Abstract
Brd4 is a member of the bromodomains and extra terminal domain (BET) family of proteins that recognize acetylated chromatin structures through their bromodomains and act as transcriptional activators. Brd4 functions as an associated factor and positive regulator of P-TEFb, a Cdk9-cyclin T heterodimer that stimulates transcriptional elongation by RNA polymerase II. Here, the crystal structures of the two bromodomains of Brd4 (BD1 and BD2) were determined at 1.5 and 1.2 Å resolution, respectively. Complex formation of BD1 with a histone H3 tail polypeptide encompassing residues 12–19 showed binding of the Nζ-acetylated lysine 14 to the conserved asparagine 140 of Brd4. In contrast, in BD2 the N-terminal linker sequence was found to interact with the binding site for acetylated lysines of the adjacent molecule to form continuous strings in the crystal lattice. This assembly shows for the first time a different binding ligand than acetylated lysine indicating that also other sequence compositions may be able to form similar interaction networks. Isothermal titration calorimetry revealed best binding of BD1 to H3 and of BD2 to H4 acetylated lysine sequences, suggesting alternating histone recognition specificities. Intriguingly, an acetylated lysine motif from cyclin T1 bound similarly well to BD2. Whereas the structure of Brd2 BD1 suggested its dimer formation, both Brd4 bromodomains appeared monomeric in solution as shown by size exclusion chromatography and mutational analyses.
Introduction
The transition from transcription initiation to productive transcription elongation depends on the phosphorylation of the C-terminal domain of the RNA polymerase II by the positive transcription elongation factor P-TEFb (1, 2). Active P-TEFb is composed of the kinase Cdk9 and its cyclin-partner cyclin T or K. In its inhibited form, P-TEFb is bound to a heterotrimeric complex formed by the small nuclear RNA 7SK and the proteins Hexim and Larp7 (3–5). This negatively regulated complex can be converted into the active form by the bromodomain (BD)3-containing protein 4 (Brd4) by a yet unknown mechanism (6–8). It is thought that Brd4 couples the P-TEFb complex to chromatin structures via binding of its bromodomains to acetylated lysines in the histone H3 and H4 tail sequences.
In eukaryotic cells, the DNA templates are condensed into chromatin structures that consist of repetitive units of nucleosomes. In each nucleosome, the DNA is wrapped around an octamer of core histones that consist of two H2A/H2B heterodimers and an H3/H4 tetramer (9). The N-terminal tail regions of histones H3 and H4 are flexible in the nucleosome and rich in lysine, arginine, and serine residues. They are thus accessible to enzymatic modifications such as acetylation, methylation, and phosphorylation (10). The variable patterns generated by these covalent modifications define the histone code, which represents a fundamental regulatory mechanism of gene expression and repression (11). Histone acetylation of lysines in H3 and H4 is mediated by acetyltransferases including Gcn5p, p300/CBP, P/CAF, and TAFII250. The selectivity of these enzymes depends on both their substrates and their environment within the cell. The acetylated histone tail sequences, in reverse, are recognized by the bromodomain, a conserved domain structure of ∼110 amino acids that was first identified in the Drosophila protein brahma (12, 13).
The bromodomain-containing protein 4 (Brd4) belongs to the BET (bromodomains and extra terminal domain) family of proteins (14). BET family proteins are defined by the presence of two bromodomains and an additional conserved ET domain (14). In vertebrates, four members of the BET family have been identified: Brd2 (15), Brd3 (16), Brd4 (initially named MCAP for mitotic chromosome-associated protein) (17), and BrdT (18) that each have distinct functions and specific domain compositions. Brd2 e.g. contains a kinase-like domain between the two bromodomains that exhibits mitogen-activated nuclear kinase activity and is involved in signal transduction (19). The bromodomains within the BET family members are thought to play an important role in the epigenetic memory of transcription and viral inheritance across cell division (20).
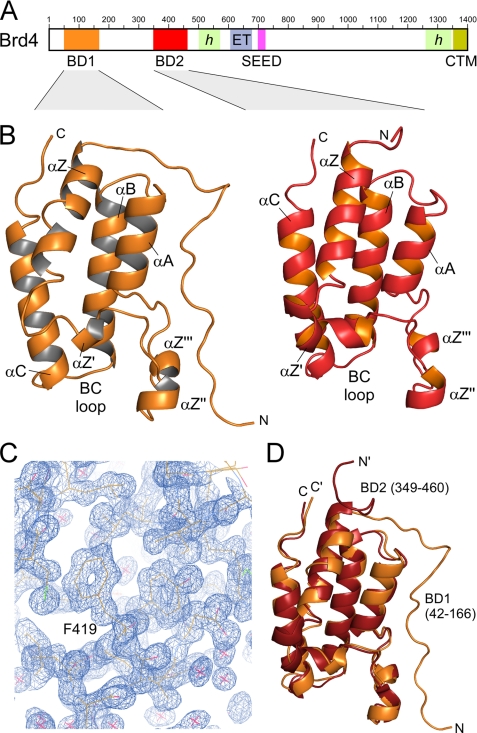
Brd4 is a ubiquitously expressed nuclear protein of 200 kDa that associates with mitotic chromosomes and is required for the regulation of cell growth (17, 21, 22). It interacts with acetylated chromatins because of its affinity for acetylated histones H3 and H4 and remains bound to chromatin during mitosis, suggesting a role in the transmission of epigenetic memory (23, 24). Moreover, dysregulation of Brd4-associated pathways was described to be involved in breast cancer progression, and its activation grade might serve as a prognostic signature (25). Brd4 contains two N-terminal bromodomains, designated as BD1 and BD2, followed by a central ET domain (26) whose function is yet unclear (Fig. 1A). A succeeding SEED motif (Ser/Glu/Asp-rich region) as well as a C-terminal motif (CTM) that was found to interact with the human papillomavirus E2 to silence the expression of human papillomavirus-encoded E6 and E7 oncoproteins (27), are likewise specific Brd4 components. To understand the molecular mechanisms of Brd4 in more detail, we determined the crystal structures of the two bromodomains BD1 (residues 42–168) and BD2 (349–464) of mouse Brd4 to a resolution of 1.5 Å and 1.2 Å, respectively. The complex structure of BD1 with a H3 tail peptide containing acetylated lysine K(ac)14 was determined displaying the mode of histone recognition. Intriguingly, the N-terminal linker sequence of BD2 interacts with the binding site for acetylated lysines to form continuous strings within the crystal showing for the first time variations in the recognition of bromodomain interaction motifs.
