Abstract
The cadA gene in Dictyostelium encodes a Ca2+-dependent cell adhesion molecule DdCAD-1 that contains two β-sandwich domains. DdCAD-1 is synthesized in the cytoplasm as a soluble protein and then transported by contractile vacuoles to the plasma membrane for surface presentation or secretion. DdCAD-1-green fluorescent protein (GFP) fusion protein was expressed in cadA-null cells for further investigation of this unconventional protein transport pathway. Both morphological and biochemical characterizations showed that DdCAD-1-GFP was imported into contractile vacuoles. Time-lapse microscopy of transfectants revealed the transient appearance of DdCAD-1-GFP-filled vesicular structures in the lumen of contractile vacuoles, suggesting that DdCAD-1 could be imported by invagination of contractile vacuole membrane. To assess the structural requirements in this transport process, the N-terminal and C-terminal domains of DdCAD-1 were expressed separately in cells as GFP fusion proteins. Both fusion proteins failed to enter the contractile vacuole, suggesting that the integrity of DdCAD-1 is required for import. Such a requirement was also observed in in vitro reconstitution assays using His6-tagged fusion proteins and purified contractile vacuoles. Import of DdCAD-1 was compromised when two of its three Ca2+-binding sites were mutated, indicating a role for Ca2+ in the import process. Spectral analysis showed that mutations in the Ca2+-binding sites resulted in subtle conformational changes. Indeed, proteins with altered conformation failed to enter the contractile vacuole, suggesting that the import signal is somehow integrated in the three-dimensional structure of DdCAD-1.
Introduction
In eukaryotes, soluble secretory proteins are typically transported through the classical ER3-Golgi transport pathway (1, 2). However, it has become evident in recent years that a growing number of soluble proteins synthesized in the cytoplasm are targeted for secretion (3, 4). This group of secretory proteins usually shares several common features, including the lack of conventional signal peptides, the absence of post-translational modifications, and the presence of free cysteines (5, 6). Many of these proteins play key roles in diverse biological processes. Among them are galectins (7, 8), interleukin-1β (9), thioredoxin (10), macrophage migration inhibitory factor (11), and fibroblast growth factor 1 and 2 (FGF-1 and FGF-2) (12, 13). Several viral proteins, such as HIV-Tat and herpes simplex VP22, are also known to be secreted via ER-Golgi-independent routes (14–16).
Multiple unconventional transport mechanisms have been discovered, and cells can utilize one or more pathways to target soluble proteins for secretion. The interleukin-1β export mechanism involves intracellular vesicles in its secretory pathway (17, 18), whereas the muscle lectin galectin-1 is externalized by the shedding of membrane vesicles (19) or through a protein transporter (20). Secretion of the growth factor FGF-2 is also likely mediated by a plasma membrane-resident transporter (21). Additionally, cell surface counter receptors are essential components in the export machinery of galectin-1 because they can provide an extracellular trap mechanism (20).
Several soluble proteins expressed by the social amoeba Dictyostelium discoideum have been found to be targeted for secretion by unconventional pathways (22, 23). At the onset of development, amoeboid cells undergo chemotactic migration to form large aggregates of ∼105 cells, which eventually culminate in the formation of fruiting bodies (24). The endogenous soluble lectin discoidin-I, which is externalized to facilitate cell-substratum adhesion (25, 26), is targeted for secretion through vesicular structures (27). Multicellularity during Dictyostelium development is maintained by the expression of several cell adhesion molecules (22). One of them is the Ca2+-dependent cell adhesion molecule DdCAD-1 that is encoded by the cadA gene. DdCAD-1 lacks the classical signal peptide and is synthesized as a soluble protein in the cytoplasm (28–30). It is imported into contractile vacuoles for transport to the plasma membrane (31). DdCAD-1 contains two distinct domains with β-sandwich architecture. Although the N-terminal domain is involved in homophilic binding, the C-terminal domain tethers the protein to a membrane anchor, thus allowing it to function as a cell adhesion molecule on the cell surface (32). Both DdCAD-1 and discoidin-I have been found to be enriched in contractile vacuoles (31), the osmoregulatory organelles that regulate water balance during the growth and the initial stages of development (33, 34). As they fuse with the plasma membrane, the contents of the contractile vacuoles are released into the medium.
In this study, we investigated the structural requirements for the import of DdCAD-1 into contractile vacuoles. Using transfectants that express DdCAD-1-GFP fusion proteins, we observe that DdCAD-1 is imported through the invagination of contractile vacuole membranes. However, the whole protein is required because neither the N-terminal domain nor the C-terminal domain alone is sufficient for transport into the contractile vacuole. Additionally, the import mechanism is dependent on Ca2+ and proper protein conformation, suggesting that the secretory targeting motif of DdCAD-1 lies in its three-dimensional structure rather than in a linear stretch of amino acids.
EXPERIMENTAL PROCEDURES
Construction of DdCAD-1-GFP, N-GFP, and C-GFP Expression Vectors and Cell Transfection
For DdCAD-1-GFP expression, a 645-bp fragment of DdCAD-1 cDNA was cloned into the HindIII and EcoRI sites of pA15/NIGFP expression vector (kindly provided by Dr. David Knecht, University of Connecticut). The N-terminal domain (N, 288 bp) and the C-terminal domain (C, 366 bp) of DdCAD-1 were amplified by PCR methods before cloning into pA15/NIGFP. Primers used in this study are listed in supplemental Fig. S1. The expression constructs were transfected into cadA-null cells by electroporation as described by Pang et al. (35). Cells were harvested at log phase, washed two times in cold H50 solution (20 mm Hepes, 50 mm KCl, 10 mm NaCl, 1 mm MgSO4, 5 mm NaHCO3, 1 mm NaH2PO4, pH 7.0), and then resuspended in H50 at 2 × 107 cells/ml. Cell samples (100 μl each) were mixed with ∼5 μg of plasmid DNA and incubated for 5 min on ice in 1-mm cuvettes. Electroporation was carried out by two consecutive pulses of 0.85 kV with a capacitance of 25 microfarads applied to the cuvette with a 5-s recovery between pulses. After 5 min of incubation on ice, the cells from each cuvette were deposited onto a 6-well tissue culture plate containing 3 ml of HL5 (0.5% Difco proteose peptone number 2, 0.5% BBL thiotone E peptone, 1% glucose, 0.5% yeast extract, 2.5 mm KH2PO4, 2.5 mm Na2HPO4, pH 6.7) in each well (35). The cells became adherent, and transfectants were then selected by sequential incubation in 2, 5, 10, and 20 μg/ml G418 (Sigma).
