Abstract
We examined activation of the human epithelial sodium channel (ENaC) by cleavage. We focused on cleavage of αENaC using the serine protease subtilisin. Trimeric channels formed with αFM, a construct with point mutations in both furin cleavage sites (R178A/R204A), exhibited marked reduction in spontaneous cleavage and an ∼10-fold decrease in amiloride-sensitive whole cell conductance as compared with αWT (2.2 versus 21.2 microsiemens (μS)). Both αWT and αFM were activated to similar levels by subtilisin cleavage. Channels formed with αFD, a construct that deleted the segment between the two furin sites (Δ175–204), exhibited an intermediate conductance of 13.2 μS. More importantly, αFD retained the ability to be activated by subtilisin to 108.8 ± 20.9 μS, a level not significantly different from that of subtilisin activated αWT (125.6 ± 23.9). Therefore, removal of the tract between the two furin sites is not the main mechanism of channel activation. In these experiments the levels of the cleaved 22-kDa N-terminal fragment of α was low and did not match those of the C-terminal 65-kDa fragment. This indicated that cleavage may activate ENaC by the loss of the smaller fragment and the first transmembrane domain. This was confirmed in channels formed with αLD, a construct that extended the deleted sequence of αFD by 17 amino acids (Δ175–221). Channels with αLD were uncleaved, exhibited low baseline activity (4.1 μS), and were insensitive to subtilisin. Collectively, these data support an alternative hypothesis of ENaC activation by cleavage that may involve the loss of the first transmembrane domain from the channel complex.
Introduction
It is well established that serine proteases activate the epithelial sodium channel (ENaC).2 Activation occurs by direct mechanisms that induce channel subunit cleavage (1, 2) as well as those that are cleavage-independent but may involve cleavage of protease-activated membrane receptors (3). Channel cleavage studies have established that cellular proteases such as furin endogenously cleave the channel α and γ subunits. Mutation of identified endogenous cleavage sites on both of these subunits diminished baseline activity, demonstrating a role for cleavage in ENaC activation.
The acute effects of ENaC cleavage have largely relied on examining the effects of the protease trypsin on the α and γ subunits. These studies have examined the effects of cleavage on wild type and furin cleavage-deficient ENaC in oocytes and epithelial cells (1, 4, 5). Although these have markedly improved our understanding of channel activation by serine proteases, they suffer from the main shortcoming that trypsin is a non-selective serine protease that can cleave after a single arginine residue (6, 7), and therefore, it only offers a limited tool for examining the mechanisms of cleavage at specific sites. Consistent with the reduced specificity for trypsin is the observation that ENaC retains its cleavage by this protease after mutation of consensus cleavage sites for furin (1).
Despite their limitations, these studies have indicated that ENaC is activated by cleavage by an increase of open probability (Po) of membrane resident-inactive channels (8). Kleyman and co-workers (9, 10) have proposed that the mechanism of the Po increase involves the release of small inhibitory fragments from the α and γ subunits and that cleavage at two points in the extracellular loop is necessary for ENaC activation. They also proposed that the resulting N- and C-terminal fragments of subunit cleavage remain associated with the active channel complex, although a direct comparison of fragment levels at the membrane was not feasible given the presence of two different tags on these fragments. Therefore, this conclusion has not been experimentally validated.
To determine the mechanisms of ENaC activation by cleavage, we focused on the acute effects of cleavage and specifically cleavage of the core α subunit that is necessary to observe channel activity. We utilized the serine protease subtilisin, which exhibits a different cleavage preference than furin. This allowed us to examine the effects of cleavage on constructs with markedly reduced endogenous cleavage. To examine ENaC cleavage, subunits were tagged with two hemagglutinin (HA) tags; that is, toward the N terminus before the currently identified cleavage sites and at the C terminus. This allowed visualization of multiple proteolytic products of the same subunit and with the same antibody.
We report that wild type channels are activated ∼6-fold after cleavage by subtilisin. Channels formed with αFD, a construct that lacks the proposed inhibitory domain contained between the two furin cleavage sites, exhibited lower rather than higher activity when compared with αWT (∼50% of WT activity). More importantly, αFD also retained the ability to be markedly activated by subtilisin. Similarly, channels formed with αFM, a construct that mutated the two furin cleavage sites but retained the proposed inhibitory segment between the two furin cleavage sites, was also activated by subtilisin and to similar levels as αFD. On the other hand, channels formed with αLD, a construct that eliminated the proposed inhibitory domain, as well as subtilisin cleavage exhibited low baseline activity and was not activated by exogenous subtilisin. In all cleavable constructs, the N-terminal fragment of α exhibited lower density at the membrane than the C-terminal α, indicating potential internalization. We propose an alternative mechanism whereby cleavage predominantly activates the channel by the loss of the first transmembrane domain from the channel complex rather than by removal of a short inhibitory domain in the extracellular loop.
EXPERIMENTAL PROCEDURES
Xenopus Oocyte
Oocytes were prepared as previously described (11). Oocytes were surgically removed and defoliculated in Ca2+-free buffer containing 1 mg/ml collagenase (type 1A, Sigma). After an overnight recovery, defoliculated oocytes were injected with cRNA for the human ENaC subunits. All subunits were cloned in the PGEM-HE oocyte expression vector as previously described (3). The 9-aa HA tag was inserted into the subunits at positions described under “Results.” All insertions, deletions, and point mutations utilized PCR with the enzyme PFU Ultra (Stratagene, La Jolla CA). All modifications were verified by sequencing the entire insert. Recordings were carried out 1–3 days after injection. Procedures to determine surface binding utilized a colorimetric assay with a horseradish peroxidase-coupled anti-HA antibody (Roche Applied Science) and were previously described (3, 12).
Oocytes incubation and recording solutions were as previously described (11). All recording were carried out in ND94 consisting of 94 mm NaCl, 2 mm KCl, 1.8 mMCaCl2, 1 mm MgCl2, and 5 mm HEPES, pH 7.4–7.5. Amiloride was obtained from Merck and was used at 10 μm to inhibit ENaC-generated currents. Commercial grade subtilisin and trypsin were obtained from Sigma and used at concentrations indicated under “Results.” Electrical recordings from oocytes have been described in detail in the references above.
Oocyte Homogenization and Western Blotting
Injected oocytes were processed as previously described (3, 13). Oocytes were biotinylated on ice using a sulfo-NHS-biotin linker (Pierce) and homogenized at 10 μl/oocyte in buffer containing 170 mm NaCl, 10 mm Tris, 5 mm EDTA, pH 7.5, and a protease inhibitor mixture (Sigma). Oocytes were first broken by triturating with 27-gauge syringes. Yolk and nuclei were pelted by centrifugation at low speed (200 × g) for 5 min. Supernatants were spun at top speed (14,000 × g) for 20 min at 4 °C to obtain the membrane pellet. This insoluble pellet represented the total (bulk) ENaC fraction and was used for further processing. For the plasma membrane fraction, homogenized proteins were solubilized with 1% Triton, pulled down with streptavidin beads (Pierce), and released by boiling into SDS sample buffer. Fractionated proteins were separated by SDS-PAGE (with or without 5% glycerol) and transferred to nitrocellulose membranes.
Blots were blocked in Tris-buffered saline containing 5% nonfat dry milk and 0.1% Tween 20. Blots were then probed with the anti-HA antibody already coupled to horseradish peroxidase (Roche Applied Science) or a commercial αENaC N terminus antibody (Affinity BioReagents, Golden CO). Surface binding was determined in intact oocytes as previously described using the same anti-HA antibody (12). The commercial anti-αENaC antibody was probed with an horseradish peroxidase-conjugated anti-rabbit secondary. Blots were washed five times, and bound antibodies were visualized by enhanced chemiluminescence Super signal ECL Dura (Pierce). All blots were repeated a minimum of three times on oocytes obtained from three different frogs and were digitally acquired using a gel documentation system (Alpha Innotech, San Leandro, CA). Signal intensity of unadjusted images was analyzed using NIH ImageJ or Alpha Innotech software.
