ICL in the Presence of TRXh1 or GRX1
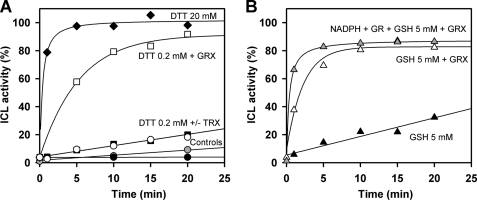
It is now well established that GRXs are very efficient glutathionyl mixed disulfide reductases, whereas TRXs are more specific for reduction of inter- or intraprotein disulfide bonds and much less efficient than GRXs to catalyze deglutathionylation reactions (6, 36, 37). Therefore, we tested the reactivation of GSSG-treated CrICL in the presence of GRX1 and TRXh1, the major cytosolic isoforms of TRX and GRX in Chlamydomonas, using DTTred as reductant. CrICL was incubated with 5 mm GSSG until no residual activity was detected and dialyzed overnight against HEPES-NaOH (pH 7.2). Reactivation was assessed in the presence of 20 mm DTTred or 0.2 mm DTTred alone or supplemented with TRXh1 or GRX1. Treatment with 20 mm DTTred fully restored protein activity within 5 min, whereas 0.2 mm DTTred only provided 20% recovery of total activity after 20 min (Fig. 4). The reactivation in the presence of TRXh1 displayed no difference with respect to 0.2 mm DTTred alone, suggesting that GSSG-treated CrICL does not contain any TRX-reducible disulfide bonds. TRXh1 also proved inefficient for reactivation of ICL when DTT was replaced by NADPH-thioredoxin reductase and NADPH. By contrast, GRX1 allowed full reactivation of protein activity within 20 min (Fig. 4). To further confirm the reactivation mediated by GRX1, we replaced DTTred by 5 mm GSH, the physiological reductant of GRX. In these conditions, we observed reactivation of CrICL to about 80% of maximal activity. The reactivation was faster in the presence of NADPH and glutathione reductase, which allows maintaining the glutathione pool reduced. These results strongly suggest that ICL inactivation in the presence of GSSG is due to the presence of one or more glutathione adducts on the protein.
FIGURE 4.
Kinetics of reactivation of glutathionylated CrICL. A, reactivation in the presence of DTTred. CrICL was inactivated by incubation with 5 mm GSSG until no residual activity was detected and dialyzed overnight against HEPES-NaOH (pH 7.2). This GSSG-inactivated ICL was incubated with no addition (black circles), with DTTred alone (0.2 mm (closed squares) or 20 mm (closed diamonds)) or with 0.2 mm DTTred in the presence of 20 μm TRXh1 (open circles) or 1 μm GRX1 (open squares). The enzyme was also treated with 20 μm TRXh1 in the presence of 0.3 mm NADPH and 0.22 μm A. thaliana NADPH-thioredoxin reductase b (gray circles). B, reactivation in the presence of GSH and GRX. GSSG inactivated CrICL was incubated with 5 mm GSH (closed triangles) or with 5 mm GSH in the presence of 1 μm GRX1 alone (open triangles) or in the presence of 0.2 mm NADPH and yeast glutathione reductase (GR) (6 μg/ml) (gray triangles). 100% corresponds to the mean maximal reactivation measured after treatment with 20 mm DTT.
Analysis of ICL Glutathionylation by Western Blot, Thiol Titration, and CD
We have previously used [35S]cysteine labeling to identify ICL among proteins undergoing S-thiolation in vivo in C. reinhardtii (11). Glutathione being, by far, the most abundant low molecular weight thiol in cells, glutathionylation is considered as the major form of S-thiolation. However, the possibility cannot be excluded that some of the radiolabeled proteins identified with this method might not be glutathionylated but cysteinylated or S-thiolated by other low molecular weight compounds synthesized from cysteine, such as the 398-Da molecule recently identified in B. subtilis (38). Another method for detection of glutathionylated proteins consists of the use of biotinylated glutathione, which allows detection by Western blot of glutathionylated proteins (39).
