Abstract
Previously, we showed that CL1-5 cells express more hypoxia-inducible factor-1α (HIF-1α) than the parental CL1 cells, which bestows CL1-5 cells a stronger invasive activity. Here, we investigated the mechanisms underlying the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells. Data showed that the transcription rate of HIF-1α gene in CL1 cells was slightly higher than that of CL1-5 cells, suggesting that the expression of HIF-1α mRNA in CL1 cells was repressed by post-transcriptional mechanisms. RNA electrophoretic mobility shift assays revealed a 61-base segment (designated as D5) within the 5′-untranslated repeat of HIF-1α mRNA, with which the CL1 cell lysates formed more prominent complexes (including complex I) than did CL1-5 cell lysates. Insertion of D5 into a reporter construct reduced the half-life of the chimeric transcripts in transfected CL1 but not CL1-5 cells; conversely, overexpression of D5-containing reporter construct in CL1 cells increased HIF-1α mRNA. We also identified the polypyrimidine tract-binding protein (PTB) as a required component of complex I. Deletion of the RNA recognition motif 1 (RRM1) or RRM3 of PTB abolished the formation of complex I. Our data showed that CL1 cells expressed more PTB than CL1-5 cells. Inhibition of PTB expression in CL1 cells decreased the formation of complex I, whereas overexpression of PTB in CL1-5 cells increased the levels of complex I, decreased the stability of HIF-1α and D5-containing chimeric mRNAs, and decreased cell invasiveness. In sum, we have identified in lung adenocarcinoma cells a mechanism that regulates HIF-1α expression by modulating HIF-1α mRNA stability.
Introduction
One of the major characteristics of cancer cells is their aberrant proliferation. For a tumor originated from avascular area, its vigorous growth results in hypoxic microenvironment, which may limit its expansion unless the supply of oxygen and nutrients is restored (1). To cope with the hypoxic stress, tumor cells elicit adaptive responses, including the improvement of glucose uptake for ATP synthesis and the induction of angiogenesis. A number of studies have reported that hypoxia-inducible factor-1 (HIF-1)2 is one of the major factors that control the cellular adaptive response to hypoxia (2–5).
HIF-1 is a heterodimeric transcription factor consisting of HIF-1α and HIF-1β subunits. HIF-1β is constitutively expressed, whereas HIF-1α is regulated by cellular oxygen concentrations (6). Under nonhypoxic conditions, HIF-1α is quickly ubiquitinated and degraded. However, the ubiquitination/proteasomal degradation of HIF-1α is inhibited under hypoxic conditions, resulting in the accumulation of HIF-1α and the increase in cellular HIF-1 (7). A number of findings have revealed the crucial role HIF-1α plays in tumor growth and expansion (8, 9). Studies have also revealed the capability of HIF-1α to enhance tumor invasion and metastasis (10, 11). To date, studies have revealed several mechanisms that up-regulate HIF-1α in cancer cells. These include (i) activation of epidermal growth factor receptor/phosphatidylinositol 3-kinase/Akt signaling pathway (12); (ii) generation of reactive oxygen species in response to hypoxia (13); (iii) inactivation of tumor suppressor genes such as Von Hippel-Lindau and p53 (14, 15); and (iv) activation of oncogene such as src (16). These mechanisms work ultimately by increasing the stability of HIF-1α protein without affecting HIF-1α mRNA levels. However, our previous study using lung adenocarcinoma cells showed that an as yet undefined mechanism may regulate HIF-1α expression at mRNA level (11).
CL1/CL1-5 is a human lung adenocarcinoma metastatic cell model. CL1 cells were cultured from a single cell clone isolated from the lung adenocarcinoma tissues of a patient with a poorly differentiated adenocarcinoma (17). As reported, CL1 cells are tumorigenic but not highly metastatic. However, after passages of culture, CL1 cells became heterogeneous, and a series of sublines with progressive invasiveness were isolated. Among these cell lines, CL1-5 subline demonstrated the highest invasive and metastatic potentials in vitro and in vivo (18). Previously, using this cell model, we described the stimulatory effect of HIF-1α to cell invasion, and showed that CL1-5 cells express much more HIF-1α protein and mRNA than CL1 cells. Here, we have investigated the molecular mechanism(s) governing the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
CL1 and CL1-5 cells were maintained in RPMI 1640 medium (Amersham Biosciences) supplemented with 10% fetal bovine serum, glutamine, penicillin, and streptomycin. For transcription shut-off assays, CL1 and CL1-5 cells were transfected with the pTet-off plasmid (BD Biosciences Clontech) and selected in the presence of 1 mg/ml G418. Stably transfected clones were picked and tested by transient transfection with a reporter construct, pTRE-DsRed2 (BD Biosciences Clontech). The CL1 and CL1-5 clones exhibiting the highest repression of the reporter construct upon the treatment of doxycycline (2 μg/ml) were designated as CL1-Tet and CL1-5-Tet and used in subsequent experiments. To generate cells overexpressing PTB or GFP, CL1-5 cells were transfected either with pCMV-SPORT6-PTB plus pCEP4 or with pcDNA3.1-GFP (a kind gift from Dr. Ling-Jun Ho) and selected with either hygromycin B (50 μg/ml) or G418 (1 mg/ml). Antibiotic-resistant cells were pooled and designated as CL1-5-PTB and CL1-5-GFP, respectively. All the cell lines were cultured at 37 °C in a humidified atmosphere containing 5% CO2.
Northern Blot Analysis, Quantitative Real-time PCR, and Western Blot Analyses
Total RNA was isolated from cells using TRIzol (Invitrogen). Reverse transcription-PCRs were performed as previously described to generate cDNA probes (11). The 5′ and 3′ primers used were as follows: DsRed2-containing transcripts, CGAGAACGTCATCACCGAG and CTTCTTCTGCATCGCGGGG; and GAPDH, CATCACCATCTTCCAGGAGC and GGATGATGTTCTGGAGAGCC. Northern blot analyses were carried out as previously described (19). Quantitative real-time PCR (RT-qPCR) was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems, CA). The 5′ and 3′ primers used were as follows: HIF-1α, AGAAAAAGATAAGTTCTGAAGGTC and GAGAAAAAAGCTTCGCTGTGTG; luciferase, GCACTCTGATTGACAAATACG and CTCGGGTGTAATCAGAATAGC; and β-actin, CCCTGGCACCCAGCAC and GCCGATCCACACGGAGTAC. All RT-qPCRs were performed in triplicate on an ABI Prism 7000 Sequence Detector System as follows: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The relative mRNA levels were calculated using the 2−ΔΔCT method, with β-actin mRNA as a normalizer. For Western blot analyses, 40 μg of whole cell lysate was resolved on SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes, and hybridized with anti-HIF-1α (BD Biosciences), anti-α-tubulin (Neo markers), anti-PTB (Abcam), and anti-β-actin (Chemicon) antibodies. Signals were obtained by enhanced chemiluminescence (Pierce).
Preparation of Luciferase Reporter Constructs and Luciferase Assays
To assess the HIF-1α gene promoter activities of CL1 and CL1-5 cells, a DNA fragment corresponding to the region from −641 to +89 of the HIF-1α gene (20), was cloned into the HindIII site of pGL3-basic vector (Promega, Madison, WI) to generate pGL3-HIF-1α-luc. Cells were transiently transfected with pGL3-HIF-1α-luc for 6 h using Effectene reagent (Qiagen). Cotransfection with a Renilla luciferase reporter served as an internal control for normalization of transient transfection. Cells were lysed 24 h after transfection and subjected to luciferase assays using the Dual-Luciferase Reporter Assay System (Promega). To examine the influence of specific regions of HIF-1α mRNA on the expression of a luciferase reporter, DNA fragments A4, D1, and D5 (Figs. 2C and 4A) were cloned into the HindIII-NcoI site of pGL3-promoter vector, and fragment B2 (Fig. 2A) was cloned into the XbaI site of pGL3-promoter vector to generate pGL3-A4, pGL3-D1, pGL3-D5, pGL3-B2, and pGL3-D5-B2, respectively. Cells were transiently transfected either with pGL3-promoter, pGL3-A4, and pGL3-D1 and subjected to luciferase assays, or with pGL3-promoter, pGL3-D5, pGL3-B2, and pGL3-D5-B2 and subjected to RT-qPCR analyses for the levels of luciferase transcript.