MALDI-TOF
Cleavage sites for subtilisin were examined with αENaC peptides using MALDI-TOF spectroscopy. Enzymatic cleavage by subtilisin was performed in the same solution used for oocyte recordings (ND94). All control and protease reactions (50 μl) were allowed to proceed for 40 min at room temperature. Reactions were terminated by the addition of 5 μl of 50% acetic acid. Before analysis, samples were purified by passage through Zip Tip C18 micro columns (Eppendorf) followed by elution in a saturated solution of cyano-4-hydroxycinnamic acid in 0.1% trifluoroacetic acid, 60% acetonitrile. Eluted samples were immediately spotted on a MALDI target and air-dried. Spectra were obtained using Biflex IV MALDI-TOF mass spectrometer (Bruker Daltonics Inc., Billerica MA). Individual spectra were obtained from 200 shots. Data were analyzed using the FindPeptide software to <0.5-dalton accuracy.
Peptide Syntheses
All peptides were synthesized by Genscript Corp. (Piscataway, NJ). Peptides were purified to >95% purity. Peptides were either directly dissolved into buffer or dissolved into DMSO at 20 mm and rediluted into buffer at the appropriate concentrations.
All biochemical experiments were repeated a minimum of three times and were carried out on oocytes harvested from separate frogs. All electrophysiological experiments were repeated as indicated in the figure legends on oocytes from a minimum of four frogs. Statistical analyses utilized Student's t test. p values <0.05 were considered statistically significant. Unless otherwise indicated, data are summarized as the means ± S.E.
RESULTS
Spontaneous Subunit Processing
To examine proteolytic processing and the fate of the processed fragments at the membrane, we introduced HA tags into each of the human ENaC subunits. Each subunit contained two HA tags which allowed detection and comparison of processed and unprocessed fragments utilizing the same antibody. This approach eliminates problems with differences in antibody sensitivity in multitagged proteins as it is expected that the two HA tags would exhibit similar binding to their antibody in fully denatured proteins. The HA tag was chosen given its low background in control oocytes and its suitability for in vivo binding (12) and immunofluorescence studies. The 9-aa HA tag was introduced into αENaC at positions 161 and 670 into βENaC at positions 138 and 641 and γENaC at positions 143 and 650. In α and γ the loop HA tag replaced existing sequences as previously used for rat ENaC (14). These constructs did not alter channel expression, biophysical properties, or amiloride sensitivity. Given that all three subunits were tagged with HA, separate experiments were carried out with one tagged and two untagged subunits to examine the effects on α, β, and γ, e.g. α2HA/β/γ, α/β2HA/γ, and α/β/γ2HA.
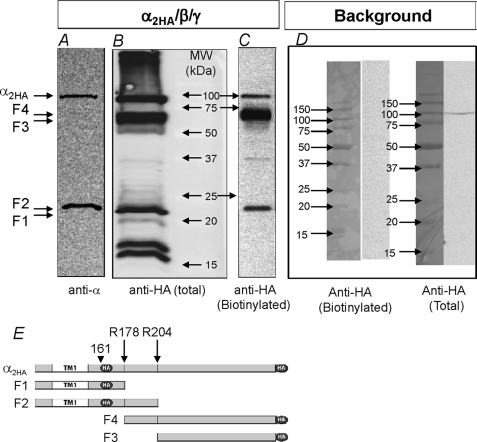
Double-tagged α is shown in Fig. 1. In this composite blot we compare the HA signal in whole cell lysate with that using a commercial N-terminal antibody as well as the differences in total versus plasma membrane-bound ENaC. Unprocessed α migrated at 80–85 kDa and was detected with both the anti-HA antibody (B) and the commercial N-terminal antibody (A). The C-terminal fragment of cleaved α containing the majority of the extracellular loop and the second transmembrane domain migrated at 60–65 kDa and was only observed in lanes probed with the anti-HA antibody. The N-terminal fragment of α containing the first transmembrane domain was detected with both antibodies and migrated at 20–25 kDa. Additional ENaC fragments were observed in the total fraction. These represent additional intracellular processing or degradation as they were not observed at the plasma membrane (C).
FIGURE 1.
Endogenous processing of α ENaC in trimeric channels. A composite Western blot of oocytes expressing α2HA/β/γ separated by SDS-PAGE on 12% acrylamide gels is shown. Total homogenates were probed with a commercial N-terminal epitope α ENaC antibody (A) or with an anti-HA antibody (B). The plasma membrane fraction was estimated from surface-biotinylated proteins probed with the anti-HA antibody (C). A background signal of non-expressing oocytes probed with the anti-HA antibody was not detected in the biotinylated pool and was very low in the total pool (D). The two furin cleavage sites are shown schematically in E at Arg-178 and Arg-204 (long arrows). The origin of the major predicted fragments observed in A, B, and C is also shown. Dark circles indicate the relative location of the HA tag. Short arrows indicate the position of the first HA tag. Full-length α migrated at ∼85 kDa, whereas the two main fragments migrated at ∼65 and 22 kDa. Full-length α was observed in all panels. The C-terminal 65-kDa α fragment did not contain the epitope recognized by the commercial antibody and was only observed when probed with the anti-HA antibody. The N-terminal 22-kDa α fragment was recognized by both antibodies. Of the intracellular processed components, only the full-length and the 22- and 65-kDa fragments were routinely observed at the plasma membrane, indicating that these fragments are amenable to biotinylation. In 12 blots no correlation was observed between the levels of the 65- and 22-kDa fragments (see “Results” and Fig. 5). This and other gels were loaded with the equivalent protein yield from 2–3 oocytes in the total lanes and 25 oocytes in the biotinylated lanes.
The data in Fig. 1 indicate that the double HA tag can be used to detect αENaC fragments at the plasma membrane. They also indicate that α is mostly cleaved under baseline conditions, although some unprocessed α is clearly evident. They also indicate that the levels of two α fragments are not similar. This difference was observed in many experiments in both the plasma membrane and total fractions (see Fig. 5 below) and likely indicates that the smaller α N-terminal fragment is degraded after subunit cleavage.
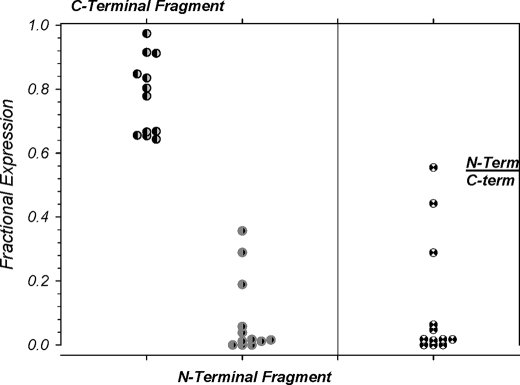
FIGURE 5.
Absence of a correlation between the N- and C-terminal αENaC proteolytic fragments. The levels of the two cleaved α fragments (22 and 65 kDa) were examined and were normalized to total plasma membrane-bound ENaC (uncleaved + 22 kDa + 65 kDa = 1.0). The levels of the C-terminal fragment were consistently high, whereas those of the N-terminal fragment were consistently low with no apparent correlation between the two. This is also evident from the calculated ratio of these two fragments, which varied between 0.55 and 0. These results along with those in Fig. 4 indicate that the N-terminal fragment is likely internalized after cleavage and is unlikely to be part of the active channel complex. n = 12.
