Abstract
Intracellular lipid accumulation in the heart is associated with cardiomyopathy, yet the precise role of triglyceride (TG) remains unclear. With exercise, wild type hearts develop physiologic hypertrophy. This was associated with greater TG stores and a marked induction of the TG-synthesizing enzyme diacylglycerol (DAG) acyltransferase 1 (DGAT1). Transgenic overexpression of DGAT1 in the heart using the cardiomyocyte- specific α-myosin heavy chain (MHC) promoter led to approximately a doubling of DGAT activity and TG content and reductions of ∼35% in cardiac ceramide, 26% in DAG, and 20% in free fatty acid levels. Cardiac function assessed by echocardiography and cardiac catheterization was unaffected. These mice were then crossed with animals expressing long-chain acyl-CoA synthetase via the MHC promoter (MHC-ACS), which develop lipotoxic cardiomyopathy. MHC-DGAT1XMHC-ACS double transgenic male mice had improved heart function; fractional shortening increased by 74%, and diastolic function improved compared with MHC-ACS mice. The improvement of heart function correlated with a reduction in cardiac DAG and ceramide and reduced cardiomyocyte apoptosis but increased fatty acid oxidation. In addition, the survival of the mice was improved. Our study indicates that TG is not likely to be a toxic lipid species directly, but rather it is a feature of physiologic hypertrophy and may serve a cytoprotective role in lipid overload states. Moreover, induction of DGAT1 could be beneficial in the setting of excess heart accumulation of toxic lipids.
Introduction
Triglyceride (TG)2 is the major energy storage form in organs. The final step of TG synthesis, the conversion of diacylglycerol (DAG) to TG, is catalyzed by diacylglycerol acyltransferase (DGAT) enzymes. DGAT1 and DGAT2 are unrelated proteins that exhibit DGAT activity (1). DGAT1 belongs to a gene family that includes ACAT1 and ACAT2 (acyl-CoA:cholesterol acyltransferases 1 and 2) (1–3), whereas DGAT2 is a member of a larger gene family whose members include acyl-CoA:monoacylglycerol acyltransferase (3). Although both enzymes catalyze the same reaction in TG synthesis, they are functionally distinguished by their differences in regulation and substrate specificity (2, 4–7). For example, DGAT1, but not DGAT2, will esterify other lipids such as retinol (2, 8). DGAT1 is widely expressed in all tissues, with high expression in white adipose tissue, skeletal muscle, heart, and intestine (1); DGAT2 is primarily expressed in the liver (1, 2).
Studies to understand the roles of DGAT1 and -2 have been performed using genetically modified mice. Investigators have studied whether DGATs regulate insulin actions and toxic effects of lipids on tissue. DGAT1 knock-out mice have reduced obesity on a high fat diet (6). Moreover, when these mice were crossed onto the agouti background they had increased insulin sensitivity (9). Transplantation of DGAT1-deficient adipose tissue into wild type (WT) mice decreased adiposity and increased insulin sensitivity (10). These experiments suggest that DGAT1 inhibition reduces obesity/lipid storage and promotes insulin actions. This has spurred pharmacologic development with the objective of inhibiting DGAT1 as a treatment for obesity.
In contrast to the beneficial effects of DGAT1 inhibition on obesity development, overexpression of DGAT1 using an AP2 promoter led to greater obesity but not insulin resistance in C57BL6 (11) but not FVB mice (12). Overexpression of DGAT1 in the liver (13) or in the skeletal muscle (14) increased TG storage but did not cause insulin resistance. Thus, in some situations greater conversion of toxic lipids, such as DAG, ceramide, or fatty acids, to inert TG may be beneficial (14, 15).
We first studied whether chronic exercise increases heart TG storage and induces lipid metabolism. Highly trained rodents and humans have skeletal muscle TG accumulation associated with up-regulation of DGAT1 (16, 17), but whether similar changes in lipid content occur in the setting of physiologic hypertrophy of the heart is unclear. We then created mice expressing DGAT1 in cardiomyocytes using the α-myosin heavy chain promoter (the transgene is referred to as MHC-DGAT1). We report the characterization of these mice and the effect of MHC-DGAT1 added to a model of cardiac lipotoxicity.
EXPERIMENTAL PROCEDURES
Exercise Protocol
All animal procedures were approved by the Institutional Animal Care and Use Committee at Columbia University. Eight-week-old FVB mice were assigned to sedentary or exercised groups. Mice were swum in tanks. The swimming protocol was a modification of the procedure used by Ryder et al. (18). Water temperature was maintained at 34–35 °C. Mice were swum for 10-min sessions twice a day separated by a 10-min break. Sessions were increased by 10 min each day until the 4th day; mice were then allowed to swim for six 30-min intervals separated by 10–15-min rest periods. After the last swim interval, mice were dried and put back in their cages for 18–20 h.
Creation of Transgenic Mice and Mouse Breeding
The MHC-DGAT1 transgene contained, from the 5′- to the 3′-end, a 5.5-kb MHC promoter, a full-length human DGAT1 cDNA and a 0.97-kb PCR fragment of human growth hormone genomic DNA containing two introns (four and five), three exons (three, four, and five), and the poly(A) signal (Fig. 2A). Transgenic mice were derived from fertilized eggs of the CBA/C57BL6 hybrid. Lines of MHC-DGAT1/FVB mice were developed after six backcrosses with the wild type FVB strain (The Jackson Laboratory). The O7 line of MHC-acyl-CoA synthetase 1 (MHC-ACS) mice has been described previously (19). Genotyping was carried out by PCR or Southern blot using genomic DNA extracted from mouse tails (14).
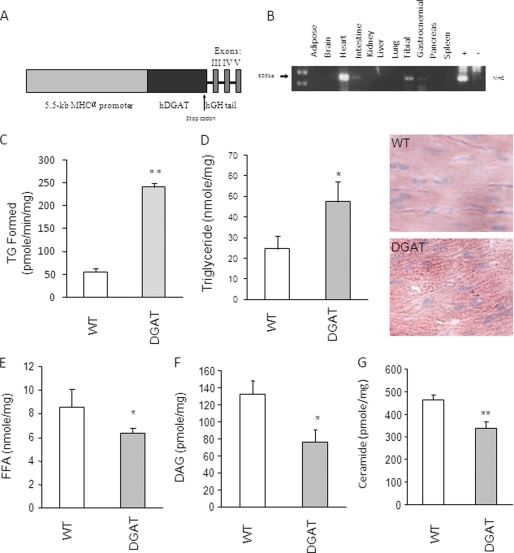
FIGURE 2.
Creation and characterization of MHC-DGAT1 transgenic mice. A, DGAT1 transgene contains (from the 5′- to 3′-end) the 5.5-kb MHC promoter, a human DGAT1 cDNA containing its own initiation and termination codons, and the genomic sequence of human growth hormone (hGH) containing the last 3 exons and 2 introns as indicated. B, tissue distribution of the DGAT1 transgene mRNA levels measured by reverse transcription and PCR amplification. C, total DGAT activity levels in membrane fractions of heart muscles from WT and transgenic mice (n = 3 in each group). D–G, TG, FFA, DAG, and ceramide contents in heart muscles isolated from WT and transgenic mice (n = 5–6). *, p < 0.05; **, p < 0.01.