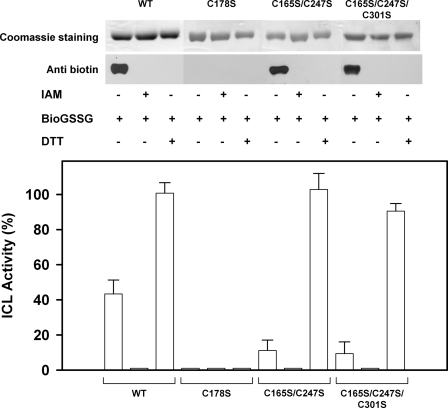
In order to confirm that CrICL effectively undergoes glutathionylation, at least in vitro, we incubated the reduced protein with 2 mm BioGSSG for 1 h and analyzed the presence of glutathione adducts by Western blot using an anti-biotin antibody. A clear signal was observed for the BioGSSG-treated ICL, which completely disappeared after DTTred treatment (Fig. 5). No signal was detected when the protein was pretreated with IAM, a specific cysteine-alkylating agent, suggesting that the absence of free cysteines prevents biotin labeling via glutathionylation. Concomitantly, we also followed protein activity during the treatments. The protein pretreated with IAM showed no activity, suggesting the presence of one or more cysteine residue(s) involved in catalysis. The treatments with BioGSSG led to a 50% inhibition of protein activity, which was fully reversed by DTTred (Fig. 5). These results confirm that purified recombinant CrICL undergoes glutathionylation in the presence of BioGSSG. In order to determine the number of cysteines modified by glutathionylation, we quantified the number of free thiols under non-denaturing conditions using 5,5′-dithiobis-2-nitrobenzoic acid before and after GSSG treatment. Although native reduced CrICL was experimentally found to contain two reactive thiols (2.13 ± 0.07), after 5 h of incubation in the presence of 5 mm GSSG, only 0.57 ± 0.02 remained accessible. This result indicates that, under these conditions, between one and two reactive thiols were lost per CrICL monomer. Moreover, CD analysis revealed that glutathionylation does not significantly affect the secondary structure of CrICL (supplemental Fig. 2). Indeed, the untreated native CrICL and the glutathionylated and fully inactive CrICL exhibited comparable CD spectra (190–260 nm).
FIGURE 5.
Analysis of CrICL glutathionylation with BioGSSG. Recombinant WT CrICL and the variants C178S, C165S/C247S, and C165S/C247S/C301S were incubated for 1 h in the presence of BioGSSG (2 mm) with or without prior incubation with 100 mm IAM. Proteins were resolved by non-reducing SDS-PAGE and transferred to nitrocellulose for Western blotting with anti-biotin antibodies. The Coomassie Brilliant Blue staining of the gel shows equal loading in each lane. The reversibility of the reaction was assessed by treatment with 20 mm DTTred for 30 min as indicated. CrICL activity was measured on aliquots after BioGSSG treatment. Activities are represented as mean percentage ± S.D. (n = 3–5) of the initial activity measured before inactivation treatments.
MALDI-TOF Mass Spectrometry on GSSG-treated ICL
We have previously observed that CrICL-treated with 5 mm GSSG for 24 h presented two glutathione adducts. In order to get more insight into the kinetics of glutathionylation and the number of glutathionylated residues, especially for shorter incubations, we incubated CrICL with 5 mm GSSG and determined the molecular mass of the protein by MALDI-TOF mass spectrometry after 1, 5, and 24 h (Table 1). After a 1-h incubation, a partial shift in molecular mass was observed, consistent with the presence of one glutathione adduct per subunit. After 5 h of incubation, the shift corresponding to one glutathione was complete, and no other glutathione adduct was observed. However, a second peak, corresponding to the presence of two molecules of glutathione on the protein, appeared after 24 h of incubation, indicating that a second cysteine can undergo glutathionylation after prolonged incubation. Upon the addition of reduced DTT, the molecular mass of CrICL shifted back to the mass of the untreated protein (data not shown).
TABLE 1.