FIGURE 1.
Structure of the two bromodomains of Brd4. A, domain organization of Brd4. Two N-terminal bromodomains (BD1 and BD2) are followed by an extra terminal domain (ET) that together constitute the BET family proteins. SEED represents a serine-, glutamate-, and aspartate-rich region. CTM denotes a conserved C-terminal motif. Green-shaded areas indicate regions of proposed helical content. The sequence numbering corresponds to mouse Brd4. B, ribbon plot representation of the Brd4 BD1 (left panel) and BD2 (right panel) crystal structures encompassing residues Ser-42 to Thr-166 and Ser-349 to Asp-460. C, electron density map of Brd4 BD2 from diffraction data recorded to 1.2 Å resolution. Displayed is the omit map at 1.0 σ of a region displaying the αB-helix in the bromodomain. D, superimposition of Brd4 BD1 (orange) and BD2 (red) yield an r.m.s.d. value of 0.83 Å for the backbone heavy atoms.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Brd4 Bromodomains
Plasmids encoding the two bromodomains of mouse Brd4 (GenBankTM accession number NM_020508) were generated by PCR-mediated amplification with primer containing BamHI and EcoRI restriction sites at the 5′- and 3′-end, respectively. For the N-terminal bromodomain (BD1), five different construct sizes with domain boundaries 27, 42, 46, 51, or 54 to 168 were tested. For the C-terminal bromodomain (BD2) constructs 346–464 and 349–464 were generated. The Brd4 BD1 and BD2 constructs were cloned into the prokaryotic expression vector pProEx-HTb (Invitrogen) for protein expression and purification. Site-directed mutagenesis of residues R445A, D449A, and A456Q were performed in a single round of PCR by modified antisense oligonucleotide primer. All clones were confirmed by DNA sequencing prior to expression.
Expression plasmids encoding Brd4 BD1 or BD2 were transformed into Escherichia coli BL21(DE3) cells (Novagen), grown at 30 °C and induced at an A600 of 0.6 to 1.0 with 0.3 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h growth. Proteins were purified by Ni-nitrilotriacetic acid affinity chromatography and gel filtration similarly as described previously (28), including cleavage of the histine tag at 4 °C over 12 h with Tev protease. Resulting protein in gel filtration buffer (50 mm HEPES pH 7.5, 150 mm NaCl, 5 mm β-mercaptoethanol) were about 98% pure as analyzed by SDS-PAGE, concentrated, and stored at −80 °C. Protein concentrations were determined by using Bradford reagent.
Polypeptides of histone tail sequences H3 and H4 with acetylated lysine residues H3–14: GG-K(ac)-APRKQ, H4–5: RG-K(ac)-GGKGL, and H4–16: GA-K(ac)-RHRKV and cyclin T1 sequences T1–390: SL-K(ac)-EYRAK and T1–404: AQ-K(ac)-RQLEN were purchased from BIOSYNTAN, Berlin, with 95% purity (HPLC grade) and dissolved in water to 20 mg/ml.
Bromodomain Crystallization
Initial screening for crystallization conditions was carried out using a Mosquito robot (TTP Labtech) with the sitting-drop method at 293 K and a concentration of 5–15 mg/ml BD1 or BD2. For the initial screening, 0.1 μl of protein solution was mixed with 0.1 μl of reservoir solution from a 100-μl reservoir in 96-well Greiner crystallization plates. Crystals of BD2 could be obtained from a PEG screen (Hampton Research). Crystal conditions were optimized to 0.1 m Tris/HCl, pH 8.5 and 25% PEG 2000 MME in Linbro crystallization plates, using the hanging drop technique. Crystals grew under these conditions within 3 days to a size of 200 × 150 × 20 μm. Crystals of BD1 grew in 3.6 m sodium formate, 10% glycerol to a size of 100 × 50 × 20 μm. Crystals of both bromodomains were exposed to octamer histone tail peptides with acetylated lysines by transferring them into a solution containing the crystallization buffer with additional H3–14 or H4–5 peptide in 20 times molar access. For cryo-protection, crystals were transferred to a solution that contained the reservoir buffer with additionally 20% ethylene glycol. After 5–10 s, crystals were flash-cooled in liquid nitrogen.
Data Collection and Processing
Native data at cryogenic temperature of 100 K were collected to 1.2 Å resolution for Brd4 BD2 and to 1.5 Å for BD1 as well as for histone peptide exposed bromodomains on beamline X10SA (PXII) of the Swiss Light Source (SLS, Villigen, Switzerland) equipped with a MAR 225 CCD detector (oscillation width per frame, 1°; 120–180 frames collected). The XDS package (29) was used to process, integrate, and scale the collected data. Brd4 BD2 crystals belong to space group P21212 and have unit cell parameters of a = 52.1, b = 73.1, c = 32.3 Å. Assuming the presence of one molecule in the asymmetric unit, the solvent content of the crystals is 30.4%, corresponding to a Matthews coefficient of 1.78 Å3 Da−1. For BD1, the space group was determined to be P212121 with a = 34.0, b = 47.4, c = 78.0 Å. A solvent content of 40% corresponds to a Matthews coefficient of 2.04 Å3 Da−1. Data collection statistics and refinement parameters are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Brd4 BD1 | Brd4 BD1 with H3-K(ac)14 peptide GGK(ac)APRKQ | Brd4 BD2 | Brd4 BD2 with H3-K(ac)14 peptide GGK(ac)APRKQ | |
|---|---|---|---|---|
| Data collection | ||||
| Beam line | SLS X10SA | SLS X10SA | SLS X10SA | SLS X10SA |
| Wavelength [Å] | 1.00 | 1.00 | 0.97642 | 0.97886 |
| Space group | P212121 | P212121 | P21212 | P21212 |
| Unit cell a, b, c [Å] α, β, γ [°] | a = 34.01, b = 47.44, c = 78.02, α = β = γ = 90° | a = 36.13, b = 47.03, c = 77.25, α = β = γ = 90° | a = 52.06, b = 73.05, c = 32.30, α = β = γ = 90° | a = 52.10, b = 73.15, c = 32.16, α = β = γ = 90° |