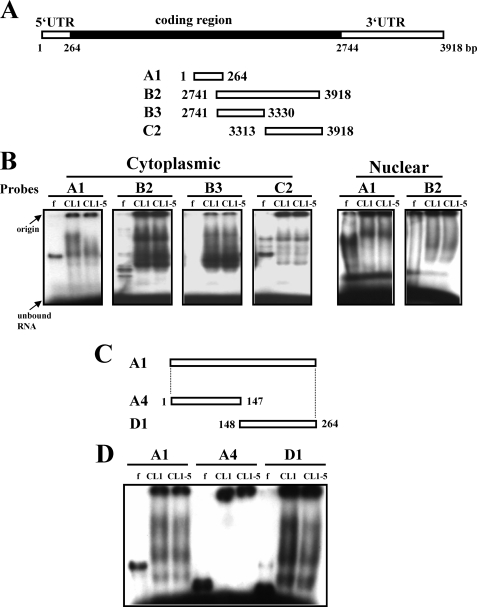
FIGURE 2.
Interactions of cellular proteins and HIF-1α transcripts. A and C, schematic representation of the HIF-1α cDNA and various transcripts derived from the 5′-UTR and 3′-UTR. These transcripts were assayed for protein binding. B, REMSA assays. CL1 and CL1-5 cells were fractionated into cytoplasmic and nuclear fractions. Ten micrograms of each fraction was mixed with 32P-radiolabeled A1, or B2, or B3, or C2 probe, and then with RNase T1. Reaction mixtures were resolved on 6% native polyacrylamide gels. Signals were visualized using FLA-2000 (Fujifilm). D, REMSA assays. Ten micrograms of cytoplasmic protein isolated from CL1 or CL1-5 cells was each mixed with radiolabeled A1, or A4, or D1 probe, and then with RNase T1. Electrophoresis was performed as described above. f, radiolabeled transcripts digested with RNase T1 without incubation with cell lysate.
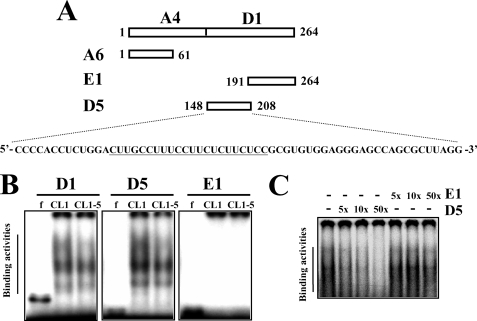
FIGURE 4.
Characterization of the protein binding region within D1. A, schematic representation of D5 and E1 regions derived from D1. A polypyrimidine tract within the D5 region is underlined. B, REMSA assays. Ten micrograms of cytoplasmic protein isolated from CL1 and CL1-5 cells was each mixed with radiolabeled D1, or D5, or E1 probe, and then with RNase T1. Electrophoresis and visualization of signals were as described in Fig. 2. C, competition assays. Cytoplasmic lysates prepared from CL1 cells were incubated with 5-, 10-, or 50-fold molar excesses of either unlabeled D5 or E1 probe on ice for 30 min, and then with radiolabeled D5 probe for another 20 min on ice. After being treated with RNase T1, reactions were subjected to REMSA.
Nuclear Run-on Assays
Five micrograms of DNA from either the β-actin-containing plasmid, or pCEP4/HIF-1α (American Type Culture Collection, ATCC) or pCEP4 vector was linearized and blotted onto nitrocellulose membranes. Nuclei from 2 × 107 cells were collected for in vitro transcription. Nascent 32P-labeled RNA was isolated using TRIzol reagent. Membranes were blocked with 100 μg of tRNA for 4 h at 65 °C, then hybridized with 4 × 106 cpm of 32P-labeled RNA in 2 ml of hybridization buffer for 72 h at 65 °C, and washed at 65 °C with wash buffer. Signals were visualized using FLA-2000 (Fujifilm).
Preparation of Radiolabeled RNA Transcripts
Total RNA prepared from CL1-5 cells was used for reverse transcription-PCRs (RT-PCRs) to generate various regions of HIF-1α cDNA. A T7 RNA polymerase promoter sequence (T7) was placed 5′ to the 5′ primers used in this study. The 5′ primers used were as follows: A, (T7)CAGGAGGATCACCCTCTTC; B, (T7)GCTTTGGATCAAGTTAACTGAG; C, (T7)AACCTGGAACATGACATTGTTAATC; D, (T7)CCCCACCTCTGGACTTGCC; and E, (T7)AGGGAGCCAGCGCTTAGG. The 3′ primers used were as follows: 1, TGGTGAATCGGTCCCCGC; 2, GCCTGGTCCACAGAAGATG; 3, AATGTCATGTTCCAGGTTTAAC; 4, GTGCGAGGCGGGAAACCCC; and 5, CCTAAGCGCTGGCTCCCT. PCR-amplified products were purified to serve as templates for synthesis of radiolabeled and nonradiolabeled RNA probes (19).
RNA Electrophoretic Mobility Shift Assays
The nuclear and cytoplasmic cell fractions were prepared as described previously (19). Ten-microgram aliquots of either nuclear or cytoplasmic fractions were incubated with 50,000 cpm of RNA probes on ice for 20 min in a 10-μl mixture. For supershift assays, 2.5-μg aliquots of cytoplasmic fractions were mixed with 1 μg each of antibodies against HuR (Santa Cruz Biotechnology, Santa Cruz, CA), PTB, or cyclin B1 (Santa Cruz Biotechnology) for 30 min, and then with 50,000 cpm of RNA probes on ice for 20 min in a 10-μl mixture. After addition of RNase T1 (1000 units/reaction), reaction mixtures were incubated at 37 °C for 20 min and electrophoresed through 6% (4% for supershift assays) nondenaturing polyacrylamide gels containing 0.3× Tris borate-EDTA at 150 V for 1.5 h at 4 °C. For cross-linking experiments, RNase-T1-treated protein-RNA mixtures were exposed to UV using a Stratalinker (Stratagene), then electrophoresed through 12% SDS-polyacrylamide gels. Gels were dried, and signals were visualized using FLA-2000 (Fujifilm).
Transcription Shut-off Assays for Measuring mRNA Half-life
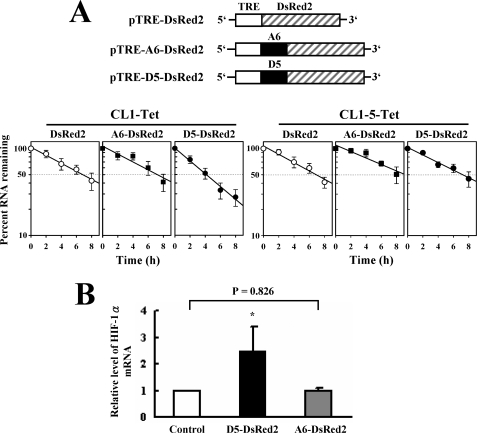
To assess the influence of specific regions of the HIF-1α mRNA on the half-life of chimeric mRNAs, PCR-amplified fragments D5 and A6 (Fig. 4A) were subcloned 5′ to the reporter gene in the tetracycline-controlled vector pTRE-DsRed2 to generate pTRE-D5-DsRed2 and pTRE-A6-DsRed2. CL1-Tet and CL1-5-Tet cells were transiently transfected with each of pTRE-DsRed2, or pTRE-D5-DsRed2, or pTRE-A6-DsRed2 for 6 h using Effectene reagent and then maintained in fresh medium for 20 h. Cotransfection with a Renilla luciferase reporter served as an internal control. Then, cells were treated with doxycycline (2 μg/ml) for 0, 2, 4, 6, and 8 h. RNA was collected, and Northern blot analyses were performed to detect expression of DsRed2-containing transcripts and GAPDH. The half-life of each transcript was measured.