Measurement of DGAT Activity
Heart muscle DGAT activity was measured in vitro in membrane fractions isolated from muscle specimens using [14C]palmitoyl-CoA as described previously (14, 20). Briefly, ventricular heart muscle was homogenized in a buffer (20 mm HEPES, pH 7.4, 1 mm CaCl2, 1 mm MgCl2), 1 mm dithiothreitol, and a mixture of protease inhibitors containing phenylmethylsulfonyl fluoride, leupeptin, and pepstatin. One-fourth volume of 30% sucrose was added to the sample immediately following homogenization. The homogenization mixture was then centrifuged at 1,500 × g for 10 min at 4 °C. The supernatant was spun at 150,000 × g for 1 h at 4 °C. The membrane pellet was homogenized and resuspended in a buffer containing 20 mm HEPES, pH 7.4, 0.25 m sucrose, 1 mm dithiothreitol, and protease inhibitors. Protein concentration was determined using the protein assay reagent kit (Bio-Rad).
To measure DGAT1 activity, 10 μg of membrane protein was used in a 200-μl reaction mixture containing 100 mm Tris, pH 7.5, 250 mm sucrose, 1 mg/ml bovine serum albumin, 150 mm MgCl2, 0.8 mm EDTA, 0.25 mm 1,2-dioleoyl-sn-glycerol, and 25 μm palmitoyl-CoA containing 0.3 μCi of 14C. The reaction was carried out at 37 °C for 5 min and stopped by adding 0.75 ml of lipid extraction solvents (chloroform/methanol in a ratio of 2:1). After adding 0.375 ml of acidic solution (1 mm H2SO4, 17 mm NaCl), the organic phase was removed and evaporated under a stream of N2. Lipids were redissolved in 20 μl of hexane and then analyzed by TLC using plates that were developed in hexane/diethyl ether/glacial acetic acid in a ratio of 70:30:1 (v/v). Chromatographic bands containing TG were cut out after iodine staining and quantified by scintillation counting.
Lipid Extraction, TG, and Free Fatty Acid (FFA) Measurements
Lipids were assessed in hearts isolated from mice fasted for 12 h and perfused with phosphate-buffered saline. To measure TG, DAG, FFA, and ceramide, lipids were first extracted from muscles using chloroform/methanol/HCl (21). Butylated hydroxytoluene (0.01%) was added in the extraction solution as an antioxidant, and [3H]triolein (0.25 μCi) was added as an internal standard for TG recovery. TG and FFA mass in lipid extracts was determined enzymatically with a colorimetric kit (TG from Sigma and FFA from Wako) and normalized to recovered standard. Blood from fasted (12 h) mice was collected from retro-orbital plexus for the measurement of plasma total cholesterol (Thermo Electron Corp.), TG, and FFAs.
DAG Kinase Activity
DAG and ceramide levels were measured using a DAG kinase-based method, in which DAG and ceramide are phosphorylated to form 32P-labeled phosphatidic acid and ceramide 1-phosphate, respectively, and then quantified (22). Lipids extracted from muscle were dried under a N2 stream and redissolved in 7.5% octyl β-d-glucoside containing 5 mm cardiolipin and 1 mm diethylenetriamine pentaacetate. The reaction was performed at room temperature for 30 min in 100 mm imidazole HCl, 100 mm NaCl, 25 mm MgCl2, 2 mm EGTA, pH 6.6, 2 mm dithiothreitol, 10 μg/100 μl DAG kinase (Sigma), 1 mm ATP, and 1 μCi/100 μl [γ-32P]ATP. The reaction was stopped by addition of chloroform/methanol (2:1, v/v) and 1% HClO4. Lipids were extracted and washed twice with 1% HClO4. Lipids were redissolved in chloroform/methanol and analyzed by TLC (Partisil K6 adsorption TLC plates, Whatman) with a mobile phase containing chloroform/methanol/acetic acid (65:15:5, v/v). The bands corresponding to phosphatidic acid and ceramide 1-phosphate were identified and silicon-scraped into a scintillation vial for radioactivity measurement. [3H]Triolein bands from the same TLC plates were also identified and quantified in the same way and were used as controls for lipid recovery.
Gene Expression
Total RNA was extracted using a TRIzol kit from Invitrogen. One μg of RNA was initially treated with DNase I (Invitrogen). The RNA samples were then reverse-transcribed using the SuperScript III First-strand Synthesis System for reverse transcription-PCR (Invitrogen). Real time amplification was performed using iQ SYBR Green Supermix (Bio-Rad). Primers used for PCR amplification are listed in supplemental Table 1. Analysis was performed using iCycler iQ Real Time Detection software (Bio-Rad).
Histology
Ventricle heart muscle was embedded in Tissue-Tek OCT compound (Sakura Finetek). Tissue sections (7-μm sections) were stained with oil red O and counterstained with hematoxylin and eosin.
Echocardiographic Analysis
Two-dimensional echocardiography was performed in conscious 3–4-month-old mice with light anesthesia using techniques described previously (Sonos 5500 System; Philips Medical Systems) (23). Two-dimensional echocardiographic images were obtained and recorded in a digital format. Images were analyzed off-line by a researcher blinded to the murine genotype. Left ventricular end diastolic dimension (LVDd) and left ventricular end systolic dimension (LVDs) were measured. Percent fractional shortening (FS), which quantifies contraction of the ventricular wall and is an indication of muscle function, was calculated as % FS = ((LVDd − LVDs)/LVDd) × 100.
Cardiac Catheterization
Hemodynamic evaluation in intact closed-chest anesthetized mice (4 months of age) was performed using a 1.4F Millar MIKRO-TIP catheter/pressure transducer (SPR-671, Millar Instruments, Inc.). Mice from the different groups were anesthetized using pentobarbital (50–100 mg/g, intraperitoneal). The Millar catheter was inserted into the right carotid artery and advanced retrograde into the left ventricle. After a stabilization period, base-line hemodynamic parameters were recorded using the PowerLab system (AD Instruments). The left ventricular end diastolic pressure and the maximal rates of rise and fall in left ventricular pressure (LVdP/dtmax and LVdP/dtmin) were determined from consecutive 10 beats and then averaged. All measurements and data analysis were performed in a blind fashion by a single investigator who had no knowledge of the genotype of the mice tested.
Mitochondrial DNA Copy Number
Mitochondrial DNA copy number was measured using quantitative real time PCR as described previously (24). Mitochondrial DNA for NADH dehydrogenases 1 (forward, acgcaaaatcttagggtaca, and reverse, tattatggctatgggtcagg) and nuclear DNA for β-actin (forward, ctgcctgacggccagg, and reverse, ggaaaagagcctcagggcat) were co-amplified within the same reaction wells.
Fatty Acid Uptake and Oxidation
Heart muscle fatty acid uptake and oxidation were measured using heart muscle slices as described previously (14, 25, 26). Hearts were removed and immediately sliced with a sharp razor transversely across the ventricles forming rings ∼0.1–0.2-mm thick and usually 20–30 mg wet weight. The time between the removal of the heart from the body and the completion of the slicing was <5 min. Isolated heart muscles were first washed for 1 h at 37 °C in Krebs-Henseleit bicarbonate buffer (10 mm HEPES, pH 7.35, 24 mm NaHCO3, 114 mm NaCl, 5 mm KCl, 1 mm MgCl2, and 2.2 mm CaCl2) containing 8 mm glucose, 32 mm mannitol, and 0.1% bovine serum albumin. The reaction was initiated by adding to the mix pre-conjugated bovine serum albumin/fatty acid solution (final concentrations: 0.2 mm palmitate with 10 mCi/ml of 9,10-[3H]palmitate and 1.25% bovine serum albumin). The fatty acid oxidation product 3H2O was determined in muscles incubated for 2 h at 37 °C. An aliquot of the completed reaction mixture was transferred into an open microtube that was then placed inside a scintillation vial containing 0.5 ml of distilled water. The scintillation vial was then capped and incubated at 50 °C for 18–24 h to reach vapor-phase equilibrium. After the transfer of 3H2O from the reaction mixture to the scintillation vial (by water equilibration), the 3H2O in the scintillation vial was scintillation-counted. The transfer and counting efficiency was calibrated using a standard solution containing 0.1 μCi of 3H2O for re-equilibration. After washing with three times ice-cold phosphate-buffered saline, the tissue was homogenized, and trapped radioactivity plus total produced 3H2O were counted as fatty acid uptake.
