Abstract
Vaccinia viral envelope protein A27 (110 amino acids) specifically interacts with heparin (HP) or heparan sulfate (HS) proteoglycans for cell surface attachment. To examine the binding mechanism, a truncated soluble form of A27 (sA27-aa; residues 21–84 of A27) with Cys71 and Cys72 mutated to Ala was used as the parent molecule. sA27-aa consists of two structurally distinct domains, a flexible Arg/Lys-rich heparin-binding site (HBS) (residues 21–32; 21STKAAKKPEAKR32) and a rigid coiled-coil domain (residues 43–84), both essential for the specific binding. As shown by surface plasmon resonance (SPR), the binding affinity of sA27-aa for HP (KA = 1.25 × 108 m−1) was approximately 3 orders of magnitude stronger than that for nonspecific binding, such as to chondroitin sulfate (KA = 1.65 × 105 m−1). Using site-directed mutagenesis of HBS and solution NMR, we identified a “KKPE” segment with a turn-like conformation that mediates specific HP binding. In addition, a double mutant T22K/A25K in which the KKPE segment remained intact showed an extremely high affinity for HP (KA = 1.9 × 1011 m−1). Importantly, T22K/A25K retained the binding specificity for HP and HS but not chondroitin sulfate, as shown by in vitro SPR and in vivo cell adhesion and competitive binding assays. Molecular modeling of the HBS was performed by dynamics simulations and provides an explanation of the specific binding mechanism in good agreement with the site-directed mutagenesis and SPR results. We conclude that a turn-like structure introduced by the KKPE segment in vaccinia viral envelope protein A27 is responsible for its specific binding to HP and to HS on cell surfaces.
Introduction
Vaccinia virus (VV)2 is the prototype of the Orthopoxvirus genus of the Poxviridae family, which infects many cell lines and animals (1, 2). The size of the viral genome is 190 kb, and it encodes more than 200 proteins. VV replicates in the cytoplasm of infected cells. The mature virion is the most abundant infectious virion particle assembled in the cytoplasmic viral factory (3). Viral envelope proteins form complexes on mature virions; for instance, viral protein A26 interacts with laminin (4). The vaccinia viral proteins A16, A21, A28, G3, G9, H2, J5, and L5 form protein complexes involved in viral fusion and viral entry (5–7). Like many viruses, such as herpesvirus, papillomavirus, paramyxovirus, and flavivirus, VV specifically interacts with glycosaminoglycans (GAGs) that determine its host tropism.
GAGs are linear polysaccharides with repeating disaccharide units predominantly found on cell surfaces and as constituents of the extracellular matrix (8, 9) that play fundamental roles in growth factor signaling, cellular differentiation, morphogenesis, pathophysiology (10–12) and proliferation, embryonic development, wound healing, and inflammation (9, 11, 13–15). GAGs interact specifically with a variety of proteins that mediate various biological processes (for details see Ref. 16 and references therein). There are four major members of the GAG family as follows: chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS) proteoglycan, and its highly sulfated analogue heparin (HP). HS is composed of N-acetyl-α-d-glucosamine as the major component, whereas the major component of HP is N-sulfo-α-d-glucosamine (16). Both CS and HS on the cell surface show a high degree of heterogeneity of the disaccharide repeating units and a variety of sulfation patterns that might influence the interaction with different VV proteins. In VV, several envelope viral proteins have been reported to interact with GAGs for specific attachment to the host cell surface, e.g. D8 binds to CS, whereas A27 and H3 interact with HS or HP (17, 18).
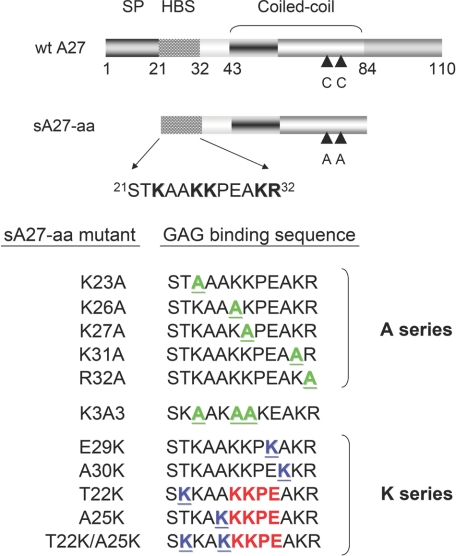
A27 consists of 110 amino acid residues that can be divided into four functional domains (Fig. 1) as follows: an N-terminal signal peptide (residues 1–20) (19); a Lys/Arg-rich domain (residues 21–32) known as the HBS (18); an α-helical coiled-coil domain (residues 43–84); and a C-terminal leucine zipper motif (residues 85–110) that interacts with viral membrane protein A17 (20). The HBS (21STKAAKKPEAKR32) is a flexible GAG-binding motif, consisting of five basic (B) and seven nonbasic (X) residues (XXBXXBBXXXBB) (see Fig. 1), whereas the rigid hydrophobic coiled-coil domain consists of four heptad repeats (abcdefg) responsible for self-assembly (21). The flexible HBS and the rigid hydrophobic coiled-coil domain are structurally and dynamically distinct, and the latter serves to stabilize the oligomeric protein structure that is essential for binding to HP via the HBS. Unfortunately, because of intrinsic self-assembly, neither an x-ray crystallographic structure nor a solution NMR structure is available. In our previous study (21), we used a truncated form of A27, sA27-aa (residues 21–84), containing both the HBS and the rigid coiled-coil domain, as the parent molecule (see Fig. 1). We demonstrated that the oligomeric structure of sA27-aa is essential for effective binding to HP and HS on cell surfaces (21). Apart from the structure/function correlation, the specific GAG binding mechanism of A27 remains unclear. In this study, in the absence of an x-ray or NMR protein structure, we used site-directed mutagenesis to examine the mechanism of the specific A27/GAG interaction.
FIGURE 1.
Illustration of the different functional domains in wild-type A27 and its truncated form sA27-aa (amino acids 21–84). Wild type A27 (wt A27) consists of the four functional domains of the signal peptide (SP, residues 1–20), the heparin binding domain (residues 21–32), the coiled-coil rigid domain (residues 43–84), and the Leu zipper domain at the C terminus (residues 84–110). sA27-aa consists of the heparin binding domain and the coiled-coil rigid domain in which two adjacent Cys residues (CC) at residues 71 and 72 are mutated to Ala. The HBS is an Arg/Lys-rich flexible domain, STKAAKKPEAKR; the basic residues are indicated in boldface. Different HBS mutants (A and K series and the K3A3 mutant) were expressed and purified; the mutated residues are shown in green (A series) or blue (K series) and are underlined. The KKPE-binding motif in the K mutants is shown in red.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
The preparation of the parent molecule sA27-aa has been described previously (22). Eleven sA27-aa mutants were constructed, consisting of five “A series” mutants in which single Lys residues were replaced by Ala, five “K series” mutants in which one or two residues were replaced by Lys residues, and a scrambled HBS sequence mutant “K3A3” (for details see Fig. 1). PCR amplification was performed using PfuUltra high fidelity DNA polymerase (Stratagene) and various oligonucleotide primers (supplemental Table S1). A PCR-based strategy was used, followed by DpnI digestion to remove the parental template, as described previously (23). To generate the K3A3 mutant, an A34L plasmid was constructed with a SacI restriction site at the end of the DNA triple coding for Leu34. The plasmid was created by BamHI/SacI double digestion, followed by K3A3 cassette insertion into the digested product using the forward primer (5′-gatcctctaaggctgctaaagcagcaaaggaggctaaacgcgagct-3′) and the reverse primer (5′-cgcgtttagcctcctttgctgctttagcagccttagag-3′). Subsequently, a second mutagenesis step was performed to restore Ala34 using the sense and antisense L34A primers (supplemental Table S1). All mutant constructs were confirmed by DNA sequencing.