Immunoprecipitation of Ribonucleoprotein Complexes
To assess the binding of PTB-containing protein complexes on the HIF-1α mRNA of CL1 and CL1-5 cells, cells were processed as described (21) with some modification. Briefly, cells were washed with ice-cold phosphate-buffered saline, transferred into a microcentrifuge tube, and followed by low speed centrifugation. Cell pellet was mixed with equal volume of polysome lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES, pH 7, 0.5% Nonidet P-40, 10 μm dithiothreitol) plus inhibitors of RNases and proteases. The mixture was centrifuged at 20,000 × g for 30 min at 4 °C, and the supernatant was used in subsequent immunoprecipitation. To prepare antibody-coated protein A beads, 15 μg of antibody was mixed with 100 μl of protein A beads (Sigma) in 150 μl of NT-2 buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm MgCl2, 0.05% Nonidet P-40) at 4 °C with gentle shaking for 16 h. The beads were then washed 5 times with NT-2 buffer, and pelleted by centrifugation. For immunoprecipitation of ribonucleoprotein complexes, the antibody-coated beads were mixed with 1 mg of cell lysate in NT-2 buffer with the final volume as 1 ml, and incubated at 4 °C with gentle shaking for 2 h. Then, the beads were washed five times with NT-2 buffer. RNAs were isolated from the precipitated ribonucleoprotein complexes as described (21) and subjected to RT-qPCR analyses.
Preparation of Constructs Expressing Partially Deleted Forms of PTB
PCRs were carried out using pCMV-SPORT6-PTB as template to generate pPTB-deRRM1, pPTB-deRRM2, and pPTB-deRRM3 constructs harboring PTB cDNA lacking of RRM1, or RRM2, or RRM3 sequence, which encode partially deleted forms of PTB with predicted molecular masses of ∼50.3, 48.1, and 47.2 kDa, respectively. The 5′ and 3′ primers used were: pPTB-deRRM1, TCCAACCACAAGGAGCTGAA and CACTCTAGAGGGGACGCC; pPTB-deRRM2, ACACGCCCAGACCTGCCT and CCTGAGCACGGGGCTCTG; and pPTB-deRRM3, CGCATCACGCTCTCGAAG and GCCGTGGACGTTCGGAAC. PCR products were purified and treated with polynucleotide kinase (Fermentas) for 1 h at 37 °C and then treated with T4 ligase (Fermentas) for 16 h at 4 °C. In addition, RT-PCR analyses were performed to examine the expression of the intact and partially deleted forms of PTB mRNAs in transfected cells. The 5′ and 3′ primers used were: CCATGGACGGCATTGTCCC and CTAGATGGTGGACTTGGAGAAGGAG.
In Vitro Invasion Assay
The cell invasiveness was determined using the growth factor-reduced Matrigel invasion system (BD Biosciences) as previously described (11). Briefly, cells were suspended in 0.5 ml of low serum medium (2% fetal bovine serum) and added to the upper chamber. The upper chamber was lodged into the lower chamber containing 0.75 ml of low serum medium. After incubating at 37 °C for the time as indicated, the cells in the upper side of the filter membrane were removed with cotton swabs. The membranes were then soaked in the fixation solution containing 10% of paraformaldehyde for 10 min. Cells that migrated to the lower side of the membranes were stained with Liu stain (Handsel Technologies, Inc., Taipei, Taiwan). The stained cells were counted in three fields under a 100× high power field.
Statistical Analysis
Data shown were the mean ± S.D. Statistical difference between two groups was determined by paired t test. A value of p < 0.05 was considered to denote statistical significance.
RESULTS
Post-transcriptional Regulation Is Involved in the Differential Expression of HIF-1α between CL1 and CL1-5 Cells
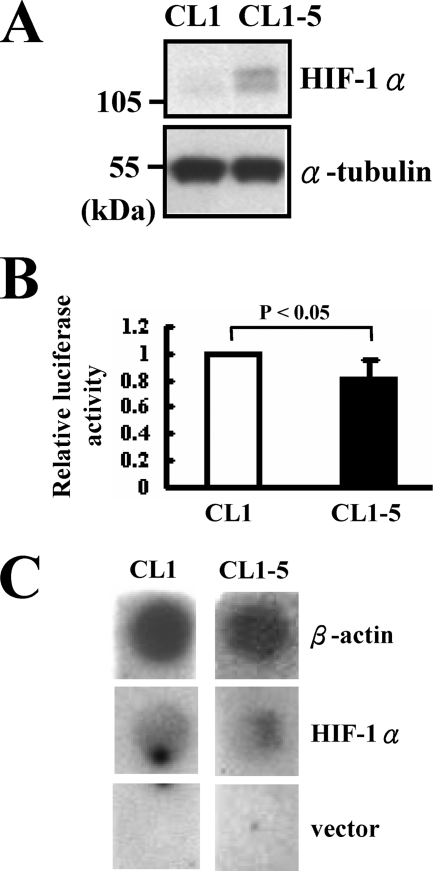
We examined the levels of HIF-1α protein in CL1 and CL1-5 cells by Western blot analyses. The results showed that CL1-5 cells expressed much more HIF-1α than CL1 cells (Fig. 1A), which is consistent with our previous report in which we reported that CL1-5 cells express much more HIF-1α mRNA than CL1 cells (11). To elucidate the mechanisms underlying the differential expression of HIF-1α in CL1 and CL1-5 cells, we examined the HIF-1α promoter activity of CL1 and CL1-5 cells. These cells were transiently transfected with a plasmid harboring a luciferase reporter under control of the HIF-1α promoter (20). Subsequent luciferase assays showed that transfected CL1 cells elicited ∼20% more luciferase activity than transfected CL1-5 cells (Fig. 1B). These data suggested that the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells was unlikely to result from altered transcription rate of the HIF-1α gene. To confirm this possibility, we performed nuclear run-on assays using nuclei prepared from CL1 and CL1-5 cells. As shown, the transcription rate of HIF-1α of CL1 cells was no lower than that of CL1-5 cells (Fig. 1C). These results indicated that one or more post-transcriptional mechanisms down-regulating the expression of HIF-1α mRNA in CL1 cells contribute to the differential expression of HIF-1α in CL1 and CL1-5 cells.
FIGURE 1.
Comparison of HIF-1α transcription in CL1 and CL1-5 cells. A, Western blot analyses. Forty micrograms of whole cell lysates prepared from CL1 and CL1-5 cells were subjected to Western blot analyses fort the detection of HIF-1α protein. α-tubulin served as an internal control. B, luciferase assays. CL1 and CL1-5 cells (3 × 105 each) were transiently transfected with pGL3-HIF-1α-luc (2 μg) plus Renilla luciferase reporter (0.1 μg). Cells were lysed 24 h after transfection and subjected to luciferase assays. The normalized luciferase activity of CL1-5 cells was presented relative to that of CL1 cells (to which a value of 1 was assigned). Data represent mean ± S.D. from three separate experiments. C, nuclear run-on assays. Nuclei from 2 × 107 CL1 and CL1-5 cells were collected and subjected to nuclear run-on assays. Signals of β-actin and HIF-1α were shown. The vector, pCEP4, was used as a negative control.
Identification of Protein Binding Activities on the 5′-UTR of HIF-1α mRNA
We performed RNA electrophoretic mobility shift assay (REMSA) analyses to examine the interactions of HIF-1α mRNA with cellular proteins of CL1 and CL1-5 cells, because the process of mRNA degradation often involves the interactions between RNA-binding proteins and certain regions of target mRNAs. CL1 and CL1-5 cells were fractionated into cytoplasmic and nuclear fractions and incubated with radiolabeled RNA transcripts corresponding to the 5′-UTR (A1) and 3′-UTR (B2, B3, and C2) of HIF-1α mRNA (Fig. 2A). Nuclear proteins of CL1 and CL1-5 cells formed complexes with A1 transcript with almost identical binding patterns, as did their binding with B2 transcript. Cytoplasmic proteins of CL1 and CL1-5 cells also exhibited identical binding patterns with each 3′-UTR transcript (B2, B3, and C2), but the former exhibited more binding activities to the A1 transcript than the later (Fig. 2B). The finding that A1 was the only segment for which protein binding activities reflected the difference in HIF-1α mRNA abundance in CL1 and CL1-5 cells led us to propose that cis-elements involved in the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells could be located within the 5′-UTR of HIF-1α mRNA. It is worth noting that the RNA probes (A1, B2, B3, and C2) used in REMSA experiments were derived from the HIF-1α mRNA of CL1-5 cells, although direct sequencing showed no difference between the 5′-UTR sequences of HIF-1α mRNAs prepared from CL1 and CL1-5 cells. REMSA assays using RNA probes derived from the HIF-1α mRNA of CL1 cells generated similar data as described above (data not shown).