Statistics
All data are presented as mean ± S.D., except if indicated. Comparisons between two groups were performed using unpaired two-tailed Student's t tests with Statistica version 6.0 (StatSoft). p value less than 0.05 was used to determine statistical significance. Multiple regression analyses were performed to show correlations between heart lipids and heart function. The forward and backward stepwise regression analyses were used to assess independent effects and contributions of different heart lipids and function. Survival was analyzed by log-rank test.
RESULTS
Exercise Increases Heart TG Stores and Induces DGAT1
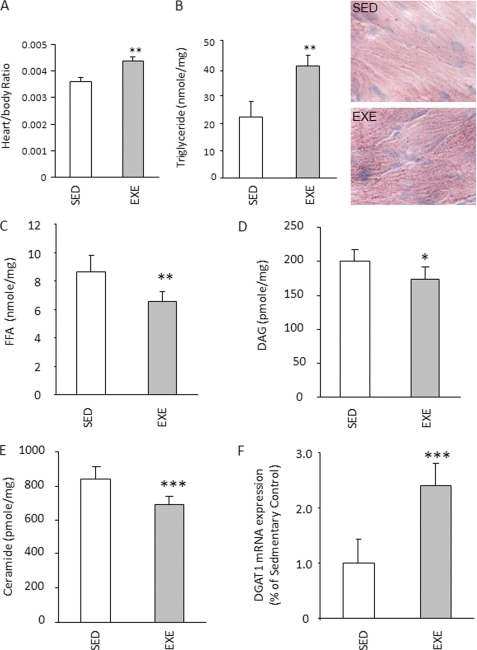
WT mice underwent a 2-week intensive swimming exercise protocol. At the end of this period, plasma cholesterol, TG, and FFA were decreased, but there was no change in plasma glucose (supplemental Table 2). Heart to body weight ratio increased ∼28%, and heart TG content doubled (Fig. 1, A and B). Other potentially toxic cardiac lipids such as FFA (∼15% less), ceramide (∼10% less), and DAG (∼10% less) were all reduced (Fig. 1, C–E). These changes in lipid metabolites were associated with an ∼3-fold increase in cardiac DGAT1 mRNA expression (Fig. 1F). Changes in gene expression with exercise suggested greater TG uptake and oxidation; lipid uptake genes CD36 and lipoprotein lipase were more than doubled, and PDK4, a regulator of glucose oxidation, and acyl-CoA oxidase and CPT1 increased about 2-fold in the exercise group (Table 1). PPARα mRNA was elevated about 3-fold in the exercise group, and mRNA for PGC1α (PPARγ coactivator 1α), a regulator of mitochondrial biogenesis, increased >3-fold. Adipose TG lipase (ATGL), the enzyme that initiates intracellular TG lipolysis (26), was also induced. mRNA levels of heart failure markers (ANF and BNP), PPARγ and -δ, and DGAT2 were not significantly altered. Therefore, similar to skeletal muscle (14), exercise training leads to more cardiac TG storage, increased expression of fatty acid oxidation genes, and marked up-regulation of the TG storage enzyme DGAT1.
FIGURE 1.
Exercise effects on heart DGAT1 expression and lipid. A, heart to body weight ratio in sedentary (SED) and exercised (EXE) FVB mice (n = 5). B–E, TG, FFA, DAG, and ceramide content in heart muscles isolated from sedentary and exercised FVB mice (n = 5). F, relative DGAT1 expression in exercise group compared with sedentary group, real time PCR (n = 5). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
TABLE 1.
mRNA expression in exercised mice
| Gene | Sedentary | Exercise |
|---|---|---|
| CD36 | 1 ± 0.17 | 3.12 ± 1.4a |
| LPL | 1 ± 0.47 | 5.6 ± 1.39 |
| PDK4 | 1 ± 0.39 | 4.0 ± 1.8b |
| AOX1 | 1 ± 0.32 | 2.24 ± 0.98b |
| CPT1 | 1 ± 0.51 | 2.17 ± 1.0a |
| PPARα | 1 ± 0.40 | 2.9 ± 0.77a |
| PGC1α | 1 ± 0.48 | 3.8 ± 1.3a |
| ATGL | 1 ± 0.38 | 2.4 ± 1.52a |
n = 5.
a p < 0.05 versus sedentary mice.
b p < 0.01 versus sedentary mice.
Creation of MHC-DGAT1 Transgenic Mice
To determine whether DGAT1 up-regulation in the heart would reproduce the metabolic effects of exercise, we generated mice with cardiac selective overexpression of DGAT1 using the MHC promoter (Fig. 2A). Although the transgene was expressed primarily in heart, a small amount of expression was also found in skeletal muscle but not in other tissues (Fig. 2B). DGAT activity was increased ∼3-fold in cardiac tissue from these mice (Fig. 2C), and this was associated with a doubling of heart TG (Fig. 2D). FFA in plasma was decreased about 10%, whereas plasma TG was not changed (supplemental Table 3). Because DGAT1 uses FFA to convert DAG to TG, we assessed the concentration of these lipids in heart tissue. There was an ∼20% reduction in FFA and ∼30% reduction in DAG (Fig. 2, E and F). Ceramide was also reduced ∼20% in MHC-DGAT1 hearts (Fig. 2G). Thus, mice overexpressing functional DGAT1 in the heart muscle were created, and the expected effect of the DGAT1 transgene, an increase in TG production, occurred.
Expression of Genes Affecting Lipid Metabolism and Heart Failure
Genes related to heart metabolism and heart failure were assessed (Table 2). We first determined whether the transfer of more lipids into TG led to reduced expression of genes regulated by PPARs and was responsible for fatty acid oxidation. Neither PPARα nor -δ expression was altered. Acyl-CoA oxidase was not changed, but CPT1 increased about 3-fold in MHC-DGAT1 transgenic mouse hearts. PDK4 was increased ∼2.5-fold in the MHC-DGAT1 mice. CD36 was up-regulated ∼3-fold, whereas lipoprotein lipase was not changed. ATGL was 4-fold higher than in the WT, whereas HSL was not changed. This gene expression pattern suggests that MHC-DGAT1 hearts both store and oxidize more lipids. ANF, BNP, and indicators of endoplasmic reticulum stress (CHOP and BIP), were not increased by the MHC-DGAT1 transgene (Table 2). Thus, the changes in metabolic gene expression and the cardiac lipid profile due to DGAT1 appeared similar to those of exercise.