Whole protein mass spectrometry analysis of GSSG treated CrICL and its variants
| 0 h |
1 h |
5 h |
24 h |
|||||
|---|---|---|---|---|---|---|---|---|
| Measured mass | No. of glutathione adducts | Measured mass | No. of glutathione adducts | Measured mass | No. of glutathione adducts | Measured mass | No. of glutathione adducts | |
| Da | Da | Da | Da | |||||
| CrICL WT | 46,870.6 | 0 | 47,173.0 (partially) | 0–1 | 47,187.5 | 1 | 47,506.0 | 2 |
| CrICL C178S | 46,861.3 | 0 | 46,869.3 | 0 | 46,887.2 | 0 | 46,885.4 | 0 |
| CrICL C247S | 46,828.2 | 0 | 47,121.3 (partially) | 0–1 | 47,155.6 | 1 | 47,137.3 | 1 |
Analysis of ICL Glutathionylation by Site-directed Mutagenesis
We used site-directed mutagenesis to investigate the molecular mechanism underlying the redox regulation of CrICL by glutathionylation and to identify the cysteine residues involved. CrICL contains four cysteine residues, which were individually replaced by serines. In addition, we also generated double and triple mutants by combining several cysteine substitutions. All proteins were produced recombinantly and purified to homogeneity. The variant lacking Cys178 (C178S) was found to be totally inactive, strongly suggesting the involvement of this cysteine in the catalytic mechanism of the enzyme. All of the other variants remained active and were further analyzed to determine the effect of 5 mm GSSG treatments on their activity. Under these conditions, the single mutants C165S, C247S, and C301S were found to behave as the WT protein and exhibited comparable kinetics of inactivation (data not shown). This suggests that these residues are either not glutathionylated or that their glutathionylation does not affect ICL activity. Similarly, the double mutant C165S/C247S and the triple mutant C165S/C247S/C301S which only retains Cys178, were inactivated by GSSG as the WT protein (data not shown).
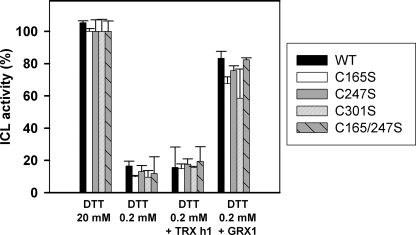
The reactivation of GSSG-treated CrICL variants was also investigated. All CrICL variants were, as above, inactivated by 5 mm GSSG treatment, extensively dialyzed against HEPES-NaOH, pH 7.2, and subsequently incubated with 20 mm DTTred or 0.2 mm DTTred alone or supplemented with TRXh1 or GRX1. As observed for the WT protein, all variants were only reactivated by treatment with either 20 mm DTTred or reduced GRX1 (Fig. 6).
FIGURE 6.
Reactivation of GSSG inactivated WT CrICL and variants by TRXh1 and GRX1. Recombinant WT CrICL and variants were inactivated by incubation with 5 mm GSSG until no residual activity was detected and dialyzed overnight against HEPES-NaOH (pH 7.2). The inactivated proteins were treated with 20 mm DTTred or 0.2 mm DTTred alone or supplemented with either 20 μm TRXh1 or 1 μm GRX1. The protein activity was determined on WT CrICL (black bar), CrICL C165S (white bar), CrICL C247S (gray bar), CrICL C301S (white bar with diagonals), and CrICL C165S/C247S (gray bar with diagonals). Activities are represented as mean percentage ± S.D. (n = 3–5) of the initial activity measured for each protein before GSSG treatment.
Altogether, these results strongly suggest that inactivation of CrICL is mediated by glutathionylation of Cys178 and that deglutathionylation of this residue is specifically catalyzed by GRX. To further confirm that Cys178 undergoes glutathionylation upon GSSG treatment, we incubated the ICL variants C178S, C165S/C247S, and C165S/C247S/C301S in the presence of 2 mm BioGSSG for 1 h. Western blot analysis revealed that the biotin signal was only detected in the mutants where Cys178 is still present, notably C165S/C247S and C165S/C247S/C301S mutants (Fig. 5). Upon DTTred treatment, the signals were completely removed, and pretreatment with IAM blocked biotin labeling via BioGSSG-mediated glutathionylation. The analysis of protein activities confirmed that BioGSSG led to inhibition. Although these results clearly show that Cys178 undergoes glutathionylation with a concomitant loss of enzyme activity, the identity of the second cysteine residue becoming glutathionylated after prolonged incubation in the presence of GSSG remained undetermined.