| Resolution range [Å]a | 19.6–1.55 (1.6–1.55) | 19.7–1.80 (1.9–1.8) | 19.5–1.20 (1.25–1.2) | 19.5–1.20 (1.25–1.2) |
| Total observations | 146 605 (13 213) | 97 615 (14 698) | 195 195 (20 535) | 365 947 (40 606) |
| Unique reflections | 18 865 (1 668) | 12 652 (1857) | 39 272 (4460) | 37 336 (4 265) |
| Rmergeb [%] | 8.2 (43.6) | 6.8 (42.9) | 8.0 (41.5) | 5.2 (23.1) |
| Multiplicity | 7.7 (7.9) | 7.7 (7.9) | 5.0 (4.6) | 9.8 (9.5) |
| Mean I/σI | 15.4 (5.1) | 16.5 (4.7) | 20.4 (4.0) | 23.4 (6.9) |
| Completeness [%] | 99.4 (99.5) | 99.4 (99.9) | 99.9 (100.0) | 95.2 (96.3) |
| Refinement | ||||
| Total number of atoms | 1177 | 1200 | 1231 | 1194 |
| Model contents | mol A: MG-S42-T166 1 glycerol 1 ethylene glycol 111 water molecules | mol A: MG-S42-T166 mol B: GGK(ac)A 1 ethylene glycol 104 water molecules | mol A: GAMG-S349-D460 3 ethylene glycol 2 β-mercaptoethanol 219 water molecules | mol A: GAMG-S349-D460 2 ethylene glycol 2 β-mercaptoethanol 223 water molecules |
| Solvent content (%) | 40 | 42 | 30 | 33 |
| Rfactorc (%) | 18.4 (23.9) | 19.6 (29.1) | 11.6 (15.3) | 11.6 (13.3) |
| Rfreed (%) | 22.0 (25.2) | 25.9 (30.8) | 14.5 (20.9) | 15.0 (14.4) |
| R.m.s. bonds [Å] | 0.026 | 0.021 | 0.030 | 0.034 |
| R.m.s. angles [Å] | 2.238 | 1.957 | 2.525 | 2.359 |
| <B> overall [Å2] | 24.6 | 41.1 | 15.5 | 15.8 |
| <B> BD [Å2] | 23.4 | 38.7 | 12.4 | 12.5 |
| <B> water [Å2] | 35.6 | 47.3 | 30.0 | 29.8 |
| Ramachandran plot | Most favored: 91.8% | Most favored: 88.4% | Most favored: 93.3% | Most favored: 94.2% |
| Allowed: 8.2% | Allowed: 11.6% | Allowed: 5.8% | Allowed: 5.8% | |
| Disallowed: 1% | ||||
| PDB accession code | 3JVJ | 3JVK | 3JVL | 3JVM |
a Values in parentheses correspond to the highest resolution shell.
bRmerge = Σ(|I − <I>|)/Σ(I), where I and <I> are the observed and mean intensities of all observations of a reflection, including its symmetry-related equivalents.
c Rfactor = Σhkl||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors of reflection hkl.
d Rfree was calculated from a randomly selected subset of the reflections (5%) that were omitted during refinement.
Structure Solution and Model Refinement
The structures of Brd4 BD1 and BD2 were solved by molecular replacement using the program Molrep (30), based on the PDB entries 2OSS and 2OUO (Filippakopoulos et al., Structural Genomics Consortium Oxford), respectively. The structures were afterward refined with Refmac5 (31). The molecular diagrams were drawn using PyMOL. Atomic coordinates and structure factors of Brd4 bromodomains have been deposited in the Protein Data Bank with accession codes 3JVJ (BD1), 3JVK (BD1 and H3-K(ac)14), 3JVL (BD2), and 3JVM (BD2 strings).
Isothermal Titration Calorimetry
Interactions of Brd4 bromodomains with acetylated lysine peptides were performed by ITC using an iTC200 microcalorimeter (MicroCal). Measurements with BD1 were carried out in 50 mm Tris buffer (pH 8.0), 100 mm NaCl, 1 mm TCEP at 25 °C. Measurements with BD2 were performed in 50 mm HEPES (pH 7.0), 150 mm NaCl, 1 mm TCEP. BD1 or BD2 were concentrated up to 90 mg/ml and stepwise injected from the syringe to the peptides placed in the measurement cell. The change in heating power was observed over the reaction time until equilibrium was reached. Data were analyzed using the software provided by the manufacturer.
Size Exclusion Chromatography
Analytical gel filtration experiments were performed using a Superdex S200 10/300 column at a flow rate of 1.0 ml/min similarly as described (32). Prior to injection of the protein samples, the column was equilibrated in 50 mm Hepes (pH 7.5), 50 mm NaCl, 1 mm TCEP buffer. Elution profiles were monitored by UV absorption at 280 nm. The column was calibrated using a gel filtration standard (Bio-Rad). Bromodomain protein samples were diluted to a concentration of 5 mg/ml each and injected onto the column at a volume of 25 μl. Gel filtration experiments were performed repeated times at 25 °C.
RESULTS
Structures of the Two Bromodomains of Brd4
The structures of Brd4 BD1 and BD2 exhibit the classical bromodomain fold that consists of four α-helices (αZ, αA, αB, αC) and two interconnecting loops (ZA and BC) at the distal site to the domain N and C terminus (33–36). The four helices form a left-handed α-helical bundle that composes the hydrophobic core of the domain while the two loops form a deep cleft that composes a recognition site for the binding to acetylated lysines within histone tail sequences (Fig. 1B). The extended loop ZA that connects helices αZ and αA comprises three short helices (αZ′, αZ″, and αZ‴, with αZ″ also called πD) whose detailed resolution in BD2 might be due to the excellent crystallographic map (Fig. 1C). The structures were determined by molecular replacement for residues Ser-42 to Thr-166 of BD1 including 111 water molecules and for residues Ser-349 to Asp-460 of BD2 including 219 water molecules. The structures were refined to 1.5 Å and 1.2 Å resolution, respectively, with excellent overall stereochemistry (Table 1). Both crystals also comprised ethylene glycol, used for cryoprotection, as well as glycerol molecules in BD1 and β-mercaptoethanol in BD2 which was covalently bound by disulfide bonds to cysteines Cys-357 and Cys-392, pointing toward the solvent. The two bromodomains of Brd4 exhibit a root mean-square deviation (r.m.s.d.) value of 0.83 Å for the backbone heavy atoms based on a sequence identity of 42.4% over 117 amino acids (Fig. 1D).
