Abstract
Zinc protoporphyrin IX (ZnPP), an endogenous heme analogue that inhibits heme oxygenase (HO) activity, represses tumor growth. It can also translocate into the nucleus and up-regulate heme oxygenase 1 (HMOX1) gene expression. Here, we demonstrate that tumor cell proliferation was inhibited by ZnPP, whereas tin protoporphyrin (SnPP), another equally potent HO-1 inhibitor, had no effect. Microarray analysis on 128 tumorigenesis related genes showed that ZnPP suppressed genes involved in cell proliferation and angiogenesis. Among these genes, CYCLIN D1 (CCND1) was specifically inhibited as were its mRNA and protein levels. Additionally, ZnPP inhibited CCND1 promoter activity through an Sp1 and Egr1 overlapping binding site (S/E). We confirmed that ZnPP modulated the S/E site, at least partially by associating with Sp1 and Egr1 proteins rather than direct binding to DNA targets. Furthermore, administration of ZnPP significantly inhibited cyclin D1 expression and progression of a B-cell leukemia/lymphoma 1 tumor in mice by preferentially targeting tumor cells. These observations show HO independent effects of ZnPP on cyclin D1 expression and tumorigenesis.
Introduction
Zinc protoporphyrin IX (ZnPP)2 is a metabolite formed in trace amounts during heme biosynthesis. In this process, the final reaction is the chelation of zinc in the protoporphyrin ring, whereas heme is formed by chelation of iron in the ring. During periods of iron insufficiency or impaired iron utilization, ZnPP formation is enhanced. Clinically, ZnPP quantification is a sensitive and specific tool for measuring iron mineral status and metabolism (1). In addition, ZnPP regulates heme catabolism through competitively inhibiting the activity of heme oxygenase (HO), the rate-limiting enzyme in the heme degradation pathway that produces carbon monoxide and biliverdin. The latter is rapidly reduced to bilirubin by biliverdin reductase. Thus, ZnPP has potential therapeutic applications in controlling exaggerated bilirubin formation leading to neonatal jaundice (2). Moreover, because the by-products of the HO reaction, carbon monoxide and bilirubin, are antioxidants, the potential effects of ZnPP have been studied in numerous diseases, including cancer, such as chronic myelogenous leukemia (3–6).
Whereas ZnPP has been largely studied in relation to its inhibition of HO activity, reports show that ZnPP could exert cellular effects independent of HO activity (7, 8). In kinetic assays, ZnPP inhibited the soluble guanylyl cyclase activity independent of HO-1 (9). In vitro studies showed that ZnPP directly interacts with human immunodeficiency virus type 1 reverse transcriptase and modulates its activity (10). We showed that ZnPP induces the expression of HMOX1 and TP53 genes and localizes to the nucleus (11). Although the underling mechanism is unknown, zinc mesoporphyrin, another analogue of heme, has been shown to induce HMOX1 expression by accelerating Bach1 protein degradation (12). Heme itself can affect gene expression by associating with transcription factors (13, 14). Therefore, we reasoned that ZnPP could potentially interact with transcriptional regulators in the nucleus and directly modulate gene expression. If this effect was targeted to genes related to cell proliferation or apoptosis, this could further explain the role of ZnPP in suppressing tumor formation.
In tumorigenesis, cyclin D1 is key to stimulate cell proliferation by enhancing G1/S transition in the cell cycle. Abundant evidence demonstrates that cyclin D1 is overexpressed in mammary, ovarian, and lung tumors among others (15–17). Cyclin D1 expression is predominantly regulated at the transcriptional level, although post-transcriptional mechanisms also exist (18, 19). The CCND1 promoter contains multiple cis-elements, including binding sites for Sp1 and Egr1 (20–23). The B-cell leukemia/lymphoma 1 (BCL1) tumor is characterized by overexpression of cyclin D1. In BCL1 tumor cells, the cyclin D1 locus is downstream of the Emu enhancer due to a chromosome translocation yet it contains the intact promoter (24). Here, we show that ZnPP interacts with transcription factors Egr1 and Sp1 and modulates their binding to the CCND1 promoter, thereby inhibiting cyclin D1 expression, cell proliferation, and BCL1 tumor progression in vivo.
EXPERIMENTAL PROCEDURES
Cell Lines and Tissue Culture
Human bone marrow K562 cells, isolated from a patient carrying chronic myelogenous leukemia, were cultured in Iscove's modified Dulbecco's medium supplemented with 10% (v/v) fetal calf serum, penicillin (100 units/ml), streptomycin (100 μg/ml), and 1.5 g/liter of sodium bicarbonate. Human hepatoma HepG2 cells were cultured in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). Both cell lines were cultured under 5% CO2 at 37 °C.
To express Sp1, cyclin D1, or Egr1, human cDNAs were cloned in a myc-pMX-puro vector and further packaged as retroviruses in human embryonic kidney 293T cells. The HepG2 cells were infected with these retroviruses and selected with 2 μg/ml of puromycin for stably expressing Sp1, cyclin D1, or Egr1. To deplete endogenous Egr1 expression, lentiviruses carrying control or shRNA targeting Egr1 mRNA (shEgr1) were produced as per the manufacturer's instructions and used to infect HepG2 cells (RHS4533-NM_001964, Thermo Fisher Scientific Open Biosystems Products, Huntsville, AL).
Experimental Design
ZnPP and SnPP (Frontier Scientific Inc, Logan, UT) were prepared as previously described (11). For controls, an equal amount of vehicle (0.5% ethanolamine, pH 7.5) was used.
Exponentially growing HepG2 or K562 cells were incubated with vehicle, ZnPP or SnPP for 6 and 12 h, respectively, then subjected to whole cell lysis as described (25) or nuclear extraction with the Pierce NE-PER® kit (78833, Pierce Biotechnology) according to the manufacturer's instructions.
To evaluate cell cycle progression, HepG2 cells were treated with vehicle, ZnPP, or SnPP for 24, 48, or 72 h after 24 h of serum starvation and analyzed by fluorescence-activated cell sorting (FACS) following 70% ethanol fixation and propidium iodide staining. FACS data were analyzed using FlowJo7 software (Tree Star, Inc., Ashland, OR).
Determination of HO Enzymatic Activity
This was measured by detecting the amount of carbon monoxide generated from cell lysates, as previously described (26).
Profiling of Genes Affected by ZnPP
HepG2 cells incubated with ZnPP or vehicle were subjected to an oligo array (GEArray System, OHS-033, SuperArray Bioscience Corporation, Frederick, MD) as per the manufacturer's protocol. The array contains 128 genes. Three independent experiments were conducted. Data analysis was performed using the GEArray Expression Analysis Suite according to the manufacturer's instructions (SuperArray Bioscience Corp., Frederick, MD).