Immunofluorescence Labeling of Cells and Laser Scanning Confocal Microscopy
Transfected cells expressing DdCAD-1-GFP, N-GFP, C-GFP, and GFP were developed in 17 mm phosphate buffer, pH 6.4, for 1–3 h at 2 × 107 cells/ml. The cell samples (1 × 106 cells) were then allowed to attach on positively charged coverslips (Fisher, catalog no. 12-545-84) for 30–60 min). Cells were then fixed in 4% paraformaldehyde, 50% HL-5, and 0.1% DMSO for 30 min at room temperature. The cells were permeabilized with 0.5% Triton X-100 and 1% paraformaldehyde for 30 min (34, 36). Nonspecific binding was blocked by incubation with 1% bovine serum albumin in MCG buffer (50 mm Mes, pH 6.4, 0.2 mm CaCl2, 2 mm MgCl2) for 30 min. Cells were labeled with the mouse anti-calmodulin mAb 6D4 (Sigma) (1:200 dilution in MCG containing 0.1% bovine serum albumin) for 1 h, washed three times with Tris-buffered saline containing 0.1% Tween 20, and then stained with Alexa-568-conjugated goat anti-mouse antibodies (1:400 dilution) (Molecular Probes, Eugene, OR) for 1 h. Coverslips were mounted in DAKO fluorescent mounting medium (DakoCytomation, Glostrup, Denmark). Confocal images were acquired using the Zeiss LSM 510 microscope.
For live cell imaging, axenically grown transfected cells were suspended in 17 mm Na/Na2 phosphate buffer at ∼2 × 105 cells/ml, and 100 μl was deposited on a concavity glass slide for 30–60 min. The medium was then replaced with 8.5 mm Na/Na2 phosphate buffer containing the styryl dye FM4-64 (Molecular Probes) at 1 mg/ml. Images were recorded between 5 and 20 min after staining to visualize both the contractile vacuoles and the plasma membrane. Time-lapse sections were collected on the Zeiss LSM510 microscope equipped with a 100× water immersion objective. Images were processed using the Zeiss LSM Image Browser software (version 4.0.0.157).
Isolation of Contractile Vacuoles and Cytosol
Contractile vacuoles were isolated according to Sesaki et al. (31) with minor modifications. Cells (2 × 109) were developed for 6 h in suspension and then homogenized in 12 ml of TM buffer (2 mm MgCl2, 10 mm Tris-HCl, pH 7.5) at room temperature. Samples (5 ml each) were layered on top of discontinuous (28 and 48% (w/w), 4 ml each) sucrose density gradients and then centrifuged for 1 h at 40,000 rpm at 4 °C in an SW 40 rotor. Contractile vacuoles enriched at the interface were collected, and aliquots were stored at −70 °C. Also, cytosol derived from cadA-null cells was obtained after centrifugation of the cell lysate at 40,000 rpm, and aliquots were stored at −70 °C. Protein concentration was determined using the bicinchoninic acid assay kit (Pierce). Proteolytic digestion of contractile vacuoles was carried out by incubating samples (containing ∼100 μg of protein) with 0.01 mg/ml proteinase K in the presence or absence of 0.05% SDS at 37 °C for 1 h. After the addition of 2 mm phenylmethylsulfonyl fluoride, samples were boiled for 10 min and subjected to SDS-PAGE, followed by Western blot analysis using either rabbit antibodies against DdCAD-1 (32) or rabbit antibodies against GFP (Molecular Probes).
Expression of His6-tagged Mutant DdCAD-1 Proteins
His-tagged wild-type DdCAD-1 (His6-WT) and, N and C domains (His6-N and His6-C) were expressed and purified as soluble native proteins as described previously (32). The Ca2+-binding site mutants were created using site-directed mutagenesis to substitute two to three residues with alanine in each Ca2+-binding site (SI, SII, and SIII) as follows: His6-SI(D39A, T81A), His6-SII(D35A, E56A), His6-SIII(E59A, D61A), and His6-S(I+II)(D35A, N38A, D39A). The primers used to generate these constructs are shown in supplemental Fig. S2. All constructs were sequenced to ascertain sequence fidelity. Protein expression was carried out in Escherichia coli strain BL21(DE3), and His-tagged proteins were purified using nickel-nitrilotriacetic acid resin (Qiagen Inc., Valencia, CA).
In Vitro Reconstitution of DdCAD-1 Import into Contractile Vacuoles
Contractile vacuoles and cytosol fractions derived from cadA-null cells were mixed at a 1:1 ratio in terms of protein amount in a 600-μl reaction sample to give a final concentration of 2 mg/ml. His6-DdCAD-1 and the different mutant forms of DdCAD-1 were added to a final concentration of 2 μm, and the mixture was incubated for 1 h at room temperature. The contractile vacuoles were pelleted at 12,000 rpm for 10 min at 4 °C, washed twice with TM buffer, and subjected to proteinase K (10 μg/ml) digestion for 1 h at 37 °C in the presence or absence of 0.1% SDS. Samples were subjected to SDS-PAGE and Western blot analysis.
45Ca2+ Overlay Assay
45Ca2+ overlay assays were carried out as described previously (32). Recombinant DdCAD-1 proteins were blotted onto nitrocellulose membrane using a slot blot apparatus (Bio-Rad). The membrane was washed for 1 h in four changes with the overlay buffer (60 mm KCl, 5 mm MgCl2, and 10 mm imidazole, pH 6.8). The blot was then incubated with 10 ml of 45Ca2+ (10 μCi/ml) in the overlay buffer for 30 min at room temperature on a platform shaker, followed by three 10-min washes with deionized water. Autoradiography was carried out by exposure of the air-dried membrane to Bioflex-MRI Films (Clonex Corp.) for 24 h at −70 °C.
Antibody-induced Cap Formation
Cells (2 × 107 cells/ml) were developed in liquid medium and collected at 3 h. Anti-GFP antiserum (1:100 dilution) was added to 1.2 × 106 cells suspended in 300 μl of 50 mm Mes buffer, pH 6.3, and incubated for 30 min at room temperature. Alexa-568-conjugated goat anti-mouse antibody was added at 1:400 dilution and rotated at room temperature for another 30 min. Next, 300 μl of cells were deposited on a positively charged coverslip (Fisher) and allowed to attach for 15 min. The coverslips were washed gently with MCG buffer, fixed with 3.7% formaldehyde, and mounted for fluorescence microscopy.
Analysis of DdCAD-1 Secretion
Axenically grown cadA null transfectants (2 × 107 cells/ml) were developed in 17 mm phosphate buffer for 6 h at room temperature. During development, 1-ml cell samples were collected every hour, and the cells were pelleted at 15,000 × g for 1 min to obtain a clear supernatant. Aliquots of the supernatant were mixed with sample buffer (1:1) and prepared for SDS-PAGE, followed by Western blot analysis.
Chemical Cross-linking of DdCAD- 1
His6-DdCAD-1 (30 μg) was cross-linked in the presence of 1 mm disuccinimidyl suberate for 30 min at room temperature. The excess disuccinimidyl suberate in the reaction sample was quenched with 50 mm Tris-HCl, pH 7.6. The cross-linked His6-DdCAD-1 was subjected to the contractile vacuole import assay.