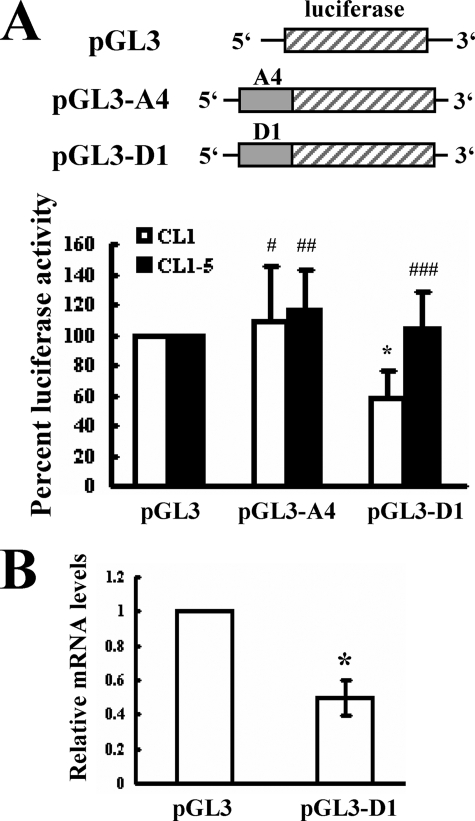
To identify the region within A1 that was recognized by cytoplasmic proteins, we subdivided A1 region into A4 and D1 fragments (Fig. 2C) and performed REMSA analyses to examine the protein binding patterns with these transcripts. As shown, cytoplasmic proteins of CL1 and CL1-5 cells only bound to the D1 transcript, and the binding pattern was identical to that seen with the A1 transcript (Fig. 2D). Subsequently, we examined the function of D1 in regulating gene expression. CL1 and CL1-5 cells were transiently transfected with pGL3-promoter, or pGL3-A4, or pGL3-D1 plasmid (see “Experimental Procedures”) together with a Renilla luciferase reporter construct (as an internal control) and subjected to luciferase analyses. As shown, compared with the luciferase activity of CL1 cells receiving pGL3-promoter, the luciferase activity of CL1 cells receiving pGL3-D1 was ∼42% lower, whereas the luciferase activity of CL1 cells receiving pGL3-A4 was not significantly changed (Fig. 3A). On the other hand, neither D1 nor A4 insertion resulted in detectable decrease of luciferase activity in transfected CL1-5 cells. Subsequent RT-qPCR analyses showed that the levels of the chimeric luciferase transcript in CL1 cells receiving pGL3-D1 was ∼50% lower than the levels of luciferase transcript in cells receiving pGL3-promoter (Fig. 3B). Thus, the binding of cytoplasmic proteins to the HIF-1α D1 region might therefore reduce the expression of HIF-1α mRNA in CL1 cells, resulting in the differential expression of HIF-1α mRNA in CL1 and CL1-5. To delineate the protein-binding site within the D1 region, we divided D1 into D5 and E1 fragments (Fig. 4A). Subsequent REMSA analyses showed that the cytoplasmic proteins of CL1 and CL1-5 cells formed multiple complexes with D1 and D5, and the binding patterns on D5 were identical to those seen with D1. There was no binding activity on E1. CL1 cell lysates formed more prominent complexes than did CL1-5 cell lysates (Fig. 4B). The specificity of the complexes formed by D5 was confirmed by the competition assays, which showed that the formation of D5-protein complexes was decreased by a 5-fold molar excess of unlabeled D5 transcript, and was completely inhibited by a 50-fold molar excess of unlabeled D5 transcript. However, a 50-fold molar excess of unlabeled E1 transcript failed to compete with D5 for protein binding (Fig. 4C). Thus, these results showed that the levels of D5-protein complexes reflected the difference in HIF-1α mRNA abundance in CL1 and CL1-5 cells and suggested that the cis-element(s) involved in the degradation of HIF-1α mRNA in CL1 cells is located within the D5 region.
FIGURE 3.
Influence of the D1 region of HIF-1α mRNA on the expression of a luciferase reporter gene. A, luciferase assays. CL1 and CL1-5 (3 × 105/6 cm-plate) were used for transient transfection with 2 μg of either pGL3-promoter (pGL3), or pGL3-A4, or pGL3-D1 plasmid together with a Renilla luciferase reporter (0.1 μg). Cells were lysed 24 h after transfection and subjected to luciferase assays. The luciferase activities of CL1 (□) and CL1-5 (■) receiving pGL3-A4 or pGL3-D1 were compared with that of cells receiving pGL3-promoter (to which a value of 100% was assigned). Data represent the mean ± S.D. from five separate experiments. *, p < 0.005; #, p = 0.276; ##, p = 0.191; ###, p = 0.359 versus controls. B, RT-qPCR analyses. CL1 cells were transiently transfected with 2 μg of either pGL3-promoter (pGL3) or pGL3-D1 plasmid together with a Renilla luciferase reporter (0.1 μg). Total RNAs were isolated 24 h after transfection. RT-qPCR analyses were performed to examine the levels of luciferase, chimeric luciferase, and β-actin mRNAs. The normalized levels of chimeric luciferase mRNA in cells receiving pGL3-D1 were compared with the normalized levels of luciferase mRNA in cells receiving pGL3 (to which a value of 1 was assigned). Data represent the mean ± S.D. from four separate analyses. *, p < 0.0006 versus cells receiving pGL3.
Influence of D5 on the Expression of Chimeric Transcripts and HIF-1α mRNA
To investigate whether the D5 region down-regulated the half-life of mRNA, we constructed pTRE-D5-DsRed2 and pTRE-A6-DsRed2 plasmids and established CL1-Tet and CL1-5-Tet cells, which constitutively express a transactivator in the absence of tetracycline. CL1-Tet and CL1-5-Tet cells were transiently transfected with pTRE-DsRed2 or pTRE-A6-DsRed2, or pTRE-D5-DsRed2, so that these cells expressed mRNA coding for a fluorescent reporter, or chimeric mRNAs containing extra D5 or A6 sequence in 5′-UTR at the absence of tetracycline. After a 20-h transcriptional pulse, cells were treated with doxycycline, and the half-lives of the three transcripts (DsRed2, D5-DsRed2, and A6-DsRed2) were determined by Northern blot analyses. As shown, in CL1-Tet cells, the half-life of D5-DsRed2 (∼4 h) was shorter than those of DsRed2 and A6-DsRed2 transcripts (∼7 h). In CL1-5-Tet cells, however, the half-life of D5-DsRed2 was no shorter than those of DsRed2 and A6-DsRed2 transcripts (Fig. 5A). By RT-qPCR analyses, the level of HIF-1α mRNA in CL1-Tet cells expressing pTRE-D5-DsRed2 was ∼2.5-fold higher than that of cells expressing pTRE-DsRed2 (Fig. 5B). Overexpression of A6-DsRed2 transcript did not affect HIF-1α mRNA expression. Taken together, these data suggested that D5 overexpression competes for the decay of HIF-1α, in turn rendering HIF-1α mRNA stable. It further indicated that interactions of the cytoplasmic proteins of CL1 with D5 could reduce the stability of HIF-1α mRNA.
FIGURE 5.
Influence of D5 on the half-life of chimeric transcripts and the expression of HIF-1α mRNA. A, transcription shot-off assays. CL1-Tet and CL1-5-Tet cells (3 × 105/6-cm plate) were transiently transfected with 5 μg of each pTRE-DsRed2, or pTRE-D5-DsRed2, or pTRE-A6-DsRed2 construct along with 0.1 μg of a Renilla luciferase reporter as a transfection control. After treated with doxycycline, cells were harvested at the times as indicated. Twenty micrograms of RNA prepared from these cells was analyzed for the levels of DsRed2, D5-DsRed2, A6-DsRed2, and GAPDH transcripts by Northern blot analyses. Signals of reporter transcripts were quantitated and normalized to that of GAPDH and to Renilla luciferase activity. The results are presented in the semilog graph. The dashed line serves to estimate the half-life values of each transcript. Values represent the mean ± S.D. from four experiments. B, RT-qPCR assays. Equal amount of cDNA (0.5 μg) from the CL1-Tet cells receiving pTRE-DsRed2, or pTRE-D5-DsRed2, or pTRE-A6-DsRed2 construct was analyzed by RT-qPCR assays for the levels of HIF-1α and β-actin mRNAs. HIF-1α signals were normalized to β-actin signals and luciferase activities. HIF-1α mRNA levels of cells receiving pTRE-D5-DsRed2 (D5-DsRed2) or pTRE-A6-DsRed2 (A6-DsRed2) construct were compared with that of cells receiving pTRE-DsRed2 (Control) construct (to which a value of 1 was assigned). Data represent the mean ± S.D. from three triplicate analyses. *, p < 0.005 versus control.