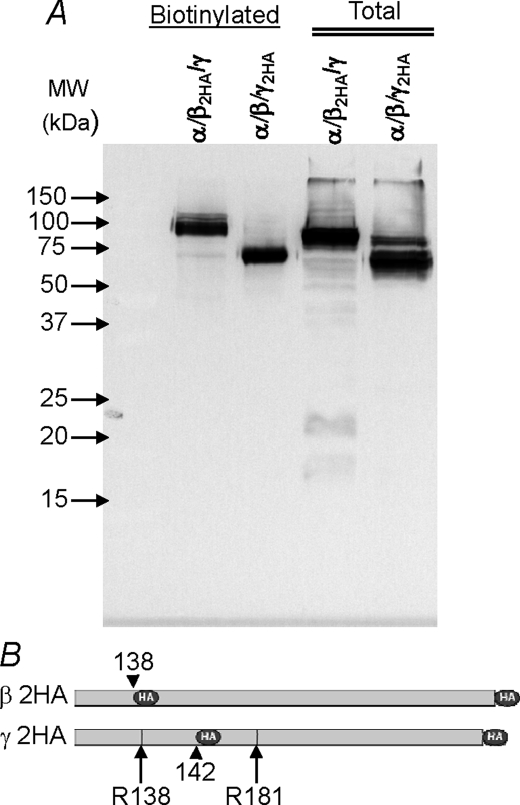
Double-tagged β and γ are shown in Fig. 2. β is uncleaved at the membrane and exhibits little intracellular processing or degradation products. Membrane bound γ is essentially all in the cleaved form at the plasma membrane. This was invariably observed when all three subunits were expressed and indicates that channel activation observed by extracellular trypsin is likely due to cleavage of unprocessed α at the oocyte membrane rather than γ (also see below). Similar to β, γ exhibited little intracellular degradation and was mostly present in the intracellular fraction in the shorter processed form.
FIGURE 2.
Endogenous processing of β and γ ENaC in trimeric channels. Shown is a Western blot of oocytes expressing trimeric α/β/γ with 2HA-tagged β or γ separated by SDS-PAGE on 12% acrylamide gels. β is unprocessed at the plasma membrane, whereas γ is essentially all in the processed shorter form. Similar results were also observed in the total compartment, although some unprocessed γ is observed. These results indicate that both β and γ are highly stable in oocytes as no significant degradation products were observed in the total (intracellular-dominated) compartment. The two proposed serine protease cleavage sites in γ are shown schematically at Arg-138 and -181 (long arrows). Dark circles and short arrows indicate the relative location of the HA tags. Data are representative of three experiments. Conditions were as described in Fig. 1. See “Results” for additional details.
Our data indicate that the effects of exogenous proteases in activating ENaC expressed in oocytes are likely mediated by processing of unprocessed α at the membrane and propose a dominant role of this subunit in channel activation. This conclusion is at odds with two recent reports indicating that γ processing is dominant in this system (5, 15). However, it is important to point out that our results are not directly comparable with those of Carattino et al. (15) as these authors did not examine plasma membrane ENaC subunit processing, and to Diakov et al. (5), who base their conclusion of the role of γ by examining processing and the effects of trypsin on dimeric α/β and α/γ channels. Notwithstanding, the role of α processing in channel activation is clearly established below.
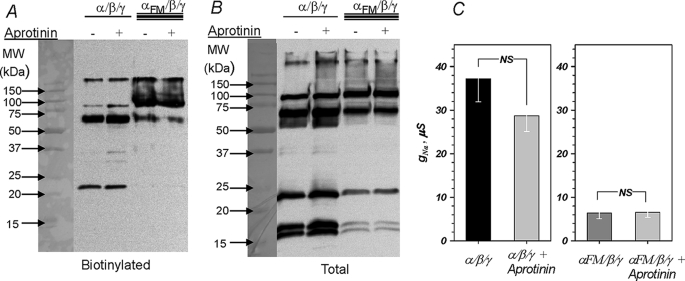
To examine the role of α and its processing in channel activation, we examined if processing can be prevented by the serine protease inhibitor aprotinin. This peptide inhibitor has been extensively used to inhibit endogenous serine proteases in epithelial preparations, including the Xenopus A6 epithelia (16, 17), and it is unknown if it can also inhibit ENaC processing in Xenopus oocytes. The effect of aprotinin on α processing is shown in Fig. 3 and indicates that aprotinin was without effects on processing in both the plasma membrane and total (cytoplasmic dominated) fractions. Consistent with this result, the amiloride-sensitive conductance was also insensitive to aprotinin, confirming the previous observation that rat ENaC activity is insensitive to aprotinin when expressed in Xenopus oocytes (18). This indicates that ENaC-cleaving oocyte-endogenous proteases are either aprotinin-insensitive at least at 100 μg/ml aprotinin or that oocytes are impermeable to this peptide inhibitor.
FIGURE 3.
Effect of αENaC cleavage on spontaneous channel activity. Western blot and the corresponding amiloride-sensitive conductance of oocytes expressing trimeric channels formed with α/β/γ or αFM/β/γ (αFM, double furin mutant containing the R178A and R204A mutations) separated on 12% acrylamide gels are shown. Spontaneous channel processing was insensitive to overnight addition of 100 μg/ml aprotinin. This was observed in both the plasma membrane and total fractions (A and B) and manifested as no significant change of the amiloride-sensitive conductance (C). Mutation of the terminal arginine in both proposed furin cleavage sites markedly reduced spontaneous α processing and the appearance of both N- and C-terminal fragments. This was accompanied by a marked and statistically significant (p < 0.01) decrease of the amiloride-sensitive conductance to 17% of control (C). NS indicates not significant. Data are representative of three experiments in A and B. n = 8 and 11 from 5 different frogs in C for the effect of aprotinin and the furin mutation, respectively.
Mutation of α ENaC Cleavage Sites and Cleavage by Subtilisin
To further examine α ENaC processing, we mutated the terminal arginine (P1) in both identified serine protease (furin) cleavage sites (Arg-178 and 204) to alanines (2). This αENaC construct is referred to as αFM. α was largely uncleaved in αFM/β/γ. In oocytes expressing wild type α and β/γ, ∼86% of α was endogenously processed at the membrane (see Figs. 3 and 5). In those expressing αFM, this effect was reversed where ∼90% of α was unprocessed at the membrane. Consistent with this observation, the 22-kDa N-terminal α fragment is nearly undetected at the membrane in cells expressing αFM. These changes in both the 65- and 22-kDa fragments between αWT and αFM further confirm our ability to detect both fragments at the plasma membrane.
Expression of αFM also markedly reduced the amiloride-sensitive conductance to ∼17% that of the control wild type (Fig. 3C), consistent with the effect of this mutation on subunit processing. These results indicate that α is processed by oocyte endogenous proteases at Arg-178 and/or Arg-204 and that this processing plays a dominant role in controlling channel activity. The importance of α processing in overall channel activity is further demonstrated below.
The above data also indicate that the acute effect of αENaC cleavage can be studied using a protease that can cleave αFM at the plasma membrane. To examine cleavage in this and other α constructs, we chose to avoid trypsin as low levels of this protease do not cleave ENaC (3), whereas high levels cause oocyte endogenous effects (19) in addition to potential multiple cleavage events that may cause nonspecific and poorly controlled effects (see Figs. 8 and 9). These shortcomings were avoided by utilizing the serine protease subtilisin. This protease is the archetypical S8 protease that also contains furin. Subtilisin favors different cleavage sites than furin (20, 21) but shares its catalytic domain with this protease (22). Thus, subtilisin would allow us to use constructs that eliminate endogenous cleavage by removing the P1 arginines but all the while retaining the ability for exogenous cleavage in a manner that is similar to that for furin.
FIGURE 8.