RESULTS
Import of DdCAD-1-GFP via Invagination of Vacuolar Membrane
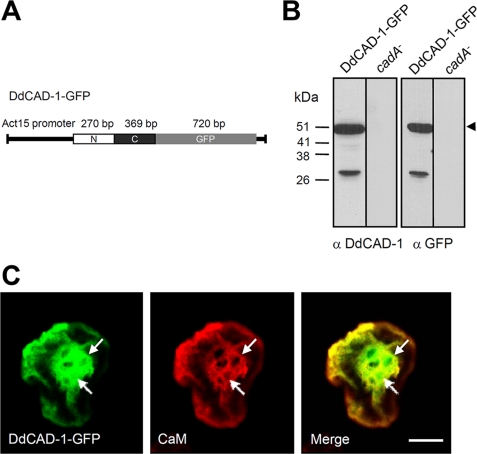
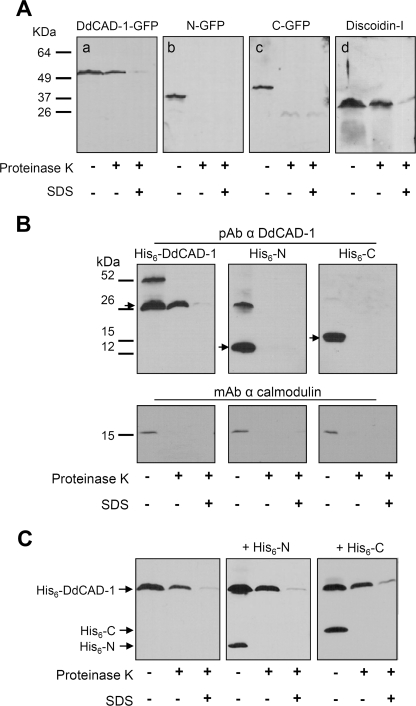
To facilitate the in vivo analysis of DdCAD-1 transport, cadA-null cells were transfected with a DdCAD-1-GFP construct (Fig. 1A). Transfectants were selected using G418 and by direct visualization under a fluorescence microscope. The expression of DdCAD-GFP was confirmed by probing protein blots with antibodies against DdCAD-1 and GFP (Fig. 1B). Both antibodies detected a protein band corresponding to the expected size of the fusion protein. Transfected cells showed uniform green fluorescence in the cytoplasm. To determine whether the fusion protein was imported by contractile vacuoles, cell samples were fixed and permeabilized to induce partial loss of the cytoplasmic GFP fusion protein. Some samples were also stained with anti-calmodulin antibodies. Calmodulin serves as a marker of contractile vacuoles because it is associated with their cytoplasmic surface (37). Confocal microscopy showed that an abundance of DdCAD-1-GFP associated with the contractile vacuole network and the cortical region (Fig. 1C). Notably, DdCAD-1-GFP was present in the lumen of contractile vacuoles, where calmodulin staining was absent.
FIGURE 1.
Construction and expression of DdCAD-1-GFP in cadA-null cells. A, schematic drawing of the DdCAD-1-GFP construct. B, Western blots of cadA-null cells transfected with plasmid DNA. Cell lysates of stable G418-resistant clones and parental cells (cadA−) were prepared from cells at 3 h of development for SDS-PAGE and protein blots, which were probed with rabbit antisera against either DdCAD-1 or GFP. The expected size of the fusion protein is indicated by an arrowhead. The lower molecular weight band probably represents a degraded product. Molecular mass markers are shown on the left. C, confocal micrographs showing the association of DdCAD-1-GFP with the contractile vacuole network. Cells were collected at 3 h of development, fixed in 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100 for 5 min. Cells were labeled with mouse anti-calmodulin (CaM) mAb (red) and subjected to laser scanning confocal microscopy. Arrows indicate contractile vacuoles. Bar, 10 μm.
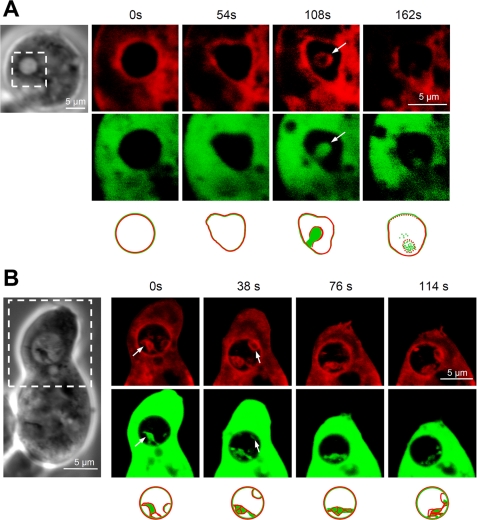
Confocal images of fixed specimens of DdCAD-1-GFP transfectants occasionally displayed a punctate staining pattern of DdCAD-1 inside contractile vacuoles. To examine the nature of these stained structures, live cells were incubated with the vital dye FM4-64 to label cellular membranes (34), followed by time lapse microscopy. The confocal series of a number of cells revealed novel vesicular structures inside their contractile vacuoles (Fig. 2, A and B). These vesicles adopted either a tubular or rounded appearance and were often filled with DdCAD-1-GFP. The membrane of these vesicles was marked by FM4-64 and showed continuity with the contractile vacuole membrane, indicating that they had originated from the contractile vacuole. These structures were relatively transient as they remained for only 2–3 min inside the lumen of contractile vacuoles and then disappeared. After that, the GFP fluorescence in the lumen became diffuse, and it was accompanied by the loss of FM4-64 staining, suggesting that the vesicles might have pinched off from the vacuolar membrane and ruptured to release the cargo in the lumen. Thus, membrane invagination might serve as a pathway by which DdCAD-1 is imported into contractile vacuoles.
FIGURE 2.
Budding of vesicles into the lumen of the contractile vacuoles. A and B, transfectants expressing DdCAD-1-GFP were collected from HL-5 medium, washed, and suspended in 17 mm phosphate buffer at ∼2 × 105 cells/ml. Live cells (100 μl) were deposited on slides for attachment. The styryl dye FM4-64 (red) was added at 1 mg/ml to visualize both the contractile vacuoles and the plasma membrane. Time-lapse sequences of confocal images were recorded between 5 and 20 min after dye addition. Confocal images of the boxed area in the light micrograph are shown. Arrows point to membrane protrusions in the contractile vacuole lumen, where DdCAD-1-GFP and FM4-64 co-localize. Schematic drawings of the contractile vacuole are shown below the confocal images.
Import of DdCAD-1 into Contractile Vacuoles Requires Both N- and C-terminal Domains
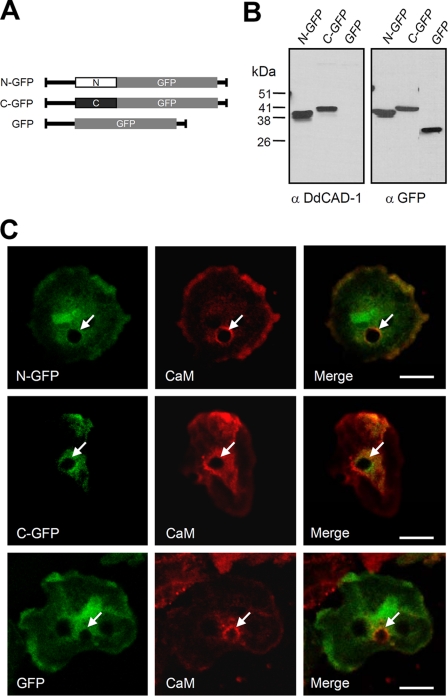
DdCAD-1 contains two distinct domains linked by a short stretch of amino acids (32, 38). To investigate the structural requirements for its import into contractile vacuoles, constructs containing GFP fused to either the N-terminal domain or the C-terminal domain were transfected into cadA-null cells (Fig. 3A). As a control, cadA-null cells were also transfected with a GFP construct. The expression of the GFP fusion proteins was confirmed by protein blot analysis (Fig. 3B). Antibodies against DdCAD-1 and GFP detected protein bands corresponding to the expected molecular size of the respective fusion protein. Transfectants expressing comparable levels of fusion protein were selected for further analysis. The two GFP fusion proteins appeared to co-localize with calmodulin to the periphery of contractile vacuoles, whereas GFP showed a more diffuse staining pattern in the cytoplasm (Fig. 3C). In all three types of transfectants, green fluorescence was not detected inside contractile vacuoles, suggesting that neither the N-terminal domain nor the C-terminal domain alone was sufficient for import into contractile vacuoles.