Purification of Recombinant Proteins
Recombinant proteins were expressed in bacteria and purified as described previously (18). All recombinant proteins carried a T7 tag at the N terminus and a hexahistidine tag at the C terminus, which did not interfere with the function of wild-type A27 (18). For heteronuclear NMR studies, transformed bacteria were grown at 37 °C in M9 medium supplemented with [15N]ammonium chloride (1 g/liter) and [13C]glucose (1 g/liter) (Cambridge Isotope Laboratories, Inc., Andover, MA) to an absorbance at 600 nm of 0.8, induced for 2 h at 37 °C with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and harvested; the proteins were then purified on a nickel-nitrilotriacetic acid affinity column, as described previously (18).
14-mer HBS Peptide Synthesis
The 14-mer HBS peptide (STKAAKKPEAKREA) that corresponds to the HS binding domain of A27 was synthesized using an automated, solid phase synthesizer 433A (Applied Biosystems, Stafford, TX) using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and PyBOP activation. The peptide was purified by high pressure liquid chromatography using a reverse phase C-18 column (Supelco, 250 × 10 mm, 5 μm) on an Agilent 1100 system. Purity and homogeneity were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
Surface Plasmon Resonance (SPR)
Real time SPR experiments were performed on a BIAcore Biosensor 3000 system (BIAcore, Uppsala, Sweden) as described previously (21). To analyze the GAG binding affinity of sA27-aa, streptavidin (SA) sensor chips were purchased from BIAcore. GAG molecules such as biotinylated HP, DS, and chondroitin sulfate A were purchased from Sigma. CS was biotinylated by the method of Osmond et al. (24) or Cain et al. (25) via oxidized cis-diol groups. For SPR analysis, biotin-HP or -CS (10 μg/ml) in running buffer (10 mm HEPES, pH 7.0, containing 0.15 m NaCl, and 0.005% Polysorbate 20) was passed over the surface of an SA sensor chip at a flow rate of 10 μl/min until the maximal amount of biotin-HP or -CS was captured. The cells were then washed with HBS buffer containing 1 m NaCl to remove unbound biotinylated GAG molecules. GAG surfaces were created with an enhancement response unit value of 250–300. Unless specified otherwise, recombinant proteins at five different concentrations (1.0, 0.5, 0.25, 0.125, and 0.0625 μm) were injected over the HP- or CS-coated surface for 180 s for association followed by 300 s of running buffer injection for dissociation. Parallel injections of analytes over a control surface were performed for background measurements. The control surface was prepared using the same procedure but without biotinylated HP or CS. After subtraction of nonspecific binding curves, the association and dissociation rate constants were derived from a 1:1 Langmuir binding model (28). The coated surface was regenerated for subsequent injections by two successive injections of 120 μl of 1 m NaCl.
NMR Spectroscopy
1H NMR spectra were recorded on a Bruker DRX500 spectrometer operating at 500.13 MHz. Spectra were collected in H2O/D2O (90:10) at a temperature of 25 °C and with a repetition time of 1.5 s. Chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate using to the water resonance calibrated at 25 °C. NOESY spectra (mixing time of 300 ms) and TOCSY spectra were recorded with a spectral width of 4.7 kHz using 512 t1 blocks of 96 scans (NOESY) or 80 scans (TOCSY), each consisting of 2048 t2 points. Two-dimensional data sets were processed using a 60° sine-squared bell window function in both dimensions and zero filling to 2048 × 1048 data points prior to Fourier transformation. All 1H-15N HSQC spectra were recorded at pH 5.0 and at 23 °C on a Bruker Avance 600 MHz spectrometer equipped with a 5-mm inverse triple resonance (1H/13C/15N) z axis gradient probe; for 15N-labeled proteins (0.8–1.0 mm), Shigemi NMR tubes (5-mm outer diameter) were used. The detailed procedures for NMR measurement, data acquisition, and analysis have been described previously (29).
In the sA27-aa/HP and K3A3/HP interaction study, 1H-15N HSQC spectra were recorded for 15N-labeled sA27-aa and K3A3 (0.6 mm) in the presence of HP using a protein/HP weight ratio of 1:5. Chemical shift perturbations were quantified using Δδ = ((0.1 ΔδN)2 + (ΔδH)2)1/2, the factor of 0.1 compensating for the fact that the 15N shift covers a range ∼10 times that of 1H.
Cell Binding Assays
For the direct cell binding assay, different amounts (1, 10, and 100 μg) of the recombinant T22K/A25K mutant and the parental sA27-aa were incubated with HeLa cells at 4 °C for 60 min.
For competition binding assay, recombinant T22K/A25K mutant and sA27-aa protein (10 μg) were preincubated with phosphate-buffered saline, soluble HP, or CS (100 μg/ml) at 4 °C for 60 min with gentle rotation, and then the mixture was added to pre-cooled HeLa cells and incubation continued at 4 °C for another 60 min. The cells were washed three times with cold phosphate-buffered saline and lysed in SDS-containing sample buffer, and cell-bound A27 protein was separated on 12% SDS-PAGE and analyzed by immunoblot analyses as described previously (29).
In the binding specificity study, three murine cell lines were used (L, gro2C, and sog9 cells; kindly provided by F. Tufaro (30, 31)). Ten milliliters of prewarmed high glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 2% penicillin/streptomycin was added to a 10-mm Petri dish. Frozen stocks (−80 °C) of the cell lines were brought to room temperature in a water bath; the thawed cells were then moved to a 1.5-ml microtube and spun down, and the supernatant was removed. 1 ml of the medium from the Petri dish was then added to the cells, and the cells were suspended by gentle repeated pipetting and spread on the Petri dish. In a few minutes, the cells became attached to the bottom of plate and appeared thick and round under the microscope. On the 4th day, cells were split onto four plates for binding studies. Different amounts of the T22K/A25K mutant were incubated with the cells at 4 °C for 1 h, and the medium was then removed, and the cells were washed. The cells were lysed and aliquots of the lysate separated by 15% SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride membrane, and monoclonal anti-T7 tag antibody (Novagen) was used at a 1:5000 dilution to detect T22K/A25K. Horseradish peroxidase-conjugated anti-mouse IgG antibody (Pierce) at a 1:5000 dilution was used as the second antibody.
HBS-HS Docking and Co-minimization Analysis
A 12-mer HBS peptide (STKAAKKPEAKR, the GAG-binding motif of A27 viral protein) was created as an object using the program Discover Studio 2.0 and typed with the CHARMm force field. The peptide conformation was generated by standard dynamic cascade with implicit solvent model of generalized born switching and by energy minimization. The x-ray structure of tetrasaccharide, ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S (Protein Data Bank entry 1T8U) bound to a sulfotransferase (32), was used as the starting substrate structure. The bound sulfotransferase was removed, and the structure of 12-mer HBS peptide was used as a template for generating the HBS·HS complex. The docking of HS with HBS was performed using a manual procedure, placing ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S in the HBS for the best fitting. The complex structure was solvated with water molecules in an orthorhombic periodic box (57.5 × 31.1 × 27.4 Å) using explicit periodic boundary as a solvation model. Appropriate counterions such as sodium and chloride ions were added to neutralize the total charge of the complex. The complex together with the explicit water molecules were subjected to molecular dynamic simulation using standard dynamics cascade. The electrostatic interactions were calculated using the Particle Mesh Ewalds method.