Quantitative Real Time RT-PCR (qRT-PCR)
After treatment of vehicle, SnPP, or ZnPP, total RNA was isolated from HepG2 cells as well as mouse spleen or liver using the RNeasy kit (Qiagen Inc., Valencia, CA). Reverse transcription was performed with the SuperscriptTM II reverse transcriptase (Invitrogen).
By using the TaqMan Gene Expression Assay (Hs00277039_ ml, Hs02758991_gl, Mm00432359_ml, and Mm99999915_gl, Applied Biosystems, Foster City, CA), the CCND1 mRNA level was analyzed and further normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level.
Detection of Proteins Modified by ZnPP
Proteins were separated using SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and blotted with appropriate primary antibodies against cyclin D1 (number 2922, Cell Signaling Technology, Inc., Danvers, MA), Sp1 (07-645, Millipore), or Egr1 (number 4152, Cell Signaling Technology Inc., Danvers, MA), followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The blotted proteins were visualized with the enhanced chemiluminescence detection kit (GE Healthcare).
Measurement of CCND1 Promoter Activity
HepG2 cells were seeded at a density of 1 × 105 cells per well in 24-well plates. Using LipofectamineTM 2000 (Invitrogen), cells were transfected with CCND1 promoters driving the Firefly luciferase reporter gene, and phRL-TK (Promega, Madison, WI) encoding Renilla luciferase was used as an internal control to normalize Firefly luciferase activity. Twenty-four hours later, cells were incubated with 5 μm SnPP, 5 μm ZnPP, or vehicle for 12 h, and then harvested for luciferase assays using the Dual-Luciferase® Reporter (DLRTM) Assay System (Promega) following the manufacturer's protocol. Firefly and R. luciferase activities were measured using a TR717 Microplate Luminometer (EG&G Berthod Technologies, Oak Ridge, TN).
Determination of ZnPP-mediated Egr1 and Sp1 Specific Binding in Vivo
HepG2 cells were seeded at a density of 3 × 106 per 100-mm dish. Twenty-four hours later, cells were incubated with 5 μm SnPP, 5 μm ZnPP or vehicle. After 6 h of incubation, cells were collected for ChIP assay using a commercially available kit (Millipore). Briefly, chromatin DNA was cross-linked to protein with formaldehyde and sheared by pulsed ultrasonication. Sheared DNA-protein complexes were incubated overnight with anti-Sp1, anti-Egr1 antibodies, or rabbit IgG. Antibody-precipitated DNA-protein complexes were reverse cross-linked and extracted with phenol/chloroform. The precipitated DNA was used as template for PCR amplification. Primers, 5′-GGGCGATTTGCATTTCTATG-3′ and 5′-AAAGATCAAAGCCCGGCAGA-3′, were used for generating PCR amplicons of the CCND1 promoter.
Evaluation of ZnPP Interaction with Protein or DNA
To analyze the ability of ZnPP to associate with cellular proteins, varying amounts of nuclear proteins were extracted from HepG2 cells incubated with 5 μm ZnPP in NER buffer (78833C, Pierce Biotechnology, Inc., Rockford, IL). The ZnPP-protein complexes were resolved on a 0.7% agarose gel and visualized under UV light taking advantage of the autofluorescent property of ZnPP.
To evaluate ZnPP binding to DNA, the DNA fragment produced from the ChIP assay (SE) was used, and another 123-bp DNA fragment, extracted from the cyclin D1 promoter using KpnI and DpnI restriction enzymes served as a control (non-SE). The fragments were incubated with 5 μm ZnPP or vehicle in binding buffer from the Gel Shift Assay System (Promega) for 20 min at room temperature followed by PAGE. Potential ZnPP-DNA complexes were visualized under UV light because of the autofluorescent properties of ZnPP. Afterward, the PAGE gel was subjected to ethidium bromide staining to further verify the potential binding of ZnPP to DNA by evaluating ZnPP-supershifted DNA bands.
Assessment of ZnPP-mediated Tumor Growth Inhibition in Vivo
As detailed previously (27), tumor cells were isolated from spleens of BCL1-bearing animals, and infected with retroviruses expressing luciferase (LUC) and green fluorescent protein. Green fluorescent protein positive cells, referred to as BCL1-gfp/luc, were sorted by FACS. Four-week-old BALB/c female mice were intraperitoneally injected with 2 × 103 BCL-gfp/luc cells. After 5 days, the mice were intraperitoneally injected with 0 (sterile saline), 40 or 80 μm/kg of ZnPP. Mice were then injected with luciferin (150 mg/kg; BioSynth, Naperville, IL) and imaged daily using the In Vivo Imaging System (IVIS, Xenogen Corp. Alameda, CA). To monitor tumor growth, photons emitted from BCL-gfp/luc cells were quantified using Living Image software (IgorPro, Xenogen Corp., Alameda, CA). All mice were handled according to the appropriate protocols approved by the Institutional Animal Care and Use Committee of the Stokes Research Institute at the Children's Hospital of Philadelphia.
Statistical Analysis
Values represent the mean ± S.D. of three experiments unless otherwise indicated. For comparison between treatment groups, the null hypothesis that there is no difference between treatment means was tested by a single factor analysis of variance for multiple groups or unpaired t test for two groups (Intsat 3, GraphPad Software, Inc., San Diego, CA). Statistical significance (*, p < 0.05) between and within groups was determined by means of the Fischer method of multiple comparisons.
RESULTS
ZnPP Inhibits Cell Proliferation
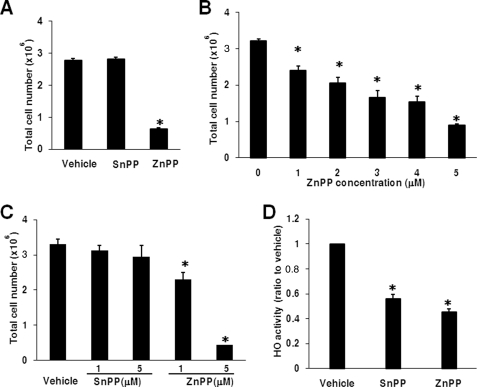
ZnPP inhibits tumor growth and accumulates in the liver after intraperitoneal administration in vivo (2, 5), hence the use of HepG2 cells. Due to its autofluorescent properties, ZnPP enhanced the reading of fluorescence or absorbance-based proliferation assays,3 obviating their use in this study, therefore, cell counting was used. Five μm ZnPP inhibited cell proliferation by 4.4-fold compared with vehicle, whereas SnPP failed to do so (Fig. 1A). The effect of ZnPP was concentration dependent (Fig. 1B). Because ZnPP inhibits proliferation of bone marrow cells (28), K562 cells were also used and 1–5 μm ZnPP significantly inhibited their proliferation (Fig. 1C). At 10 μm, the number of trypan blue positive cells increased after incubation with ZnPP suggesting toxicity.3 Corroborating the action of ZnPP on cell proliferation was not due to the inhibition of HO activity, HO enzymatic suppression by ZnPP and SnPP was 58 and 42%, respectively (p = 0.145 ZnPP versus SnPP) (Fig. 1D).