MALDI-TOF Analysis Confirms Glutathionylation of Cys178 and Cys247
We have determined the molecular mass of different CrICL variants by MALDI-TOF mass spectrometry after incubation with 5 mm GSSG for 1, 5, and 24 h (Table 1). The C247S variant presented a complete shift in molecular mass corresponding to one glutathione adduct after 5 h of incubation, but prolonged incubation did not induce any additional glutathionylation, strongly suggesting that Cys247 is the second cysteine residue undergoing glutathionylation. The same results were obtained with the C165S/C247S and C165S/C247S/C301S variants, whereas the C301S variant behaved like WT CrICL (data not shown). The analysis of C178S treated by GSSG showed that after 5 h, no glutathionylation occurs. This result coupled with those obtained above confirms that Cys178 is indeed the first residue modified by glutathionylation (Table 1). However, after 24 h of incubation, the C178S mutant did not present the peak corresponding to glutathionylation of Cys247. This could be linked to the fact that glutathionylation of Cys247 occurs subsequently to Cys178 modification. On the other hand, we cannot exclude the possibility that in the C178S variant, a conformational change occurs, thus modifying the microenvironment surrounding Cys247.
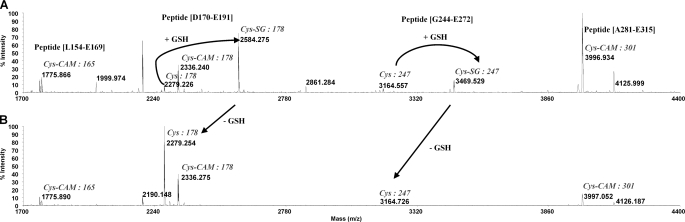
In order to have a direct confirmation of the identity of the two cysteine residues modified by glutathionylation after prolonged incubation with 5 mm GSSG, we have used MALDI-TOF peptide mass fingerprints. With standard trypsin digestions, several cysteine-containing peptides could not be detected. Therefore, GSSG-treated CrICL was initially alkylated with IAM, and we tested different proteases under acidic (Glu-C) and alkaline conditions (trypsin and chymotrypsin). Finally, Glu-C digestions revealed a clear mass increase of ∼305 Da for peptides Asp170–Glu191 and Gly244–Glu272 containing Cys178 and Cys247, respectively (Fig. 7A). By contrast, the peptides containing Cys165 and Cys301 were only found to be carbamidomethylated. As shown in Fig. 7B, the 305-Da mass increases were fully reversed by DTTred treatment, thereby confirming that Cys178 and Cys247 are indeed the two glutathionylated residues of CrICL.
FIGURE 7.
Peptide mass fingerprinting of CrICL reveals that Cys178 and Cys247 are glutathionylated. WT CrICL was incubated with 5 mm GSSG for 24 h and dialyzed overnight against HEPES-NaOH (pH 7.2). Peptide mass fingerprints of Glu-C digestions after alkylation of cysteines with iodoacetamide (+57 Da) showed a glutathione adduct (+305 Da) on peptides Asp170–Glu191 and Gly244–Glu272 (A). For both peptides, the 305-Da mass increase was reversed by DTTred treatment (B). The peptides containing Cys165 and Cys301 were found to be exclusively carbamidomethylated.