RESULTS
Specific and Nonspecific GAG Binding
To differentiate nonspecific and specific GAG binding in vitro, we determined the binding affinity of vaccinia viral protein A27 for HP and CS using SPR. Biotin-tagged GAG molecules were immobilized on the SA sensor chip, and sA27-aa or mutants were passed over the immobilized GAGs. Fig. 2A shows SPR sensorgrams obtained at five different protein concentrations for sA27-aa and immobilized HP. The values of the kinetic binding constants kon, koff, and KA (= kon/koff) were 1.71 × 103 m−1 s−1, 13.7 × 10−6 s−1, and 1.25 × 108 m−1, respectively (Table 1). However, for nonspecific GAG binding, such as the sA27-aa/CS interaction, a lower KA value (∼105 m−1) was observed. The binding affinities of the specific and nonspecific binding differed by approximately 3 orders of magnitude. Note that the dissociation rate constant koff for the nonspecific interaction was also 3 orders of magnitude higher than that for the specific interactions, suggesting that, in the case of nonspecific binding, the viral protein was rather easily dissociated from GAGs. For comparison, we also measured the specific binding to CS and nonspecific binding to HP of another VV envelope protein, D8, and we found similar binding affinities that differed at least by 2 orders of magnitude (Table 1). The specific interactions showed a characteristic slow association and rather slow dissociation binding mechanism, implying that the local structure conformation of the GAG binding domain might be critical for binding specificity.
FIGURE 2.
SPR analyses of GAG binding of sA27-aa and different mutants. Biotinylated HP was captured by streptavidin onto an SA sensor flow chip. A, sA27-aa at five different protein concentrations (1.0, 0.5, 0.25, 0.125, and 0.0625 μm) was injected onto the immobilized HP, and the association and dissociation kinetic interactions were monitored as a function of protein concentration in real time. B, representative sensorgrams for sA27-aa and the five A series mutants at a protein concentration of 10 μm. C, representative sensorgrams for sA27-aa and three positive control K series mutants at a protein concentration of 2.5 μm.
TABLE 1.
Binding constants (KA), association rate constant (kon), and dissociation rate constant (koff) of specific and nonspecific GAG binding affinity of vaccinia viral envelope proteins sA27-aa and D8
Binding constants (KA) were calculated from the ratio kon/koff. Data represent the means ± S.D. of at least four independent experiments at five different protein concentrations.
| Specific | kon (×103m−1s−1) | koff (×10−6s−1) | KA (×108m−1) |
|---|---|---|---|
| sA27-aa, HP | 1.71 ± 0.42 | 13.7 ± 2.3 | 1.25 ± 0.80 |
| D8, CS | 11.9 ± 1.5 | 36.8 ± 26.6 | 3.23 ± 0.20 |
| Nonspecific | kon (×103m−1s−1) | koff(×10−3s−1) | KA (×105m−1) |
|---|---|---|---|
| sA27-aa, CS | 2.58 ± 0.04 | 15.6 ± 0.7 | 1.65 ± 0.40 |
| D8, HP | 1.45 ± 0.04 | 1.60 ± 0.35 | 9.06 ± 0.17 |
| T22K/A25K, CS | 3.57 ± 0.02 | 5.64 ± 0.18 | 6.33 ± 0.30 |
Each Basic Residue in HBS Is Critical for HP Binding
As mentioned in the Introduction, the HBS, containing five positively charged amino acids, is responsible for the interaction with HP. To identify the residues involved in the interaction with HP, we constructed and purified a series of sA27-aa mutants (K23A, K26A, K27A, K31A, and R32A) in which each basic residue in HBS was systematically replaced with Ala (Fig. 1). These HBS mutants were denoted the A series, and HP affinities were determined by SPR and compared with those of the parent sA27-aa (Fig. 2B). As shown in Table 2, the effective binding constant (KA) for HP for each mutant was 2 orders of magnitude lower than that for the parent molecule, whereas no significant change was observed in the association rate constant kon. In fact, some of these HBS mutants had an HP binding affinity similar to that of the nonspecific binding of D8 to HP (Table 1). Although it has been reported that Arg residues might bind HP more tightly than Lys residues in acidic fibroblast growth factor at physiological ionic strength (33), in the case of sA27-aa we found no substantial difference in GAG binding affinity when either Arg or Lys was mutated (Table 2). Because the binding affinities were of the same order of magnitude in the A series, we believe that the basic residues contribute equally to the binding of HP.
TABLE 2.
Heparin binding constants (KA), association rate constant (kon), and dissociation rate constant (koff) of sA27-aa mutants determined by SPR
Binding constants (KA) were calculated from the ratio kon/koff. Data represent the means ± S.D. of at least four independent experiments at five different protein concentrations.
| kon (×103m−1s−1) | koff (×10−3s−1) | KA (×106m−1) | |
|---|---|---|---|
| A series | |||
| K23A | 1.60 ± 0.22 | 2.20 ± 0.14 | 0.72 ± 0.07 |
| K26A | 2.00 ± 0.27 | 1.70 ± 0.71 | 1.18 ± 0.53 |
| K27A | 1.10 ± 0.19 | 1.90 ± 0.13 | 0.58 ± 0.05 |
| K31A | 1.20 ± 0.04 | 1.20 ± 0.15 | 1.00 ± 0.12 |
| R32A | 0.97 ± 0.03 | 1.40 ± 0.13 | 0.69 ± 0.07 |
| Scrambled | |||
| K3A3 | 2.56 ± 0.36 | 0.46 ± 0.03 | 5.57 ± 0.78 |
| K series | |||
| E29K | 26.5 ± 16.0 | 1.92 ± 0.11 | 13.80 ± 1.80 |
| A30K | 12.8 ± 0.63 | 3.76 ± 0.10 | 3.40 ± 0.23 |
| kon (×103m−1s−1) | koff (×10−6s−1) | KA (×109m−1) | |
|---|---|---|---|
| T22K | 3.88 ± 0.15 | 2.21 ± 0.10 | 1.75 ± 0.50 |
| A25K | 2.55 ± 0.10 | 3.40 ± 0.12 | 0.75 ± 0.20 |
| kon (×103m−1s−1) | koff (×10−7s−1) | KA (×1011m−1) | |
|---|---|---|---|
| T22K/A25K | 26.7 ± 2.3 | 1.39 ± 0.01 | 1.90 ± 0.40 |
“KKPE” Segment Is Required for HP Binding
We proposed that the ordering of the amino acids in the HBS might be critical for specific HP binding. To test this hypothesis, we created an sA27-aa mutant, denoted K3A3, in which the HP-binding sequence was scrambled. In this mutant, the first eight amino acids (SKAAKAAKEAKR), including the three basic residues Lys22, Lys25, and Lys28, were displaced compared with the original HBS sequence (STKAAKKPEAKR), although the remaining residues were unchanged. In the scrambled mutant, the original KKPE-binding motif in the central region was replaced with “AAKE,” but the total number of charged residues remained the same (see Fig. 1). If sA27-aa/HP binding were nonspecific, the resulting binding affinity would be sensitive to the number of charged residues accessible in HBS, rather than the HBS sequence itself, and one would expect that K3A3 would have an HP binding affinity comparable with that of the parent sA27-aa. On the other hand, if the sA27-aa/HP interaction were specific, then binding would be dependent not only on the total number of charged residues but also on the sequence. In fact, K3A3 showed a rather weak HP binding affinity (KA = 5.57 × 106 m−1) (Table 2), 2 orders of magnitude less than that of the parent molecule, suggesting that the latter is the case. Although the binding association rate constant kon was similar to that for specific HP binding, the dissociation rate constant koff increased substantially, similar to those for nonspecific GAG interactions (see Table 1). Based on these observations, we believe that the order of the HBS amino acids is critical for the HP binding affinity and that the KKPE segment plays an important role in determining binding specificity.