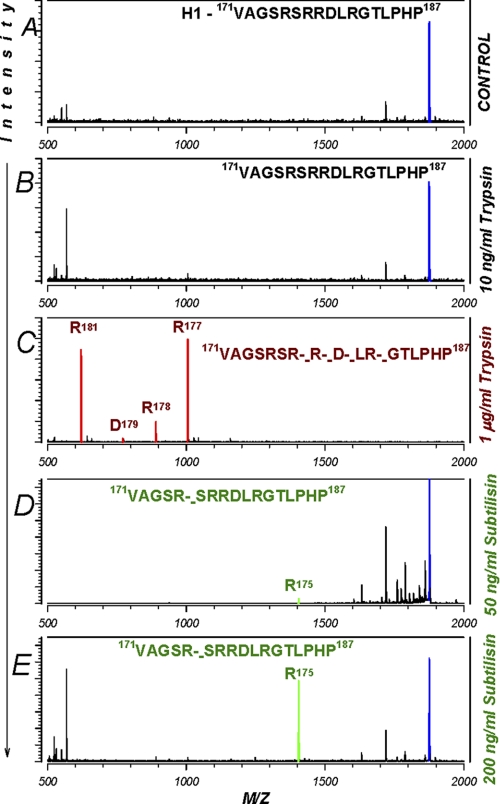
Cleavage of human α H1 peptide by trypsin and subtilisin. A, cleavage of a peptide encompassing the first furin cleavage site on α was examined. Only fragments originating from the parental peptide were labeled. All other fragments were unlabeled and originated from the added trypsin or subtilisin. The corresponding cleavages in the original peptide are shown by broken dashes. B, trypsin at 10 ng/ml was without effect. C, at 1 μg/ml, trypsin caused multiple cleavages indicating the non-specificity of this protease. These sites and the corresponding peaks are shown in red. D and E, subtilisin, on the other hand, caused a single cleavage at Arg-175 at both 50 and 200 ng/ml (shown in green). Thus, subtilisin cleaves ENaC once within three aa upstream of the first furin cleavage site. This result explains the ability of this protease to cleave the αFM, which lacks Arg-178 but retains the sequences at Arg-175. Data are representative of three separate experiments.
FIGURE 9.
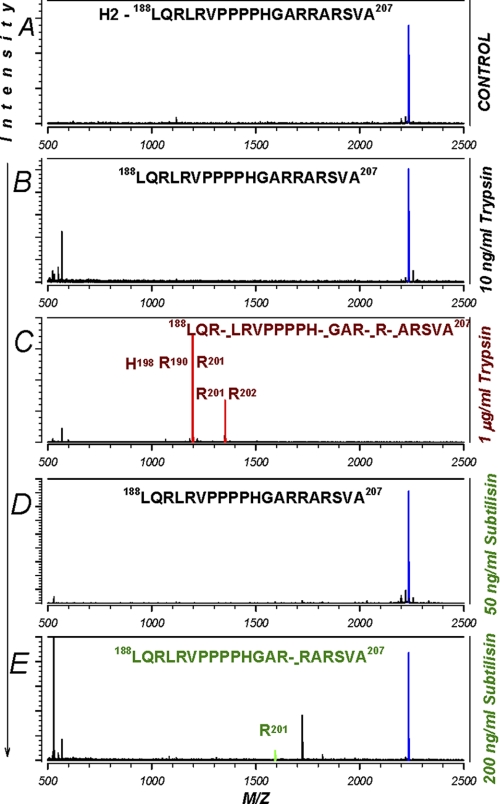
Cleavage of human α H2 peptide by trypsin and subtilisin. A, cleavage of a peptide encompassing the second furin cleavage site on α was examined. B, trypsin at 10 ng/ml was without effect. C, as above, trypsin caused multiple cleavages at 1 μg/ml. D and E, subtilisin, on the other hand, was without effect at 50 ng/ml, indicating that this and lower levels of subtilisin likely activate ENaC via a single cleavage effect at Arg-175. See the Fig. 8 legend for additional details. Data are representative of three separate experiments.
The biochemical effects of subtilisin on αWT and αFM cleavage are shown in Fig. 4. Endogenous processing of wild type α was mostly complete in trimeric channels (Fig. 4A). By contrast, αFM was largely in the unprocessed form. Uncleaved α in both of these constructs could be cleaved with subtilisin treatment at either 10 or 50 ng/ml for 15 min. The increase of the cleaved 65 kDa α fragment was easy to visualize in αFM given the low baseline levels of this fragment. To visualize the effect on αWT, we examined the disappearance of the uncleaved form of this subunit.
FIGURE 4.
Subtilisin cleaves wild type and furin mutant α. Shown is a Western blot of trimeric channels with WT or FM α separated on 12% acrylamide gels. Oocytes were untreated or treated for 25 or 15 min with the protease subtilisin at 10 or 50 ng/ml, respectively. A, subtilisin at 50 ng/ml cleaved both αWT and αFM at the plasma membrane with no effect on the total fraction, which contained 90–95% intracellular proteins. This effect was not blocked by 1 μg/ml soybean trypsin inhibitor (STI) ruling out a contamination by trypsin or trypsin like proteases. No effect of subtilisin was observed in the total compartment. B, shown is the dose-dependent decrease of full-length plasma membrane-bound wild type α with subtilisin treatment (n = 4). C, shown is the dose-dependent increase in the levels of the 65-kDa C-terminal processed α in the furin mutant construct (n = 4). p values were calculated from paired measurements of band intensities in treated and untreated groups. See Fig. 6 for the parallel electrophysiological data.
On average, 50 ng/ml subtilisin caused cleavage of ∼64% of plasma membrane uncleaved full-length αWT (Fig. 4B). This effect was insensitive to soybean trypsin inhibitor at 20-fold excess, ruling out a contamination of this protease by trypsin. Subtilisin did not affect fragment levels in the total fraction, which is dominated by the intracellular compartment (Fig. 4), consistent with a membrane-only effect of this protease.
Subtilisin also caused processing of plasma membrane-bound αFM (Fig. 4). These effects are summarized in Fig. 4C, where the data are normalized to the levels of αFM cleaved by 50 ng/ml subtilisin (65-kDa fragment). On average, 50 ng/ml subtilisin caused a 6-fold increase in the levels of the 65-kDa C-terminal fragment in αFM. Similar to wild type, the effects of subtilisin on αFM were not discernable in the total protein fraction. α processed by subtilisin in the furin mutant migrated at an indistinguishable size from endogenously cleaved wild type α and from subtilisin-cleaved wild type α. This indicates that subtilisin cleaves at sites that overlap or are in close vicinity to Arg-178 and Arg-204.
Interestingly, no consistent changes were observed in the plasma membrane levels of the N-terminal 22-kDa fragment after subtilisin treatment in either αWT or αFM. This disjoint in the levels of these two fragments after acute cleavage is consistent with the lack of a correlation observed with spontaneous baseline cleavage. This result indicates that the N-terminal fragment likely dissociates from the channel complex. Given that the 22-kDa N-terminal fragment would contain the proposed ubiquitination sites on the α subunit, this result would also be consistent with rapid internalization of this fragment and of ubiquitinated proteins in general (23).
The interpretation that the 22-kDa fragment is possibly internalized and may not be necessary to form an activated channel is further supported by the data examining the spontaneous levels of the two α fragments at the membrane. As shown in Fig. 5, the 22- and 65kDa α fragments averaged 8.2 and 77.9% of total α detected at the plasma membrane (the remaining ∼14% was uncleaved α). More importantly, no correlation was observed between these two fragments, and the ratio of their plasma membrane levels (N-term/C-term) varied between 0.55 and 0. In these experiment, the range of the plasma membrane levels of the N-terminal fragment was large, a result that could not be explained by small potential differences in the antigenicity of the HA tag between the N-terminal and C-terminal fragment of α. Furthermore, any such potential antigenic differences would be expected to be minimal as (a) antibody binding is carried out on denatured proteins, and (b) the N-terminal fragment is easily detectable in the bulk fraction (see Figs. 1, 3, and 4). This variability in the levels of the N-terminal fragment was also unlikely because of marked differences in the ability to biotinylate this fragment as it is easily detectable in many experiments at the plasma membrane (see Figs. 1C and 3A). Thus, this variability likely indicates that this fragment is not stable at the plasma membrane and that it is not necessary to be part of the activated cleavage complex (see “Discussion”).