FIGURE 3.
Expression of N-GFP and C-GFP fusion proteins in transfected cells. A, schematic drawings of GFP fusion protein constructs. B, Western blots of transfectants probed with antibodies against DdCAD-1 and GFP. C, cells were collected at 3 h of development and then fixed and labeled for confocal microscopy. Arrows indicate contractile vacuoles. Bars, 10 μm. CaM, calmodulin.
Surface Expression and Secretion of DdCAD-1-GFP
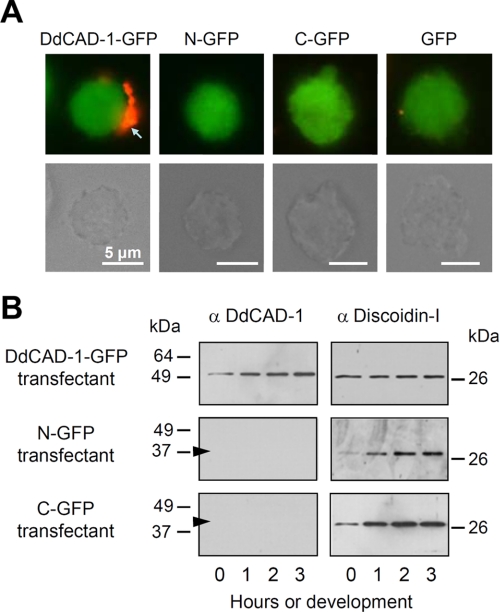
Our previous work showed that DdCAD-1 binds to an anchoring protein on the luminal surface of contractile vacuoles for presentation on the cell surface, whereas the unbound protein is targeted for secretion (23, 31). To determine whether N-GFP and C-GFP were capable of reaching the cell membrane independent of the contractile vacuole route, live cells were treated with antibodies against GFP to induce antigen clustering, which would eventually lead to the formation of “caps.” Caps were only observed in transfectants expressing DdCAD-1-GFP, indicating that it was associated with the cell surface (Fig. 4A). In contrast, neither N-GFP nor C-GFP was detectable on the surface of their respective transfectants.
FIGURE 4.
Differential cell surface expression and secretion of the DdCAD-1 fusion proteins in transfectants. A, fluorescence micrographs showing antibody-induced cap formation of DdCAD-1-GFP on live cells. cadA-null cells expressing the different GFP fusion proteins were incubated with anti-GFP antibodies for 30 min. After washing, cells were incubated at room temperature for another 30 min with a secondary antibody to induce “cap” formation (red). The corresponding light micrographs are shown in the lower panels. Bars, 5 μm. B, DdCAD-1 secretion during development. Transfectants were developed in 17 mm phosphate buffer at 2 × 107 cells/ml, and the conditioned media were collected at 1-h intervals for Western blot analysis using rabbit antibodies against DdCAD-1 or discoidin-I. The arrowheads indicate the position of N-GFP (38 kDa) and C-GFP (41 kDa), respectively.
To determine whether fusion proteins were secreted into the medium, cells were developed in phosphate buffer, and samples were collected at hourly intervals. After the removal of cells, the supernatants were subjected to SDS-PAGE and Western blot analysis. Among the three fusion proteins, only DdCAD-1-GFP was found in the conditioned medium (Fig. 4B). Neither N-GFP nor C-GFP was detected in the medium. As a positive control, protein blots were probed with antibodies against the endogenous lectin discoidin-I, which is also targeted for secretion through the contractile vacuole. Discoidin-I was found in the conditioned media of all three transfectants (Fig. 4B), indicating that this unconventional secretory pathway functioned normally in these transfectants.
In Vitro Import Analysis of GFP Fusion Protein
To further investigate the import of DdCAD-1-GFP, contractile vacuoles were isolated from transfectants and then subjected to Western blot analysis. Consistent with our morphological observation, DdCAD-1-GFP, N-GFP, and C-GFP co-purified with contractile vacuoles (Fig. 5A). If fusion proteins were present in the lumen, they should be protected from protease digestion unless the vacuolar membrane was disrupted by detergent. To test this, contractile vacuoles isolated from different transfectants were subjected to proteinase K digestion in the presence or absence of SDS, followed by protein blot analysis (Fig. 5A). As a positive control, blots were probed with antibodies against discoidin-I. Whereas N-GFP and C-GFP were completely degraded in the absence of SDS, DdCAD-1-GFP and discoidin-I were reduced by only 40–50%, indicating that a substantial amount of these two proteins was protected from proteolysis. In the presence of detergent, both DdCAD-1 and discoidin-I were reduced by >90% (Fig. 5A). The data provided biochemical evidence demonstrating that only DdCAD-1-GFP and discoidin-I were able to enter the contractile vacuole.
FIGURE 5.
Import of DdCAD-1 fusion proteins into contractile vacuoles. A, association of GFP fusion proteins with purified contractile vacuoles. Transfectants were developed for 6 h in liquid culture and then homogenized. The post-nuclear supernatant was fractionated on a 28 and 48% step sucrose density gradient by centrifugation at 40,000 rpm for 1 h at 4 °C. Contractile vacuoles banded in the interface were collected and subjected to digestion with proteinase K in the presence or absence of 0.05% SDS for 1 h at 37 °C. Panels a–c were probed with rabbit antibody against GFP (Molecular Probes). Panel d was probed with rabbit antibody against discoidin-I, which served as a positive control. B, in vitro import of His-tagged DdCAD-1 fusion proteins into contractile vacuoles. Contractile vacuoles (2 mg/ml) and cytosol (2 mg/ml) fractions derived from cadA-null cells were mixed together in a 600-μl reaction volume. His6-DdCAD-1, His6-N, or His6-C was added to a final concentration of 2 μm, and the assay was carried out as described under “Experimental Procedures.” Samples were subjected to SDS-PAGE, and the protein blots were probed with rabbit antibodies against either DdCAD-1 (29) (upper panels) or anti-calmodulin mAb (Sigma) (lower panels). The arrows indicate the position of the His-tagged proteins. A higher molecular weight band representing the dimer form of His6-DdCAD-1 and His6-N samples was frequently observed. pAb, polyclonal antibody. C, in vitro import assays of the His6-DdCAD-1 in the presence of the individual domains. His6-N or His6-C (4 μm) was added to the import assay samples, and the mixture was incubated at room temperature for 30 min. Then His6-DdCAD-1 was added to a final concentration of 2 μm, and the mixture was incubated for 1 h at room temperature. The contractile vacuoles were pelleted and subjected to proteinase K digestion in the presence or absence of detergent, and the protein blots were probed with rabbit anti-DdCAD-1 antibody.