FIGURE 1.
ZnPP but not SnPP inhibits the proliferation of HepG2 and K562 cells. Cell counts from HepG2 incubated with 5 μm SnPP or ZnPP are shown in A. In B, HepG2 cell counts after 0–5 μm ZnPP incubations are shown. In C, cell counts in K562 after incubation with vehicle, SnPP, or ZnPP are shown. In D, HO activity in HepG2 cells treated with 5 μm ZnPP or SnPP for 12 h are shown. *, p < 0.05 versus vehicle.
ZnPP Inhibits the Expression of Cyclin D1
Because ZnPP inhibits cell proliferation and localizes to the nucleus (11), could it modulate the expression of key genes involved in cell proliferation and consequently affect tumorigenesis? Among 128 cancer-related genes detected using a microarray, incubation with 5 μm ZnPP for 6 h resulted in the decrease of 26, many closely involved in cell proliferation, by more than 2-fold (Table 1). However, we did not see modulation of genes involved in apoptosis,4 arguing against cell death by incubation.
TABLE 1.
Genes inhibited by ZnPP in HepG2 cells
| UniGene | RefSeq number | Symbol | Description | -Fold inhibition |
|---|---|---|---|---|
| Hs.396530 | NM_000601 | HGF | Hepatocyte growth factor | 7.2 |
| Hs.507621 | NM_002019 | FLT-1 | Fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 5.0 |
| Hs.160562 | NM_000618 | IGF1 | Insulin-like growth factor 1 | 4.2 |
| Hs.369675 | NM_001146 | ANGPT1 | Angiopoietin 1 | 4.1 |
| Hs.132966 | NM_000245 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | 3.7 |
| Hs.239818 | NM_006219 | PIK3CB | Phosphoinositide-3-kinase, catalytic, β polypeptide | 3.5 |
| Hs.523852 | NM_053056 | CCND1 | Cyclin D1 | 2.9 |
| Hs.533683 | NM_000141 | FGFR2 | Fibroblast growth factor receptor 2 | 2.5 |
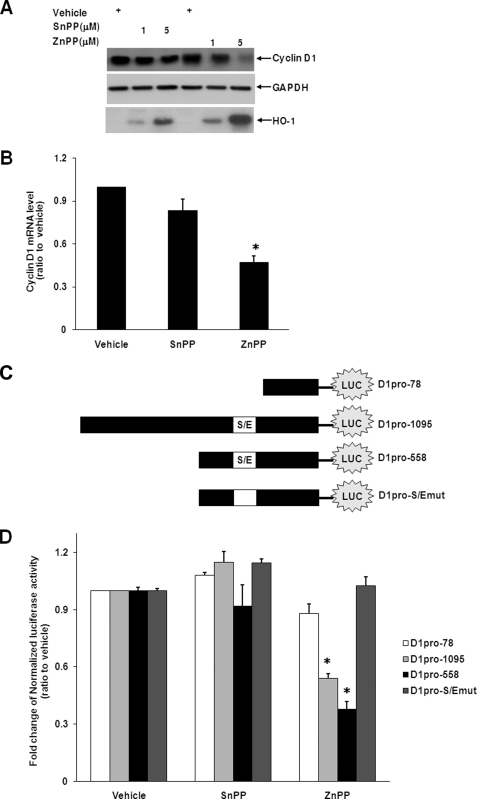
Among the genes inhibited by ZnPP, cyclin D1 is well documented to enhance cell proliferation in tumorigenesis (29–31). ZnPP decreased cyclin D1 protein levels at both 1 and 5 μm (Fig. 2A). This was not due to generalized inhibition of gene expression because ZnPP increased HO-1 protein levels, consistent with our previous data (11). As shown by qRT-PCR, cyclin D1 mRNA levels were also suppressed significantly by incubation with ZnPP but not by SnPP (Fig. 2B) although they both equally inhibited HO activity (Fig. 1). Because ZnPP decreases cyclin D1 protein and mRNA levels, we predicted that it would regulate cyclin D1 gene promoter activity. The promoter region of the CCND1 locus contains several transcription factor binding sites, including the Sp1 and Egr1 overlapping binding site (S/E) (23). Previously, we showed that ZnPP enhanced nuclear protein binding to the Egr1 targeting sequence, but not to Sp1 or AP-1 consensus sequences on the HMOX1 gene (11). We now speculate that ZnPP could regulate the promoter activity of CCND1 by modulating nuclear protein targeting to the S/E site on the cyclin D1 promoter. To this end, a luciferase construct containing the full-length CCND1 promoter (D1pro-1095), deletion mutants harboring the S/E site (D1pro-558), and lacking the S/E site (D1pro-78) were transfected into HepG2 cells (Fig. 2C). As shown in Fig. 2D, ZnPP decreased the promoter activities of D1pro-1095 and D1pro-558, especially D1pro-1095 by more than 2-fold. In contrast, it did not alter D1pro-78 activity. These data suggest that ZnPP regulates cyclin D1 gene expression by modulating the S/E site of the cyclin D1 promoter. Furthermore, by using direct mutagenesis, the S/E site in D1pro-558 was mutated (D1pro-S/Emut) and introduced into HepG2 cells (Fig. 2C). The luciferase analysis showed that contrary to D1pro-1095 and D1pro-558, D1pro-S/Emut did not demonstrate any inhibition of ZnPP on CCND1 promoter activity (Fig. 2D), suggesting that the S/E site is essential for ZnPP-mediated suppression of cyclin D1 gene expression.
FIGURE 2.
ZnPP inhibits cyclin D1 expression. A representative Western blot of cyclin D1 in HepG2 cells after 12-h SnPP or ZnPP incubations is shown in A. Equal loading is demonstrated with the housekeeping gene GAPDH. Representative qRT-PCR of cyclin D1 mRNA is shown in B. The GAPDH mRNA levels were used to normalize the cyclin D1 signal. In C, constructs used in the luciferase reporter assays are shown. The promoter activity was shown in D. *, p < 0.05 versus vehicle control.