DISCUSSION
In C. reinhardtii, isocitrate lyase was previously identified both as a putative TRX target retained on a monocysteinic TRX affinity column (10) and as a protein undergoing S-thiolation in vivo (11). The aim of the present study was to clarify the molecular mechanism of CrICL redox regulation using a combination of biochemical and biophysical methods. The results clearly show that purified CrICL can be inactivated by glutathionylation and reactivated by GRX, whereas TRX does not appear to regulate ICL activity, and no inter- or intramolecular disulfide bond could be formed under any of the conditions tested. These results suggest that the TRX-mediated increase of ICL activity previously observed in total extracts of C. reinhardtii (10) was probably indirect. This apparent activation could be due to the presence of TRX-reducible GRXs in the extract (40, 41) or other enzymes catalyzing deglutathionylation, such as sulfiredoxins (42, 43). However, the possibility cannot be completely ruled out that a disulfide bond could be formed in CrICL in vivo under specific conditions or by interaction with partner proteins not present in our in vitro assays. It has been previously suggested that a number of putative TRX targets, identified by their ability to bind to monocysteinic TRX affinity columns, could represent S-thiolated proteins (1). These proteins could either exhibit a double regulation by TRX and by S-thiolation or, alternatively, could be only S-thiolated and artifactually retained on TRX columns. The latter hypothesis is consistent with the ability of TRXs to catalyze deglutathionylation at high non-physiological concentrations (6) and with the very high local concentration of immobilized TRX on the column matrix. Moreover, the deglutathionylation activity of TRXs might be significantly favored by removal of the second active site cysteine. The results presented here on CrICL provide the first clear demonstration that a glutathionylated protein that is not regulated by TRX can be retained on TRX affinity columns and erroneously considered as a TRX target. There is increasing evidence about numerous interactions between the TRX and the glutathione/GRX systems (1). Several GRXs have been shown to be reduced by thioredoxin reductases (6, 44, 45). Conversely, some TRXs appear to be reduced by glutathione and/or GRX (40, 41), and inactivation of NADPH-thioredoxin reductase genes in Arabidopsis revealed that an alternative reduction system allows TRX reduction in a glutathione-dependent manner, probably implicating a GRX (46). Moreover, TRXs from humans and higher plants have been shown to be regulated by glutathionylation (47–49).
We have shown that Cys178 is the most reactive cysteine of CrICL and that it becomes glutathionylated rapidly in the presence of GSSG or H2O2 plus GSH. However, a second cysteine, Cys247, was found to undergo glutathionylation after prolonged incubation with GSSG. The glutathionylation of this residue does not appear to affect ICL activity and is most likely artifactual because it is only observed under non-physiological oxidative conditions. Similarly, it has been reported for human TRX that one cysteine is the primary target of glutathionylation but that long incubation with GSSG also leads to artifactual glutathionylation of several other cysteine residues (47). We have previously observed that Cys165 could be glutathionylated after prolonged incubation in the presence of GSSG (11). However, we observed here that the glutathionylation of this residue is artifactual because it only occurs after tryptic digestion and most likely arises from a transfer of glutathione adduct between the peptide containing Cys178 and the one containing Cys165. Consequently, after tryptic digestion of CrICL treated with GSSG overnight, Cys165 and Cys247 appear to be glutathionylated, whereas the peptide containing Cys178 is not detected. This explanation is supported by the fact that the isomerization is only observed in peptide mass fingerprints under alkaline conditions without carbamidomethylation of cysteines. When tryptic digestions are performed after prior cysteine alkylation by IAM, Cys165 is only carbamidomethylated. All of these considerations underline that great caution must be taken when analyzing redox regulation mechanisms in vitro, especially in the case of glutathionylation.
In Chlamydomonas ICL, the primary target of glutathionylation and oxidation is the catalytic cysteine 178. This cysteine is strictly conserved in all known ICL, consistent with its crucial role in catalysis (29, 30, 50, 51). This cysteine was proposed to be required for deprotonation of a carboxylate of succinate during the catalytic reaction (52). The strict conservation of this residue suggests that it is likely to be glutathionylated in all organisms containing glutathione. Despite this conservation, higher plant ICLs were not identified among putative TRX targets or S-thiolated proteins by proteomic analyses (2, 53). This is most probably linked to the abundance of the protein. Indeed, ICL is very abundant in Chlamydomonas extracts from cultures grown in the presence of acetate, whereas in higher plants, ICL is not detectable in mature leaves, the enzyme being only present in earlier developmental stages (54).