NMR Analysis of the KKPE-binding Motif
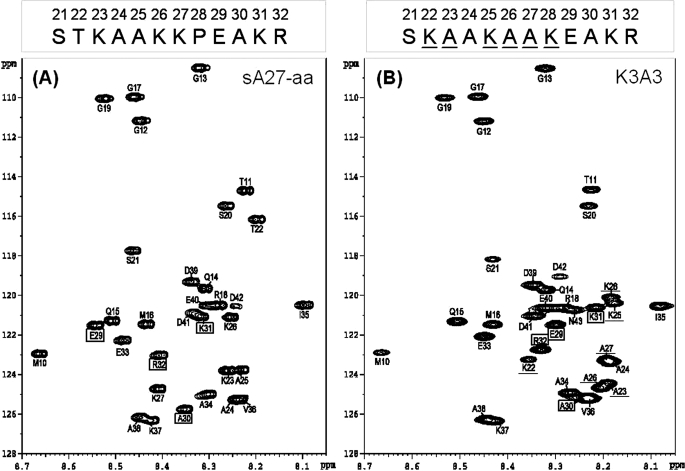
The 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum provides a reliable measure of self-assembly of sA27-aa (21). In the native state of the protein, the HSQC spectrum displays only the cross-peaks arising from the flexible residues Ser21–Asp42, and the residues that are involved in self-assembly are silent due to rapid relaxation (21). To determine possible structural requirements for HP binding specificity, we recorded 1H/15N HSQC spectra for the parent sA27-aa and K3A3 (Fig. 3, A and B). Comparison showed distinct chemical shift changes for a few residues, such as Glu29, Ala30, Lys31, and Arg32, although for all others (for example, Glu33, Ala34, and Ile35), chemical shifts were basically unchanged. The three-dimensional molecular structure of soluble proteins can be determined by NMR spectroscopy using 1H-1H NOE measurements; however, mainly due to protein self-assembly, in sA27-aa NOEs were too weak to be analyzed. Instead, we assessed the residual secondary structural information from the 1HN, 13Cα, and 13CO chemical shifts. By comparing these chemical shifts to the values reported for a random coil peptide (34), a structural propensity index was calculated using (δ-δrandom)/(δβ-δrandom), where δ is the experimental chemical shift and δβ and δrandom the chemical shift values reported in the literature for a β-turn or a random coil, respectively (34). A residue is considered likely to form a β-motif if the 1HN propensity index falls between 0.8 and 1 and to be a random coil if it is less than 0.2, whereas for indices between 0.3 and 0.8, the relative tendency to form a β-motif is proportional to the propensity index. Similarly, a residue is considered likely to form a β-motif if the 13Cα or 13CO propensity index falls between −0.8 and −1.0 and to be random coil if it is less than −0.2. This analysis is particularly useful for extracting secondary structural information for partially structured domains. In the HBS of sA27-aa (Fig. 3A), most of the chemical shift propensity indices were less than 0.5, with only a few residues in the region of Lys27-Pro28-Glu29-Ala30, having their 1HN indices between 0.5 and 0.8 and some 13Cα/13CO indices outside the range of −0.5 to +0.5 (Fig. 3, C–E), indicating a turn-like character in the secondary structure. In contrast, no secondary structure was indicated in the same region of K3A3.
FIGURE 3.
NMR structural analyses of sA27-aa and K3A3. A, two-dimensional 1H-15N HSQC NMR spectra of the parent sA27-aa. B, HSQC spectrum of the scrambled HBS sequence mutant K3A3 at pH 5.0. The respective GAG-binding sequences are shown above the spectrum with the mutated residues underlined. Due to protein self-assembly, only the residues of the flexible HBS were observable (for details, see the text). The respective sequential assignments were achieved on the basis of three-dimensional HNCA and HN(CO)CA NMR experiments (43), and the inter- and intra-residue chemical shift correlations were determined (supplemental Table S2). Residues are boxed if a substantial chemical shift difference was found in the spectra. Single letter abbreviations for amino acids are used. The numbering is based on the sequence of wild type A27. C, 1HN; D, 13Cα; and E, 13CO chemical shift propensity index analysis of the GAG-binding sequences of sA27-aa and K3A3. The chemical shift index was determined as (δ-δrandom)/(δβ-δrandom), where δ is the experimental chemical shift and δβ and δrandom the chemical shift values reported in the literature for a β-structure and random coil, respectively. In the KKPE-binding motif of sA27-aa, the propensity index is around +0.5, suggesting a β-turn conformation, whereas in K3A3, no such structure is seen. The sequence numbering for sA27-aa (single letter code) is shown on the horizontal axis. F, two-dimensional 1H-15N HSQC NMR spectra of sA27-aa interacting with HP, respectively. Spectrum of 0.6 mm [15N]sA27-aa protein without HP (red) is overlaid with the spectrum of the protein after the addition of HP (blue). In sA27-aa, for residues such as Ala25, Lys26, Lys27, Glu29, and Ala30, chemical shift perturbations are greater than 0.01 ppm, suggesting that these residues are involved in specific HP binding. For other residues, namely Ser21, Glu33, Ala34, chemical shifts were unperturbed, suggesting that these are not involved in the HP binding. Insets showed expanded regions for some of these residues. G, two-dimensional 1H-15N HSQC NMR spectra of K3A3 interacting with HP, respectively. Spectrum of 0.6 mm [15N]K3A3 protein without HP (red) is overlaid with the spectrum of the protein after the addition of HP (blue). Weight ratios of protein to HP are 1:0 (red), and 1:5 (blue).
To further characterize the local structure of HBS, we recorded NMR spectra for a synthetic 14-mer peptide, STKAAKKPEAKREA, a copy of the HBS sequence. Resonance assignments for this HBS domain were made from inspection of the homonuclear two-dimensional NOESY and TOCSY spectra collected in H2O/D2O (90:10) at 298 K (supplemental Figs. S1 and S2). The 1HN/1Hα cross-peaks in the two-dimensional chemical shift correlation spectra were clearly distinct, which made the sequential assignments straightforward (Fig. 4A and Table 3). In the NOESY spectrum, strong sequential (i, i + 1) inter-residue NOE cross-peaks were seen between 1Hδ of Pro28 and 1Hγ of Lys26 and between 1Hδ of Pro28 and 1Hβ of Lys27. Interestingly, (i, i + 2) NOE signals were also detected between 1Hγ of Pro28 and 1Hα of Ala30 and between 1Hδ of Pro28 and 1Hγ of Lys26 (Fig. 4B). Together with the NOE signal determined from the 14-mer HBS peptide and the β-turn conformation resolved from the 1HN, 13Cα, and 13CO chemical shift propensity analyses (Fig. 3, C–E), we interpret these data as suggesting that Lys26-Lys27-Pro28-Glu29-Ala30 adopts a characteristic turn-like backbone conformation. Because both the 1HN and the 1Hα chemical shifts derived from the 14-mer HBS peptide are consistent with those observed for the parental sA27-aa (see Table 3), this indicated that the molecular conformations in the center part of HBS arising from the two molecules are closely related.
FIGURE 4.
Chemical shift resonance assignments and structural analysis of the 14-mer peptide (STKAAKKPEAKREA), by two-dimensional TOCSY and NOESY spectroscopy. A, 1H chemical shift assignments were successfully determined using the inter- and intra-residual correlations in TOCSY and NOESY spectra. All 1H chemical shifts are tabulated in Table 3. B, inter-residual NOE signals were found between 1Hδ of Pro28 and 1Hγ of Lys26, 1Hδ of Pro28 and 1Hβ of Lys27, 1Hγ of Pro28 and 1Hα of Lys26, and 1Hδ of Pro28 and 1Hα of Lys27.
TABLE 3.