ZnPP Facilitates the Competition of Egr1 with Sp1 in Targeting the S/E Site
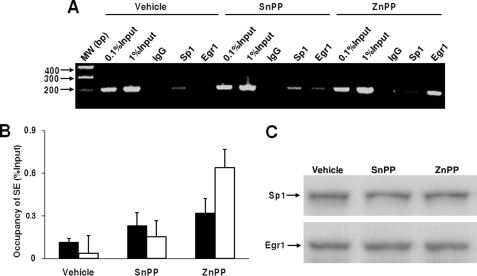
Because ZnPP incubation was associated with enhanced levels of nuclear proteins targeting the Egr1 binding site (11) and inhibited the promoter activity of CCND1 via the S/E site, we speculated that ZnPP could associate with nuclear proteins that target this site. Therefore, we performed ChIP assays using a fragment including the S/E site generated from the cyclin D1 locus (referred as SE below). Incubation with ZnPP increased binding of the Egr1 protein to the CCND1 locus by 12.5-fold, whereas it decreased the binding of Sp1 to this region by more than 2-fold. In contrast, incubation with vehicle or SnPP showed preferential Sp1 binding and incubation with SnPP minimally increased Sp1 and Egr1 protein binding (Fig. 3, A and B). Despite these observations, neither ZnPP nor SnPP incubation altered Egr1 or Sp1 protein levels (Fig. 3C), indicating that the effect of ZnPP on the S/E site was not due to changes in Egr1 or Sp1 abundance.
FIGURE 3.
ZnPP enhances Egr1 but decreases Sp1 binding at the S/E site in the CCND1 promoter. In A, a representative ChIP assay of ZnPP-mediated Sp1 and Egr1 binding to the cyclin D1 promoter is shown. HepG2 cells were treated with vehicle, 5 μm SnPP or ZnPP for 6 h. The SE DNA fragment, generated from the CCND1 promoter and harboring the SE site, was PCR amplified with chromatin DNA precipitated by IgG or anti-Sp1 or Egr1 antibodies. In B, quantitation was achieved by subtracting the IgG signal from the experimental value and divided by the signal of the input. Empty bars represent samples incubated with anti-Egr1 antibodies; solid bars represent samples incubated with anti-Sp1 antibodies. In C, a representative Western blot of Egr1 and Sp1 immunoreactive signals from the cells used for the ChIP assay is shown.
ZnPP Interacts with SP1 and Egr1 Complexes Rather Than Modulate a DNA Target
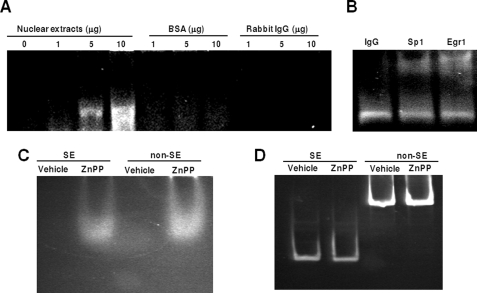
Others show that ZnPP interacts with human immunodeficiency virus type 1 reverse transcriptase protein in vitro (10). Because ZnPP preferentially enhanced Egr1 but decreased Sp1 binding at the S/E site, we speculated that ZnPP inhibits cyclin D1 gene expression through direct interaction with transcription factor complexes, specifically those containing Egr1 and/or Sp1. Therefore, increasing amounts of nuclear proteins extracted from HepG2 cells were incubated with 5 μm ZnPP. The ZnPP-protein complexes were separated on agarose gels. Because ZnPP is autofluorescent, altered migration of nuclear proteins with fluorescence can be used as an index of ZnPP/protein binding. As shown in Fig. 4A, ZnPP interacted with nuclear extracts from HepG2 cells, but failed to show any specific association with either bovine serum albumin or IgG. Furthermore, the interaction of protein and ZnPP was enhanced with an increasing amount of nuclear protein, indicating that ZnPP associates with nuclear proteins in a dose-dependent manner.
FIGURE 4.
ZnPP associates with transcription factors Sp1 and Egr1 but not DNA. In A, increasing amounts of nuclear proteins extracted from HepG2 cells were incubated with 5 μm ZnPP, and then separated on an agarose gel. The protein complexes bound to ZnPP, shown as retention on the gel, were visualized under UV light. Varying amounts of bovine serum albumin (BSA) or rabbit IgG incubated with ZnPP served as controls. In B, the nuclear proteins were incubated with IgG, or anti-Sp1 and Egr1 antibodies and further incubated with ZnPP. The ZnPP-protein complexes were separated on the agarose gel and visualized under UV light. In C, the DNA fragments SE and non-SE were incubated with 5 μm ZnPP or vehicle, respectively, and then separated using PAGE. The ZnPP complex was visualized under UV light. In D, after the ZnPP bands shown in C were photographed, the gel was further stained with ethidium bromide and visualized under UV light to evaluate the migration of free DNA and DNA bound by ZnPP.
To verify that transcription factors Egr1 and Sp1 interacted with ZnPP, the complexes were incubated with IgG, anti-Sp1, or anti-Egr1 antibody, separated on the agarose gel, and visualized under UV light. As shown, anti-Sp1 and anti-Egr1 antibodies further retarded the migration of ZnPP-proteins complexes, whereas incubation with IgG had no effect (Fig. 4B). These results suggest that ZnPP interacts with the endogenous Sp1 and Egr1 proteins. Of note, we used agarose gels in these experiments because ZnPP-protein complexes were unable to migrate out of the wells in polyacrylamide gels.3 Agarose gels have large pores, which suggests that the ZnPP-protein complexes are large and may involve multimers. This remains to be determined systematically.
Previously, others have shown that protoporphyrin interacts with DNA in vitro (32). Because ZnPP affects the ability of transcription factors to bind to the S/E site of the CCND1 locus, we wondered if this may be due to DNA conformational changes resulting from direct interaction between ZnPP and DNA. By using the same approach as described above, a DNA fragment harboring the S/E site (SE), which was previously analyzed in the ChIP assay, was incubated with ZnPP. For the control, a DNA fragment lacking the S/E site (non-SE) was used, which was isolated from D1pro-1095 by restriction enzyme digestion with KpnI or DpnI. As shown in Fig. 4C, equal fluorescence was observed with SE and non-SE DNA fragments. Ethidium bromide staining of the gel showed that the SE or non-SE DNA fragments did not migrate differentially after ZnPP incubation and no additional bands appeared in either sample (Fig. 4D). This suggests that ZnPP does not specifically interact with DNA targets harboring S/E sites.