Homology of BET Family Bromodomains
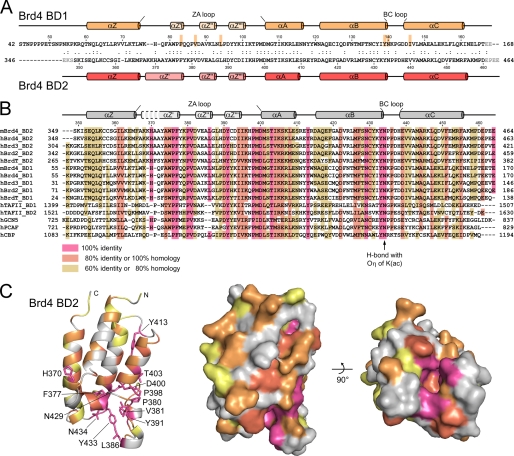
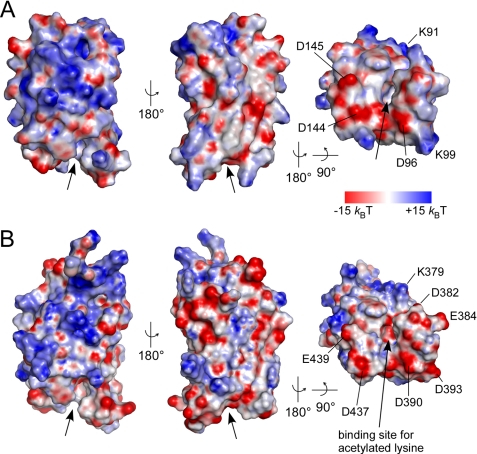
From a structure-based sequence alignment of BET family bromodomains, it is becoming apparent that the highest degree of sequence conservation is found in the ZA loop between helices αZ' and αA as well as for the YN motif in the BC loop (Fig. 2). These residues form the structural basis for loop formation and acetylated lysine recognition, whereas interspersed charged residues e.g. at positions 379, 382, 384, and particularly 393 of BD2 determine the adjacent electrostatic surface that might contribute to the specificity for histone recognition (Fig. 2, B and C). A large variability in the electrostatic surface potential of bromodomains was indeed observed from a comparison of PCAF and CBP bromodomains (37), Brd2 BD1 (38) and the TAFII250 double bromodomain module (34). A representation of the electrostatic surface potential showed an overall polar domain surface for both Brd4 bromodomains (Fig. 3). Although the two domains exhibit different theoretical isoelectric points with pI 8.8 for BD1 and pI 5.8 for BD2, the surface potential at the proximate binding sites for acetylated lysines appeared rather similar. A tight accumulation of acidic residues occurred in helices αZ″ and αZ‴ of the ZA loop in BD2, whereas BD1 exhibits with Lys-99 in αZ‴ a partly basic surface (Fig. 3, A and B). These electrostatic patches might contribute to the specificity for the recognition of acetylated lysines motifs based on their neighboring sequence compositions.
FIGURE 2.
Structure-based alignment of BET family bromodomains and sequence conservation. A, secondary structure display and sequence alignment of Brd4 bromodomains. Residues of BD1 that interact with acetylated lysine are marked. B, sequence alignment of bromodomains from the BET family and structure-based alignment to other known bromodomains. The secondary structure elements as determined for Brd4 BD2 are printed on top. C, display of the sequence conservation on the structure of Brd4 BD2. The ribbon plot and surface depiction reveal the accumulation of conserved residues (colored red) at the recognition cavity for acetylated lysines.
FIGURE 3.
Electrostatic surface potential of the two Brd4 bromodomains. Displayed is the electrostatic surface potential from −15 kBT (red) to +15 kBT (blue) for BD1 (A) and BD2 (B). The binding site for acetylated lysines is indicated by arrows. Acidic residues on helices αZ″ to αA and the BC loop form negatively charged surfaces surrounding the interaction cleft of both bromodomains (right panel).
Brd4 Bromodomain Binding to Acetylated Lysine Sequences
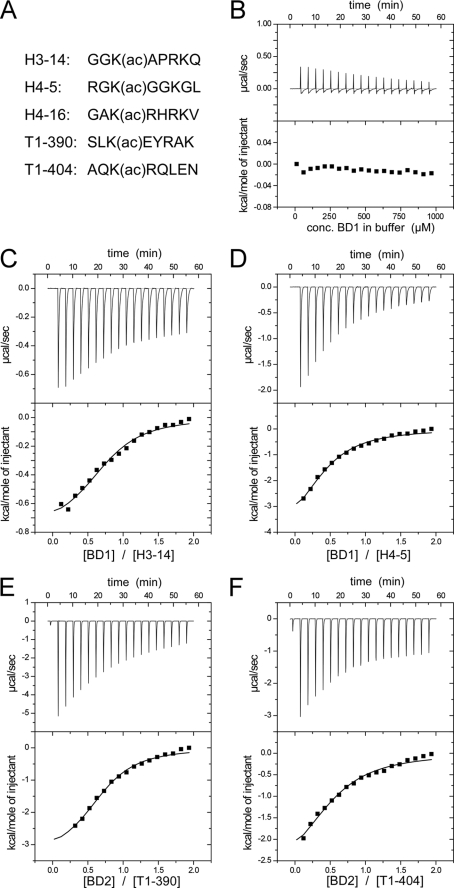
We next tested for binding to acetylated lysine motifs within different histone tail sequences. To this end three histone octamer peptides were probed containing an acetylated lysine either at position 14 of H3, at lysine 5 of H4 or at lysine 16 of H4 (Fig. 4A). The selection was based on previous immunoprecipitation analysis that suggested Brd4 similarly active for binding to modified lysines at all three different positions (23). Using isothermal titration calorimetry, we determined the dissociation constants and the thermodynamic parameters of the mutual interactions (Table 2). High concentrations of the bromodomains were titrated in successive steps to the peptides placed in the measurement cell (Fig. 4, B–F). The binding isotherms showed best binding of Brd4 BD1 to the H3-K(ac)14 peptide with a Kd of 118 μm. The second bromodomain Brd4 BD2 bound similarly well to H4-K(ac)5 and H4-K(ac)16 acetylated lysine sequences, whereas the mutual interactions were about 3-fold weaker. These observations suggest differences in the substrate recognition for the dual bromodomain protein.