1H chemical shift assignments of synthesized 14-mer, a copy of HBS sequence STKAAKKPEAKREA, in the absence of HP
For comparison, the 1HN and 1Hα chemical shifts that extracted from recombinant sA27-aa protein are given in parentheses.
| Residue | HN | Hα | Hβ | Others |
|---|---|---|---|---|
| Thr22 | 8.666 (8.190) | 4.408 (4.374) | 4.204 | Hγ 1.228 |
| Lys23 | 8.445 (8.262) | 4.309 (4.336) | 1.816, 1.750 | Hγ 1.430, 1.430 |
| Hδ 1.690, 1.690 | ||||
| Hϵ 2.997, 2.997 | ||||
| Hζ 7.540 | ||||
| Ala24 | 8.334 (8.234) | 4.270 (4.306) | 1.356 | |
| Ala25 | 8.280 (8.219) | 4.280 (4.296) | 1.375 | |
| Lys26 | 8.279 (8.253) | 4.298 (4.325) | 1.779, 1.742 | Hγ 1.443, 1.443 |
| Hδ 1.690, 1.690 | ||||
| Hϵ 2.997, 2.997 | ||||
| Hζ 7.540 | ||||
| Lys27 | 8.413 (8.404) | 4.573 (4.608) | 1.816, 1.740 | Hγ 1.466, 1.483 |
| Hδ 1.706, 1.723 | ||||
| Hϵ 3.011 | ||||
| Hζ 7.540 | ||||
| Pro28 | 4.399 (4.436) | 2.296, 2.019 | Hγ 1.889 | |
| Hδ 3.837, 3.654 | ||||
| Glu29 | 8.530 (8.546) | 4.242 (4.276) | 2.018, 1.944 | Hγ 2.349, 2.331 |
| Ala30 | 8.357 (8.351) | 4.299 (4.325) | 1.374 | |
| Lys31 | 8.314 (8.311) | 4.280 (4.318) | 1.800, 1.760 | Hγ 1.412, 1.412 |
| Hδ 1.685, 1.685 | ||||
| Hϵ 2.992, 3.008 | ||||
| Hζ 7.525 | ||||
| Arg32 | 8.417 (8.404) | 4.334 (4.354) | 1.834, 1.779 | Hγ 1.632, 1.632 |
| Hδ 3.213, 3.213 | ||||
| Hϵ 7.223 | ||||
| Hη 6.670 | ||||
| Glu33 | 8.477 (8.480) | 4.316 (4.306) | 2.073, 1.928 | Hγ 2.349, 2.330 |
| Ala34 | 8.036 (8.318) | 4.113 (4.346) | 1.338 |
Identification of Specific HP Binding by 1H-15N HSQC
In addition to analyzing protein self-assembly, HSQC spectroscopy provides a simple and powerful method for identifying specific protein/GAG interactions. Presuming the sA27-aa/HP interaction to be specific, the chemical shifts of HBS residues involved in specific binding should be perturbed, whereas those of HBS residues not involved in specific binding should not be perturbed. Comparison of the 1H-15N HSQC spectrum of sA27-aa recorded in the presence of HP (at a protein to HP weight ratio of 1:5) with that recorded in the absence of HP showed that HBS residues were perturbed to different degrees. In particular, for residues in the “AKKPEA” segment critical for HP binding, the chemical shifts were perturbed the most (>0.01 ppm). Among these, only Glu29 resonances shifted downfield in 1HN dimension, although all others shifted upfield in both dimensions (see Fig. 3F). For other residues, such as Lys23, Ala24, Lys31, and Arg32, the chemical shifts were less perturbed, although for the remaining residues (namely Ser21, Glu33, and Ala34), chemical shifts were basically unperturbed, suggesting that these residues were not involved in HP binding. An increase in the weight ratio to 1:10 resulted in a higher degree of chemical shift perturbation in the AKKPEA segment, following the same trend as seen in Fig. 3F, but much less for other residues. Under these conditions, however, the overall intensity of signals in the spectrum dropped substantially due to sample precipitation (data not shown). In the case of K3A3, the resonances of all HBS residues were unperturbed in the presence of HP (at a protein to HP weight ratio of 1:5), see Fig. 3G. In summary, we have determined that the AKKPEA sequence in the central part of HBS is responsible for the specific binding of sA27-aa to HP.
T22K/A25K Mutant Binds Tightly to HP
We postulated that the turn-like structure of the “KKPEA” segment was required for HP binding. To check this, we constructed Lys mutants, denoted the K series, to determine the importance of the KKPEA segment. The K series consisted of five mutants, four single and one double Lys mutants (T22K, A25K, E29K, A30K, and T22K/A25K). T22K, A25K, and T22K/A25K were treated as positive controls, in which Thr22 and/or Ala25 was substituted with Lys (see Fig. 1). By adding more basic residue(s) while leaving the KKPEA-binding motif intact, we expected to achieve a higher GAG binding affinity without loss of GAG binding specificity. E29K and A30K were treated as negative controls. Using SPR (Fig. 2C), as expected, the KA values of the positive controls for HP were high, in the range of 109–1011 m−1 (Table 2). Of these mutants, the double mutant T22K/A25K had the highest HP binding affinity (KA = 1.9 × 1011 m−1). The two negative controls did not show high binding affinity (see Table 2).
To determine whether T22K/A25K showed tight HP binding at the expense of specificity, we compared its affinity for CS to that of sA27-aa and D8 protein and found a low binding affinity (KA = 6.33 × 105 m−1) in the same range as that for nonspecific GAG binding (Table 1). Because the 1H-15N HSQC spectrum of the double mutant T22K/A25K was very similar to that of sA27-aa (supplemental Fig. S3), we believe that the central region of the HBS of the double mutant T22K/A25K adopts a turn-like conformation similar to that seen in the parent molecule.
T22K/A25K Retains Binding Specificity for HS on Cell Surface
To further examine HP binding specificity in vivo, we performed an inhibition assay and two cell binding assays using two types of cell.
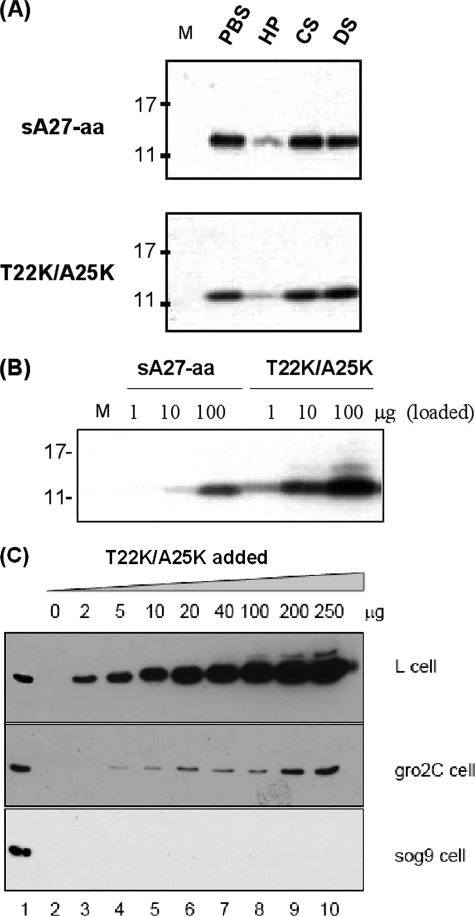
In the competitive inhibition assay, sA27-aa and T22K/A25K were incubated with HP, CS, or DS, and then the protein/GAG mixture was added to cultured HeLa cells to see if it bound to the cell surface-bound protein by Western blot analysis. As shown in Fig. 5A, this competition assay clearly demonstrated that HP, but not CS or DS, inhibited the binding of both sA27-aa and T22K/A25K to the cells, showing that the binding of the T22K/A25K mutant was specific for HP.
FIGURE 5.