Overexpression of Sp1 or Depletion of Egr1 Diminishes ZnPP-inhibited Cell Proliferation
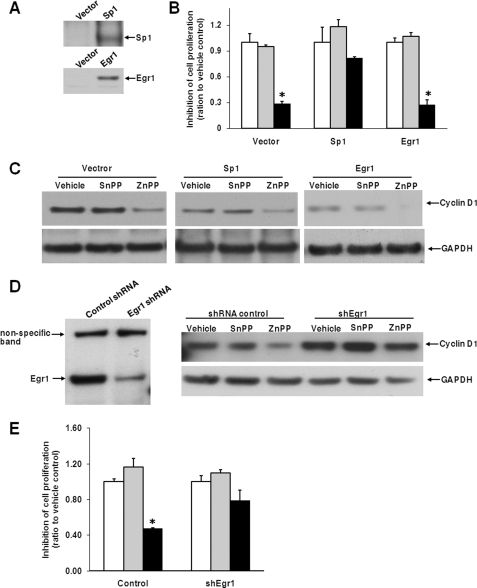
To further explore the role of Egr1 and Sp1 in the function of ZnPP, we introduced Egr1 and Sp1 into HepG2 cells using a retroviral system (Fig. 5A). As shown in Fig. 5B, overexpression of Sp1 diminished ZnPP-inhibited cell proliferation (p = 0.1 for ZnPP versus vehicle). On the other hand, exogenous Egr1 did not enhance ZnPP-suppressed cell proliferation compared with control. Also, Sp1 overexpression dampened the inhibition of cyclin D1 by ZnPP, whereas this was not affected by overexpression of Egr1 (Fig. 5C). This could be due to a high level of endogenous Egr1 already maximizing the effect of ZnPP. To address this issue, we further depleted endogenous Egr1 expression by using a shRNA lentiviral model. Compared with shRNA control, shEgr1 successfully decreased the expression of Egr1, and compromised the inhibition of cyclin D1 mediated by ZnPP (Fig. 5D). Additionally, the suppression of cell proliferation by ZnPP was diminished by silencing Egr1 (Fig. 5E; p = 0.1 for ZnPP versus vehicle). The exogenous Sp1 or Egr1 protein levels were not changed by ZnPP incubation.5 In summary, these data demonstrate that Egr1 and Sp1 play critical roles in the function of ZnPP.
FIGURE 5.
Sp1 and Egr1 modify ZnPP-inhibited cell proliferation. HepG2 cells were infected with retroviruses expressing Sp1, Egr1, or empty vector as control, and analyzed with Western analysis of Sp1 and Egr1 protein (A). These cells were further subjected to a cell proliferation assay (B) and Western blot analysis (C) as described in the legends to Figs. 1A and 3A. In D and E, HepG2 cells were infected with shRNA control or shEgr1 lentiviruses and selected with puromycin. Puromycin-resistant cells were subjected to Western analysis with anti-Egr1 and anti-cyclin D1 antibodies (D), and cell proliferation was evaluated (E). Equal loading is demonstrated with the housekeeping gene GAPDH. *, p < 0.05 versus vehicle. Clear bars, vehicle; gray bars, SnPP; and black bars, ZnPP, at 5 μm concentration.
Cyclin D1 Overexpression Alone Does Not Block ZnPP-mediated Inhibition of HepG2 Cell Proliferation
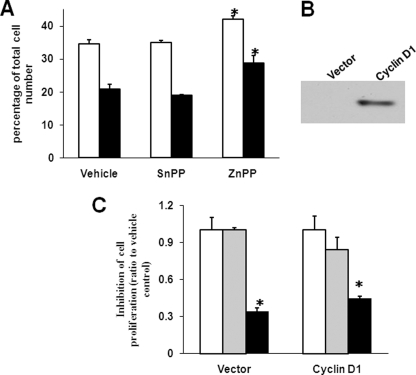
Because ZnPP significantly suppresses the expression of cyclin D1, which promotes cell cycle progression, we analyzed the cell cycle profile upon ZnPP incubation. Cell cycle progression was not affected after 24 h of ZnPP incubation.3 However, 48 h of ZnPP incubation significantly increased the number of cells at the G1 phase, whereas a 72-h incubation resulted in accumulation of cells at the G2 phase, suggesting that ZnPP delays the cell cycle at the G1/S and G2/M transitions (Fig. 6A). These data corroborate the role of cyclin D1 as a major regulator of the G1/S transition and the G2/M checkpoint (33). However, the possibility remains that other ZnPP targets may impede the cell cycle as well, particularly at the G2/M transition. Thus, cyclin D1 was overexpressed in HepG2 cells using a retroviral system (Fig. 6B). This was expected to rescue ZnPP-repressed HepG2 cell proliferation if cyclin D1 was the major target of ZnPP action. However, ZnPP still inhibited cell proliferation as shown in the vector control (p = 0.052 for ZnPP versus vehicle; Fig. 6C). This indicates that cyclin D1 overexpression alone cannot fully rescue HepG2 cells from ZnPP-mediated inhibition, suggesting that ZnPP may affect other targets involved in cell proliferation as well as cyclin D1. Of note, the exogenous cyclin D1 protein level was not affected by ZnPP in cells overexpressing cyclin D1.5
FIGURE 6.
Cyclin D1 overexpression alone does not block ZnPP-mediated suppression of HepG2 cell proliferation. By using propidium iodide staining by FACS, HepG2 cells were analyzed for G1 phase (empty bars) and G2 phase (solid bars) following serum starvation and incubation with 5 μm ZnPP for 2 or 3 days (A). In B, a representative Western blot of the cyclin D1 immunoreactive protein in control or cyclin D1 overexpressing cells is shown. In C, cyclin D1-overexpressing and control HepG2 cells were subjected to cell proliferation assays after incubation with 5 μm ZnPP for 72 h. *, p < 0.05 versus vehicle. Clear bars, vehicle; gray bars, SnPP; and black bars, ZnPP, at 5 μm concentration.