FIGURE 4.
Binding analyses of Brd4 BD1 and BD2 bromodomains to acetylated lysine peptides by isothermal titration calorimetry. A, peptide sequences used within this study. B, control measurement of a Brd4 BD1 titration in buffer solution. C, ITC measurements of BD1 to H3-K(ac)14 revealed a dissociation constant of 118 μm. D, interaction of BD1 with H4-K(ac)5. E, binding of BD2 to acetylated cyclin T1 peptides K(ac)390. F, binding of BD2 to CycT1-K(ac)404. The thermodynamic parameter and dissociation constants are listed in Table 2.
TABLE 2.
Thermodynamic analysis of Brd4 bromodomain binding to acetylated lysine containing peptides by ITC
All measurements were performed at 25 °C.
| Titration scheme | Kd | ΔH | TΔS | Molar ratio n |
|---|---|---|---|---|
| μm | kcal/mol | kcal/mol | ||
| Brd4 BD1ato | ||||
| H3-K(ac)14 | 118 ± 28 | −0.59 ± 0.022 | 4.77 | 1.33 |
| H4-K(ac)5 | 325 ± 72 | −1.99 ± 0.067 | 2.76 | 2.35 |
| H4-K(ac)16 | 308 ± 142 | −0.76 ± 0.062 | 4.02 | 1.47 |
| Brd4 BD2ato | ||||
| H3-K(ac)14 | 327 ± 75 | −1.68 ± 0.059 | 3.07 | 2.31 |
| H4-K(ac)5 | 107 ± 23 | −1.81 ± 0.060 | 3.61 | 1.91 |
| H4-K(ac)16 | 133 ± 25 | −0.63 ± 0.019 | 4.64 | 1.43 |
| CycT1-K(ac)390 | 110 ± 21 | −2.35 ± 0.070 | 3.04 | 1.44 |
| CycT1-K(ac)404 | 319 ± 93 | −1.69 ± 0.080 | 3.07 | 1.84 |
a Brd4 BD1 encompasses residues 42–166 and Brd4 BD2 residues 349–460.
A recent study showed lysine acetylation in the central part of cyclin T1 to regulate the equilibrium between active and inactive P-TEFb (39). Because Brd4 was reported to associate with P-TEFb we included two cyclin T1 peptides T1-K(ac)390 and T1-K(ac)404 in the binding analysis (Fig. 4, A, E, and F). Unexpectedly, Brd4 BD2 exhibited a Kd of 110 μm to T1-K(ac)390 that is just as tight as the interaction to histone H4 sequences (Table 2). BD1 however precipitated during the measurement course prohibiting the determination of binding parameters. Overall, the rather weak binding affinities determined for Brd4 to acetylated lysine sequences are in line with whose of other non-tandem bromodomains (37).
Structural Basis of Brd4-BD1 Binding to H3-K(ac)14
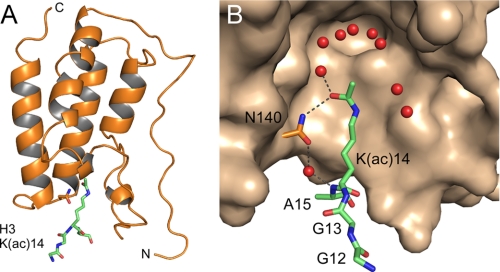
Histone tail peptides were exposed to the bromodomain crystals by soaking experiments. As a result we now recorded additional electron density in the binding site for acetylated lysines of Brd4 BD1 for all three peptides, but best resolution was achieved for the H3-K(ac)14 peptide (Fig. 5A). The modified lysine is encircled by a shell of water molecules that surround the methyl group toward the cavity formed by conserved residues Phe-83, Met-105, Met-132, and Cys-136. Intermolecular hydrogen bonds are formed only between Oη of K(ac) and asparagine 140 of the BC loop that is highly conserved in all bromodomains (Fig. 5B). A water-mediated H-bond is in addition formed between the hydroxy group of the asparagine site chain and the backbone amide of the following alanine residue in the histone peptide. This interaction network largely reflects previous results of the histone acetyltransferase Gcn5 bound to a histone H4-acetylated lysine peptide (35), whereas hydrophobic contacts of the modified lysine prevail over the interaction in the complex structure of an acetylated Tat peptide to the PCAF bromodomain (40). Additional contacts within a shell of 3.6 Å to the acetylated lysine were mediated by Leu-94, Tyr-139, and Ile-146 of Brd4. The H3 arginine residue three positions away from the critical lysine could unfortunately not be observed in the crystal density. This position is suggested to determine the course of the ligand binding chain by the formation of ionic interactions with the backbone hydroxy groups of the YN motif in the BC loop and its preceding residue Ile-138.
FIGURE 5.
Structural basis of H3-K(ac)14 binding to Brd4 BD1. A, complex structure of the bromodomain 42–166 with the peptide GGK(ac)A from H3 (green). B, hydrogen bonds between the H3 peptide and the essential asparagine Asn-140 in BD1 define the interaction network. Several water molecules (colored red) in the binding cavity of the bromodomain surround the acetyl moiety.
Unusual Binding of the N Terminus to the Adjacent Recognition Site for Acetylated Lysines
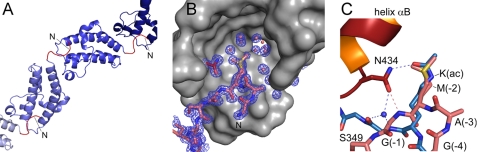
Surprisingly, the density observed upon histone peptide addition to Brd4 BD2 corresponded to the N-terminal linker sequence GAMG-S349 that remained after Tev protease cleavage but not to any of the peptides. This linker sequence was bound to each preceding bromodomain molecule forming thus continuous strings within the crystal (Fig. 6A). The side chain of methionine sticks into the binding pocket of the adjacent molecule and is surrounded by a shell of water molecules. Because of the excellent crystallographic resolution of 1.2 Å two different conformations of the Cϵ methyl group could be observed, affecting successively also the Sδ and Cγ atom position that together fill the space of the acetyl group of the modified lysine (Fig. 6B).
FIGURE 6.