Identification of PTB as a Component of D5-Proteins Complexes
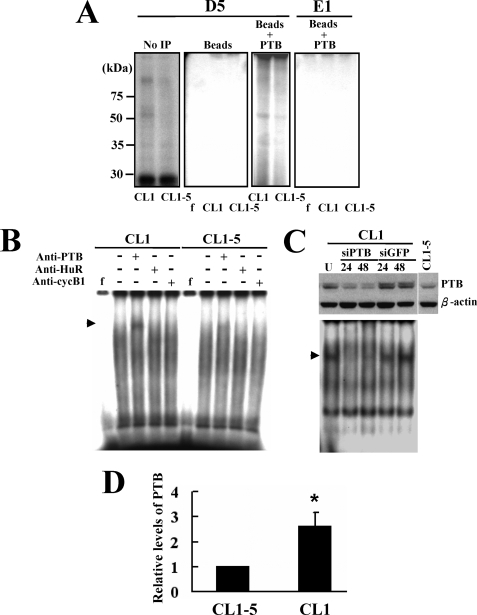
We performed UV cross-linking experiments to investigate the proteins binding to the D5 transcript. Our results revealed several complexes formed by the proteins of CL1 and CL1-5 cells. One of the major D5-protein complexes of which the levels reflected the difference in HIF-1α mRNA abundance in CL1 and CL1-5 cells showed a molecular size slightly higher than 50 kDa (Fig. 6A). Considering that D5 contains a pyrimidine-rich region (Fig. 4A), we thought that the D5-binding proteins might contain the 58-kDa PTB. So, we performed UV cross-linking experiments and used an anti-PTB antibody to precipitate the D5-protein complexes. Subsequent analysis by SDS-PAGE showed that the molecular size of the D5-protein complexes immunoprecipitated by anti-PTB antibody was slightly higher than 50 kDa (Fig. 6A). These data indicated that PTB binds to the D5 region of HIF-1α mRNA. Accordingly, we conducted supershift assays using antibodies against PTB, HuR (an RNA-binding protein), and cyclin B1, and found that only the anti-PTB antibody supershifted the complexes formed by the cytoplasmic proteins of CL1 cells and, if any, by the cytoplasmic proteins of CL1-5 cells (Fig. 6B). No supershifted signal was detected in the reaction mixture containing anti-PTB antibody and D5 probe only (data not shown). Targeted repression of PTB in CL1 cells specifically decreased the formation of one of the D5-protein complexes. For our convenience, this complex was designated as complex I. These data indicated the requirement of PTB for the formation of complex I of D5-protein complexes (Fig. 6C). Interestingly, Western blot analyses showed that CL1 cells expressed ∼2.6-fold more PTB than CL1-5 cells (Fig. 6, C and D).
FIGURE 6.
Identification of the PTB as a component of the D5-protein complexes. A, detection of PTB-D5 associations by immunoprecipitation. Cell lysates prepared from CL1 and CL1-5 cells were mixed with radiolabeled probes (either D5 or E1) and then subjected to UV cross-linking. Complexes were either resolved directly by electrophoresis through SDS-PAGE gels (No IP), or first immunoprecipitated using protein A beads coated with anti-PTB antibody (Beads + PTB) or with protein A beads (Beads) and then resolved by electrophoresis through SDS-PAGE gels. f, radiolabeled transcripts digested with RNase T1 without incubation with cell lysate. B, supershift assays. Cytoplasmic proteins of CL1 and CL1-5 cells were mixed with antibodies against PTB, HuR, or cyclin B1 for 30 min on ice, prior to the addition of radiolabeled D5 probe. After addition of RNase T1, reaction mixtures were resolved on 4% native polyacrylamide gels. Signals were visualized as described above. The arrowhead indicates the supershifted band. f, radiolabeled transcripts digested with RNase T1 without incubation with cell lysate. C, Western blot and REMSA analyses. CL1 cells (1 × 106) were either treated with transfection reagents (U, untreated) or transfected with 100 nm of siRNAPTB (siPTB) (Qiagen, catalogue no. SI00301490) or siRNAGFP (siGFP) (Dharmacon, catalogue no. D-001300-01-20) for 6 h using Effectene reagent. Cells were harvested 24 and 48 h after transfection. Forty micrograms of whole cell lysates prepared from these cells and CL1-5 cells were subjected to Western blot analyses for the levels of PTB (upper). Ten micrograms of cytoplasmic fractions was subjected to REMSA assays with D5 transcript (lower) as described above. The arrowhead indicates the position of complex I. D, Western blot analyses. Forty micrograms of whole cell lysates prepared from CL1 and CL1-5 cells were subjected to Western blot analyses for the cellular levels of PTB. The PTB levels in CL1 cells were compared with that of CL1-5 cells (to which a value of 1 was assigned). Data represent the mean ± S.D. from three separate analyses. *, p < 0.05 versus CL1-5 cells.
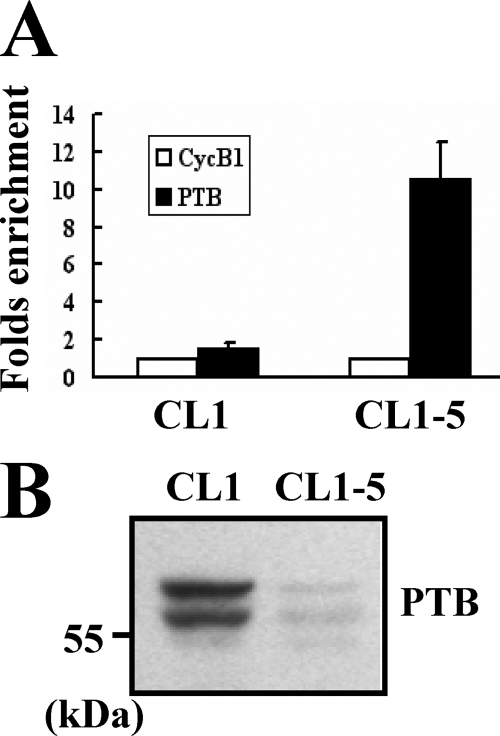
To further examine the binding of PTB-containing protein complexes on HIF-1α mRNA in CL1 and CL1-5 cells, we performed immunoprecipitation experiments using anti-PTB or control antibody to pull down the ribonucleoprotein complexes in equal amounts of lysates prepared from CL1 and CL1-5 cells. RNAs were extracted from these immunoprecipitates and subjected to RT-qPCR analyses for HIF-1α and β-actin mRNAs. In the lysate of CL1-5 cells, RT-qPCR assays revealed an enrichment of ∼10.5-fold in HIF-1α mRNA in the immunoprecipitate using anti-PTB antibody compared with that in the immunoprecipitate using a control antibody (Fig. 7A). In the lysate of CL1 cells, the -fold enrichment of HIF-1α mRNA was ∼1.5-fold. Because Western blot analysis showed that the immunoprecipitate of CL1 cells contains more PTB than that of CL1-5 cells (Fig. 7B), our data showed that the PTB-containing protein complexes can bind HIF-1α mRNA and that the ribonucleoprotein complexes, which contain more PTB, have less remaining HIF-1α mRNA.
FIGURE 7.
Further examination of the binding of PTB-containing protein complexes on HIF-1α mRNA. A, ribonucleoprotein immunoprecipitation and RT-qPCR assays. One milligram of lysates prepared from CL1 or CL1-5 cells (1 × 107) was incubated with protein A beads precoated with 15 μg of either anti-PTB (PTB) or anti-cyclin B1 (CycB1) antibody to precipitate ribonucleoprotein complexes and to extract RNAs from the complexes as described under “Experimental Procedures.” RNAs were used in subsequent RT-qPCR assays for HIF-1α and β-actin mRNAs. HIF-1α signals were normalized to β-actin signals. The normalized HIF-1α signals obtained from the ribonucleoprotein complexes pulled down by anti-PTB antibody were compared with those pulled down by anti-cyclin B1 antibody (to which a value of 1 was assigned). Data represent the mean ± S.D. from three triplicate analyses. B, Western blot analyses. Forty micrograms of the immunoprecipitates prepared from CL1 and CL1-5 cells were subjected to Western blot analyses for the levels of PTB.