Cell binding assays. sA27-aa and the double mutant T22K/A25K (10 μg) were added to cells, and bound protein was measured by Western blot analysis. A, competitive binding assay. HP, CS, or DS (all 100 μg/ml) was used to compete with the protein for binding to the HeLa cell surface. Lane M, protein standard markers. B, direct binding of different amounts of sA27-aa and T22K/A25K to HeLa cells. Phosphate-buffered saline (PBS) was used as a negative control. C, binding of different amounts of T22K/A25K to L, gro2C, and sog9 cells. The GAG expression phenotypes for the three cells are as follows: L (HS+/CS+), gro2C (HS−/CS+), and sog9 (HS−/CS−).
In the first direct cell binding experiment, different amounts (1, 10, or 100 μg) of sA27-aa and T22K/A25K were added to cultured HeLa cells, and bound protein was measured by Western blotting. As shown in Fig. 5B, the sA27-aa control bound to the cells, consistent with previous findings (18). However, under the same conditions, much more T22K/A25K was bound, reflecting the higher binding affinity of T22K/A25K determined in the SPR analyses (Table 2). To confirm the binding specificity, we carried out additional cell binding assays using different amounts (0–250 μg) of T22K/A25K and three different cell lines as follows: gro2C cells that lack mature HS; sog9 cells that lack both HS and CS (30, 35); and L cells with both HS and CS on the cell surface. As shown in Fig. 5C, on increasing the amount of T22K/A25K, a significant amount of T22K/A25K bound to L cells. In contrast, the amount bound to gro2C cells was very low. This substantial reduction in the amount of bound protein was interpreted as resulting from the lack of mature HS on the gro2C cell surface, the small amount of bound protein being possibly due to nonspecific binding to molecules other than HP, such as CS. This observation is consistent with the nonspecific weak binding detected by SPR. Using sog9 cells that lack both HS and CS, no T22K/A25K was bound. Taking these results together with the in vitro SPR measurements, we conclude that the T22K/A25K double mutant has a high HP binding affinity without any loss of HP binding specificity.
Computer Modeling Analysis of HBS
To resolve the local molecular structure of the HBS, we carried out a computer modeling analysis of a 12-mer HBS peptide in the absence and presence of HS using Discovery Studio software. As shown in Fig. 6A, in the absence of HS, the HBS backbone was seen as an elongated, extended segment. Notably, a turn-like local conformation was observed in the KKPE segment. Because of this turn-like conformation, the side chain of acidic residue Glu29 sits between the side chains of Lys26 and Lys31 in a co-planar parallel manner. Similarly, the side chain of Ala30 sits between the side chains of Lys27 and Arg32. In fact, the side chains of these basic residues are well organized, forming a “branch” pattern on the same plane that allows interaction with HS. It can be seen that the positively charged groups of Lys/Arg side chains are accessible to sulfate groups on HS molecules, allowing multiple ionic interactions. In the presence of HS, the cooperative HBS/HS binding structure is demonstrated by the docking of an α-helical type HS molecule, exemplified by an HS tetrasaccharide (32) as follows: unsaturated 2-O-sulfate uronic acid (ΔUA2S), 2-N-sulfo, 6-O-sulfate glucosamine (GlcNS6S), 2-O-sulfate iduronic acid (IdoA2S), and 2-N-sulfo, 6-O-sulfate glucosamine (GlcNS6S) (ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S). The HBS·HS complex revealed a perfect match between the α-helical HS and the branched side chain pattern (Fig. 6B). On simulating more accurately the binding event by introducing solvent molecules in the calculation, the complex structure resulted in tighter binding through a series of hydrogen bonds, in particular those of Lys26, Lys27, Lys31, and Arg32 (Fig. 6C). These side chains behave as buckles that reinforce the ionic interaction with HS, suggesting a slight modification of the side chain conformations upon HS binding that is responsible for the tight binding in the specific interaction of A27 with HS (Fig. 6C). This phenomenon explains the slow association and even slower dissociation phenomena observed by SPR. As mentioned above, both Glu29 and Ala30 play pivotal roles in recruiting the side chains of Lys26/Lys31 and Lys27/Arg32. As indicated by the SPR data (Table 2), the HP binding affinities for mutants E29K and A30K were 2 orders of magnitude lower than those of other mutants in the K series. Interestingly, although the association rate constants kon increased 7–15-fold, the dissociation rate constants koff decreased by at least 500-fold. We interpret these changes in kinetic binding constants as being primarily due to a disruption of the local turn-like conformation needed for HS binding. Molecular dynamics simulations of nonspecific GAG binding are in progress in our laboratory to obtain more detailed information. An important point is that the extent of ionic interaction and hydrogen bonding formation in the HBS·CS complex was much lower than that in the HBS·HS complex. We therefore believe that the turn-like conformation provided by the KKPE segment allows a specific binding of vaccinia viral envelope protein A27 to HS, despite the random coil secondary structure of the HBS.
FIGURE 6.
Dynamic simulation modeling of HBS in vaccinia viral protein sA27-aa. The backbone of HBS is shown as a blue solid ribbon in which the KKPE segment forms a turn-like conformation. To distinguish the positively charged residues, Lys26, Lys27, Lys31, and Arg32 are colored green. A, molecular dynamics simulation of the HBS in the absence of HS. The HBS molecule is shown using ball and stick representation (white, hydrogen atom; red, oxygen atom; blue, nitrogen atom; gray, carbon atom). For clarity, Lys26, Lys27, Glu29, Ala30, Lys31, and Arg32 residues are highlighted in yellow. B, docking of HS with the HBS without any solvent molecules. A tetrasaccharide, unsaturated 2-O-sulfate uronic acid (ΔUA2S), 2-N-sulfo-,6-O-sulfate glucosamine (GlcNS6S), 2-O-sulfate iduronic acid (IdoA2S), and 2-N-sulfo-, 6-O-sulfate glucosamine (GlcNS6S) (ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S), was used as a GAG-binding substrate; its x-ray structure has been determined elsewhere (32). The HS molecule is shown in stick form wrapped around the HBS, revealing close multiple ionic interactions. C, molecular dynamics simulation of HBS in the presence of HS, including solvent molecules. The structures of both the HBS and HS show slight modification.
DISCUSSION
Viruses make use of diverse GAG molecules on the cell surface in determining host restriction and tropism. Dengue virus envelope protein has sequentially distant basic residues that are closely packed into a basic patch, forming a long cleft, capable of interacting with HP (36). In the case of VV, the envelope protein D8 interacts specifically with CS (18), although others, such as A27 (or sA27-aa) and H3, bind to HS (17), facilitating cell surface attachment and viral entry. Secondary structure analysis of the HBS backbone structure of sA27-aa by solution-state NMR revealed an unstructured flexible random coil (23). In our previous study (21), we proposed a molecular model that illustrates the cooperative structural and functional relationship of two distinct domains, a flexible, unstructured, extended coil and a rather rigid α-helical, coiled-coil domain. However, the specific mechanism by which A27 binds to HBS remained unclear until now. In this study, we used a combination of site-directed mutagenesis, NMR, and SPR techniques to elucidate the specific binding mechanism of sA27-aa. We first measured the binding affinities for specific protein/GAG interactions (such as sA27-aa/HP and D8L/CS) and nonspecific protein/GAG interactions using SPR. The specific and nonspecific GAG binding affinities differed by approximately 3 orders of magnitude. In addition, the specific interactions showed a characteristic slow association and even slower dissociation binding behavior, suggesting that the local molecular structure might be critical for specific binding. Unfortunately, due to protein self-assembly, neither NMR nor x-ray structures are available. In the absence of structural information, we expressed two series of HBS mutants and a scrambled HBS sequence mutant, K3A3, in an attempt to identify local structure required for specific HP binding. The mutant K3A3 showed loss of HP binding affinity and specificity. Using solution-state NMR spectroscopy, we identified a turn-like twisted KKPE segment as being critical for specific binding. The K series mutants, in which Lys residue(s) were added to HBS whereas the KKPE segment remained intact, were found to have an extremely high binding affinity for HP. Furthermore, as demonstrated by in vitro SPR assays and in vivo cell adhesion and competition binding assays, one of the K series mutants, T22K/A25K, had an affinity of 1.9 × 1011 m−1 and still retained the binding specificity for HP and HS on the cell surface. Taking these results together, we therefore conclude that the turn-like structure provided by the KKPE segment is required in the vaccinia viral envelope protein A27 for specific HP binding.