Binding of the N-terminal linker sequence GAMG-S349KI to Brd4 BD2. A, in the protein crystal BD2 bromodomains are successively linked by their N-terminal tail sequence GAMGS (colored red), which leads to formation of continuous strings. B, electron density omit map of the bromodomain binding site for Nζ-acetyl-lysines contoured at 1 σ. The methionine side chain sticks into the binding pocket for acetylated lysines of a neighboring molecule and interacts with Asn-434 of Brd4 BD2. The terminal methyl group (Cϵ) of methionine adopts two different conformations with an approximate 6:4 population ratio. The moiety is surrounded by a shell of water molecules at the base of the recognition cavity. C, superimposition of the N-terminal linker sequence GAMGS bound to its proximate bromodomain from Brd4 BD2 (red) with the complex of a histone H4 tail peptide comprising acetylated lysine 16 with the bromodomain Gcn5 (blue; 1E6I; Ref. 35). The hydrogen bond network is displayed as dashed lines. Note that the backbone atoms of the GAMG sequence are roped into the binding pocket leading to the formation of a direct hydrogen bond between Asn-434 and the G(-1) amide group that is otherwise mediated by a water molecule upon binding to acetylated lysines.
Because the shorter methionine side chain inserts as deeply into the binding site as the acetylated lysine does, the backbone of the GAMG peptide is much deeper pulled into the bromodomain recognition cleft as in case of the histone sequence. Thus, a direct hydrogen bond is now formed between the amide group of the succeeding glycine and the highly conserved asparagine 434 that is otherwise only indirectly mediated by an additional water molecule as seen in BD1. A superimposition of the binding mode observed here with the one of the Gcn5 bromodomain structure in complex with an acetylated histone H4 peptide (35) reveals similarities in the positioning of the interacting moiety but also differences in the routing of the linker sequences (Fig. 6C). A glutamine or threonine residue directly surrounded by small residues might be able to form similar interaction networks as the acetylated lysine.
Size Exclusion Chromatography of Brd4 Bromodomains
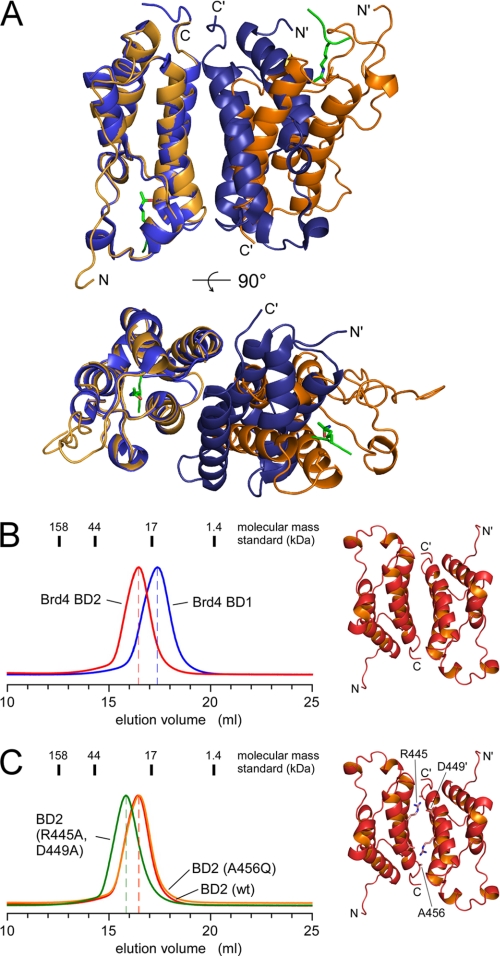
The Brd2 BD1 bromodomain was recently shown to form an intact homodimer presenting two acetyl-lysine-binding pockets in close proximity (38). The crystallographic assembly of the BD1 bromodomains of Brd2 and Brd4, however, appeared very different with opposing binding sites in the crystal mates, despite a sequence identity of 79% over 112 residues (Fig. 7A). To further analyze the elution profiles of the two Brd4 bromodomains we performed analytical gel filtration experiments using a Superdex S200 10/300 column. Strikingly, the two domains of a calculated molecular weight of 14.2 and 13.9 kDa for BD1 and BD2, respectively, showed markedly different elution volumes at 16.4 and 17.3 ml (Fig. 7B). Whereas the apparent molecular mass of 16 kDa for BD1 as derived from an equilibration to the molecular mass standard is in accordance with a globular monomeric protein, BD2 elutes at an apparent mass of 21 kDa, tolerable with both monomer and dimer structures. We, therefore, introduced two mutations in the interface of the crystal contacts that were designed to break these interactions and to impair a putative dimer formation. The double mutant R445A/D449A on helix αC was aimed to eliminate the mutual electrostatic interactions of Arg-445 from one molecule with Asp-449 from the opposite one (Fig. 7C, right panel). The A456Q mutation at the C terminus of helix αC instead was designed to introduce a sterical hindrance in the proposed dimer interface because of the increased residue size.
FIGURE 7.
Size exclusion chromatography of Brd4 bomodomains. A, overlay plot of the Brd2 BD1 dimer (1X0J, Ref. 38; blue) with the crystal mates of Brd4 BD1 (3JVJ, this study; orange). The bromodomains on the left side are aligned to each other, whereas the orientation of the bromodomains at the right side varies significantly. B, elution profiles of Brd4 BD1-(51–168) and Brd4 BD2-(349–464) separated by the elution volume in 50 mm Hepes buffer (pH 7.5), 50 mm NaCl, and 1 mm TCEP. Despite similar molecular masses of 14.2 kDa (BD1) and 13.9 kDa (BD2) the second bromodomain eluted significantly earlier suggesting an increased molecular size. C, mutations of residues R445A/D449A (green line), or A456Q (orange line) in Brd4 BD2, that contribute to the crystallographic dimer interface, did not change the elution profile toward smaller volumes, suggesting a monomeric state in solution.
Gel filtration analysis of these two BD2 mutants compared with the wild-type protein, however, did not show a retardation of the protein elution peak that would indicate the change to smaller domain volumes (Fig. 7C). Instead, the double mutant (R445A/D449A) even increased its apparent size, possibly because of a destabilization of helix αC. We, therefore, conclude that BD2 is a monomer in solution as is BD1. In addition, we tested binding of the two bromodomains for the formation of a BD1-BD2 heterodimer. Using size exclusion chromatography and isothermal titration calorimetry we could not observe any indication for such heterodimerization of the dual bromodomains.