A cell-free reconstitution assay was employed to examine the selective import of DdCAD-1 into contractile vacuoles. His6-tagged fusion proteins were used in these assays to avoid potential steric hindrance due to the bulky GFP. Contractile vacuoles and cytosol were isolated from cadA-null cells and then incubated with one of the three fusion proteins, His6-DdCAD-1, His6-N, and His6-C. At the end of the incubation period, the contractile vacuoles were washed and then subjected to proteinase K digestion in the presence or absence of detergent. Western blot analysis showed that although all three proteins bound to contractile vacuoles, only His6-DdCAD-1 was protected from proteolysis in the absence of detergent, indicating the selective import of His6-DdCAD-1 (Fig. 5B). The blots were also probed with antibodies against calmodulin. Calmodulin was digested completely in the absence of detergent, demonstrating the effectiveness of proteinase K in the removal of proteins associated with the surface of these vacuoles (Fig. 5B).
Because both His6-N and His6-C were able to bind contractile vacuoles, an important question was whether they would compete for the DdCAD-1-binding sites on contractile vacuoles. Import assays were carried out for His6-DdCAD-1 in the presence of a 2-fold higher concentration of either His6-N or His6-C (Fig. 5C). In all cases, comparable levels of His6-DdCAD-1 were found inside the contractile vacuoles, although both His6-N and His6-C bound efficiently to the contractile vacuole. The fact that they did not interfere with the import of His6-DdCAD-1 suggests that both His6-N and His6-C bound nonspecifically or to components that are not involved in DdCAD-1 transport.
Requirement of Ca2+ in the Import of DdCAD-1 into Contractile Vacuole
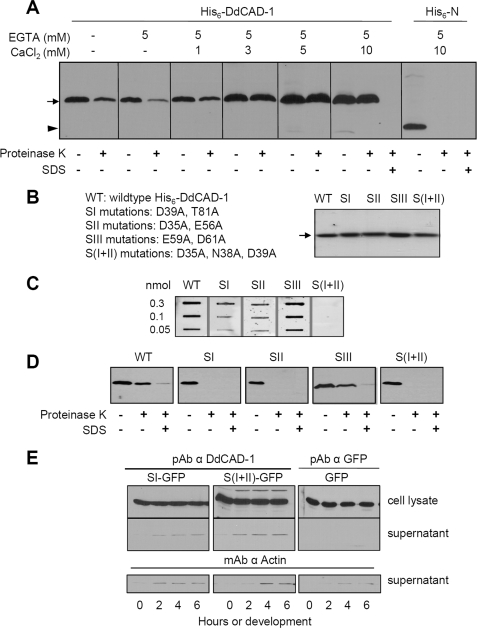
DdCAD-1 is a Ca2+-binding protein that mediates cell-cell adhesion via homophilic binding in a Ca2+-dependent manner (29, 32). To assess the role of Ca2+ in DdCAD-1 transport, the import assay was performed in the presence of EGTA. DdCAD-1 import was reduced by ∼80% in the presence of 5 mm EGTA (Fig. 6A). The inhibitory effects of EGTA could be reversed by the addition of Ca2+. Furthermore, the efficiency of DdCAD-1 import was enhanced as the Ca2+ level increased (Fig. 6A). In contrast, Ca2+ did not have any effect on His6-N in the import assay.
FIGURE 6.
Role of Ca2+ on DdCAD-1 import into contractile vacuole. A, effects of EGTA on DdCAD-1 import into contractile vacuoles. DdCAD-1 import assays were performed in the presence of 5 mm EGTA plus different concentrations of CaCl2. Samples were subjected to SDS-PAGE, and protein blots were probed with anti-DdCAD-1 antibody. In a separate experiment, import of His6-N was tested in the presence of 5 mm EGTA and 10 mm CaCl2. The arrow indicates the position of His6-DdCAD-1, and the arrowhead indicates the position His6-N. B, Coomassie-stained gel of purified His-tagged wild-type (WT) and mutant DdCAD-1 containing mutations in one of the Ca2+-binding sites (SI, SII, SIII, and S(I + II)). C, binding of 45Ca2+ to His6-DdCAD-1 and Ca2+-binding site mutant DdCAD-1. Protein samples were blotted onto nitrocellulose membrane. The blot was incubated with 10 ml of 45Ca2+ (10 μCi/ml) for 30 min at room temperature. After washing extensively with water, the blot was exposed to x-ray film at −70 °C for 24 h. D, in vitro import of His-tagged wild-type (WT) and mutant (SI, SII, SIII, and S(I + II)) DdCAD-1 into contractile vacuoles. Protein blots of import assay samples were probed with anti-DdCAD-1 antibody. E, secretion of mutant DdCAD-1 during development. Transfectants expressing SI- or S(I + II) mutant DdCAD-1 were developed in 17 mm phosphate buffer at 2 × 107 cells/ml, and the conditioned media were collected at 2-h intervals for Western blot analysis using rabbit antibodies against DdCAD-1. The same samples were also probed with mAb against actin. The very low level of protein detected in the medium was probably due to cell lysis, as indicated by the actin blots. pAb, polyclonal antibody.
DdCAD-1 consists of three Ca2+-binding pockets (32). Site I (SI) involves the residues Asp-39, Thr-81, Asn-84, and Phe-41 in the N-terminal domain, whereas site II (SII) consists of Asp-35, Asn-38, Glu-56, and Ser-87. Site III (SIII) is unique as it is situated at the interface of the two domains, including Glu-59 and Asp-61 from the N-terminal domain and Thr-179, Gln-181, and Asn-202 from the C-terminal domain. To further assess the requirement of Ca2+ in DdCAD-1 transport, mutations were introduced in the Ca2+-binding sites of DdCAD-1, with SI mutant containing D39A and T81A, SII mutant containing D35A and E56A, SIII mutant containing E59A and D61A, and S(I + II) mutant containing D35A, D39A, and N38A. His-tagged mutant proteins were expressed and purified from bacteria (Fig. 6B). Their ability to bind Ca2+ was assessed using the 45Ca2+ overlay assay (Fig. 6C). His6-SI and His6-SII showed substantial reduction in 45Ca2+ binding, whereas His6-SIII exhibited a similar level of bound 45Ca2+ as the wild-type protein, indicating that the side chains of residues Glu-59 and Asp-61 do not contribute significantly to the Ca2+-binding affinity. Binding of 45Ca2+ was not observed in the His6-S(I + II) mutant.
Import assays showed that the Ca2+-binding site mutants that displayed reduced binding of Ca2+ failed to enter the contractile vacuole, whereas His6-SIII was imported into the contractile vacuole at a level similar to that of the wild-type protein (Fig. 6D). Therefore, the import of DdCAD-1 into contractile vacuoles likely depends on the function of Ca2+-binding sites SI and SII. Consistent with this observation, cadA-null transfectants expressing the SI and S(I + II) mutant DdCAD-1 (see supplemental Fig. S3) did not secrete significant amounts of these mutant proteins (Fig. 6E).