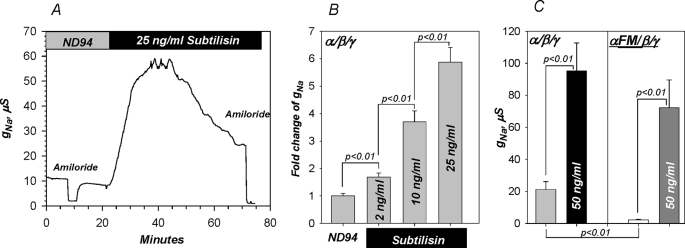
The electrophysiological effects of subtilisin are shown in Fig. 6. This protease markedly activated WT as well as FM α. The effects were dose-dependent, with higher levels causing more rapid and higher activation. At 50 ng/ml, subtilisin activation peaked at 15 min leading to a ∼6-fold activation of WT α and ∼35-fold activation of FM α. Activation rebounded at times longer than 20 min (at 50 ng/ml), a process previously described and attributed to Nedd4-2-mediated internalization of cleaved ENaC (4). The potential complication of this rebound was avoided by limiting our analyses to the initial period of channel activation; i.e. the first 15 min after 50 ng/ml subtilisin.
FIGURE 6.
Activation of ENaC by subtilisin cleavage of α. A, subtilisin caused marked time-dependent activation of the amiloride-sensitive conductance in α/β/γ-expressing oocytes. Stimulation was dose-dependent with activation observed at levels as low as 2 ng/ml (B). Activation peaked at 15 min at 50 ng/ml subtilisin. In this group of experiments 50 ng/ml subtilisin activated αWT by ∼6-fold and αFM by ∼35-fold. C, both activated channels reached similar absolute values, indicating the same mechanism of activation (p < 0.01 between the conductance of α/β/γ and αFM/β/γ before subtilisin and p > 0.05 after subtilisin). This effect is consistent with the biochemical data summarized in Fig. 4 and the in vitro cleavage data shown below. Subtilisin did not affect oocyte endogenous currents (control oocytes are not shown, but note the absence of changes of amiloride-sensitive currents before and after subtilisin). n = 5 for B and n = 10 for C.
Activation of conductance by subtilisin was also insensitive to 20-fold excess soybean trypsin inhibitor (not shown), consistent with the biochemical data in Fig. 5. Activated amiloride-sensitive whole cell conductances in αWT and αFM reached the same absolute levels. In combination with the Western blot data indicating cleavage products of indistinguishable sizes, these results indicate that subtilisin activated ENaC by cleaving the α subunit at or near the same sites as those cleaved by oocyte endogenous proteases. This is verified below.
The ability of subtilisin to cleave plasma membrane-bound ENaC with no detectable effects on the total pool, one that is dominated by intracellular ENaC protein, indicates a surface membrane-only effect and activation of mostly inactive membrane resident channels. Such a mechanism would be consistent with that observed for ENaC activation by a variety of serine proteases in oocytes as well as epithelial cells (3, 8, 24). To further confirm this mechanism for subtilisin we examined its effects on ENaC surface expression in intact oocytes. Experiments have been previously described (12) and utilized expression of HA-tagged α in trimeric channels and examined binding of a horseradish peroxidase-coupled anti-HA antibody.
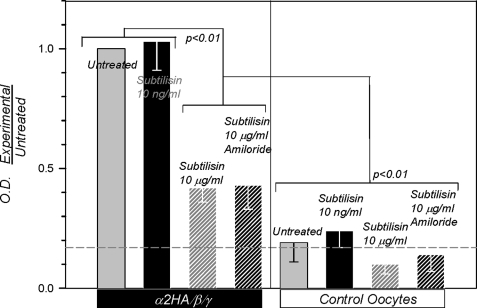
Subtilisin at 10 ng/ml was without effect on surface expression, indicating activation of membrane resident channels (Fig. 7), consistent with the data above. At 10 μg/ml, subtilisin decreased binding consistent with surface subunit degradation. This effect could not be modified by amiloride, indicating that changes of Na+ transport per se have no effect on surface binding. All of these maneuvers were without effect on background binding. These results indicate that subtilisin can specifically activate membrane resident ENaC at ng/ml levels and that it is well suited for examining the role of cleavage in channel activation.
FIGURE 7.
Subtilisin activates membrane resident ENaC. Binding studies demonstrate the absence of changes to channel density at the membrane in α/β/γ-expressing oocytes in response to ng levels of subtilisin. At much higher levels of this protease, binding intensity decreased consistent with channel subunit protein digestion and loss of HA signal. As expected, amiloride at 10 μm was without effects on anti-HA binding. The horizontal dashed line indicates the average background binding. Data were obtained from oocytes batches (25 each) from four different frogs. OD, outer diameter.
The above biochemical and electrophysiological data indicate that subtilisin cleaved near the endogenously cleaved sites at Arg-178 and/or Arg-204. This segment between Arg-178 and Arg-204 maps at or near the exposed finger domain of the crystallized ASIC1 (25). Therefore, it is expected to be solution-exposed and amenable to extracellular cleavage. To determine the sites of subtilisin cleavage within this segment, we examined cleavage of peptides with sequences derived from α Arg-178 to Arg-204 using MALDI-TOF. This technique allows us to precisely calculate the sizes of the cleaved products and consequently determine the cleavage sites and the cleavage preference for subtilisin. Given the size of this segment, it was divided into two peptides shown in Figs. 8 and 9 that encompassed each furin cleavage site.
In vitro cleavage of peptide H1 spanning Val-171—Pro-187 and encompassing the first cleavage site is shown in Fig. 8. This peptide was uncleaved by 10 ng/ml trypsin, confirming our previous result that this level of trypsin activates ENaC via an indirect, cleavage-independent mechanism (3). At higher trypsin levels, this peptide is cleaved at multiple sites. At 1 μg/ml trypsin cleavage occurred after most basic amino acids with no requirement for additional specific sequences. This indicates that trypsin is a poor tool to study ENaC cleavage given this potential non-specificity. By extrapolation, 10–100 μg/ml trypsin (levels that are commonly used to activate ENaC) could cause additional cleavage throughout solution-accessible segments of the extracellular loop further confounding the interpretation of experiments with this protease.
Cleavage of H1 by subtilisin occurred at a single position after Arg-175. This was observed with 50 ng/ml subtilisin and to a higher level with 200 ng/ml concentrations of this protease. This result is not surprising given the cleavage preference for subtilisin, which favors non-acidic residues at P2-P4 (20, 21), and the observed size of the subtilisin-cleaved α, which is indistinguishable from that cleaved by oocyte endogenous proteases at Arg-178 or Arg-204. This shift in cleavage by three aa explains the ability of subtilisin to cleave the αFM. Thus, subtilisin presents an ideal tool to study the acute effects of cleavage as it allows us to cleave channels with mutated endogenous cleavage sites.
Shown in Fig. 9 is peptide H2. This peptide encompassed the second furin cleavage site and spanned Leu-188—Ala-207. Like H1, this peptide was also uncleaved by 10 ng/ml trypsin but was cleaved at multiple sites at higher concentrations (1 μg/ml). Subtilisin was without effects on H2 at 50 ng/ml and was slightly cleaved after Arg-201 at 200 ng/ml. As the cleavage and activation effects are observed with subtilisin levels as low as 2 ng/ml, these results indicate that subtilisin does not likely cleave in this segment.
These results demonstrate that subtilisin is more specific than trypsin, that it can cleave near the sites used by endogenous proteases, and that it favors non-acidic residues at P2-P4. These results are consistent with activation after a single cleavage in the segment between the two proposed furin cleavage sites. However, these data do not rule out additional cleavage outside this segment in either αWT or αFM. This is tested below as well as the role of cleavage per se, and this segment activates the channel.