DISCUSSION
The crystal structures of the two bromodomains of Brd4 add another component to the emerging picture of P-TEFb regulation. Yet, it remains unresolved how Brd4 activates transcription elongation at a molecular level. Crystal structures of the Cdk9/CycT1 heterodimer and the activating CycT1/Tat/TAR complex gave first insights into the molecular basis of P-TEFb regulation (41, 42). Because the lentiviral Tat protein was shown to displace the cellular P-TEFb regulator Hexim1 from a mutual binding site on CycT1, Hexim1 is supposed to bind to a similar surface on the first cyclin box repeat of CycT1 (28, 43, 44). In contrast, Brd4 bromodomains were initially described to interact with a region in CycT1 that is located C-terminal to the cyclin box repeat, whereas a later report showed the far C terminus of Brd4 to be required and sufficient for P-TEFb activation (6–8). Using heteronuclear NMR spectroscopy at high protein concentrations an interaction of the central part of CycT1-(316–566) with either Brd4 bromodomain could not be observed (data not shown). A very recent study, however, described lysine acetylation of CycT1 at four distinct sites in its central part (residues 380–404) by the histone acetyltransferase p300 and suggested such modification to support P-TEFb activation (39). A combined interaction of the C terminus of Brd4 and its two N-terminal bromodomains might therefore be possible to generate the specificity and binding affinity that is required for P-TEFb recognition and activation.
The binding analysis performed here on acetylated lysine recognition by BD1 and BD2 showed mutually specific interactions to H3 and H4 histone tail sequences, respectively (Fig. 4). A sequential cross-talk between histone H3-S10 phosphorylation and H4-K16 acetylation was indeed recently shown to generate a histone code that mediates transcription elongation (45). Likewise, inducible gene expression of primary response genes that encode e.g. proinflammatory cytokines was shown to depend on the recruitment of P-TEFb to acetylated H4 histones by Brd4 (46). It is intriguing in that respect that Brd4 BD2 was also found to bind similarly well to the acetylated lysine motif Lys-390 from cyclin T1. Such a modification of P-TEFb could function in direct targeting of Brd4 to P-TEFb for the transition to transcription elongation. Alternatively, the release of Brd4 from acetylated histones could be stimulated by cyclin T1 acetylation to proceed with RNA polymerase II transcription elongation.
The complex structure of Brd4 BD1 bound to the acetylated H3-K(ac)14 peptide supports previous findings on Gcn5 and Rsc4 in the recognition of histone tail sequences (35, 47). A shell of water molecules is lining the base of the recognition groove in the bromodomain, shielding all direct contacts between the acetyl moiety and its recognition domain, besides a hydrogen bond to the conserved asparagine in the BC loop. Differences in the recognition mode of the bromodomain for acetylated lysine observed in structures determined from x-ray crystallography and NMR spectroscopy may arise from the incorporation of these surrounding water molecules (35, 37, 40, 47, 48). The specificity for the modified histone tail sequence, however, is defined at surfaces adjacent to the binding groove that interact with residues ±3 amino acids apart from the acetylated lysine (49). Overall, the few hydrogen bonds and the sparse hydrophobic contacts formed may account for the low affinity between bromodomains and histones that on the other hand allow cooperative binding and synergistic effects in the recognition of the epigenetic code.
The interaction of the N-terminal tail sequence GAMGS with a neighboring bromodomain observed for BD2 must truly be considered a crystallographic artifact; however, it shows for the first time a residue other than the acetylated lysine to occupy the bromodomain recognition site (Fig. 6). A glutamine or threonine residue that would be directly surrounded by small residues might insert as deeply into the binding cleft and undergo similar hydrogen bond interactions and hydrophobic contacts as the modified lysine. Such sequence composition could potentially serve as an autoregulation motif for bromodomains that would stabilize the binding groove in the absence of acetylated lysines. A model for bromodomain regulation by binding to an intramolecular acetylated lysine has been recently proposed for Rsc4 whose N-terminal bromodomain was found to be occupied by a modified lysine, about 28 residues ahead of the bromodomain (47). It is therefore intriguing that the 20 residues preceding the canonical bromodomain fold in BD1 loop backwards to the acetylated lysine recognition site on the other side of the molecule (Fig. 5). The preceding sequence MSTTQAQAQPQPANAA (26–41) is rich in glutamine residues that are interspersed with alanines and prolines. Whereas we did not succeed in crystallizing a Brd4 BD1 construct (27–168) that encompasses this sequence, such interactions could serve to stabilize the binding site in the absence of a bona fide ligand. On a speculative level, these residues might interact with the acetylated lysine recognition site, thus forming an autoregulation motif in Brd4.
An increase in binding affinity and specificity to histones could result from oligomerization of the bromodomains. Indeed, the N-terminal bromodomain of Brd2 was recently shown to form an intact homodimer; thus, presenting two acetyl-lysine-binding pockets in close proximity (38). Together with a negatively charged secondary binding pocket, produced at the dimer interface between the two bromodomains, this unique feature was suggested to selectively recognize acetylated H4 histone tail motifs. A similar dimerization interface has been proposed for Brd4 because of the high sequence similarity to Brd2. However, we could not observe any dimer formation either as homo- or hetero-oligomerization of Brd4. These results are in agreement with recent structural data from NMR spectroscopy (50). The mechanistic activity that determines the readout of the histone code and its translation into the different phases of transcription initiation, elongation, and subsequent splicing of premature mRNA has yet to be determined. Other cofactors as the recently described acetylated RelA that coactivates NF-κB might be required to generate the specificity for gene activation (51). The structures of the dual Brd4 bromodomains and their binding specificities to acetylated lysine sequences add another component to the regulation factors of eukaryotic transcription elongation.
Acknowledgments
We thank Diana Ludwig for excellent technical assistance, Antje Schulte for initial help with crystallization, André SchÖnichen for advice on ITC measurements, and M. Weyand, C. Steegborn, N. Schrader, E. Hofmann, and I. Vetter for crystallographic data collection. We are grateful to Matija Peterlin and Matjaz Barboric, University of California at San Francisco, for providing Brd4 cDNA.
This work was supported by Grant GE-976/5 from the Deutsche Forschungsgemeinschaft (to M. G.).