Effect of Conformation on the Import of DdCAD-1 into Contractile Vacuole
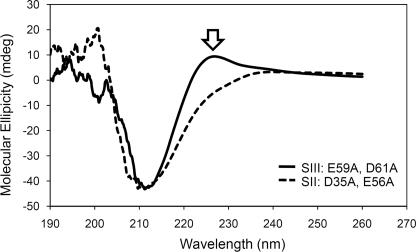
CD analysis was carried out to determine whether mutations in the Ca2+-binding sites had any effect on the conformation of DdCAD-1, because loss of Ca2+ binding could lead to subtle conformational changes, which might in turn influence the import process. A comparison between the CD spectra of the Ca2+-binding impaired mutant His6-SII and Ca2+-binding competent mutant His6-SIII showed a loss of the positive peak between 220 and 230 nm in the His6-SII spectra (Fig. 7). In contrast, the His6-SIII mutant, which showed no impairment in its import into contractile vacuoles, exhibited a CD spectrum similar to that of the wild-type protein (see supplemental Fig. S4). Because aromatic residues are known to contribute to the positive signal in the far UV region of CD spectra (39, 40), the data suggest that the mutations in SI, SII, and S(I + II) might have caused conformation perturbations around Trp-37, which is situated close to the Ca2+-binding sites SI and SII (32).
FIGURE 7.
Conformational changes in mutant DdCAD-1. The CD spectra of 40 μm His6-SII(D35A, E56A) (dashed line) and His6-SIII(E59A, D61A) (solid line) were plotted together for comparison. The open arrow indicates loss of the positive peak between 220 and 230 nm.
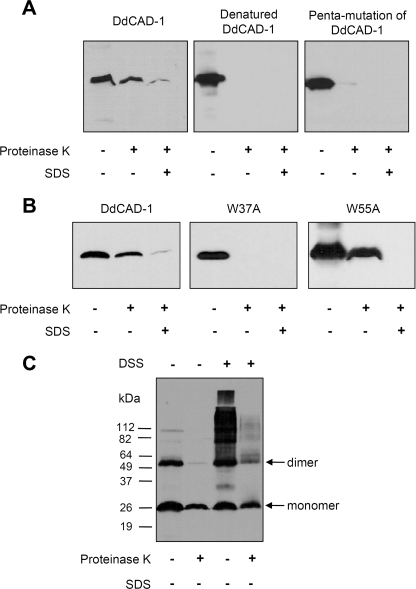
To investigate whether conformation plays a role in DdCAD-1 import, we first examined the import of denatured His6-DdCAD-1 and a His6-tagged mutant DdCAD-1 containing five mutations (N58A, E59A, D61A, D201A, and N202A), which displayed severe distortions in secondary structure (see supplemental Fig. S4F). The results showed that neither of them was able to enter the contractile vacuole (Fig. 8A). Next, we used a His6-DdCAD-1 mutant protein with Trp-37 substituted with Ala (W37A) because it is situated close to SI and SII. The CD spectrum of His6-DdCAD-1 (W37A) showed a reduced positive peak between 220 and 230 nm similar to those of the SI and SII mutant proteins (see supplemental Fig. S4). Import assays showed that the W37A mutant protein failed to enter the contractile vacuole (Fig. 8B). As a control, Trp-55, which is situated further from the SI and SII, was substituted with Ala. The W55A mutation did not cause any adverse effect in import assays, although it showed similar changes in the CD spectra. Therefore, the integrity of the SI and SII Ca2+-binding sites are crucial to DdCAD-1 import.
FIGURE 8.
Effects of DdCAD-1 conformational changes on the import process. A, His6-DdCAD-1 was denatured by boiling for 10 min followed by quick cooling on ice. The denatured form of His6-DdCAD-1 and the penta-mutant (N58A, E59A, D61A, D201A, and N202A) protein were subjected to the in vitro import assay. B, effects of the tryptophan mutations, W37A and W55A, on DdCAD-1 import into contractile vacuoles were examined. Protein blots were probed with anti-DdCAD-1 antibody. C, import of DdCAD-1 dimers into contractile vacuoles. His6-DdCAD-1 was cross-linked using disuccinimidyl suberate (DSS) and then subjected to the import assay. Protein blots were probed with anti-DdCAD-1 antibody.
Our previous studies suggest that Trp-37 is involved in the homophilic interactions of DdCAD-1 (32). This observation suggested the possibility that DdCAD-1 might be imported into contractile vacuoles as a dimer. To test this hypothesis, we cross-linked His6-DdCAD-1 with disuccinimidyl suberate before the import assay. Import studies showed that a substantial level of dimer and cross-linked oligomers was able to enter the vacuole (Fig. 8C). However, the monomeric form of DdCAD-1 appeared to be imported more efficiently.
DISCUSSION
In Dictyostelium, the contractile vacuoles serve as the major vehicle for the transport of DdCAD-1 to the plasma membrane for either secretion or cell surface presentation. Our in vivo analysis using time-lapse microscopy has revealed transient membrane budding into the lumen of contractile vacuoles. DdCAD-1 docked on the contractile vacuoles is mobilized to fill these invaginations, which are then pinched off to become vesicles inside the lumen. DdCAD-1 is released upon the burst of these vesicles. In many ways, the export of DdCAD-1 via contractile vacuole is similar to the budding of vesicles in yeast vacuoles (41) or the formation of multivesicular bodies in murine macrophages (42). In yeast, the tubular structures facilitate that lateral sorting of proteins and lipids into vesicles inside the vacuole (41). In macrophages, the multivesicular bodies are the primary vehicle for the transport of interleukin-1β trapped inside the exosomes (42). However, both vesicles and tubules are only transient structures in the lumen of contractile vacuoles in Dictyostelium cells, and multivesicular structures are rarely observed. It is possible that the water collected in the contractile vacuole creates a hypo-osmotic environment that can cause the rapid rupture of vesicles soon after they bud off into the lumen.
To further investigate the structural mechanisms involved in DdCAD-1 import into contractile vacuoles, individual domains were transfected into cadA-null cells. Both in vivo and in vitro analyses show that neither the N-terminal domain nor the C-terminal domain of DdCAD-1 alone is sufficient for import into the contractile vacuole. Because the docking of His6-DdCAD-1 to contractile vacuoles is not affected by the presence by either His6-N or His6-C, the import signal is likely integrated in the three-dimensional structure of DdCAD-1.