ZnPP Inhibits the Tumor Growth in Vivo
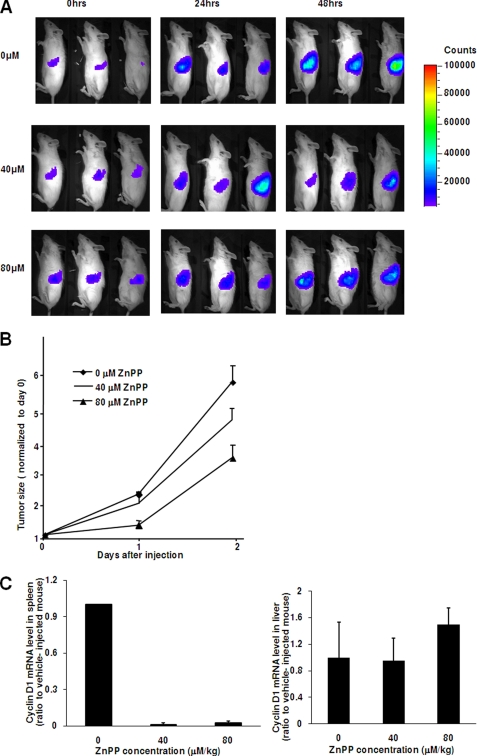
Although cyclin D1 overexpression alone cannot restore HepG2 cell proliferation inhibited by ZnPP, we speculated that ZnPP may have an inhibitory effect on tumors where cyclin D1 is the driving force for the origin and progression of tumor. To this end, an in vivo BCL1 model was used (24). As described previously, BCL1 originated spontaneously from aged mice (27). In these cells, the CCND1 locus is downstream of the Emu enhancer due to a chromosome translocation yet it contains the intact promoter. This results in constitutively high expression of cyclin D1 and consequently leads to tumorigenesis (24). Following intravenous injection, BCL1-gfp/luc cells were successfully engrafted into the animals (Fig. 7A). By measuring the amount of light emitted from the BCL1-gfp/luc cells, tumor growth was shown to be exponential in the saline-injected controls but decreased 2-fold following injection with both 80 and 40 μm/kg of ZnPP, albeit at a lower efficiency with the 40 μm/kg dose (Fig. 7B). Collectively, these data demonstrate that ZnPP inhibits BCL1 tumor progression in a dose-dependent manner. In the spleen where a majority of the tumor cells were engrafted, CCND1 expression was inhibited by more than 40-fold with both 40 and 80 μm/kg of ZnPP as compared with control (Fig. 7C). Intraperitoneal administration of ZnPP leads to its accumulation in the liver (2). We therefore evaluated cyclin D1 gene expression from the liver where the ZnPP concentration would be highest. Surprisingly, ZnPP did not significantly affect cyclin D1 expression in the liver (1.1 ± 0.33-fold for ZnPP versus control at 40 μm/kg and 1.49 ± 0.25-fold at 80 μm/kg, n = 6 in each group). This suggests that intraperitoneal injection of ZnPP specifically targets tumor cells rather than normal cells. Overall, these data demonstrate that ZnPP inhibits cyclin D1 expression and growth in engrafted BCL1 tumor cells in vivo.
FIGURE 7.
ZnPP inhibits tumor growth and cyclin D1 expression in the BCL1 tumor in vivo. In A and B, BCL1-gfp/luc recipients were injected with 0 (saline), 40, or 80 μm/kg of ZnPP followed by injection of luciferin and daily imaging. In A, representative sequential images of mice are shown in each group. Mice are in the lateral position with the left side up. In B, tumor growth was quantitated using light emission from BCL1-gfp/luc tumor cells. Values are the mean ± S.E. of six animals. In C, total RNA isolated from the spleen (left panel) or liver (right panel) of mice after inoculation with ZnPP was evaluated by qRT-PCR. The cyclin D1 expression level was normalized to GAPDH mRNA level. Values are the mean ± S.E. of six animals.
DISCUSSION
ZnPP is an endogenous metalloporphyrin formed under various clinical conditions (28, 34). The role of ZnPP in inhibiting tumor progression has been ascribed to its ability to inhibit HO activity (3–5). However, we show that ZnPP inhibits the proliferation of hepatoma and leukemia cells and cyclin D1 expression. This could not be entirely explained by decreased HO activity because another similarly potent metalloporphyrin, SnPP, did not exert the same effect. Moreover, an S/E site on the cyclin D1 promoter is responsible for ZnPP-mediated inhibition of cyclin D1 gene expression, and ZnPP interacts with transcription factors Sp1 and Egr1 to modulate their ability to bind to the S/E site of CCND1. Last, ZnPP administration significantly inhibits cyclin D1 expression from engrafted BCL1-gfp/luc cells but not normal cells, and further limits tumor progression of BCL1 in vivo.
In erythrocytes, ZnPP is normally present at a concentration of 0.5 μm with a ratio of 1:40,000 to heme. In pathological states, such as anemia and leukemia, ZnPP concentrations can increase to 5 μm (34). A previous study showed that ZnPP fails to stimulate the expression of globin genes in K562 cells, whereas heme did (7), suggesting a role for ZnPP in modifying cell differentiation in leukemia. In the current study, we showed that 1 to 5 μm doses of ZnPP suppress cell proliferation in K562 and HepG2 cells in part by inhibiting cyclin D1 expression. Corroborating our findings, the overexpression of cyclin D1 is known to play a role in hepatoma and leukemia development by stimulating cell proliferation (35, 36). Additionally, we showed that ZnPP significantly inhibited the expression of a number of genes critically related to cell proliferation and angiogenesis (Table 1). Among them, hepatocyte growth factor receptor (MET) is an protooncogene, and the hepatocyte growth factor/Met pathway is considered a novel target in cancer therapy (37). The vascular endothelial growth factor receptor (FLT1) plays a critical role in tumorigenesis and its inhibitors have been used in cancer therapy (38). Therefore, enhanced ZnPP formation could be an endogenous self-defense system that is important in pathological conditions, such as leukemia. Nonetheless, there may be other important ZnPP-targeted genes that we have not yet explored.
In addition to enhanced ZnPP production in leukemia, ZnPP inhibits tumor progression. In fact, micelles of styrene maleic acid containing 10 μm ZnPP damaged oncogenic cells derived from patients with chronic myelogenous leukemia (6). Here, we confirm that ZnPP clearly suppresses cyclin D1 expression in BCL1 tumor cells and limits tumor progression. Perhaps the effect of ZnPP antagonizes the influence of the Emu enhancer on the cyclin D1 locus in BCL1 growth. Cyclin D1 overexpression is an essential hallmark of BCL1 pathogenesis as described in the literature (24). For instance, cyclin D1 is expressed at high levels in almost all cases of mantle cell leukemia/lymphoma and plays an important role in the progression of cells through the G1 phase of the cell cycle and thus serves as an important prognostic factor (39). Herein, we used a BCL1 tumor model to help elucidate the role of ZnPP on cyclin D1 expression and its effects on tumor proliferation in vivo. On the other hand, suppression of BCL1 tumor progression would likely not be solely controlled by the expression of a single gene as with numerous other types of cancers. However, ZnPP significantly impacts cell proliferation and nearly eradicates CCND1 gene expression in this model at two different doses, suggesting that a correlation between ZnPP-decreased cyclin D1 expression and retarded tumor growth exists. Moreover, the possibility remains that other ZnPP-targeted genes may also contribute to tumor suppression. Importantly, despite accumulation of ZnPP in the liver of normal mice (2), intraperitoneal injection of ZnPP affected the expression of cyclin D1 predominantly in tumor cells found in spleen but not in the normal cells of liver. This could be important for clinic application.