In CrICL, alkylation of Cys178 or its substitution by serine resulted in an inactive enzyme. Moreover, we have shown that CrICL is very sensitive to oxidative inactivation mediated by H2O2 and that glutathionylation of Cys178 can efficiently and reversibly protect the enzyme from irreversible inactivation. Indeed, the enzyme was not only glutathionylated in the presence of 5 mm GSSG, a concentration far above the physiological concentration of oxidized glutathione in vivo, but also under the physiologically relevant concentration of GSH in the presence of H2O2. This strongly suggests that H2O2-mediated irreversible inactivation of ICL is linked to oxidation of Cys178 to sulfenic acid, followed by overoxidation to sulfinic and/or sulfonic acid forms. In the presence of GSH, the sulfenic acid form would be converted to a glutathionylated thiol, thereby providing protection from further oxidation. A similar mechanism of sulfenic acid-mediated glutathionylation has been demonstrated for human protein tyrosine phosphatase 1B (32, 55) and for several chloroplastic proteins, such as glyceraldehyde-3-phosphate dehydrogenase (33) and 1-Cys-methionine sulfoxide reductase B (56). Therefore, under conditions of enhanced production of reactive oxygen species, ICL would be inactivated by glutathionylation of Cys178 and reactivated by GRX once reactive oxygen species have been scavenged.
Besides providing protection against oxidative inactivation, glutathionylation of ICL may also constitute a mechanism of regulation of ICL activity, allowing modulation of the glyoxylate cycle, depending on the intracellular redox state. Citrate and isocitrate enzymatic conversions are considered as important metabolic branch points in plant metabolism (57). Isocitrate dehydrogenase catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate, an important energy yielding step of the tricarboxylic acid cycle. Isocitrate is also used by the glyoxylate cycle, which allows net production of succinate from two molecules of acetyl-CoA, thereby supporting biosynthetic processes. The control of the isocitrate branch point through the regulation of isocitrate dehydrogenase and/or ICL activities has been suggested to provide the cellular metabolism with flexibility in the utilization of isocitrate (57). Under oxidative stress conditions, glutathionylation may constitute a mechanism regulating the partitioning of isocitrate between the two cycles. Transient inactivation of ICL by glutathionylation could allow favoring of the energy-yielding tricarboxylic acid cycle versus the carbon-conserving glyoxylate cycle, thereby providing the cell with the energy required for oxidative stress defense. After regeneration of the intracellular redox state, GRXs could ensure reactivation of ICL. Such a mechanism could allow fine tuning of these two cycles according to the relative need of the cell for energy versus biosynthetic precursors. Further studies will be required to determine the functional significance of ICL glutathionylation in vivo and its importance in the regulation of cell metabolism under oxidative stress.
Acknowledgments
We thank Myroslawa Miginiac-Maslow for critical reading of the manuscript and helpful suggestions. We are grateful to Loïc Martin and Pascal Kessler for help with CD experiments.
This work was supported by Agence Nationale de la Recherche Grant ANR-08-BLAN-0153.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- TRX
- thioredoxin
- GRX
- glutaredoxin
- ICL
- isocitrate lyase
- CrICL
- C. reinhardtii ICL
- BioGSSG
- biotinylated oxidized glutathione
- WT
- wild type
- DTT
- dithiothreitol
- DTTred
- reduced dithiothreitol
- IAM
- iodoacetamide
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- PTS
- peroxisomal targeting signal.