Further analysis has revealed a role for Ca2+ because the import of DdCAD-1 is abrogated by EGTA. Mutations that disrupt the Ca2+-binding capability of DdCAD-1 also inhibit its import. Also, Ca2+ enhances the binding of DdCAD-1 to contractile vacuoles as well as its entry into the lumen. DdCAD-1 is a Ca2+-binding protein. However, the Ca2+-binding affinity of DdCAD-1 is relatively low (Kd 12–15 μm) (32). Because the intracellular free Ca2+ concentration is estimated to be ∼56 nm in aggregation-competent cells (43), most of the intracellular DdCAD-1 probably exists in the Ca2+-free form. On the other hand, contractile vacuoles are enriched in Ca2+ pumps (44, and they constitute a highly efficient acidic Ca2+ store (45). It is conceivable that the release of Ca2+ from contractile vacuoles may serve as an attractant to DdCAD-1. Moreover, the negatively charged surface of membranes is known to determine the targeting of proteins with polybasic clusters (46, 47). The bound Ca2+ would increase the overall positive charge on the surface of DdCAD-1. The positively charged regions as well as the His6 tag would promote electrostatic interactions with acidic lipids and facilitate the binding of DdCAD-1 to the contractile vacuole membrane. Structural studies have shown that Ca2+-binding interfacial membrane proteins, such as annexin V, complex with phospholipid head groups via Ca2+ bridges (48). Such a phenomenon may explain the nonspecific binding of the C- and N-terminal fusion proteins.
Because neither the C-terminal domain nor the N-terminal domain competes with DdCAD-1 for binding to the contractile vacuole, it is likely that DdCAD-1 binds to a specific docking element in the midst of negatively charged lipids on the vacuolar membrane. In addition to Ca2+ pumps, an abundance of the ubiquitous Ca2+ regulator, calmodulin, is associated with the surface of contractile vacuoles (36, 37). Calmodulin is also found on endosomal and lysosomal membranes (49, 50) and is known to play a role in the endosome-mediated transport system (51). Interestingly, bioinformatic analysis shows that DdCAD-1 contains a putative binding site for calmodulin in the C-terminal domain, suggesting that the Ca2+-bound form of calmodulin may serve as a potential docking partner for DdCAD-1. Indeed, direct interaction between DdCAD-1 and calmodulin has been detected in far Western blots, whereas pharmacological inhibition of calmodulin function by W7 leads to a reduction in DdCAD-1 binding to contractile vacuoles.4
DdCAD-1 contains three Ca2+-binding pockets, and the NMR solution structures of DdCAD-1 have shown that binding of Ca2+ leads to more ordered side chain packing in the Ca2+-binding sites and stabilizes the whole structure of DdCAD-1 (32). An analysis of the CD spectra of the Ca2+-binding site mutants suggests conformational perturbations in regions that contain tryptophan. A tryptophan (Trp-37) is situated in the loop structure connecting SI and SII. Substitution of Trp-37 with alanine results in alterations in CD spectra similar to those observed for the SI and SII mutants. The W37A mutation as well as mutations in either the SI or SII Ca2+-binding site results in the loss of DdCAD-1 import into contractile vacuoles, implicating a role for the three-dimensional structure in DdCAD-1 transport. It is possible that the Ca2+-binding sites may serve a dual function. Although contributions by Ca2+ to the surface charge can influence the docking of DdCAD-1 to contractile vacuoles, conformational changes induced by Ca2+ binding may regulate its import into the lumen.
In both animal and plant cells, several unconventional pathways are known to utilize transporters to import or export soluble proteins across cellular membranes (52). Matrix components of peroxisomes are transported in folded form from the cytoplasm into the peroxisomal matrix (53, 54). Other pathways that can accommodate folded substrates include the bacterial twin-arginine translocation pathway (55, 56), the ΔpH-dependent pathway of plant plastids (57), the cytoplasm-to-vacuole targeting pathway of Saccharomyces cerevisiae (58), and the direct translocation of proteins, such as FGF-2 and galectin-1, across the plasma membrane in mammalian cells (20, 21). Among them, only the peroxisomal and twin-arginine translocation pathways involve targeting signals, whereas the others do not seem to require a generalized signal for protein transport (59, 60). Similarly, there has been no targeting motif detected in the primary sequence of DdCAD-1, and the import signal is likely integrated in its three-dimensional structure.
The export of soluble proteins may involve more than one unconventional pathway (3, 4). Our previous in vitro studies suggest that the translocation of DdCAD-1 across the contractile vacuole membrane may involve a membrane transporter (31). Exogenously added ATP and an ATP regeneration system enhance the import of DdCAD-1, suggesting the involvement of specific ATP-dependent transporter(s) in the contractile vacuole membrane. Also, the import of DdCAD-1 is inhibited by 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole, a specific inhibitor of vacuolar-type H+-ATPase (61, 62), implicating a role for the V-ATPase in the import mechanism. Among other secreted proteins in Dictyostelium, the acyl-CoA-binding protein (AcbA), which is required for the terminal differentiation of prespore cells, is also externalized through an unconventional protein secretion pathway. Interestingly, this pathway involves GRASP (the Golgi reassembly stacking protein) and a membrane transporter (63). However, the identity of the transporter for AcbA remains to be elucidated. It is therefore evident that multiple unconventional pathways are employed by eukaryotic cells to target soluble proteins for surface expression and secretion. The combination of genetic and biochemical analyses in Dictyostelium should provides a useful model for the future dissection of these pathways.
Acknowledgments
We thank Drs. David Isenman and Mitsu Ikura for invaluable advice. We thank Eric Huang for excellent technical support and members of the Siu laboratory for discussion.
This work was supported in part by Canadian Institutes of Health Research Operating Grant FRN-6140.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