TABLE 2.
mRNA expression in 3 month-old male WT, MHC-Dgat1, MHC-ACS1, and MHC-DGAT1/MHC-ACS1 mice
n = 6. Gly-6-Pase is glucose-6-phosphate dehydrogenase.
| mRNA | WT | DGAT | ACS | ACSXDGAT |
|---|---|---|---|---|
| Lipid and blood metabolic genes | ||||
| PPARα | 1 ± 0.17 | 0.94 ± 0.11 | 0.38 ± 0.08a | 0.67 ± 0.16a,b |
| PPARδ | 1 ± 0.28 | 1.17 ± 0.37 | 0.67 ± 0.13a | 0.55 ± 0.12 |
| PPARγ | 1 ± 0.14 | 1.28 ± 0.07 | 0.41 ± 0.15a | 0.74 ± 0.17a,b |
| ACO1 | 1 ± 0.32 | 0.75 ± 0.25 | 1.21 ± 0.41 | 0.57 ± 0.29 |
| CPT1 | 1 ± 0.33 | 3.15 ± 1.06a | 1.27 ± 0.26 | 1.83 ± 0.62 |
| PDK4 | 1 ± 0.22 | 2.56 ± 0.17a | 0.66 ± 0.37 | 0.52 ± 0.29 |
| CD36 | 1 ± 0.28 | 3.13 ± 0.37c | 2.14 ± 0.56a | 1.56 ± 0.30 |
| LPL | 1 ± 0.10 | 1.02 ± 0.24 | 0.74 ± 0.20 | 0.86 ± 0.18 |
| ATGL | 1 ± 0.19 | 4.2 ± 0.33c | 1.51 ± 0.48 | 3.37 ± 0.68a,b |
| HSL | 1 ± 0.28 | 1.08 ± 0.09 | 0.56 ± 0.14a | 0.82 ± 0.14b |
| Heart failure genes | ||||
| ANF | 1 ± 0.21 | 0.7 ± 0.32 | 1.91 ± 0.6a | 0.91 ± 0.42b |
| BNP | 1 ± 0.28 | 0.57 ± 0.42 | 3 ± 0.57a | 1.57 ± 0.30b |
| Endoplasmic reticulum stress genes | ||||
| BIP | 1 ± 0.15 | 0.87 ± 0.28 | 1.09 ± 0.18 | 1.22 ± 0.27 |
| CHOP | 1 ± 0.33 | 0.98 ± 0.27 | 0.86 ± 0.23 | 0.65 ± 0.21 |
| Apoptosis genes | ||||
| BCL2 | 1 ± 0.32 | 1.26 ± 0.25 | 0.43 ± 0.18a | 0.79 ± 0.26b |
| BAX | 1 ± 0.24 | 0.86 ± 0.28 | 0.75 ± 0.37 | 1.21 ± 0.33 |
| Caspase9 | 1 ± 0.19 | 0.97 ± 0.22 | 2.03 ± 0.31a | 2.29 ± 0.49a |
| ROS genes | ||||
| Catalase | 1 ± 0.28 | 3.27 ± 0.81c | 0.24 ± 0.07a | 0.56 ± 0.12b |
| Gly-6-Pase | 1 ± 0.17 | 0.82 ± 0.27 | 5.76 ± 1.73c | 0.71 ± 0.19d |
| GPX1 | 1 ± 0.35 | 0.88 ± 0.28 | 0.38 ± 0.16a | 0.62 ± 0.20b |
a p < 0.05 versus WT.
b p < 0.05 versus ACS.
c p < 0.01 versus WT.
d p < 0.01 versus ACS.
Effects of MHC-DGAT1 Expression on Heart Lipids in MHC-ACS Mice
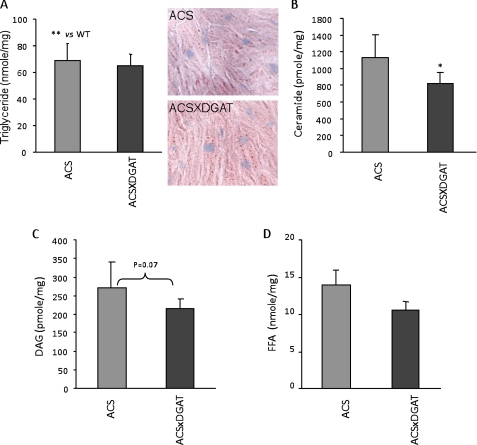
Because the MHC-DGAT1 transgene reduced ceramide and DAG, we next studied whether the MHC-DGAT1 transgene would affect heart lipids, gene expression, or function in a model of lipotoxicity because of increased heart trapping of fatty acids. Heart TG levels in MHC-ACS mice are even greater than those of MHC-DGAT1 (compare Fig. 3A versus Fig. 2D). Surprisingly, TG levels in the double transgenic mice were similar to both of the single transgenic lines (Fig. 3A). Compared with MHC-ACS single transgenic mice, the double transgenic mice had reduced ceramide and a trend for decreased DAG but no changes in FFA levels in cardiac tissue (Fig. 3, B–D).
FIGURE 3.
Comparison of cardiac lipids in MHC-ACS and MHC-ACSXMHC-DGAT1 mice. A–D, TG, FFA, DAG, and ceramide concentrations in heart muscles isolated from MHC-ACS and MHC-ACSXMHC-DGAT1 transgenic mice (n = 8–11).
MHC-DGAT1 Improves MHC-ACS Cardiomyopathy
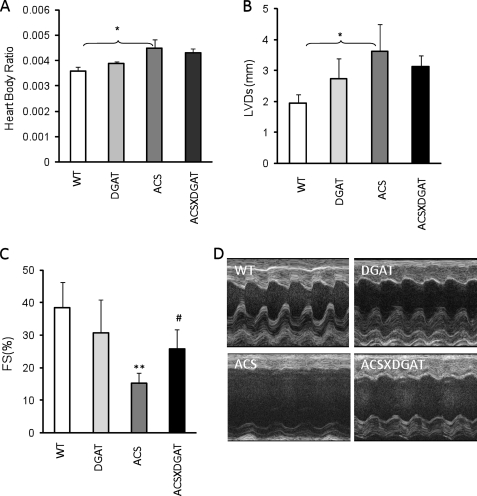
Heart/body ratio was not changed in MHC-DGAT1 transgenic mice, compared with the WT (Fig. 4A). Despite increased TG content, cardiac function of MHC-DGAT1 mice evaluated by both echocardiography and direct catheterization was normal; neither FS nor LVDs were altered (Fig. 4, B–D). Similarly cardiac catheterization showed no evidence of heart dysfunction in MHC-DGAT1 mice (supplemental Fig. 1, A–C).
FIGURE 4.
Cardiac function in MHC-DGAT1 and MHC-DGAT1/MHC-ACS mice. A, heart to body weight ratio in WT, MHC-DGAT1, MHC-ACS, and MHC-DGAT1/MHC-ACS transgenic mice (n = 5–7). Echocardiography showed left ventricular systolic dimension (B) and fractional shortening (C). Photographs are shown of echocardiograms (D) (n = 5–7). **, p < 0.01 MHC-ACS versus WT or MCH-DGAT1; #, p < 0.05 MHC-ACS versus MHC-ACS/MHC-DGAT1.
In contrast and as has been reported previously (19), MHC-ACS mice had cardiac dysfunction, but this was reduced in the MHC-ACSXMHC-DGAT1 double transgenic mice. Echocardiography of 4-month-old male mice showed improved FS (15% FS in MHC-ACS versus 24% FS in MHC-DGAT1XMHC-ACS, see Fig. 4, B–D). Cardiac catheterization revealed that MHC-DGAT1XMHC-ACS hearts had improved systolic contractile function (LVdP/dtmax is MHC-ACS 3761 mm Hg and MHC-DGAT1XMHC-ACS 6550 mm Hg, mean values), improved diastolic function (LVdP/dtmin is MHC-ACS −3333 mm Hg and MHC-DGAT1XMHC-ACS −5087 mm Hg, mean values), and decreased left ventricular end diastolic pressure compared with MHC-ACS hearts (supplemental Fig. 1, D–F).
Expression of Genes in MHC-DGAT1XMHC-ACS Hearts
Crossing MHC-DGAT1 onto MHC-ACS improved heart failure markers despite minor effects on lipid metabolic genes (Table 2). ANF and BNP were increased in MHC-ACS mice, but consistent with the functional results, these heart failure markers were decreased by the MHC-DGAT1 transgene. ATGL expression was about 2-fold higher in MHC-DGAT1XMHC-ACS mice compared with MHC-ACS transgenic mice. PPARα and PPARγ mRNA levels in MHC-DGAT1XMHC-ACS mice were more than 2-fold greater than those in MHC-ACS mice. There were no obvious changes in genes responsible for lipid oxidation. Compared with WT hearts, both MHC-ACS and MHC-DGAT1XMHC-ACS hearts had reduced expression of PDK4, suggestive of greater glucose oxidation, which might be related to the developing heart failure.