REFERENCES
- 1.Michelet L., Zaffagnini M., Massot V., Keryer E., Vanacker H., Miginiac-Maslow M., Issakidis-Bourguet E., Lemaire S. D. (2006) Photosynth. Res. 89, 225–245 [DOI] [PubMed] [Google Scholar]
- 2.Montrichard F., Alkhalfioui F., Yano H., Vensel W. H., Hurkman W. J., Buchanan B. B. (2009) J. Proteomics 72, 452–474 [DOI] [PubMed] [Google Scholar]
- 3.Rouhier N., Lemaire S. D., Jacquot J. P. (2008) Annu. Rev. Plant Biol. 59, 143–166 [DOI] [PubMed] [Google Scholar]
- 4.Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009) Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 5.Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaffagnini M., Michelet L., Massot V., Trost P., Lemaire S. D. (2008) J. Biol. Chem. 283, 8868–8876 [DOI] [PubMed] [Google Scholar]
- 7.Gallogly M. M., Starke D. W., Leonberg A. K., Ospina S. M., Mieyal J. J. (2008) Biochemistry 47, 11144–11157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallogly M. M., Starke D. W., Mieyal J. J. (2009) Antioxid. Redox Signal. 11, 1059–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemaire S. D., Guillon B., Le Maréchal P., Keryer E., Miginiac-Maslow M., Decottignies P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michelet L., Zaffagnini M., Vanacker H., Le Maréchal P., Marchand C., Schroda M., Lemaire S. D., Decottignies P. (2008) J. Biol. Chem. 283, 21571–21578 [DOI] [PubMed] [Google Scholar]
- 12.Kornberg H. L., Krebs H. A. (1957) Nature 179, 988–991 [DOI] [PubMed] [Google Scholar]
- 13.Graham I. A. (2008) Annu. Rev. Plant Biol. 59, 115–142 [DOI] [PubMed] [Google Scholar]
- 14.Schnarrenberger C., Martin W. (2002) Eur. J. Biochem. 269, 868–883 [DOI] [PubMed] [Google Scholar]
- 15.Vanni P., Giachetti E., Pinzauti G., McFadden B. A. (1990) Comp. Biochem. Physiol. B 95, 431–458 [DOI] [PubMed] [Google Scholar]
- 16.Petridou S., Foster K., Kindle K. (1997) Plant Mol. Biol. 33, 381–392 [DOI] [PubMed] [Google Scholar]
- 17.Rúa J., Soler J., Busto F., de Arriaga D. (2002) Fungal Genet. Biol. 35, 223–234 [DOI] [PubMed] [Google Scholar]
- 18.Jacquot J. P., Rivera-Madrid R., Marinho P., Kollarova M., Le Maréchal P., Miginiac-Maslow M., Meyer Y. (1994) J. Mol. Biol. 235, 1357–1363 [DOI] [PubMed] [Google Scholar]
- 19.Stein M., Jacquot J. P., Jeannette E., Decottignies P., Hodges M., Lancelin J. M., Mittard V., Schmitter J. M., Miginiac-Maslow M. (1995) Plant Mol. Biol. 28, 487–503 [DOI] [PubMed] [Google Scholar]
- 20.Chang A., Scheer M., Grote A., Schomburg I., Schomburg D. (2009) Nucleic Acids Res. 37, D588–D592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durrant I. (1990) Nature 346, 297–298 [DOI] [PubMed] [Google Scholar]
- 22.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 23.Sicard-Roselli C., Lemaire S., Jacquot J. P., Favaudon V., Marchand C., Houée-Levin C. (2004) Eur. J. Biochem. 271, 3481–3487 [DOI] [PubMed] [Google Scholar]
- 24.Marchand C., Le Maréchal P., Meyer Y., Decottignies P. (2006) Proteomics 6, 6528–6537 [DOI] [PubMed] [Google Scholar]
- 25.Serrano J. A., Bonete M. J. (2001) Biochim. Biophys. Acta 1520, 154–162 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe S., Takada Y. (2004) Microbiology 150, 3393–3403 [DOI] [PubMed] [Google Scholar]
- 27.Taylor K. M., Kaplan C. P., Gao X., Baker A. (1996) Biochem. J. 319, 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaves R. S., Herrero P., Ordiz I., Angeles del Brio M., Moreno F. (1997) Gene 198, 165–169 [DOI] [PubMed] [Google Scholar]
- 29.Robertson A. G., Nimmo H. G. (1995) Biochem. J. 305, 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehman A., McFadden B. A. (1997) Curr. Microbiol. 35, 267–269 [DOI] [PubMed] [Google Scholar]
- 31.Diehl P., McFadden B. A. (1994) J. Bacteriol. 176, 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett W. C., DeGnore J. P., König S., Fales H. M., Keng Y. F., Zhang Z. Y., Yim M. B., Chock P. B. (1999) Biochemistry 38, 6699–6705 [DOI] [PubMed] [Google Scholar]