Cardin and Weintraub (37) examined the amino acid sequences of HP-binding proteins and reported that certain amino acids could fold into either α-helices or β-sheets, generating a linear array of cations that facilitates the interaction with anions on a sulfated polysaccharide (38). In addition, the HP-binding proteins were found to contain two binding consensus sequences, namely XBBXBX and XBBBXXBX, in which “B” is a basic amino acid (Arg, Lys, or His) and “X” any type of amino acid. In both consensus sequences, the basic residue(s) are sandwiched between noncharged residues. It should be noted that the double Lys mutant T22K/A25K reported here contained an HP-binding motif (XBBXBBBXXXBB), which is a superimposition of the two binding consensuses. Here, we interpret the extremely high binding affinity for HP of the double mutant T22K/A25K as being due to the consecutive HP-binding motifs. Although HP binding affinity is in principle dependent on the positively charged residues within the HBS, the specificity of T22K/A25K for HP was primarily due to the KKPE peptide segment present in the HBS. Only with the KKPE segment intact can we generate an sA27-aa mutant that has both extremely high HP binding affinity as well as HP binding specificity. Analogous HP-binding consensus sequences “DPKR” and/or “KKDPE” are found in several HP-binding proteins, such as fibroblast growth factor (39, 40), purpurin (22), and retinol-binding protein (41, 42). Given the sequence homology, we postulate that an “APBB” or “BBAPA” motif, in which “P” stands for a Pro residue and “A” for one of the acidic amino acids (Asp or Glu), might regulate HP binding specificity.
The binding affinity of a synthesized 14-mer HBS peptide to HP was considered to be weak with a low binding constant (KD = 0.35 ± 0.10 mm) determined from chemical shift perturbations in HP NMR titration experiments (supplemental Fig. S4). Chemical shifts assigned for the synthesized HBS 14-mer as well as the HBS in recombinant sA27-aa showed small perturbations in the presence of HP. We ascribed these small perturbations to the fact that the mechanism of HP binding was enthalpy-driven, inducing no substantial but small change in local conformation. Indeed, in our previous thermodynamic studies using isothermal titration calorimetry (21), we determined thermodynamic parameters for the interaction of sA27-aa with HP and found a negative enthalpic term of −2150 cal mol−1 and a small positive entropic term of 15.4 cal K−1 mol−1, indicating that sA27-aa/HBS binding to HP is enthalpy-driven. This is consistent with the formation of energetically favorable electrostatic interactions between the positively charged side chains of HBS and the negatively charged sulfate groups of HP. Similar thermodynamic phenomena were found in other HP-binding proteins, such as human immunodeficiency virus, type 1, TAT protein and fibroblast growth factor (26, 27). In sA27-aa, the small increase in entropy suggested the local conformation in HBS was only slightly changed (discussed below). This may explain why the chemical shift perturbations observed by NMR were rather small.
The structural information from our molecular modeling of HBS provides the basis for designing highly potent inhibitors of viral attachment to the cell surface. As indicated (Fig. 6), the arrangement of side chains restricts the size and the shape of the substrate such that only an α-helical structure of a particular dimension is able to fit. A tetrasaccharide, ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S, can fit well into this binding pocket forming strong specific cooperative ionic interactions. A number of residues in the HBS are found within hydrogen bonding distance of the carboxyl groups in the tetrasaccharide molecule, in particular, between 1Hζ of Lys26 and O-1 of 4GlcNS6S, 1HN of Ala30 and O-6 of 3IdoA2S, 1HN of Lys31 and O-6 of 3IdoA2S, and 1Hζ s of Arg32 and O-6 of 1ΔUA2S. These hydrogen bonds reinforce the molecular interactions between HBS and HS. We speculate that HS binding induces a mild side chain rearrangement in the HBS of vaccinia viral protein A27. To determine whether HS binding would induce a conformational change in sA27-aa, we performed chemical shift analysis of a mixture of a 14-mer HBS peptide and HP. Most chemical shifts were basically unchanged, the 1H chemical shifts differing by 0.05 ppm or a little more for only a few resonances, i.e. the 1HN resonances of Ala25, Lys26, Lys27, and Ala30. The upfield shifts might imply that the secondary structure was altered slightly due to the presence of HS. Of these residues, Lys27 showed the most significant change (see Table 3 and Table 4), in agreement with the chemical shift perturbation data observed by HSQC spectra (see Fig. 3F). In addition, there were slight modifications in the arrangement of the side chains of Lys and Arg residues. As indicated by the HBS·HS complex model, a slight distortion was observed in the Lys27 side chain, causing it to approach that of Glu29, and as a result of a slight backbone distortion, Lys26 and Lys31 cooperatively form a “molecular click” that locks in the HS molecule. The interplay of Glu29, Lys26, and Lys31 is critical for specific HS binding. This might explain why E29K had an even higher association rate constant than T22K and A25K and yet a fast dissociation rate constant comparable with nonspecific binding (see Tables 1 and 2).
TABLE 4.
1H chemical shift assignments of synthesized 14-mer, a copy of HBS sequence STKAAKKPEAKREA, in the presence of HP
| Residue | HN | 1Hα | Hβ | Others |
|---|---|---|---|---|
| Thr22 | 8.635 | 4.420 | 4.239 | Hγ 1.228 |
| Lys23 | 8.382 | 4.291 | 1.845, 1.773 | Hγ 1.456, 1.448 |
| Hδ 1.701, 1.701 | ||||
| Hϵ 3.022, 3.022 | ||||
| Hζ 7.494 | ||||
| Ala24 | 8.292 | 4.271 | 1.384 | |
| Ala25 | 8.190 | 4.280 | 1.384 | |
| Lys26 | 8.197 | 4.280 | 1.813, 1.753 | Hγ 1.466, 1.466 |
| Hδ 1.701, 1.701 | ||||
| Hϵ 3.012, 3.012 | ||||
| Hζ 7.494 | ||||
| Lys27 | 8.297 | 4.583 | 1.824, 1.793 | Hγ 1.488, 1.488 |
| Hδ 1.713, 1.713 | ||||
| Hϵ 3.022, 3.022 | ||||
| Hζ 7.494 | ||||
| Pro28 | 4.402 | 2.303, 1.965 | Hγ 2.038 | |
| Hδ 3.832, 3.656 | ||||
| Glu29 | 8.488 | 4.258 | 2.050, 1.956 | Hγ 2.380, 2.380 |
| Ala30 | 8.303 | 4.291 | 1.394 | |
| Lys31 | 8.256 | 4.290 | 1.824, 1.766 | Hγ 1.436, 1.436 |
| Hδ 1.692, 1.692 | ||||
| Hϵ 3.022, 3.011 | ||||
| Hζ 7.494 | ||||
| Arg32 | 8.333 | 4.331 | 1.854, 1.782 | Hγ 1.641, 1.630 |
| Hδ 3.215, 3.215 | ||||
| Hϵ 7.207 | ||||
| Hη 6.670 | ||||
| Glu33 | 8.415 | 4.342 | 2.101, 1.947 | Hγ 2.376, 2.376 |
| Ala34 | 8.036 | 4.127 | 1.344 |
Acknowledgments
We thank Liane Wang (Institute of Chemistry, Academia Sinica) for preparation of the documents and Dr. Andrew Atkinson (Institute of Genetics and Molecular and Cellular Biology, France) for careful reading of the manuscript. The NMR spectra were obtained at the High Field Nuclear Magnetic Resonance Center which is supported by the National Research Program for Genomic Medicine.