Reduced expression of antioxidant genes might promote myocardial dysfunction (27). The expression of anti-oxidant genes catalase and glutathione peroxidase (GPX1) was reduced to about 20 and 30% of WT control in MHC-ACS mice and partially recovered in MHC-DGAT1XMHC-ACS mice. The expression of glucose-6-phosphate dehydrogenase, another antioxidant gene, increased about 5-fold in MHC-ACS mice and went back to normal level in double transgenic mice (Table 2).
MHC-DGAT1 Reduced Cardiac Apoptosis and Increased Mitochondria in MHC-ACS Mice
Histologic examination of the hearts by terminal dUTP nick-end labeling showed reduced apoptosis in the double transgenic hearts (Fig. 5, A and B). MHC-ACS mice have altered expression of apoptosis-related genes, and these were partially normalized with the MHC-DGAT1 cross. BCL2 (B cell leukemia/lymphoma 2) was decreased in MHC-ACS transgenic mice and increased with the MHC-DGAT1 transgene (Table 2). Caspase 9 was doubled in MHC-ACS mice, and this was not changed by the MHC-DGAT1 transgene. Another apoptosis-related gene BAX (BCL2-associated X protein) was not changed.
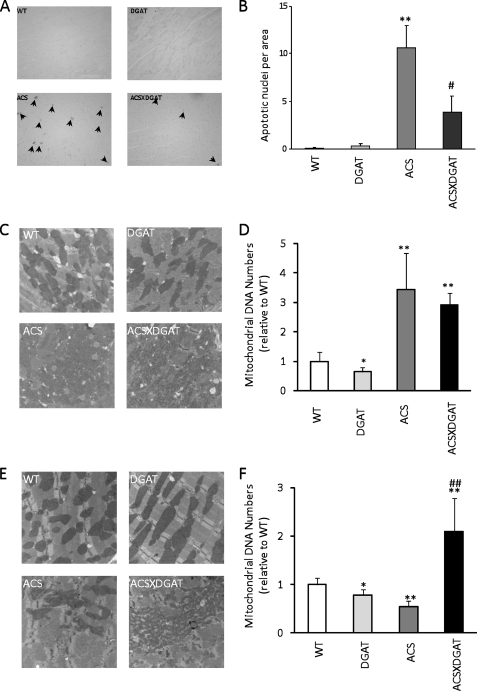
FIGURE 5.
Apoptosis and mitochondrial analysis. A, histologic examination of the hearts by terminal dUTP nick-end labeling staining for WT, MHC-DGAT1, MHC-ACS, and MHC-DGAT1X MHC-ACS transgenic mice (A) and quantification of the apoptotic cells (B) (n = 3). Data are shown as mean ± S.D. *, p < 0.05; **, p < 0.01; #, p < 0.05; ##, p < 0.01 versus MHC-ACS. Arrows indicate apoptotic cells. C, electron micrograph; D, quantification of mitochondria in 4-month-old mice. E, electron micrograph; F, and quantification of mitochondria in 6-month-old mice.
The reduction in apoptosis suggested that the MHC-DGAT1XMHC- ACS had improved mitochondrial function. Mitochondrial DNA copy number was slightly decreased (∼20%) in MHC-DGAT1 transgenic mouse hearts but was tremendously increased in 4-month-old MHC-ACS and MHC-DGAT1XMHC-ACS mice (about 3-fold compared with WT and 4-fold compared with MHC-ACS) (Fig. 5, C and D). In some surviving 6-month-old MHC-ACS mice, mitochondria number was significantly decreased, whereas age-matched MHC-DGAT1XMHC-ACS mice had the same number mitochondria as 4-month-old mice (Fig. 5, E and F). Therefore, the lipid alterations due to DGAT1 expression appeared to preserve mitochondria.
Greater Fatty Acid Oxidation and Reduced Toxic Lipids Correlate with Improved Survival
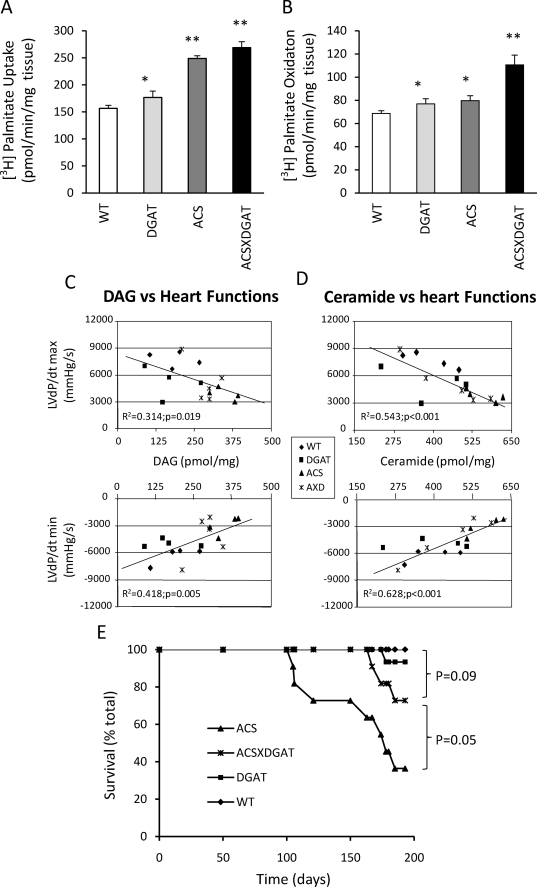
To determine whether the histologic suggestion of improved mitochondrial function was associated with changes in lipid oxidation, we measured fatty acid uptake and oxidation in these mice. Fatty acid uptake in MHC-DGAT1 heart muscle increased about 13% and in MHC-ACS heart increased about 59% (Fig. 6A). Fatty acid oxidation in MHC-DGAT1 mice also increased about 12%, almost equal to the increased fatty acid uptake. However, in MHC-ACS, fatty acid oxidation only increased about 16%, much less than the lipid uptake. In MHC-DGAT1XMHC-ACS mice, the fatty acid uptake and oxidation rose in parallel again (72 versus 61%). Thus, MHC-ACS hearts acquired amounts of fatty acids that were disproportionate to their oxidative requirements.
FIGURE 6.
Fatty acid uptake and oxidation, correlation of lipids and heart function, and survival rates. A and B, wild type and transgenic mice (MHC-DGAT1, MHC-ACS, and MHC-DGAT1XMHC-ACS) (n = 4) heart slices were incubated with [3H]palmitate acid as described under “Experimental Procedures.” C, positive correlation between heart muscle DAG levels with left ventricle contractile function (mm Hg/s), (adjusted R2 = 0.314; p = 0.019) and with diastolic function (mm Hg/s) (adjusted R2 = 0.418; p = 0.005). D, positive correlation between heart muscle ceramide levels with left ventricle contractile function (mm Hg/s) (adjusted R2 = 0.543; p < 0.001) and with diastolic function (mm Hg/s) (adjusted R2 = 0.628; p ≤ 0.001). E, wild type (WT, n = 10) and transgenic mice (WT, n = 10; MHC-DGAT, n = 15; MHC-ACS, n = 11; and MHC-ACSXMHC-DGAT1, n = 11) were followed for 180 days, and incidence of spontaneous death was recorded as a function of time. *, p < 0.5; **, p < 0.01.