- 33.Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S. D. (2007) FEBS J. 274, 212–226 [DOI] [PubMed] [Google Scholar]
- 34.Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 35.Dixon D. P., Fordham-Skelton A. P., Edwards R. (2005) Biochemistry 44, 7696–7703 [DOI] [PubMed] [Google Scholar]
- 36.Jung C. H., Thomas J. A. (1996) Arch. Biochem. Biophys. 335, 61–72 [DOI] [PubMed] [Google Scholar]
- 37.Chrestensen C. A., Starke D. W., Mieyal J. J. (2000) J. Biol. Chem. 275, 26556–26565 [DOI] [PubMed] [Google Scholar]
- 38.Lee J. W., Soonsanga S., Helmann J. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X. H., Bedhomme M., Veyel D., Zaffagnini M., Lemaire S. D. (2009) Mol. Plant 2, 218–235 [DOI] [PubMed] [Google Scholar]
- 40.Koh C. S., Navrot N., Didierjean C., Rouhier N., Hirasawa M., Knaff D. B., Wingsle G., Samian R., Jacquot J. P., Corbier C., Gelhaye E. (2008) J. Biol. Chem. 283, 23062–23072 [DOI] [PubMed] [Google Scholar]
- 41.Gelhaye E., Rouhier N., Jacquot J. P. (2003) FEBS Lett. 555, 443–448 [DOI] [PubMed] [Google Scholar]
- 42.Findlay V. J., Townsend D. M., Morris T. E., Fraser J. P., He L., Tew K. D. (2006) Cancer Res. 66, 6800–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. W., Mieyal J. J., Rhee S. G., Chock P. B. (2009) J. Biol. Chem. 284, 23364–23374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandes A. P., Fladvad M., Berndt C., Andrésen C., Lillig C. H., Neubauer P., Sunnerhagen M., Holmgren A., Vlamis-Gardikas A. (2005) J. Biol. Chem. 280, 24544–24552 [DOI] [PubMed] [Google Scholar]
- 45.Marteyn B., Domain F., Legrain P., Chauvat F., Cassier-Chauvat C. (2009) Mol. Microbiol. 71, 520–532 [DOI] [PubMed] [Google Scholar]
- 46.Reichheld J. P., Khafif M., Riondet C., Droux M., Bonnard G., Meyer Y. (2007) Plant Cell 19, 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casagrande S., Bonetto V., Fratelli M., Gianazza E., Eberini I., Massignan T., Salmona M., Chang G., Holmgren A., Ghezzi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9745–9749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelhaye E., Rouhier N., Gérard J., Jolivet Y., Gualberto J., Navrot N., Ohlsson P. I., Wingsle G., Hirasawa M., Knaff D. B., Wang H., Dizengremel P., Meyer Y., Jacquot J. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelet L., Zaffagnini M., Marchand C., Collin V., Decottignies P., Tsan P., Lancelin J. M., Trost P., Miginiac-Maslow M., Noctor G., Lemaire S. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Britton K. L., Abeysinghe I. S., Baker P. J., Barynin V., Diehl P., Langridge S. J., McFadden B. A., Sedelnikova S. E., Stillman T. J., Weeradechapon K., Rice D. W. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1209–1218 [DOI] [PubMed] [Google Scholar]
- 51.Nimmo H. G., Douglas F., Kleanthous C., Campbell D. G., MacKintosh C. (1989) Biochem. J. 261, 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma V., Sharma S., Hoener zu Bentrup K., McKinney J. D., Russell D. G., Jacobs W. R., Jr., Sacchettini J. C. (2000) Nat. Struct. Biol. 7, 663–668 [DOI] [PubMed] [Google Scholar]
- 53.Dixon D. P., Skipsey M., Grundy N. M., Edwards R. (2005) Plant Physiol. 138, 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J. Z., Gomez-Pedrozo M., Baden C. S., Harada J. J. (1993) Mol. Gen. Genet. 238, 177–184 [DOI] [PubMed] [Google Scholar]
- 55.Barrett W. C., DeGnore J. P., Keng Y. F., Zhang Z. Y., Yim M. B., Chock P. B. (1999) J. Biol. Chem. 274, 34543–34546 [DOI] [PubMed] [Google Scholar]
- 56.Tarrago L., Laugier E., Zaffagnini M., Marchand C., Le Maréchal P., Rouhier N., Lemaire S. D., Rey P. (2009) J. Biol. Chem. 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popova T. N., Pinheiro de Carvalho M. A. (1998) Biochim. Biophys. Acta 1364, 307–325 [DOI] [PubMed] [Google Scholar]