Interactions of PTB with D5, but Not with B2, Down-regulated the Expression of Chimeric Transcripts
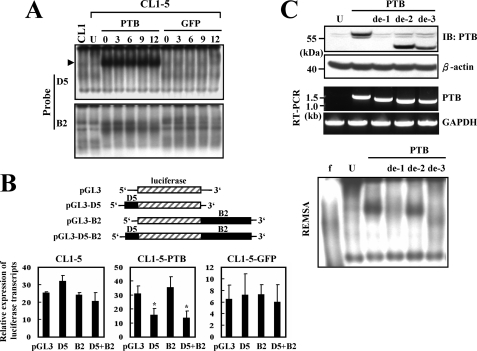
Since it was reported that PTB can bind to the 3′-UTR of HIF-1α mRNA in HeLa cells (21), we set out to examine whether overexpression of PTB in CL1-5 cells affected protein-binding activities on D5 and B2 (3′-UTR) regions of HIF-1α mRNA. Transfected CL1-5 cells were treated with an inhibitor of de novo RNA synthesis, actinomycin D, for 0, 3, 6, 9, and 12 h, and subsequent REMSA analyses showed that overexpression of PTB increased the formation of D5-protein complexes (complex I) in transfected CL1-5 cells (Fig. 8A). On the other hand, overexpression of PTB also increased protein-binding activities on B2, but those activities were readily detected in untransfected CL1-5 cells. Taken together, these data indicated that PTB participates in the protein binding on 5′-UTR and 3′-UTR of HIF-1α mRNA. Subsequently, we investigated whether binding of PTB on D5 or B2 decreased gene expression. D5 or B2 was inserted 5′ or 3′ to the luciferase cDNA in pGL3-promoter vector to generate pGL3-D5, pGL3-B2, and pGL3-D5-B2 reporters (Fig. 8B). We established CL1-5 cells overexpressing either PTB (CL1-5-PTB) or GFP (CL1-5-GFP). These cells were transfected with pGL3-promoter, pGL3-D5, pGL3-B2, and pGL3-D5-B2 reporters. Transfected cells were either left untreated or treated with actinomycin D for 6 h, and then subjected to RT-qPCR assays to quantitate the levels of those four transcripts, i.e. luciferase, D5-luciferase, luciferase-B2, and D5-luciferase-B2 (Fig. 8B). As shown, in actinomycin D-treated CL1-5 cells, the levels of luciferase, D5-luciferase, luciferase-B2, and D5-luciferase-B2 transcripts were ∼25.3-, 32.1-, 24.3-, and 20.5-fold of that seen in untreated CL1-5 cells. In actinomycin D-treated CL1-5-PTB cells, the levels of the four transcripts mentioned above were ∼30.7-, 15.5-, 35.2-, and 13.6-fold of that in untreated CL1-5-PTB cells. In actinomycin D-treated CL1-5-GFP cells, the levels of those four transcripts were ∼6.6-, 7.3-, 7.4-, and 6-fold of that in untreated CL1-5-GFP cells. These data presented the inhibitory effect of the D5 insert, but not B2, to the expression of chimeric luciferase transcript in CL1-5-PTB, but not in CL1-5 and CL1-5-GFP cells. Thus, our data indicated that PTB down-regulates the expression of chimeric mRNAs by binding to the D5 region in the 5′-UTR. Furthermore, the data obtained by using the D5-luciferase-B2 transcript, which to a certain extent mimics the HIF-1α mRNA, suggested that binding of PTB to the D5 region in the 5′-UTR may down-regulate the expression of HIF-1α mRNA.
FIGURE 8.
Binding of PTB-containing protein complexes on D5 but not B2 down-regulated gene expression. A, REMSA assays. CL1-5 cells (1 × 106/10-cm plate) were either treated with transfection reagents (U, untreated) or transiently transfected with 2 μg of each pCMV-SPORT6-PTB (PTB) or pcDNA3.1-GFP (GFP) construct along with 0.1 μg of a Renilla luciferase reporter as a transfection control. Twenty-four hours after transfection, cells were treated with actinomycin D (1 μg/ml) for 0, 3, 6, 9, and 12 h. Cytoplasmic fractions were isolated from these cells and subjected to REMSA assays using radiolabeled D5 (upper panel) and B2 (lower panel) transcripts as probes. Lysates of CL1 cells were used in the assays as references. The arrowhead indicates the position of complex I. B, RT-qPCR analyses. Schematic representation of pGL3, pGL3-D5, pGL3-B2, and pGL3-D5-B2 constructs is shown. CL1-5, CL1-5-PTB, and CL1-5-GFP cells (3 × 105/6-cm plate) were transiently transfected with 2 μg of either pGL3-promoter (pGL3), or pGL3-D5 (D5), or pGL3-B2 (B2), or pGL3-D5-B2 (D5 + B2) construct along with 0.1 μg of a Renilla luciferase reporter as a transfection control. Twenty-four hours after transfection, cells were treated with actinomycin D (1 μg/ml) for 0 and 6 h. Cells were harvested for RNA preparation and subsequent RT-qPCR analyses to quantitate the levels of luciferase and β-actin transcripts. Luciferase signals were normalized to β-actin signals and Renilla luciferase activities. Data represent the relative levels of luciferase mRNA in cells receiving 6-h actinomycin D treatment comparing with that of cells without actinomycin D treatment (to which a value of 1 was assigned). Data represent the mean ± S.D. from three triplicate analyses. *, p < 0.05 versus CL1-5-PTB cells receiving pGL3. C, Western blot, RT-PCR, and REMSA analyses. CL1-5 cells were either treated with transfection reagents (U, untreated) or transiently transfected with 2 μg of each pCMV-SPORT6-PTB (PTB), or pPTB-deRRM1 (de-1), or pPTB-deRRM2 (de-2), or pPTB-deRRM3 (de-3). Cells were harvested 24 h after transfection for the preparation of whole cell and cytoplasmic lysates and RNAs. Western blot analyses were carried out to detect the intact and partially deleted forms of PTB. A representative blot was shown (upper). RT-PCR analyses were performed to detect the various forms of PTB mRNAs (middle). The sizes of PTB mRNAs generated by pCMV-SPORT6-PTB, pPTB-deRRM1, pPTB-deRRM2, and pPTB-deRRM3 are 1675, 1465, 1405, and 1381 bases, respectively. REMSA assays using radiolabeled D5 probe were performed as described in Fig. 2 (lower).
PTB contains four RNA recognition motifs (RRMs) separated by three linkers (22). To identify the RRM(s) responsible for the PTB-triggered formation of D5-protein complexes, we generated three constructs coding for PTB with deleted RRM1, or RRM2, or RRM3, respectively. For unknown reasons, we were not able to generate the RRM4-deleted construct. So, CL1-5 cells were transiently transfected with these constructs and then subjected to Western blot, RT-PCR, and REMSA analyses. Because the RRM1 contains the recognition site of the anti-PTB antibody, the RRM1-deleted PTB could not be detected by Western blot analyses (Fig. 8C). However, RT-PCR analyses showed the overexpression of the intact and the three partially deleted forms of PTB mRNA in transfected CL1-5 cells, whereas the endogenous PTB mRNA could not be detected (Fig. 8C). REMSA assays showed that, compared with untreated CL1-5 cells, overexpression of intact PTB increased the formation of D5-proteins complex I (Fig. 8C), whereas overexpression of RRM1-deleted PTB did not. Overexpression of RRM2-deleted PTB also induced strong binding activities on D5, and the mobility of the complexes was similar to that of complex I. Overexpression of RRM3-deleted PTB also increased the formation of D5-protein complexes, although the binding activities were much weaker than the activities induced by the overexpression of intact PTB, and the mobility of the complexes was significantly different from that of complex I. Therefore, RRM1 and RRM3 might play an important role in the PTB-mediated D5 binding and complex I formation.