This work was supported by National Science Council Grant 97-2113-M-001-020-MY3 and by the Academia Sinica.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, Figs. S1–S4, and additional text and references.
- VV
- vaccinia virus
- HS
- heparan sulfate
- HP
- heparin
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- GAG
- glycosaminoglycan
- HBS
- heparin-binding site
- SPR
- surface plasmon resonance
- HSQC
- heteronuclear single quantum coherence
- NOE
- nuclear Overhauser enhancement
- NOESY
- nuclear Overhauser enhancement spectroscopy
- TOCSY
- total correlated spectroscopy
- ΔUA2S
- unsaturated 2-O-sulfate uronic acid
- GlcNS6S
- 2-N-sulfo, 6-O-sulfate glucosamine
- IdoA2S
- 2-O-sulfate iduronic acid
- sA27aa
- soluble form of A27
- SA
- streptavidin.
REFERENCES
- 1.Goebel S. J., Johnson G. P., Perkus M. E., Davis S. W., Winslow J. P., Paoletti E. (1990) Virology 179, 247–266, 517–563 [DOI] [PubMed] [Google Scholar]
- 2.Moss B. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., pp. 2905–2946, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Condit R. C., Moussatche N., Traktman P. (2006) Adv. Virus Res. 66, 31–124 [DOI] [PubMed] [Google Scholar]
- 4.Ching Y. C., Chung C. S., Huang C. Y., Hsia Y., Tang Y. L., Chang W. (2009) J. Virol. 83, 6464–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izmailyan R. A., Huang C. Y., Mohammad S., Isaacs S. N., Chang W. (2006) J. Virol. 80, 8402–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senkevich T. G., Ojeda S., Townsley A., Nelson G. E., Moss B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols R. J., Stanitsa E., Unger B., Traktman P. (2008) J. Virol. 82, 10247–10261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson R. L., Busch S. J., Cardin A. D. (1991) Physiol. Rev. 71, 481–539 [DOI] [PubMed] [Google Scholar]
- 9.Lindahl U., Lidholt K., Spillmann D., Kjellén L. (1994) Thromb. Res. 75, 1–32 [DOI] [PubMed] [Google Scholar]
- 10.Kjellén L., Lindahl U. (1991) Annu. Rev. Biochem. 60, 443–475 [DOI] [PubMed] [Google Scholar]
- 11.Perrimon N., Bernfield M. (2000) Nature 404, 725–728 [DOI] [PubMed] [Google Scholar]
- 12.Salmivirta M., Lidholt K., Lindahl U. (1996) FASEB J. 10, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 13.Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 14.Liu J., Thorp S. C. (2002) Med. Res. Rev. 22, 1–25 [DOI] [PubMed] [Google Scholar]
- 15.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U. (2002) Nat. Rev. Cancer 2, 521–528 [DOI] [PubMed] [Google Scholar]
- 16.Capila I., Linhardt R. J. (2002) Angew. Chem. Int. Ed. Engl. 41, 391–412 [DOI] [PubMed] [Google Scholar]
- 17.Lin C. L., Chung C. S., Heine H. G., Chang W. (2000) J. Virol. 74, 3353–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung C. S., Hsiao J. C., Chang Y. S., Chang W. (1998) J. Virol. 72, 1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi T., Oie M., Ichihashi Y. (1994) Virology 202, 844–852 [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez D., Rodriguez J. R., Esteban M. (1993) J. Virol. 67, 3435–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho Y., Hsiao J. C., Yang M. H., Chung C. S., Peng Y. C., Lin T. H., Chang W., Tzou D. L. (2005) J. Mol. Biol. 349, 1060–1071 [DOI] [PubMed] [Google Scholar]
- 22.Berman P., Gray P., Chen E., Keyser K., Ehrlich D., Karten H., LaCorbiere M., Esch F., Schubert D. (1987) Cell 51, 135–142 [DOI] [PubMed] [Google Scholar]
- 23.Lin T. H., Chia C. M., Hsiao J. C., Chang W., Ku C. C., Hung S. C., Tzou D. L. (2002) J. Biol. Chem. 277, 20949–20959 [DOI] [PubMed] [Google Scholar]
- 24.Osmond R. I., Kett W. C., Skett S. E., Coombe D. R. (2002) Anal. Biochem. 310, 199–207 [DOI] [PubMed] [Google Scholar]
- 25.Cain S. A., Baldock C., Gallagher J., Morgan A., Bax D. V., Weiss A. S., Shuttleworth C. A., Kielty C. M. (2005) J. Biol. Chem. 280, 30526–30537 [DOI] [PubMed] [Google Scholar]
- 26.Hakansson S., Caffrey M. (2003) Biochemistry 42, 8999–9006 [DOI] [PubMed] [Google Scholar]
- 27.Pantoliano M. W., Horlick R. A., Springer B. A., Van Dyk D. E., Tobery T., Wetmore D. R., Lear J. D., Nahapetian A. T., Bradley J. D., Sisk W. P. (1994) Biochemistry 33, 10229–10248 [DOI] [PubMed] [Google Scholar]
- 28.Adamson A. W. (1960) Physical Chemistry of Surfaces, pp. 457–481, Interscience Publishers, Inc., New York [Google Scholar]
- 29.Hsiao J. C., Chung C. S., Chang W. (1998) J. Virol. 72, 8374–8379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruenheid S., Gatzke L., Meadows H., Tufaro F. (1993) J. Virol. 67, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banfield B. W., Leduc Y., Esford L., Schubert K., Tufaro F. (1995) J. Virol. 69, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon A. F., Edavettal S. C., Krahn J. M., Munoz E. M., Negishi M., Linhardt R. J., Liu J., Pedersen L. C. (2004) J. Biol. Chem. 279, 45185–45193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromm J. R., Hileman R. E., Caldwell E. E., Weiler J. M., Linhardt R. J. (1995) Arch. Biochem. Biophys. 323, 279–287 [DOI] [PubMed] [Google Scholar]
- 34.Wishart D. S., Sykes B. D. (1994) Methods Enzymol. 239, 363–392 [DOI] [PubMed] [Google Scholar]
- 35.Uyama T., Ishida M., Izumikawa T., Trybala E., Tufaro F., Bergström T., Sugahara K., Kitagawa H. (2006) J. Biol. Chem. 281, 38668–38674 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y., Maguire T., Hileman R. E., Fromm J. R., Esko J. D., Linhardt R. J., Marks R. M. (1997) Nat. Med. 3, 866–871 [DOI] [PubMed] [Google Scholar]
- 37.Cardin A. D., Weintraub H. J. (1989) Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 38.Margalit H., Fischer N., Ben-Sasson S. A. (1993) J. Biol. Chem. 268, 19228–19231 [PubMed] [Google Scholar]
- 39.Gospodarowicz D., Baird A., Cheng J., Lui G. M., Esch F., Bohlen P. (1986) Endocrinology 118, 82–90 [DOI] [PubMed] [Google Scholar]
- 40.Esch F., Baird A., Ling N., Ueno N., Hill F., Denoroy L., Klepper R., Gospodarowicz D., Böhlen P., Guillemin R. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6507–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newcomer M. E., Liljas A., Sundelin J., Rask L., Peterson P. A. (1984) J. Biol. Chem. 259, 5230–5231 [PubMed] [Google Scholar]
- 42.Newcomer M. E., Jones T. A., Aqvist J., Sundelin J., Eriksson U., Rask L., Peterson P. A. (1984) EMBO J. 3, 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kay L. E. (1997) Biochem. Cell Biol. 75, 1–15 [DOI] [PubMed] [Google Scholar]