S. Sriskanthadevan, T. Lee, and C.-H. Siu, unpublished data.
- ER
- endoplasmic reticulum
- FGF
- fibroblast growth factor
- GFP
- green fluorescence protein
- N
- N-terminal domain
- C
- C-terminal domain
- mAb
- monoclonal antibody
- Mes
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Palade G. E. (1975) Science 189, 347–358 [DOI] [PubMed] [Google Scholar]
- 2.Rothman J. E. (1994) Nature 372, 55–63 [DOI] [PubMed] [Google Scholar]
- 3.Nickel W., Rabouille C. (2009) Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 4.Nickel W., Seedorf M. (2008) Annu. Rev. Cell Dev. Biol. 24, 287–308 [DOI] [PubMed] [Google Scholar]
- 5.Cleves A. E. (1997) Curr. Biol. 7, R318–R320 [DOI] [PubMed] [Google Scholar]
- 6.Nickel W. (2003) Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 7.Pohlschröder M., Hartmann E., Hand N. J., Dilks K., Haddad A. (2005) Annu. Rev. Microbiol. 59, 91–111 [DOI] [PubMed] [Google Scholar]
- 8.Cleves A. E., Cooper D. N., Barondes S. H., Kelly R. B. (1996) J. Cell Biol. 133, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orci L., Tagaya M., Amherdt M., Perrelet A., Donaldson J. G., Lippincott-Schwartz J., Klausner R. D., Rothman J. E. (1991) Cell 64, 1183–1195 [DOI] [PubMed] [Google Scholar]
- 10.Rubartelli A., Bajetto A., Allavena G., Wollman E., Sitia R. (1992) J. Biol. Chem. 267, 24161–24164 [PubMed] [Google Scholar]
- 11.Flieger O., Engling A., Bucala R., Lue H., Nickel W., Bernhagen J. (2003) FEBS Lett. 551, 78–86 [DOI] [PubMed] [Google Scholar]
- 12.Engling A., Backhaus R., Stegmayer C., Zehe C., Seelenmeyer C., Kehlenbach A., Schwappach B., Wegehingel S., Nickel W. (2002) J. Cell Sci. 115, 3619–3631 [DOI] [PubMed] [Google Scholar]
- 13.Mignatti P., Morimoto T., Rifkin D. B. (1992) J. Cell Physiol. 151, 81–93 [DOI] [PubMed] [Google Scholar]
- 14.Denny P. W., Gokool S., Russell D. G., Field M. C., Smith D. F. (2000) J. Biol. Chem. 275, 11017–11025 [DOI] [PubMed] [Google Scholar]
- 15.Elliott G., O'Hare P. (1997) Cell 88, 223–233 [DOI] [PubMed] [Google Scholar]
- 16.Mann D. A., Frankel A. D. (1991) EMBO J. 10, 1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrei C., Dazzi C., Lotti L., Torrisi M. R., Chimini G., Rubartelli A. (1999) Mol. Biol. Cell 10, 1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubartelli A., Cozzolino F., Talio M., Sitia R. (1990) EMBO J. 9, 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper D. N., Barondes S. H. (1990) J. Cell Biol. 110, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seelenmeyer C., Wegehingel S., Tews I., Künzler M., Aebi M., Nickel W. (2005) J. Cell Biol. 171, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schäfer T., Zentgraf H., Zehe C., Brügger B., Bernhagen J., Nickel W. (2004) J. Biol. Chem. 279, 6244–6251 [DOI] [PubMed] [Google Scholar]
- 22.Siu C. H., Harris T. J., Wang J., Wong E. (2004) Semin. Cell Dev. Biol. 15, 633–641 [DOI] [PubMed] [Google Scholar]
- 23.Sriskanthadevan S., Ivanov I., Yang C., Siu C. H. (2007) in Recent Research Developments in Cell Biology (Pandalai S. G. ed) Vol. 3, 9–21, Research Signpost, Trivandrum [Google Scholar]
- 24.Aubry L., Firtel R. (1999) Annu. Rev. Cell Dev. Biol. 15, 469–517 [DOI] [PubMed] [Google Scholar]
- 25.Crowley T. E., Nellen W., Gomer R. H., Firtel R. A. (1985) Cell 43, 633–641 [DOI] [PubMed] [Google Scholar]
- 26.Springer W. R., Cooper D. N., Barondes S. H. (1984) Cell 39, 557–564 [DOI] [PubMed] [Google Scholar]
- 27.Barondes S. H., Haywood-Reid P. L., Cooper D. N. (1985) J. Cell Biol. 100, 1825–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brar S. K., Siu C. H. (1993) J. Biol. Chem. 268, 24902–24909 [PubMed] [Google Scholar]
- 29.Wong E. F., Brar S. K., Sesaki H., Yang C., Siu C. H. (1996) J. Biol. Chem. 271, 16399–16408 [DOI] [PubMed] [Google Scholar]
- 30.Yang C., Brar S. K., Desbarats L., Siu C. H. (1997) Differentiation 61, 275–284 [DOI] [PubMed] [Google Scholar]
- 31.Sesaki H., Wong E. F., Siu C. H. (1997) J. Cell Biol. 138, 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Z., Sriskanthadevan S., Huang H., Siu C. H., Yang D. (2006) Nat. Struct. Mol. Biol. 13, 1016–1022 [DOI] [PubMed] [Google Scholar]
- 33.Gerisch G., Heuser J., Clarke M. (2002) Cell Biol. Int. 26, 845–852 [DOI] [PubMed] [Google Scholar]
- 34.Heuser J., Zhu Q., Clarke M. (1993) J. Cell Biol. 121, 1311–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang K. M., Lynes M. A., Knecht D. A. (1999) Plasmid 41, 187–197 [DOI] [PubMed] [Google Scholar]
- 36.Zhu Q., Clarke M. (1992) J. Cell Biol. 118, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Q., Liu T., Clarke M. (1993) J. Cell Sci. 104, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 38.Lin Z., Huang H., Siu C. H., Yang D. (2004) J. Biomol. NMR 30, 375–376 [DOI] [PubMed] [Google Scholar]
- 39.Andersson D., Carlsson U., Freskgård P. O. (2001) Eur. J. Biochem. 268, 1118–1128 [DOI] [PubMed] [Google Scholar]
- 40.Clark P. L., Liu Z. P., Zhang J., Gierasch L. M. (1996) Protein Sci. 5, 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller O., Sattler T., Flötenmeyer M., Schwarz H., Plattner H., Mayer A. (2000) J. Cell Biol. 151, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu Y., Franchi L., Nunez G., Dubyak G. R. (2007) J. Immunol. 179, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 43.Yumura S., Furuya K., Takeuchi I. (1996) J. Cell Sci. 109, 2673–2678 [DOI] [PubMed] [Google Scholar]
- 44.Moniakis J., Coukell M. B., Janiec A. (1999) J. Cell Sci. 112, 405–414 [DOI] [PubMed] [Google Scholar]
- 45.Malchow D., Lusche D. F., Schlatterer C., De Lozanne A., Müller-Taubenberger A. (2006) BMC Dev. Biol. 6, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaughlin S., Murray D. (2005) Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 47.Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 48.Swairjo M. A., Concha N. O., Kaetzel M. A., Dedman J. R., Seaton B. A. (1995) Nat. Struct. Biol. 2, 968–974 [DOI] [PubMed] [Google Scholar]
- 49.Enrich C., Jäckle S., Havel R. J. (1996) Hepatology 24, 226–232 [DOI] [PubMed] [Google Scholar]
- 50.Nielsen T. B., Field J. B., Dedman J. R. (1987) J. Cell Sci. 87, 327–336 [DOI] [PubMed] [Google Scholar]
- 51.Pryor P. R., Mullock B. M., Bright N. A., Gray S. R., Luzio J. P. (2000) J. Cell Biol. 149, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teter S. A., Klionsky D. J. (1999) Trends Cell Biol. 9, 428–431 [DOI] [PubMed] [Google Scholar]
- 53.Brocard C. B., Jedeszko C., Song H. C., Terlecky S. R., Walton P. A. (2003) Traffic 4, 74–82 [DOI] [PubMed] [Google Scholar]
- 54.Nickel W. (2005) Traffic 6, 607–614 [DOI] [PubMed] [Google Scholar]
- 55.Cline K., McCaffery M. (2007) EMBO J. 26, 3039–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pugsley A. P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 12058–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark S. A., Theg S. (1997) Mol. Biol. Cell 8, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J., Scott S. V., Oda M. N., Klionsky D. J. (1997) J. Cell Biol. 137, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schatz G., Dobberstein B. (1996) Science 271, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 60.Wickner W., Schekman R. (2005) Science 310, 1452–1456 [DOI] [PubMed] [Google Scholar]
- 61.Mellman I., Fuchs R., Helenius A. (1986) Annu. Rev. Biochem. 55, 663–700 [DOI] [PubMed] [Google Scholar]
- 62.Padh H., Lavasa M., Steck T. L. (1989) Biochim. Biophys. Acta 982, 271–278 [DOI] [PubMed] [Google Scholar]
- 63.Kinseth M. A., Anjard C., Fuller D., Guizzunti G., Loomis W. F., Malhotra V. (2007) Cell 130, 524–534 [DOI] [PubMed] [Google Scholar]