Previously, we demonstrated that increased nuclear protein binding to the EBS site on the HMOX1 gene leads to enhanced HO-1 expression in the presence of ZnPP (11). Others have shown that enhanced Egr1 binding to the EBS site without Sp1 competition elevates cyclin D1 promoter activity (23). However, this mechanism could not account for regulation of the CCND1 gene by ZnPP in heptoma cell lines. Despite equal levels of Sp1 and Egr1 proteins, ZnPP preferentially facilitated Egr1 but interrupted Sp1 binding to the S/E site. This corroborates with the competitive nature of Egr1 and Sp1 transcription factors as to their binding to the S/E site. As previously shown, the molecular mechanism guiding the interplay between Egr1 and Sp1 is mainly controlled by competing for binding to the overlapping cis-acting S/E site (40–43). Decreased Sp1 or enhanced Egr1 binding could lead to either transcriptional activation or repression, respectively. For example, Egr1 acts as a negative regulator and represses Sp1-mediated activation of some genes including protein-tyrosine phosphatase 1B, Met, Epstein-Barr virus C, and β1-adrenergic receptor (40, 42, 44, 45). In another circumstance, Egr1 competes with Sp1 protein for an overlapping region in the promoter of platelet-derived growth factor A and functions as a positive activator (46). By altering the interplay between Sp1 and Egr1, ZnPP could provide a means to manipulate gene expression relevant to tumorigenesis. Consistent with this hypothesis, gene expression of Met and Flt-1, all regulated by the interplay between Sp1 and Egr1 on the S/E site (40, 47), are also inhibited by ZnPP in this study.
Porphyrins bind DNA directly at purine-rich regions. For example, cationic porphyrins inhibit telomerase activity by binding a G-rich telomeric DNA fragment. This suppresses gene expression of c-myc and K-Ras by stabilizing a purine-rich G-quadruplex DNA structure (48, 49). Although the S/E site of cyclin D1 is GC-rich, ZnPP did not preferentially bind to this region. However, we cannot exclude that ZnPP may associate with the chromatin structure surrounding the S/E site, facilitating Egr1 but interrupting Sp1 targeting on the S/E site. This remains to be explored.
Heme is a cofactor for transcription factors Bach1 and Rev-erbα, and heme in the Rev-erbα complex can be displaced by its analogues Ga(III)protoporphyrin IX chloride (GaPP) and Fe(III) mesoporphyrin IX chloride (FePP), which are structurally similar to ZnPP (13, 14). Increased ZnPP under certain pathological circumstances might modulate the activity of these transcription factors by replacing heme. Furthermore, HO-1 protein, which can be dramatically up-regulated by ZnPP, translocates into the nucleus and acts as a self-regulator under oxidative stress (50, 51). Thus, ZnPP-enhanced HO-1 protein may also play a role in ZnPP-mediated transcriptional regulation although preliminary observations do not yet confirm this.
In summary, we show that ZnPP inhibits tumor cell proliferation by suppressing cyclin D1 gene expression, at least in part. Moreover, ZnPP directly interacts with transcription factor complexes of Sp1 and/or Egr1 and modulates their ability to target the CCND1 locus. We also demonstrate that ZnPP inhibits BCL1 tumor progression, very likely through inhibition of cyclin D1 expression. We speculate that ZnPP plays a role in the regulation of other SE containing genes in normal and pathological states.
This work was supported, in whole or in part, by National Institutes of Health Grant HL70285-01 (to P. A. D.).
P. La, unpublished data.
C. J. Wright, unpublished data.
A. P. Fernando, unpublished data.
- ZnPP
- zinc protoporphyrin IX
- HO
- heme oxygenase
- qRT
- quantitative real time
- BCL1
- B-cell leukemia/lymphoma 1
- FACS
- fluorescence-activated cell sorting
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- S/E
- Sp1 and Egr1 overlapping binding site
- SnPP
- tin protoporphyrin
- shRNA
- short hairpin RNA
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Labbé R. F. (1992) Clin. Chem. 38, 2167–2168 [PubMed] [Google Scholar]
- 2.Rodgers P. A., Seidman D. S., Wei P. L., Dennery P. A., Stevenson D. K. (1996) Pediatr. Res. 39, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 3.Fang J., Sawa T., Akaike T., Greish K., Maeda H. (2004) Int. J. Cancer 109, 1–8 [DOI] [PubMed] [Google Scholar]
- 4.Fang J., Sawa T., Akaike T., Akuta T., Sahoo S. K., Khaled G., Hamada A., Maeda H. (2003) Cancer Res. 63, 3567–3574 [PubMed] [Google Scholar]
- 5.Hirai K., Sasahira T., Ohmori H., Fujii K., Kuniyasu H. (2007) Int. J. Cancer 120, 500–505 [DOI] [PubMed] [Google Scholar]
- 6.Mayerhofer M., Gleixner K. V., Mayerhofer J., Hoermann G., Jaeger E., Aichberger K. J., Ott R. G., Greish K., Nakamura H., Derdak S., Samorapoompichit P., Pickl W. F., Sexl V., Esterbauer H., Schwarzinger I., Sillaber C., Maeda H., Valent P. (2008) Blood 111, 2200–2210 [DOI] [PubMed] [Google Scholar]
- 7.Palma J. F., Gao X., Lin C. H., Wu S., Solomon W. B. (1994) Blood 84, 1288–1297 [PubMed] [Google Scholar]
- 8.Ny L., Andersson K. E., Grundemar L. (1995) Br. J. Pharmacol. 115, 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serfass L., Burstyn J. N. (1998) Arch. Biochem. Biophys. 359, 8–16 [DOI] [PubMed] [Google Scholar]
- 10.Argyris E. G., Vanderkooi J. M., Venkateswaran P. S., Kay B. K., Paterson Y. (1999) J. Biol. Chem. 274, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 11.Yang G., Nguyen X., Ou J., Rekulapelli P., Stevenson D. K., Dennery P. A. (2001) Blood 97, 1306–1313 [DOI] [PubMed] [Google Scholar]