We hypothesized that the functional differences between MHC-ACS and MHC-DGAT1XMHC-ACS mice might relate to differences in cardiac content of toxic lipids other than TG. We thus evaluated the relationship between specific lipid species in the heart and catheterization-derived measures of cardiac function in WT, transgenic, and doubly transgenic mice. Both DAG and ceramide correlated inversely with LVdP/dtmax and positively with LVdP/dtmin (Fig. 6, C and D). This suggests that these lipid species or their metabolites play an important role in lipotoxicity in the heart.
The improved cardiac function in the MHC-DGAT1XMHC-ACS mice was associated with reduced mortality compared with single transgenic MHC-ACS mice (Fig. 6E). MHC-ACS mice began to die at ∼3 months of age, and 40% of the animals died by 5 months. The double transgenic line had markedly improved survival with only ∼15% of the mice dying by 5 months. Thus, overexpression of DGAT1 in the setting of lipotoxicity reduced mortality.
DISCUSSION
We studied how cardiac TG is modulated during exercise and whether overexpression of DGAT1 affects heart function in normal and lipotoxic hearts. In contrast to pathologic hypertrophied hearts that have greater glucose and reduced fatty acid oxidation, we show that physiologic hypertrophy due to swimming leads to increased heart TG and induction of DGAT1 expression. We created a new line of transgenic mice overexpressing DGAT1 specifically in cardiomyocytes, which increased heart TG and reduced heart concentrations of both DAG and ceramide, potentially toxic lipids. MHC-DGAT1 transgenic mice had normal cardiac function. Thus, increasing heart TG stores either with exercise or via DGAT1 expression was not harmful.
DGAT1 is the rate-limiting enzyme required for the final step of TG synthesis in the heart, the conversion of DAG to TG. Studies in the C57/BL6 strain of mice showed that overexpression of DGAT1 led to no greater insulin resistance in adipose tissue despite more obesity (11). Similarly transgenic expression of DGAT1 in skeletal muscle (14) led to greater insulin sensitivity despite increased tissue concentrations of TG. Skeletal muscle overexpression of DGAT1 led to a phenotype that was similar to that found with chronic exercise, greater TG stores, greater insulin sensitivity, and also greater fatty acid oxidation (14, 24). The heart response to exercise was similar to that of skeletal muscle. After 2 weeks of exercise training, hypertrophied mouse hearts had greater TG content that was associated with a marked up-regulation of DGAT1. Although excess heart accumulation of lipid is associated with lipotoxicity in genetically modified mice (19, 28–31) and is thought to underlie heart dysfunction in obese and diabetic humans (31, 32), the greater TG storage associated with up-regulation of DGAT1 appears to be a beneficial metabolic adaptation to allow greater exercise capacity.
Metabolic adaptation in skeletal muscle to chronic exercise leads to increased storage but also oxidation of TG (14, 33). A similar process occurs in the heart; physiologic hypertrophy is associated with increased lipoprotein lipase expression (34, 35) and greater fatty acid oxidation (36). In addition, we show that DGAT1 is markedly up-regulated in the heart with exercise and is likely responsible for the greater TG storage. As discussed below, DGAT1 overexpression might play a role in the greater fatty acid oxidation as MHC-DGAT1 hearts had greater expression of PPAR downstream genes involved in fatty acid oxidation.
The MHC promoter leads to cardiomyocyte-specific expression of genes and has been used to assess specific genetic alterations in heart metabolic pathways. This promoter led to a modest ∼3-fold increase in DGAT activity in the heart and the expected changes in heart lipids. TG was increased and DAG reduced. The increase in cardiac TG did not alter heart function. Thus, a modest increase in heart TG is not toxic. The data should be contrasted with that of the ATGL knock-out mouse that develops massive enlargement of heart due to a marked increase in heart TG that leads to mechanical dysfunction (28).
Both DAG and ceramide are thought to alter insulin actions (37, 38) and would be expected to shift cellular metabolism to fatty acids. Indeed, the increase in PDK4 in MHC-DGAT1 hearts is consistent with this. A similar change in gene expression was found in muscles of mice expressing DGAT1 via the skeletal muscle MCK promoter, and this was associated with greater fatty acid oxidation in isolated skeletal muscle (24). ACO1 and CD36, other PPAR regulated genes, were increased with cardiomyocyte DGAT1 expression. The reason responsible for this is not immediately obvious; however, it should be noted that the DGAT1 transgenic mice also have increased expression of ATGL, an enzyme whose expression provides ligands and activates PPARs in other organs (39).
Excess heart accumulation of lipid is associated with lipotoxicity in genetically modified mice (19, 28–30). The toxic effects of lipids have been ascribed to a variety of causes. A widely held view of cardiac lipotoxicity is that excess TG and excess fatty acid oxidation are causal. Our study does not support this. Excess heart TG storage in the MHC-DGAT1 transgenic mice was associated with normal cardiac function. MHC-ACS mice develop lipotoxicity due to greater trapping of FFA within cardiomyocytes, cardiac hypertrophy, and heart failure, resulting in premature death (19). DGAT1 expression improved cardiac function in MHC-ACS mice. Expression of heart failure markers ANF and BNP was reduced. In addition, cardiac function assessed both by echocardiography and invasive hemodynamic monitoring was improved. Surprisingly, MHC-DGAT1 did not increase TG when crossed onto the MHC-ACS background. A likely reason for this is that up-regulation of ATGL and the fatty acid oxidation pathway balanced the greater lipid uptake. Nevertheless, the MHC-DGAT1 transgene improved survival when crossed onto the lipotoxic MHC-ACS background. Although our data suggest that MHC-DGAT1 leads to similar changes as exercise, it should be noted that when we exercised the MHC-ACS mice, it led to increased death, an outcome reflecting the underlying cardiac toxicity in these animals.
The mechanism of cardiac myocyte loss and resultant heart failure in MHC-ACS mice likely involves both necrotic and apoptotic pathways. In the MHC-ACS mice, DNA fragmentation and cytochrome c release are consistent with induction of apoptotic pathways. Moreover, lipid accumulation is associated with increased cardiac ceramide content in MHC-ACS mice. This signaling lipid may contribute to the induction or amplification of apoptotic pathways (19). MHC-DGAT1 reduced apoptosis, presumably due to a reduction in toxic lipids, DAG and ceramide.
One likely beneficial effect of MHC-DGAT1 is increased fatty acid oxidation. Why this happens is unknown. However, reduction of ceramide (27, 40), some other toxic lipid, or reactive oxygen species (see below) could have prevented mitochondrial death. Myocardial antioxidants inhibit or delay the oxidative damage to subcellular proteins, carbohydrates, lipids, and DNA. Although the exact mechanisms are not fully understood, it is possible that some endogenous antioxidants may reduce apoptotic cell death by up-regulating the anti-death gene BCL-2 (27, 41). Similarly, peroxidases such as GPX1 or catalase may protect the myocardium against ischemia-reperfusion injury (42) (43–45). In our MHC-DGAT1 transgenic mice, catalase increased about 3–4-fold in heart tissue compared with WT and normalized the depressed catalase expression in the MHC-ACS model. Thus, the up-regulation of catalase could have prevented oxidant damage to cells or mitochondria.
Glucose-6-phosphate dehydrogenase-derived NADPH, which is a co-factor for NADPH oxidase, enhances superoxide anion generation and elevates oxidative stress in diabetes, heart failure, and angiotensin II-induced hypertrophy of smooth muscle (46). Elevated glucose-6-phosphate dehydrogenase activity and NADPH levels are found in diabetes (47). Also, there is a 10-fold increase in myocardial glucose-6-phosphate dehydrogenase expression and a 2-fold increase in glucose-6-phosphate dehydrogenase activity in pacing-induced heart failure compared with normal hearts (48). In MHC-ACS mice, glucose-6-phosphate dehydrogenase mRNA was increased about 5-fold compared with WT and MHC-DGAT mice. This increase was normalized in double transgenic mice.