Mechanism of Channel Activation; Loss of the N Terminus Versus Removal of a Short Inhibitory Domain
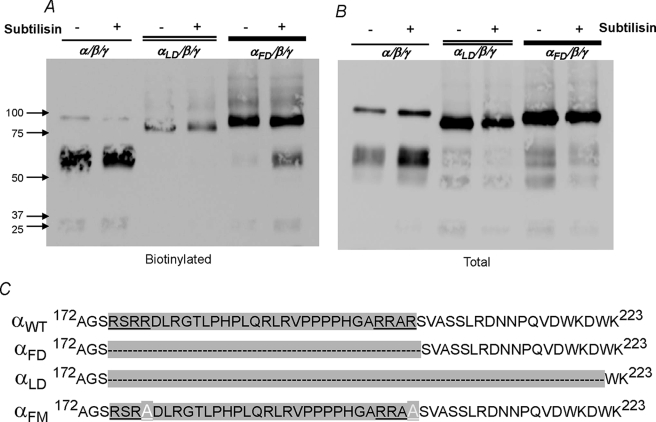
As mentioned above, it has been proposed that channel activation requires obligatory removal of the tract in α between the two furin cleavage sites (10). To directly test this hypothesis we removed the equivalent tract in human α. This construct is referred to as αFD and eliminates aa Arg-175–Arg-204. As shown in Fig. 10, this construct expresses to similar protein levels as αWT (and also αFM) in both fractions. This construct was also poorly cleaved by oocyte endogenous proteases, similar to that observed for αFM.
FIGURE 10.
Spontaneous processing and the effects of subtilisin on α lacking the inhibitory tract between the two furin cleavage sites. Western blots of oocytes expressing trimeric αWT, αLD, and αFD channels are shown. A and B represent the plasma membrane and total fractions. To visualize the small differences in the size of these different constructs, proteins were separated on 5% acrylamide gels with 5% added glycerol to increase gel strength. Subtilisin caused a 3-fold decrease of uncleaved (full-length) αWT consistent with its effect summarized above (Fig. 4B). This was only observed in the biotinylated fraction. Subtilisin also cleaved αFD at the plasma membrane, accompanied by increased density of the 65-kDa C-terminal fragment. The αLD construct, which extends the furin deletion by 17 aa, was not spontaneously cleaved at the plasma membrane and was furthermore insensitive to subtilisin. This construct expressed at a lower density at the plasma membrane despite normal levels in the total compartment (see “Results”). The amino acid sequences of these three α constructs in the vicinity of the two furin cleavage sites are shown in C. Also shown is the sequence of αFM. Data are representative of three experiments. See Fig. 11 for the electrophysiological effects. Oocytes were injected with twice the cRNA of αLD to enhance expression and plasma membrane levels.
Extracellular subtilisin retained the ability to cleave αFD as evident by the increase of the 65-kDa α fragment after subtilisin treatment. This effect was only observed in the plasma membrane fraction, indicating that subtilisin either cleaved at a new site not previously accessible before deletion of aa 175–204 or that subtilisin cleaved at a second site outside, but in close proximity to the sequences between the two furin sites. Nonetheless, the combination of subtilisin and αFD construct now provides a direct test for the two hypotheses of channel activation by cleavage. This is examined below in electrophysiological experiments.
To determine the potential site of action of subtilisin on αFD, we examined the amino acid sequences in the vicinity of the two furin sites for the general motif of basic-XX-basic for furin-like proteases and for AGSR-like sequences shown in Fig. 8 to be cleaved by subtilisin. Two sites were identified at KDWK and SSLR, respectively (Fig. 10C). We hypothesized that subtilisin may either normally cleave at one of these sites or that they may become more exposed to the actions of this protease after deletion of 30 aa from the loop in the αLD construct leading to enhanced proteolysis. This hypothesis was tested by generating an α construct that eliminated these sites. This construct is referred to as αLD and extends the deleted amino acids of αFD by 17. As shown in Fig. 10, αLD was uncleaved at the plasma membrane and was also uncleavable by subtilisin.
An interesting result was that αLD expressed at the plasma membrane at lower levels than any of the other α constructs. This effect was not observed in the total fraction indicating that the segment between Ser-205—Asp-221 may affect channel trafficking or stability. Notwithstanding this effect, αLD now provides a construct that is uncleaved and lacks the proposed inhibitory domain that can be used to directly address the mechanism of channel activation by cleavage.
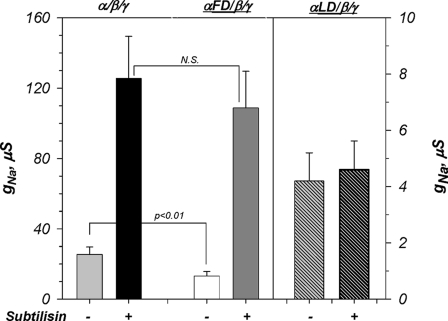
The electrophysiological data for these α constructs are shown in Fig. 11. The amiloride-sensitive whole cell conductance of αFD was elevated as compared with αFM but remained significantly lower than that of αWT. More importantly, αFD activity remained nowhere close to that of subtilisin-activated αWT. If the segment between the two furin cleavage sites, which is deleted in αFD, is assumed to be the main mechanism of channel activation, it would be expected that αFD would exhibit a conductance that is even higher than that of subtilisin-activated αWT. This follows, as expression of αFD would then be equivalent to a 100% cleavage of plasma membrane αWT by subtilisin, as all of these channels would be missing this segment. As shown in Fig. 11, this is clearly not the case. Furthermore, if channel activation primarily occurred by removing the tract between Arg-178 and Arg-204, then αFD would be expected to be insensitive to activation by cleavage. Again, this is not the case, as subtilisin caused an ∼8-fold activation of αFD. Thus, cleavage cannot activate ENaC primarily by removing a short inhibitory domain from α. Rather, we propose that this occurs by an alternative mechanism that involves the loss of the N terminus and TM1 of this subunit and the channel complex. This is consistent with the above data indicating much lower levels of the 22-kDa N-terminal fragment at plasma membrane. This is also consistent with the result that αLD, which eliminates subtilisin cleavage altogether, forms a non-activated, low baseline activity channel. These results would be also consistent with the data that the exact site of channel cleavage and the number of cleavage sites within the tract of aa 174–222 do not play a critical role in the magnitude of activation. These data do not completely rule out an inhibitory effect of the tract between Arg-178 and Arg-204; rather, they do clearly indicate that such an effect is not the dominant or obligatory mechanism of channel activation by cleavage.
FIGURE 11.
ENaC is markedly activated by subtilisin cleavage of α lacking the tract between the two furin cleavage sites. Electrophysiological data correspond to the biochemical data summarized in Fig. 10. Spontaneous activity of αFD was significantly lower than that of αWT but higher than that of αFM (see Fig. 6C). Channels formed with either αWT or αFD were activated to similar levels after 50 ng/ml subtilisin (N.S. indicates p > 0.05). Channels formed with αLD exhibited low activity due to the absence of cleavage as well as its decreased plasma membrane levels (see Fig. 10). Subtilisin was without effect on channels containing αLD. Note the difference in the y axis between αWT/αFD and αLD. n = 12, 14, and 8 from 5 different frogs for αWT, αFD, and αLD, respectively. See “Results” for additional details.
DISCUSSION
We examined the mechanism of ENaC activation by extracellular serine proteases. Our experiments utilized subunits with two homomeric tags to examine channel cleavage and the S8 serine protease subtilisin. We demonstrate that α subunit cleavage is critical to channel activation. We also demonstrate that channels formed with an α lacking the tract between the two furin cleavage sites retain the ability to be fully activated by cleavage. Conversely, we demonstrate that further elimination of cleavage in this construct results in a low activity channel that can no longer be fully activated. These results support an alternative mechanism that αENaC cleavage activates the channel by removal of the N terminus and TM1.