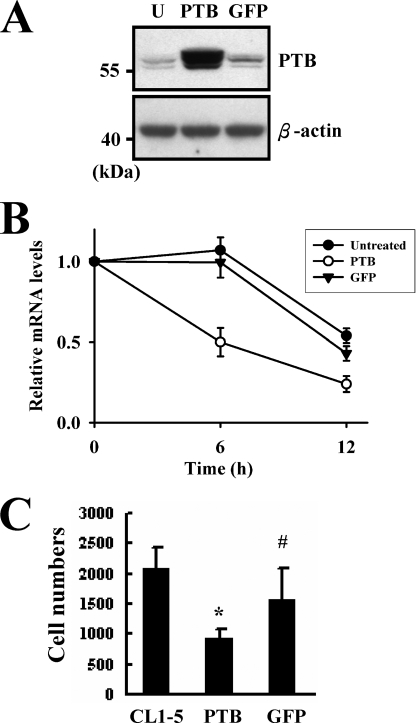
PTB Is Involved in Regulating the Expression of HIF-1α mRNA and Cell Invasiveness
Based on the finding that the ribonucleoprotein complexes, which contain more PTB, have less remaining HIF-1α mRNA (Fig. 7), we tested if PTB regulates HIF-1α mRNA levels. Because PTB also participates in RNA synthesis, inhibition of PTB expression may not necessarily cause the increase of HIF-1α mRNA. Thus, to examine if PTB regulates the stability of HIF-1α mRNA, we overexpressed PTB in CL1-5 cells (Fig. 9A) instead of inhibiting PTB expression in CL1 cells. Transfected cells were treated with actinomycin D, for 0, 6, and 12 h and then subjected to RT-qPCR assays to depict the elimination of endogenous HIF-1α mRNA (Fig. 9B). These data showed that overexpression of PTB caused a 50 and 76% decrease in HIF-1α mRNA by 6 and 12 h of actinomycin D treatment. In comparison, HIF-1α mRNA levels in untreated and GFP-expressing CL1-5 cells remained intact 6 h after actinomycin D treatment, and decreased ∼50% after another 6 h. These data indicated that PTB participates in regulating the stability of HIF-1α mRNA in vivo. Next, we examined the in vitro invasive activities of CL1-5, CL1-5-PTB, and CL1-5-GFP cells to examine if PTB regulates cell invasiveness. The results showed that the numbers of migrated CL1-5, CL1-5-PTB, and CL1-5-GFP cells were 2069 ± 374, 903 ± 186, and 1559 ± 530, respectively. Overexpression of GFP seemed to decrease cell invasiveness, but it did not reach statistical significance. On the other hand, overexpression of PTB decreased ∼56% of invasive activity (Fig. 9C). Given that HIF-1α can stimulate cell invasiveness (11), our data suggested that PTB might negatively regulate the invasive activity of CL1-5 cells at least by down-regulating HIF-1α expression.
FIGURE 9.
PTB regulated the stability of HIF-1α mRNA and cell invasiveness. CL1-5 cells (1 × 106/10-cm plate) were either treated with transfection reagents (U, untreated) or transiently transfected with 2 μg of each pCMV-SPORT6-PTB (PTB) or pcDNA3.1-GFP (GFP) construct along with 0.1 μg of a Renilla luciferase reporter as a transfection control. Twenty-four hours after transfection, cells were treated with actinomycin D (1 μg/ml) for 0, 6, and 12 h. A, Western blot analyses. Forty micrograms of whole cell lysates prepared from cells harvested 24 h after transfection was analyzed for the levels of PTB. A representative blot is shown. B, RT-qPCR assays. RNAs were isolated from cells treated with actinomycin D for 0, 6, and 12 h for the preparation of cDNAs. Equal amount of cDNA (0.5 μg) from these cells was analyzed by RT-qPCR assays for the levels of HIF-1α and β-actin mRNAs. HIF-1α signals were normalized to β-actin signals and Renilla luciferase activities. HIF-1α mRNA levels of cells receiving 6-h and 12-h actinomycin D treatment were compared with that of cells without actinomycin D treatment (to which a value of 1 was assigned). Data represent the mean ± S.D. from three triplicate analyses. C, in vitro invasion assays. CL1-5, CL1-5-PTB (PTB), and CL1-5-GFP (GFP) cells (5 × 104) were seeded on the filter membranes separately for 16 h at 37 °C and processed as described under “Experimental Procedures.” Each bar represents the mean ± S.D. from three separate experiments. *, p < 0.001; ##, p = 0.09 versus CL1-5 cells.
DISCUSSION
HIF-1α is expressed in many types of cancers. It plays a crucial role in the processes of tumor growth, expansion, invasion, and metastasis. Elucidation of the mechanisms governing the up-regulation of HIF-1α expression in cancer cells is expected to reveal important molecules that regulate this process, which may provide us more information regarding the development of invasive phenotype and reveal potential therapeutic targets for the treatment of cancers. Although many other studies reveal mechanisms that regulate HIF-1α expression at protein level, our studies using human lung adenocarcinoma cells have revealed that modulation of mRNA stability represents another mechanism controlling HIF-1α expression.
That the higher HIF-1α mRNA expression in CL1-5 cells than in CL1 cells is not via transcriptional up-regulation is evidenced by the finding that the transcription rate of HIF-1α in CL1-5 cells is slightly lower than that in CL1 cells. In addition, we found no difference in the copy number of HIF-1α gene between CL1 and CL1-5 cells (data not shown). Thus, we explored the post-transcriptional mechanisms mediating the repressed expression of HIF-1α mRNA in CL1 cells. Using gel mobility-shift analyses plus in vivo functional assays, we have revealed that a 61-base segment (D5) within the 5′-UTR of HIF-1α mRNA, which, while inserted 5′ directly to a luciferase gene, can reduce the half-life of the D5-containing chimeric mRNAs in CL1 cells but not in CL1-5 cells (Fig. 5A). In addition, overexpression of a D5-containing chimeric transcript in CL1 cells, which presumably dilutes the protein-binding activities on the D5 region of endogenous HIF-1α mRNA, increases the expression of HIF-1α mRNA (Fig. 5B). These data reveal that interactions between D5 and its binding proteins play an important role in decreasing the stability of HIF-1α mRNA in CL1 cells. Given that CL1-generated cytoplasmic proteins form more complexes with D5 than CL1-5-generated proteins (Fig. 4), and that D5 fails to decrease the stability of the D5-containing chimeric transcripts in transfected CL1-5 cells, our findings further indicate that the expression of certain protein(s) that bind D5 plays a critical role in the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells.
We have identified the PTB as a critical component of one of the D5-protein complexes (i.e. complex I) by showing that (i) an anti-PTB antibody immunoprecipitated D5-protein complexes (Fig. 6A) and (ii) repression of PTB expression in CL1 cells decreased complex I formation (Fig. 6C), whereas overexpression of PTB in CL1-5 cells increased complex I formation (Fig. 8, A and C). PTB, also known as heterogeneous nuclear ribonucleoprotein I, is an RNA-binding protein involved in many aspects of RNA metabolism, including alternative splicing, translocation, polyadenylation, translation, etc. (Ref. 22 and references therein). In our studies, although PTB is identified from the investigation of protein-binding activities on 5′-UTR of HIF-1α mRNA, REMSA assays reveal that PTB also participates in the protein-binding activities on the 3′-UTR of this message (Fig. 8A). These data are consistent with the other reports showing the binding capability of PTB on the 5′-UTR and/or 3′-UTR of several mRNAs, including HIF-1α (21, 23–26). However, there is a big difference in the consequence of PTB binding on the UTRs of mRNAs. It is reported that, by binding to the 3′-UTR, PTB stabilizes such mRNAs as those encoding insulin, CD40 ligand, phosphoglycerate kinase 2, and inducible nitric-oxide synthase (23–26). However, a recent paper reported that PTB binds to the 3′-UTR of HIF-1α mRNA in HeLa cells to enhance translation, but the binding activity doesn't affect mRNA abundance (21). On the other hand, an internal ribosome entry site has been identified in the 5′-UTR of HIF-1α mRNA, and it has been proposed that PTB binding to the internal ribosome entry site can stimulate HIF-1α translation (27, 28). Therefore, PTB had not been found to regulate the stability of HIF-1α mRNA. Here, by showing that (i) an anti-PTB antibody immunoprecipitated the ribonucleoprotein complexes, which contained HIF-1α mRNA, (ii) overexpression of PTB in CL1-5 cells decreased the stability of HIF-1α mRNA (Fig. 9B), and (iii) the expression of a luciferase reporter harboring either D5 or D5 plus B2, but not B2 insert alone, was down-regulated in CL1-5 cells overexpressing PTB (Fig. 8B), we suggest that PTB may act as a destabilizing factor of HIF-1α mRNA by mediating protein binding on the D5 region of the message. Accordingly, our findings provide a possible explanation to address why, in the ribonucleoprotein complex immunoprecipitation assays, the remaining HIF-1α mRNA in the PTB-containing complexes isolated from CL1 cells is much less than that from CL1-5 cells. It could be because HIF-1α mRNA is degraded by the D5 binding, PTB-containing protein complexes in CL1 cells, whereas the complexes in CL1-5 cells have less PTB and less binding on D5, hence more remaining HIF-1α mRNA. Taken together, we propose that (i) PTB may bind to different sites on mRNAs and form complexes with different sets of proteins to elicit various biological effects, including stabilization, degradation, and translation of mRNAs, and (ii) the fates of PTB-bound mRNAs could be cell type- and binding site-specific.