High rates of myocardial fatty acid utilization are postulated to cause “lipotoxic” cardiomyopathy (49). In contrast, greater lipid uptake and oxidation are characteristics of exercise-trained muscles and was a protective metabolic phenotype in exercised rat heart (50). Fatty acid uptake and oxidation were increased in all our genetically modified mouse hearts, but their function differed. In MHC-DGAT1 mice, both fatty acid uptake and oxidation increased; a similar situation occurred in the MHC-DGAT1/MHC-ACS transgenics. However, in MHC-ACS mice, the increase in fatty acid uptake was out of proportion to fatty acid oxidation. Although this might have been a primary cause of the heart dysfunction, it is also possible that it was secondary to a lipid-induced mitochondrial defect. De novo synthesis of ceramide requires palmitate (51). We hypothesize that consumption of extra fatty acids via the introduction of the MHC-DGAT1 transgene increased utilization of palmitic acid.
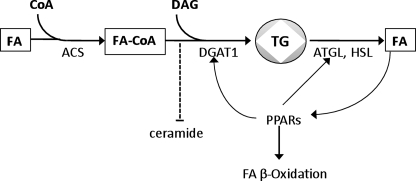
Our studies and others (12, 52) illustrate a link between TG storage via DGAT1, induction of ATGL, and greater expression of PPAR downstream genes (Fig. 7). Both ATGL and DGAT1 are PPAR-responsive genes, and recycled stored lipids might be especially potent agonists that promote fatty acid oxidation.
FIGURE 7.
DGAT1, ATGL, and PPAR regulation. Increased TG droplet storage induced by DGAT1 is associated with greater ATGL expression. Subsequent hydrolysis of TG provides agonists that lead to greater expression of PPAR downstream genes and greater fatty acid oxidation.
Both diabetes and obesity are associated with greater heart TG stores and decreased cardiac function (53). It has been presumed that either TG stores or greater fatty acid oxidation are detrimental to the heart (19, 28, 54). In part, this view is supported by studies in isolated perfused hearts that show that greater fatty acid oxidation in the setting of limited oxygen leads to more heart dysfunction or ischemic damage (54). In contrast, studies in humans (55, 56) and in rodents (56) demonstrate that continued cardiac use of fatty acid is associated with normal response to increased afterload. Perhaps this need for a continuing source of energetic lipids is the reason that high fat diets are not detrimental in the setting of heart failure (57). Our data show that processes associated with greater TG storage and fatty acid oxidation can be protective. Toxic effects of accumulated lipids that might affect mitochondrial function (58) or activate apoptosis pathways are more likely culprits. Although our studies focused on the heart, it is not unlikely that similar lipotoxic pathways are activated in other tissues. Lipotoxicity is implicated as the cause of a number of abnormalities associated with diabetes and obesity, including skeletal muscle insulin resistance, nonalcoholic hepatosteatosis, and pancreatic β-cell dysfunction (45, 53). Future studies will determine whether DGAT1 activation is a viable therapeutic strategy for all lipotoxic diseases.
This work was supported, in whole or in part, by National Institutes of Health Grants HL45095 and HL73029 from the NHLBI (to I. J. G.) and DK 064989 (to J. S.). This work was also supported by Burroughs Wellcome Foundation Grant 1005935 (to J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Fig. 1.
- TG
- triglyceride
- ACS
- acyl-CoA synthetase
- ATGL
- adipose TG lipase
- BNP
- brain-type natriuretic peptide
- CPT1
- carnitine palmitoyltransferase-1
- DAG
- diacylglycerol
- DGAT1
- diacylglycerol acyltransferase 1
- FFA
- free fatty acids
- FS
- fraction shorting
- FVB
- Fv1b allele
- LVDd
- left ventricular end diastolic dimension
- LVDs
- left ventricular end systolic dimension
- LV dP/dtmax
- maximal rates of rise in left ventricular pressure
- LV dP/dtmin
- maximal rates of fall in left ventricular pressure
- MHC
- myosin heavy chain
- PDK
- pyruvate dehydrogenase kinase
- PPAR
- peroxisome proliferator-activated receptor
- WT
- wild type.
REFERENCES
- 1.Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., Erickson S. K., Farese R. V., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yen C. L., Monetti M., Burri B. J., Farese R. V., Jr. (2005) J. Lipid Res. 46, 1502–1511 [DOI] [PubMed] [Google Scholar]
- 3.Oelkers P., Behari A., Cromley D., Billheimer J. T., Sturley S. L. (1998) J. Biol. Chem. 273, 26765–26771 [DOI] [PubMed] [Google Scholar]
- 4.Lardizabal K. D., Mai J. T., Wagner N. W., Wyrick A., Voelker T., Hawkins D. J. (2001) J. Biol. Chem. 276, 38862–38869 [DOI] [PubMed] [Google Scholar]
- 5.Meegalla R. L., Billheimer J. T., Cheng D. (2002) Biochem. Biophys. Res. Commun. 298, 317–323 [DOI] [PubMed] [Google Scholar]
- 6.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr. (2000) Nat. Genet. 25, 87–90 [DOI] [PubMed] [Google Scholar]
- 7.Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V., Jr. (2004) J. Biol. Chem. 279, 11767–11776 [DOI] [PubMed] [Google Scholar]
- 8.Wongsiriroj N., Piantedosi R., Palczewski K., Goldberg I. J., Johnston T. P., Li E., Blaner W. S. (2008) J. Biol. Chem. 283, 13510–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H. C., Smith S. J., Ladha Z., Jensen D. R., Ferreira L. D., Pulawa L. K., McGuire J. G., Pitas R. E., Eckel R. H., Farese R. V., Jr. (2002) J. Clin. Invest. 109, 1049–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H. C., Jensen D. R., Myers H. M., Eckel R. H., Farese R. V., Jr. (2003) J. Clin. Invest. 111, 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H. C., Stone S. J., Zhou P., Buhman K. K., Farese R. V., Jr. (2002) Diabetes 51, 3189–3195 [DOI] [PubMed] [Google Scholar]
- 12.Chen N., Liu L., Zhang Y., Ginsberg H. N., Yu Y. H. (2005) Diabetes 54, 3379–3386 [DOI] [PubMed] [Google Scholar]
- 13.Monetti M., Levin M. C., Watt M. J., Sajan M. P., Marmor S., Hubbard B. K., Stevens R. D., Bain J. R., Newgard C. B., Farese R. V., Sr., Hevener A. L., Farese R. V., Jr. (2007) Cell Metab. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 14.Liu L., Zhang Y., Chen N., Shi X., Tsang B., Yu Y. H. (2007) J. Clin. Invest. 117, 1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schenk S., Horowitz J. F. (2007) J. Clin. Invest. 117, 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda S., Miyazaki H., Nakatani T., Kai Y., Kamei Y., Miura S., Tsuboyama-Kasaoka N., Ezaki O. (2002) Biochem. Biophys. Res. Commun. 296, 395–400 [DOI] [PubMed] [Google Scholar]