Role of a Subunit Cleavage
The range of ENaC activation between channels formed with mostly uncleaved and those with mostly cleaved α is nearly 35-fold (Fig. 6C). This represents an enormous capacity for changes in ENaC activity at the plasma membrane and demonstrates the critical role of α subunit cleavage in the control of activity. This dominant role of α does not rule out the currently proposed role for γ subunit cleavage (5, 15). These studies cannot be directly compared with those of ours as Diakov et al. (5) examined expression of channels with two subunit combinations, whereas Carattino et al. (15) did not show any comparable biochemical data. Moreover, in our studies γ was maximally endogenously cleaved, and therefore, it could not be rate-limiting to channel activation. This represents a unique advantage of the oocyte system as it allows the separate examination of the role of α. In the absence of such an advantage it would have been difficult to experimentally cleave these two subunits separately.
It is also important to note that we do not discount the role of γ cleavage in vivo. Indeed, it is well documented that the endogenous levels of γ cleavage change with maneuvers that alter sodium transport in the rat kidney (26, 27). However, in these studies the antibodies used to detect α are N-terminal and only recognize the uncleaved form of this subunit. Therefore, a comparison of the in vivo changes to α and γ processing, and the link to changes to Na+ transport is not currently feasible and requires the development of additional tools. It is tempting to propose that cleavage of both subunits can markedly affect activity and that the role or the relative importance of cleavage of each subunit may differ for ENaC expressed in different epithelial tissue. However, it is also possible that cleavage of these two subunits may represent a redundant mechanism to control activity. Such a mechanism would be consistent with our proposed hypothesis that the loss of the first transmembrane domain of α leads to the activation of ENaC (see below) and that cleavage on α in multiple positions can lead to the same activation.
Cleavage by Subtilisin
We used the protease subtilisin to cleave ENaC. This protease is available in large quantities that allow its use as a widespread tool to cleave ENaC. In oocytes, subtilisin also has many advantages including that it has no effects on endogenous currents. In contrast, trypsin at concentrations used to cleave ENaC activates oocyte endogenous Ca2+- dependent currents (19). Although these currents are transient in nature, they nonetheless preclude an accurate determination of the time course of ENaC activation. More importantly, these currents originate from changes to the intra-oocyte Ca2+ concentration, and it is unknown if these changes can also alter ENaC activity, membrane density, or the response to cleavage. It is well known that ENaC activity and membrane density are sensitive to protein kinase C and specifically to Ca2+-dependent isoforms of protein kinase C, and in many systems it is also sensitive to the intracellular Ca2+ concentration (11, 28–30). Therefore, it would not be altogether surprising if the initial Ca2+ transient also affected the subsequent response of ENaC to trypsin. Thus, subtilisin presents a better tool to examine ENaC activation by cleavage in oocytes.
A second equally important issue with trypsin is the potential to cleave after single arginine residues. Indeed, this protease is used in digesting and fingerprinting proteins. We demonstrate in peptides with sequences derived from αENaC that trypsin cleaves at multiple sites, mostly after single arginines. This does not necessarily indicate that all of these arginines are exposed in functional plasma membrane-embedded trimeric channels. However, it is difficult to envision how all arginines, except those at the two furin cleavage sites, would be buried within the channel complex. Therefore, a protease with increased specificity such as subtilisin can serve as a more appropriate surrogate for furin cleavage.
We conclude from our in vitro experiments that subtilisin cleaves α once between aa Val-171 and Ala-207 after AGSR. Based on the data from αLD, we also determine that subtilisin can only cleave the native α between aa 175 and 221. Thus, all subtilisin cleavage in the native channel (and whether this occurs at one or two sites) is limited to this segment of the extracellular loop of α. Two possibilities exist. The first is that subtilisin cleaves at two positions in all the α constructs except αFD, which lacks one site, and αLD, which lacks both sites. In this case one would conclude that cleavage in these sites is likely redundant as they lead to the same final outcome of channel activation. In this case, this redundancy would argue against a specific inhibitory role of the segment between the two furin cleavage sites.
The second possibility is that subtilisin only cleaves at Arg-175 as demonstrated from the MALDI-TOF experiments but that the deletion of 30 aa between 175 and 204 in the αFD exposes an additional cleavage site between 205 and 221 and increases its affinity for cleavage by subtilisin. This second scenario would be expected to cause a small shift in the size of the 65-kDa-cleaved α between αWT/αFM and αFD. This is not observed in Fig. 10. However, this potential 2-kDa change in size in a 65-kDa protein is likely within the limit of resolution of mini gel electrophoresis. This explanation would indicate that cleavage is redundant and also argues against a specific inhibitory role of the segment between the two furin cleavage sites. Thus, in both cases the loss of the segment between the two furin cleavage sites is not likely to be an obligatory mechanism of ENaC activation by α cleavage. Such built-in redundancy into channel cleavage would certainly be consistent with the ability of multiple diverse proteases such as trypsin, subtilisin, elastase, prostasin, furin, and plasmin to activate ENaC in different systems with a similar final outcome (9, 24, 31–35).
Mechanism of Activation: Loss of TM1 Versus Short Inhibitory Domain
The N terminus of cleaved α exhibited much lower plasma membrane levels than those of the C terminus and at times was undetectable at the plasma membrane (see Fig. 5). This difference is unlikely because of differences in the recognition of the HA antibody to its antigenic site between the first and second part of αENaC as appreciable N-terminal signal is detectable in the intracellular fraction (see “Results”) and as these experiments utilize denaturing electrophoresis. Moreover, it is also unlikely that the differences in the levels of these two fragments at the plasma membrane is because of the inability or poor efficiency of biotinylation of the N-terminal fragment as this fragment is detectable in many experiments (Figs. 1 and 3), and one would have to invoke that the accessibility of primary amines on this fragment is a variable factor to yield the result obtained in Fig. 5. Moreover, both fragments contain multiple primary amines suitable for modification by the biotinylating reagent.
The absence of a correlation between the levels of these two fragments is rather consistent with internalization and degradation of the N-terminal fragment. The N-terminal fragment contains lysines that are likely ubiquitinated after binding of the ubiquitin ligase to the C-terminal PY motifs (23). In this case, it would be expected that the N terminus would be rapidly internalized and degraded, exactly as observed. Such a result further argues against a role of this fragment as a component of the active channel complex. Our attempts to determine whether the N terminus could be trapped at the membrane before potential internalization by examining shorter (5 min) cleavage times were unsuccessful as they yielded poor cleavage efficiency (not shown). This is consistent with the time course of activation by subtilisin shown in Fig. 6, which indicates a very linear initial phase that would severely limit activation and presumably the levels of cleaved fragments within the first 5 min.
How can the loss of the N terminus and TM1 lead to channel activation? Recent data from the crystal structure of ASIC1, a homologous ion channel, may shed insight into such a mechanism. In the trimeric form of ASIC1 it was found that TM1 of the individual subunits contributes much less than TM2 to the overall conduction pore (36). In this case the loss of TM1 may be a favorable process. From our results we find that channels formed with α/γ, which do not contain TM1 and TM2 from β, are active without cleavage (not shown). A possible mechanism explaining the activity of α/γ channels is the absence of the additional transmembrane domains from the third subunit. This effect would be mimicked in trimeric channels by cleavage of α and γ and the loss of TM1s from both of these subunits. In the case of oocytes, this effect in trimeric channels may simply just depend on the loss of TM1 from α, as γ is already maximally cleaved at the plasma membrane and likely missing its TM1. Channel activation after the loss of TM1 may then occur if this domain causes a mismatch, possibly a hydrophobic mismatch in the length or the pore of the channel, leading to a decrease of Po to levels that make the channel inactive. In this case any cleavage of αENaC at an externally accessible cleavage site within the vicinity of the furin cleavage sites would cause the same channel activation. Such mechanisms of hydrophobic mismatch leading to effects on Po have been proposed to affect the gating of many ions channels including gramicidin (37–42).