The comparison of the D5 binding and D5-protein complex I formation activities induced by overexpression of either intact or partially deleted forms of PTB shows that RRMs 1, 2, and 3 may contribute differently to the D5-binding activity of PTB. Among the three RRMs, RRM2 seems to be the least required domain for PTB to bind D5. Unlike the inhibitory effect of RRM1 deletion to the PTB-triggered D5 binding and complex I formation, RRM3-deleted PTB could form D5-protein complexes whose mobility is different from that of complex I (Fig. 8C). It is possible that the difference in the molecular weight of the intact and the RRM3-deleted forms of PTB accounts for the difference in the mobility of those complexes. It is also possible that the composition of the D5-binding protein complexes recruited by the RRM3-deleted PTB is different from that recruited by the intact PTB, hence the changes in the mobility of these complexes. Whether RRM1 and RRM3 exert different binding preference to D5 requires further investigation, although our data suggest that RRM1 and RRM3 are critical for the PTB-triggered formation of D5-proteins complex I, which suggests that at least RRM1 and RRM3 may play an important role in the PTB-triggered degradation of HIF-1α mRNA. The effect of these partially deleted forms of PTB on the expression of HIF-1α mRNA is currently under investigation. Notably, CL1 cells express more PTB than CL1-5 cells, the regulation of PTB expression may therefore play a role in the differential expression of HIF-1α mRNA in CL1 and CL1-5 cells. Nonetheless, exactly how PTB enhances the degradation of HIF-1α mRNA in CL1 cells remains to be elucidated.
Finally, HIF-1α has been found to play a stimulatory role in the invasiveness of CL1 and CL1-5 cells and is highly expressed in CL1-5 cells (11). Because CL1-5 cells are evolved from CL1 cells, our findings support the notion that alterations of post-transcriptional control of HIF-1α mRNA expression could play an important role in the evolvement of highly invasive and metastatic subpopulations from primary lung adenocarcinoma cells. In addition, our data suggest that, by regulating the expression of HIF-1α, PTB may regulate the cell invasiveness in CL1 and CL1-5 cells. Indeed, overexpression of PTB in CL1-5 cells decreased cell invasive activity (Fig. 9C). Thus, our data suggest that the regulation of PTB expression may play a role in the development of invasive and metastatic phenotypes. Further studies of the expression of PTB, and characterization of the PTB-containing, D5-binding protein complexes using our cell model are expected to provide critical insight into the development of invasive and metastatic phenotypes and the post-transcriptional regulation of HIF-1α expression in lung adenocarcinoma cells. In turn, this information could lead to improved treatment strategies.
Acknowledgment
We thank Dr. Myriam Gorospe for valuable suggestions and critical reading of the manuscript.
This work was supported by the National Science Council (Grant 92-2320-B-341-002 to S. L.) of Taiwan.
- HIF-1
- hypoxia-inducible factor-1
- siRNA
- small interfering RNA
- PTB
- polypyrimidine tract-binding protein
- GFP
- green fluorescence protein
- CMV
- cytomegalovirus
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- REMSA
- RNA electrophoretic mobility shift assay
- UTR
- untranslated repeat
- RRM
- RNA recognition motif
- RT-qPCR
- quantitative real-time PCR
- RT-PCR
- reverse transcription-PCR.
REFERENCES
- 1.Fidler I. J., Ellis L. M. (1994) Cell 79, 185–188 [DOI] [PubMed] [Google Scholar]
- 2.Graeber T. G., Osmanian C., Jacks T., Housman D. E., Koch C. J., Lowe S. W., Giaccia A. J. (1996) Nature 379, 88–91 [DOI] [PubMed] [Google Scholar]
- 3.Semenza G. L. (2000) Crit. Rev. Biochem. Mol. Biol. 35, 71–103 [DOI] [PubMed] [Google Scholar]
- 4.Guillemin K., Krasnow M. A. (1997) Cell 89, 9–12 [DOI] [PubMed] [Google Scholar]
- 5.Wenger R. H., Gassmann M. (1997) Biol. Chem. 378, 609–616 [PubMed] [Google Scholar]
- 6.Jiang B. H., Semenza G. L., Bauer C., Marti H. H. (1996) Am. J. Physiol. 271, C1172–C1180 [DOI] [PubMed] [Google Scholar]
- 7.Semenza G. L. (2001) Curr. Opin. Cell Biol. 13, 167–171 [DOI] [PubMed] [Google Scholar]
- 8.Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 9.Zagzag D., Zhong H., Scalzitti J. M., Laughner E., Simons J. W., Semenza G. L. (2000) Cancer 88, 2606–2618 [PubMed] [Google Scholar]
- 10.Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., Semenza G. L. (2003) Cancer Res. 63, 1138–1143 [PubMed] [Google Scholar]
- 11.Shyu K. G., Hsu F. L., Wang M. J., Wang B. W., Lin S. (2007) Exp. Cell Res. 313, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 12.Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M. M., Simons J. W., Semenza G. L. (2000) Cancer Res. 60, 1541–1545 [PubMed] [Google Scholar]
- 13.Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. (2000) J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 14.Maxwell P. H., Weisener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 15.Blagosklonny M. V. (1999) BioEssays 21, 704–709 [DOI] [PubMed] [Google Scholar]
- 16.Jiang B. H., Agani F., Passaniti A., Semenza G. L. (1997) Cancer Res. 57, 5328–5335 [PubMed] [Google Scholar]
- 17.Yang P. C., Luh K. T., Wu R., Wu C. W. (1992) Am. J. Respir. Cell Mol. Biol. 7, 161–171 [DOI] [PubMed] [Google Scholar]
- 18.Chu Y. W., Yang P. C., Yang S. C., Shyu Y. C., Hendrix M. J., Wu R., Wu C. W. (1997) Am. J. Respir. Cell Mol. Biol. 17, 353–360 [DOI] [PubMed] [Google Scholar]
- 19.Lin S., Wang W., Wilson G. M., Yang X., Brewer G., Holbrook N. J., Gorospe M. (2000) Mol. Cell. Biol. 20, 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer N. V., Leung S. W., Semenza G. L. (1998) Genomics 52, 159–165 [DOI] [PubMed] [Google Scholar]
- 21.Galbán S., Kuwano Y., Pullmann R., Jr., Martindale J. L., Kim H. H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J. O., Lewis S. M., Holcik M., Gorospe M. (2008) Mol. Cell. Biol. 28, 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auweter S. D., Allain F. H. (2008) Cell. Mol. Life Sci. 65, 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tillmar J., Welsh N. (2002) Mol. Med. 8, 263–272 [PMC free article] [PubMed] [Google Scholar]
- 24.Kosinski P. A., Laughlin J., Singh K., Covey L. R. (2003) J. Immunol. 170, 979–988 [DOI] [PubMed] [Google Scholar]
- 25.Xu M., Hecht N. B. (2007) Biol. Reprod. 76, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 26.Pautz A., Linker K., Hubrich T., Korhonen R., Altenhöfer S., Kleinert H. (2006) J. Biol. Chem. 281, 32294–32302 [DOI] [PubMed] [Google Scholar]
- 27.Lang K. J., Kappel A., Goodall G. J. (2002) Mol. Biol. Cell 13, 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schepens B., Tinton S. A., Bruynooghe Y., Beyaert R., Cornelis S. (2005) Nucleic Acids Res. 33, 6884–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]