- 12.Hou W., Shan Y., Zheng J., Lambrecht R. W., Donohue S. E., Bonkovsky H. L. (2008) Biochim. Biophys. Acta 1779, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 14.Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., Igarashi K. (2001) EMBO J. 20, 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stendahl M., Kronblad A., Rydén L., Emdin S., Bengtsson N. O., Landberg G. (2004) Br. J. Cancer 90, 1942–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbieri F., Lorenzi P., Ragni N., Schettini G., Bruzzo C., Pedullà F., Alama A. (2004) Oncology 66, 310–315 [DOI] [PubMed] [Google Scholar]
- 17.Ikehara M., Oshita F., Ito H., Ohgane N., Suzuki R., Saito H., Yamada K., Noda K., Mitsuda A., Kameda Y. (2003) Oncol. Rep. 10, 137–139 [PubMed] [Google Scholar]
- 18.Hashemolhosseini S., Nagamine Y., Morley S. J., Desrivières S., Mercep L., Ferrari S. (1998) J. Biol. Chem. 273, 14424–14429 [DOI] [PubMed] [Google Scholar]
- 19.Lin S., Wang W., Wilson G. M., Yang X., Brewer G., Holbrook N. J., Gorospe M. (2000) Mol. Cell. Biol. 20, 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiozawa T., Miyamoto T., Kashima H., Nakayama K., Nikaido T., Konishi I. (2004) Oncogene 23, 8603–8610 [DOI] [PubMed] [Google Scholar]
- 21.Matsumura I., Kitamura T., Wakao H., Tanaka H., Hashimoto K., Albanese C., Downward J., Pestell R. G., Kanakura Y. (1999) EMBO J. 18, 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee R. J., Albanese C., Stenger R. J., Watanabe G., Inghirami G., Haines G. K., 3rd, Webster M., Muller W. J., Brugge J. S., Davis R. J., Pestell R. G. (1999) J. Biol. Chem. 274, 7341–7350 [DOI] [PubMed] [Google Scholar]
- 23.Yan Y. X., Nakagawa H., Lee M. H., Rustgi A. K. (1997) J. Biol. Chem. 272, 33181–33190 [DOI] [PubMed] [Google Scholar]
- 24.Callanan M., Leroux D., Magaud J. P., Rimokh R. (1996) Crit. Rev. Oncog. 7, 191–203 [DOI] [PubMed] [Google Scholar]
- 25.Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., Diehl J. A. (2006) Mol. Cell 24, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vreman H. J., Mahoney J. J., Van Kessel A. L., Stevenson D. K. (1988) Clin. Chem. 34, 2562–2566 [PubMed] [Google Scholar]
- 27.Edinger M., Cao Y. A., Verneris M. R., Bachmann M. H., Contag C. H., Negrin R. S. (2003) Blood 101, 640–648 [DOI] [PubMed] [Google Scholar]
- 28.Lutton J. D., Abraham N. G., Drummond G. S., Levere R. D., Kappas A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1432–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alao J. P. (2007) Mol. Cancer 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy P. G., Thompson A. M. (2006) Breast 15, 718–727 [DOI] [PubMed] [Google Scholar]
- 31.Petty W. J., Dragnev K. H., Dmitrovsky E. (2003) Lung Cancer 41, Suppl. 1, S155–S161 [DOI] [PubMed] [Google Scholar]
- 32.Tong A. J., Tong C. Y., Yang Q. Y. (2003) Spectrochim. Acta A Mol. Biomol. Spectrosc. 59, 2967–2970 [DOI] [PubMed] [Google Scholar]
- 33.Stacey D. W. (2003) Curr. Opin. Cell Biol. 15, 158–163 [DOI] [PubMed] [Google Scholar]
- 34.Iyer J. K., Shi L., Shankar A. H., Sullivan D. J., Jr. (2003) Mol. Med 9, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aref S., Mabed M., El-Sherbiny M., Selim T., Metwaly A. (2006) Hematology 11, 31–34 [DOI] [PubMed] [Google Scholar]
- 36.Xu J. M., Wen J. M., Zhang M., Lü G. L., Wu L. Z., Wang W. S. (2004) Zhonghua Bing Li Xue Za Zhi 33, 26–30 [PubMed] [Google Scholar]
- 37.Sattler M., Salgia R. (2007) Curr. Oncol. Rep. 9, 102–108 [DOI] [PubMed] [Google Scholar]
- 38.Homsi J., Daud A. I. (2007) Cancer Control 14, 285–294 [DOI] [PubMed] [Google Scholar]
- 39.Ravandi-Kashani F., O'Brien S., Manshouri T., Lerner S., Sim S., Dodd K., Kantarjian H., Freireich E., Keating M., Albitar M. (2000) Leuk. Res. 24, 469–474 [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Liu Y. (2003) Am. J. Physiol. Renal Physiol. 284, F1216–F1225 [DOI] [PubMed] [Google Scholar]
- 41.Nenoi M., Ichimura S., Mita K., Yukawa O., Cartwright I. L. (2001) Cancer Res. 61, 5885–5894 [PubMed] [Google Scholar]
- 42.Bahouth S. W., Beauchamp M. J., Vu K. N. (2002) Mol. Pharmacol. 61, 379–390 [DOI] [PubMed] [Google Scholar]
- 43.Huang R. P., Fan Y., Ni Z., Mercola D., Adamson E. D. (1997) J. Cell. Biochem. 66, 489–499 [PubMed] [Google Scholar]
- 44.Fukada T., Tonks N. K. (2001) J. Biol. Chem. 276, 25512–25519 [DOI] [PubMed] [Google Scholar]
- 45.Nilsson T., Zetterberg H., Wang Y. C., Rymo L. (2001) J. Virol. 75, 5796–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silverman E. S., Khachigian L. M., Lindner V., Williams A. J., Collins T. (1997) Am. J. Physiol. Heart Circ. Physiol. 273, H1415–H1426 [DOI] [PubMed] [Google Scholar]
- 47.Akuzawa N., Kurabayashi M., Ohyama Y., Arai M., Nagai R. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 377–384 [DOI] [PubMed] [Google Scholar]
- 48.Hurley L. H., Von Hoff D. D., Siddiqui-Jain A., Yang D. (2006) Semin. Oncol. 33, 498–512 [DOI] [PubMed] [Google Scholar]
- 49.Dixon I. M., Lopez F., Estève J. P., Tejera A. M., Blasco M. A., Pratviel G., Meunier B. (2005) ChemBioChem 6, 123–132 [DOI] [PubMed] [Google Scholar]
- 50.Lin Q. S., Weis S., Yang G., Zhuang T., Abate A., Dennery P. A. (2008) Free Radic. Biol. Med. 44, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q., Weis S., Yang G., Weng Y. H., Helston R., Rish K., Smith A., Bordner J., Polte T., Gaunitz F., Dennery P. A. (2007) J. Biol. Chem. 282, 20621–20633 [DOI] [PubMed] [Google Scholar]