- 18.Ryder J. W., Kawano Y., Galuska D., Fahlman R., Wallberg-Henriksson H., Charron M. J., Zierath J. R. (1999) FASEB J. 13, 2246–2256 [DOI] [PubMed] [Google Scholar]
- 19.Chiu H. C., Kovacs A., Ford D. A., Hsu F. F., Garcia R., Herrero P., Saffitz J. E., Schaffer J. E. (2001) J. Clin. Invest. 107, 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y. H., Zhang Y., Oelkers P., Sturley S. L., Rader D. J., Ginsberg H. N. (2002) J. Biol. Chem. 277, 50876–50884 [DOI] [PubMed] [Google Scholar]
- 21.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 22.Preiss J. E., Loomis C. R., Bell R. M., Niedel J. E. (1987) Methods Enzymol. 141, 294–300 [DOI] [PubMed] [Google Scholar]
- 23.Goossens G. H. (2008) Physiol. Behav. 94, 206–218 [DOI] [PubMed] [Google Scholar]
- 24.Liu L., Shi X., Choi C. S., Shulman G. I., Klaus K., Nair K. S., Schwartz G. J., Zhang Y., Goldberg I. J., Yu Y. (2009) Diabetes 58, 2516–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volk M. E., Millington R. H., Weinhouse S. (1952) J. Biol. Chem. 195, 493–501 [PubMed] [Google Scholar]
- 26.Cabrero A., Alegret M., Sánchez R. M., Adzet T., Laguna J. C., Vázquez M. (2001) Diabetes 50, 1883–1890 [DOI] [PubMed] [Google Scholar]
- 27.Dhalla N. S., Elmoselhi A. B., Hata T., Makino N. (2000) Cardiovasc. Res. 47, 446–456 [DOI] [PubMed] [Google Scholar]
- 28.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 29.Yagyu H., Chen G., Yokoyama M., Hirata K., Augustus A., Kako Y., Seo T., Hu Y., Lutz E. P., Merkel M., Bensadoun A., Homma S., Goldberg I. J. (2003) J. Clin. Invest. 111, 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son N. H., Park T. S., Yamashita H., Yokoyama M., Huggins L. A., Okajima K., Homma S., Szabolcs M. J., Huang L. S., Goldberg I. J. (2007) J. Clin. Invest. 117, 2791–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park T. S., Yamashita H., Blaner W. S., Goldberg I. J. (2007) Curr. Opin. Lipidol. 18, 277–282 [DOI] [PubMed] [Google Scholar]
- 32.Szczepaniak L. S., Victor R. G., Orci L., Unger R. H. (2007) Circ. Res. 101, 759–767 [DOI] [PubMed] [Google Scholar]
- 33.Goodpaster B. H., He J., Watkins S., Kelley D. E. (2001) J. Clin. Endocrinol. Metab. 86, 5755–5761 [DOI] [PubMed] [Google Scholar]
- 34.Flavell D. M., Wootton P. T., Myerson S. G., World M. J., Pennell D. J., Humphries S. E., Talmud P. J., Montgomery H. E. (2006) J. Mol. Med. 84, 126–131 [DOI] [PubMed] [Google Scholar]
- 35.Goldberg D. I., Rumsey W. L., Kendrick Z. V. (1984) Metabolism 33, 964–969 [DOI] [PubMed] [Google Scholar]
- 36.Goodwin G. W., Taegtmeyer H. (2000) Am. J. Physiol. Heart Circ. Physiol. 279, H1490–H1501 [DOI] [PubMed] [Google Scholar]
- 37.Yu C., Chen Y., Cline G. W., Zhang D., Zong H., Wang Y., Bergeron R., Kim J. K., Cushman S. W., Cooney G. J., Atcheson B., White M. F., Kraegen E. W., Shulman G. I. (2002) J. Biol. Chem. 277, 50230–50236 [DOI] [PubMed] [Google Scholar]
- 38.Holland W. L., Brozinick J. T., Wang L. P., Hawkins E. D., Sargent K. M., Liu Y., Narra K., Hoehn K. L., Knotts T. A., Siesky A., Nelson D. H., Karathanasis S. K., Fontenot G. K., Birnbaum M. J., Summers S. A. (2007) Cell Metab. 5, 167–179 [DOI] [PubMed] [Google Scholar]
- 39.Reid B. N., Ables G. P., Otlivanchik O. A., Schoiswohl G., Zechner R., Blaner W. S., Goldberg I. J., Schwabe R. F., Chua S. C., Jr., Huang L. S. (2008) J. Biol. Chem. 283, 13087–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suematsu N., Tsutsui H., Wen J., Kang D., Ikeuchi M., Ide T., Hayashidani S., Shiomi T., Kubota T., Hamasaki N., Takeshita A. (2003) Circulation 107, 1418–1423 [DOI] [PubMed] [Google Scholar]
- 41.Maulik N., Engelman R. M., Rousou J. A., Flack J. E., 3rd, Deaton D., Das D. K. (1999) Circulation 100, II369–II375 [DOI] [PubMed] [Google Scholar]
- 42.Woo Y. J., Zhang J. C., Vijayasarathy C., Zwacka R. M., Englehardt J. F., Gardner T. J., Sweeney H. L. (1998) Circulation 98, II255–II260 [PubMed] [Google Scholar]
- 43.Marklund S. L., Westman N. G., Lundgren E., Roos G. (1982) Cancer Res. 42, 1955–1961 [PubMed] [Google Scholar]
- 44.Sekili S., McCay P. B., Li X. Y., Zughaib M., Sun J. Z., Tang L., Thornby J. I., Bolli R. (1993) Circ. Res. 73, 705–723 [DOI] [PubMed] [Google Scholar]
- 45.Triana J. F., Li X. Y., Jamaluddin U., Thornby J. I., Bolli R. (1991) Circ. Res. 69, 731–747 [DOI] [PubMed] [Google Scholar]
- 46.Gupte S. A. (2008) Curr. Opin. Investig. Drugs 9, 993–1000 [PubMed] [Google Scholar]
- 47.Gupte R. S., Floyd B. C., Kozicky M., George S., Ungvari Z. I., Neito V., Wolin M. S., Gupte S. A. (2009) Free Radic. Biol. Med. 47, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupte R. S., Vijay V., Marks B., Levine R. J., Sabbah H. N., Wolin M. S., Recchia F. A., Gupte S. A. (2007) J. Card Fail. 13, 497–506 [DOI] [PubMed] [Google Scholar]
- 49.Burkart E. M., Sambandam N., Han X., Gross R. W., Courtois M., Gierasch C. M., Shoghi K., Welch M. J., Kelly D. P. (2007) J. Clin. Invest. 117, 3930–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burelle Y., Wambolt R. B., Grist M., Parsons H. L., Chow J. C., Antler C., Bonen A., Keller A., Dunaway G. A., Popov K. M., Hochachka P. W., Allard M. F. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1055–H1063 [DOI] [PubMed] [Google Scholar]
- 51.Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 52.Alsted T. J., Nybo L., Schweiger M., Fledelius C., Jacobsen P., Zimmermann R., Zechner R., Kiens B. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E445–E453 [DOI] [PubMed] [Google Scholar]
- 53.Schaffer J. E. (2003) Curr. Opin. Lipidol. 14, 281–287 [DOI] [PubMed] [Google Scholar]
- 54.Lopaschuk G. D., Barr R., Thomas P. D., Dyck J. R. (2003) Circ. Res. 93, e33–e37 [DOI] [PubMed] [Google Scholar]
- 55.Chandler M. P., Kerner J., Huang H., Vazquez E., Reszko A., Martini W. Z., Hoppel C. L., Imai M., Rastogi S., Sabbah H. N., Stanley W. C. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1538–H1543 [DOI] [PubMed] [Google Scholar]
- 56.Yamashita H., Bharadwaj K. G., Ikeda S., Park T. S., Goldberg I. J. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E705–E713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okere I. C., Chess D. J., McElfresh T. A., Johnson J., Rennison J., Ernsberger P., Hoit B. D., Chandler M. P., Stanley W. C. (2005) Clin. Exp. Pharmacol. Physiol. 32, 825–831 [DOI] [PubMed] [Google Scholar]
- 58.Boudina S., Abel E. D. (2007) Circulation 115, 3213–3223 [DOI] [PubMed] [Google Scholar]