Are channels inhibited by the fragment contained between the two furin cleavage sites? Kleyman and co-workers (10) have demonstrated that a synthetic peptide derived from this fragment inhibits ENaC activity in the μm range in oocytes. This fragment is present in αFM but absent from αFD. Both of these channels exhibited lower baseline activity than αWT despite the fact that the αFD construct eliminated this presumed inhibitory segment. Comparing these two constructs, we find that oocytes expressing αFD exhibited higher conductance than those expressing αFM (Figs. 6 and 11; ∼10 microsiemens higher activity). However, αFD was further activated by more than 8-fold (by an additional 100 microsiemens) by cleavage to levels indistinguishable from those of αWT, indicating that an alternative mechanism accounts for the majority of protease activation of ENaC. We propose that this mechanism involves the loss of the N terminus and TM1 from the channel based in part on the result that αLD, a construct that eliminates cleavage altogether, exhibits very low activity that is similar to that observed with αFM.
The differences in the activity of αFM and αFD may indicate that the segment between the two furin cleavage sites may exhibit a secondary lesser effect on activity in addition to the predominant effect observed by removing the N terminus and TM1. Alternatively, an effect of this segment on activity may be only important in the artificial situation in uncleaved channels missing the tract between aa 175–204 (e.g. αFM versus αFD). In this case the ability of the peptide with sequences originating from this segment to inhibit ENaC may be due to other unrelated mechanisms such as interaction with other parts of the channel that are different than those that aa 175–204 interacted with when they were part of the native channel. In support of such an alternative indirect inhibitory mechanism is the observation that the efficiency of inhibition by this peptide differs by nearly 100-fold between oocytes and mammalian cells expressing ENaC (10), indicating that this is not likely because of inhibition of an intrinsic channel property. These possibilities remain to be tested and represent future experimental challenges to a better understanding of channel regulation.
This work was supported, in whole or in part, by National Institutes of Health Grant DK55626. This work was also supported by the John R. Oishei Foundation.
- ENaC
- epithelial sodium channel
- HA
- hemagglutinin
- aa
- amino acids
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- TM
- transmembrane domain
- WT
- wild type.
REFERENCES
- 1.Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 2.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 3.Bengrine A., Li J., Hamm L. L., Awayda M. S. (2007) J. Biol. Chem. 282, 26884–26896 [DOI] [PubMed] [Google Scholar]
- 4.Kabra R., Knight K. K., Zhou R., Snyder P. M. (2008) J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 5.Diakov A., Bera K., Mokrushina M., Krueger B., Korbmacher C. (2008) J Physiol 586, 4587–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedstrom L. (1996) Biol. Chem. 377, 465–470 [PubMed] [Google Scholar]
- 7.Kam C. M., Hernandez M. A., Patil G. S., Ueda T., Simmons W. H., Braganza V. J., Powers J. C. (1995) Arch. Biochem. Biophys. 316, 808–814 [DOI] [PubMed] [Google Scholar]
- 8.Caldwell R. A., Boucher R. C., Stutts M. J. (2004) Am. J. Physiol. Cell Physiol. 286, C190–C194 [DOI] [PubMed] [Google Scholar]
- 9.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 10.Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 11.Awayda M. S. (2000) J. Gen. Physiol. 115, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awayda M. S., Shao W., Vukojicic I., Bengrine A. (2006) Ion Channels Methods and Protocols 337, 101–115 [DOI] [PubMed] [Google Scholar]
- 13.Bengrine A., Li J., Awayda M. S. (2007) FASEB J. 21, 1189–1201 [DOI] [PubMed] [Google Scholar]
- 14.Firsov D., Schild L., Gautschi I., Mérillat A. M., Schneeberger E., Rossier B. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattino M. D., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 17.Vallet V., Pfister C., Loffing J., Rossier B. C. (2002) J. Am. Soc. Nephrol. 13, 588–594 [DOI] [PubMed] [Google Scholar]
- 18.Adachi M., Kitamura K., Miyoshi T., Narikiyo T., Iwashita K., Shiraishi N., Nonoguchi H., Tomita K. (2001) J. Am. Soc. Nephrol. 12, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 19.Durieux M. E., Salafranca M. N., Lynch K. R. (1994) FEBS Lett. 337, 235–238 [DOI] [PubMed] [Google Scholar]
- 20.Ruan B., London V., Fisher K. E., Gallagher D. T., Bryan P. N. (2008) Biochemistry 47, 6628–6636 [DOI] [PubMed] [Google Scholar]
- 21.Gosalia D. N., Salisbury C. M., Ellman J. A., Diamond S. L. (2005) Mol. Cell. Proteomics 4, 626–636 [DOI] [PubMed] [Google Scholar]
- 22.Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) EMBO J 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldwell R. A., Boucher R. C., Stutts M. J. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol. 288, L813–L819 [DOI] [PubMed] [Google Scholar]
- 25.Stockand J. D., Staruschenko A., Pochynyuk O., Booth R. E., Silverthorn D. U. (2008) IUBMB Life 60, 620–628 [DOI] [PubMed] [Google Scholar]
- 26.Frindt G., Palmer L. G. (2009) Am. J. Physiol. Renal Physiol. 297, F1249–F1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masilamani S., Kim G. H., Mitchell C., Wade J. B., Knepper M. A. (1999) J. Clin. Invest. 104, R19–R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Awayda M. S., Ismailov, Berdiev B. K., Fuller C. M., Benos D. J. (1996) J. Gen. Physiol. 108, 49–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Awayda M. S., Platzer J. D., Reger R. L., Bengrine A. (2002) Am. J. Physiol. Cell Physiol. 283, C1122–C1132 [DOI] [PubMed] [Google Scholar]
- 30.Frindt G., Palmer L. G., Windhager E. E. (1996) Am. J. Physiol. 270, F371–F376 [DOI] [PubMed] [Google Scholar]
- 31.García-Caballero A., Dang Y., He H., Stutts M. J. (2008) J. Gen. Physiol. 132, 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris M., Firsov D., Vuagniaux G., Stutts M. J., Rossier B. C. (2007) J. Biol. Chem. 282, 58–64 [DOI] [PubMed] [Google Scholar]
- 33.Nesterov V., Dahlmann A., Bertog M., Korbmacher C. (2008) Am. J. Physiol. Renal Physiol. 295, F1052–F1062 [DOI] [PubMed] [Google Scholar]
- 34.Svenningsen P., Bistrup C., Friis U. G., Bertog M., Haerteis S., Krueger B., Stubbe J., Jensen O. N., Thiesson H. C., Uhrenholt T. R., Jespersen B., Jensen B. L., Korbmacher C., Skøtt O. (2009) J. Am. Soc. Nephrol. 20, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svenningsen P., Uhrenholt T. R., Palarasah Y., Skjoedt K., Jensen B. L., Skott O. (2009) Am. J. Physiol. Regul. Integr. Comp. Physiol., in press [DOI] [PubMed] [Google Scholar]
- 36.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 37.Andersen O. S., Bruno M. J., Sun H., Koeppe R. E., 2nd (2007) Methods Mol. Biol. 400, 543–570 [DOI] [PubMed] [Google Scholar]
- 38.Kelkar D. A., Chattopadhyay A. (2007) Biochim. Biophys. Acta 1768, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 39.Lee K. J. (2005) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72, 031917 [Google Scholar]
- 40.Lee K. J. (2006) Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 73, 021909 [Google Scholar]
- 41.Liu W., Caffrey M. (2005) J. Struct. Biol. 150, 23–40 [DOI] [PubMed] [Google Scholar]
- 42.Lundbaek J. A., Andersen O. S. (1999) Biophys. J. 76, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]