The atomic coordinates and structure factors (codes 3JVJ, 3JVK, 3JVL, and 3JVM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BD
- bromodomain
- Cdk
- cyclin-dependent kinase
- CycT1
- cyclin T1
- P-TEFb
- positive transcription elongation factor b
- ITC
- isothermal titration calorimetry
- NMR
- nuclear magnetic resonance
- r.m.s.d.
- root mean-square deviation
- BET
- bromodomains and extra terminal domain
- CREB
- cAMP-response element-binding protein
- CBP
- CREB-binding protein.
REFERENCES
- 1.Peterlin B. M., Price D. H. (2006) Mol. Cell 23, 297–305 [DOI] [PubMed] [Google Scholar]
- 2.Saunders A., Core L. J., Lis J. T. (2006) Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 3.He N., Jahchan N. S., Hong E., Li Q., Bayfield M. A., Maraia R. J., Luo K., Zhou Q. (2008) Mol. Cell 29, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krueger B. J., Jeronimo C., Roy B. B., Bouchard A., Barrandon C., Byers S. A., Searcey C. E., Cooper J. J., Bensaude O., Cohen E. A., Coulombe B., Price D. H. (2008) Nucleic Acids Res. 36, 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markert A., Grimm M., Martinez J., Wiesner J., Meyerhans A., Meyuhas O., Sickmann A., Fischer U. (2008) EMBO Rep. 9, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang M. K., Mochizuki K., Zhou M., Jeong H. S., Brady J. N., Ozato K. (2005) Mol. Cell 19, 523–534 [DOI] [PubMed] [Google Scholar]
- 7.Yang Z., Yik J. H., Chen R., He N., Jang M. K., Ozato K., Zhou Q. (2005) Mol. Cell 19, 535–545 [DOI] [PubMed] [Google Scholar]
- 8.Bisgrove D. A., Mahmoudi T., Henklein P., Verdin E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13690–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 11.Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 12.Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. (1992) Nucleic Acids Res. 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamkun J. W., Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. (1992) Cell 68, 561–572 [DOI] [PubMed] [Google Scholar]
- 14.Wu S. Y., Chiang C. M. (2007) J. Biol. Chem. 282, 13141–13145 [DOI] [PubMed] [Google Scholar]
- 15.Beck S., Hanson I., Kelly A., Pappin D. J., Trowsdale J. (1992) DNA Seq. 2, 203–210 [DOI] [PubMed] [Google Scholar]
- 16.Thorpe K. L., Gorman P., Thomas C., Sheer D., Trowsdale J., Beck S. (1997) Gene 200, 177–183 [DOI] [PubMed] [Google Scholar]
- 17.Dey A., Ellenberg J., Farina A., Coleman A. E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. (2000) Mol. Cell Biol. 20, 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M. H., Numata M., Shimane M. (1997) Genomics 45, 529–534 [DOI] [PubMed] [Google Scholar]
- 19.Denis G. V., Green M. R. (1996) Genes Dev. 10, 261–271 [DOI] [PubMed] [Google Scholar]
- 20.Chua P., Roeder G. S. (1995) Mol. Cell Biol. 15, 3685–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maruyama T., Farina A., Dey A., Cheong J., Bermudez V. P., Tamura T., Sciortino S., Shuman J., Hurwitz J., Ozato K. (2002) Mol. Cell Biol. 22, 6509–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farina A., Hattori M., Qin J., Nakatani Y., Minato N., Ozato K. (2004) Mol. Cell Biol. 24, 9059–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanno T., Kanno Y., Siegel R. M., Jang M. K., Lenardo M. J., Ozato K. (2004) Mol. Cell 13, 33–43 [DOI] [PubMed] [Google Scholar]
- 25.Crawford N. P., Alsarraj J., Lukes L., Walker R. C., Officewala J. S., Yang H. H., Lee M. P., Ozato K., Hunter K. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6380–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y. J., Umehara T., Inoue M., Saito K., Kigawa T., Jang M. K., Ozato K., Yokoyama S., Padmanabhan B., Güntert P. (2008) Protein Sci. 17, 2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbate E. A., Voitenleitner C., Botchan M. R. (2006) Mol. Cell 24, 877–889 [DOI] [PubMed] [Google Scholar]
- 28.Schulte A., Czudnochowski N., Barboric M., Schönichen A., Blazek D., Peterlin B. M., Geyer M. (2005) J. Biol. Chem. 280, 24968–24977 [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 30.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 31.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32.Schönichen A., Alexander M., Gasteier J. E., Cuesta F. E., Fackler O. T., Geyer M. (2006) J. Biol. Chem. 281, 5084–5093 [DOI] [PubMed] [Google Scholar]
- 33.Dhalluin C., Carlson J. E., Zeng L., He C., Aggarwal A. K., Zhou M. M. (1999) Nature 399, 491–496 [DOI] [PubMed] [Google Scholar]
- 34.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. (2000) Science 288, 1422–1425 [DOI] [PubMed] [Google Scholar]
- 35.Owen D. J., Ornaghi P., Yang J. C., Lowe N., Evans P. R., Ballario P., Neuhaus D., Filetici P., Travers A. A. (2000) EMBO J. 19, 6141–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson B. P., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2000) J. Mol. Biol. 304, 355–370 [DOI] [PubMed] [Google Scholar]
- 37.Zeng L., Zhang Q., Gerona-Navarro G., Moshkina N., Zhou M. M. (2008) Structure 16, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura Y., Umehara T., Nakano K., Jang M. K., Shirouzu M., Morita S., Uda-Tochio H., Hamana H., Terada T., Adachi N., Matsumoto T., Tanaka A., Horikoshi M., Ozato K., Padmanabhan B., Yokoyama S. (2007) J. Biol. Chem. 282, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 39.Cho S., Schroeder S., Kaehlcke K., Kwon H. S., Pedal A., Herker E., Schnoelzer M., Ott M. (2009) EMBO J. 28, 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mujtaba S., He Y., Zeng L., Farooq A., Carlson J. E., Ott M., Verdin E., Zhou M. M. (2002) Mol. Cell 9, 575–586 [DOI] [PubMed] [Google Scholar]
- 41.Baumli S., Lolli G., Lowe E. D., Troiani S., Rusconi L., Bullock A. N., Debreczeni J. E., Knapp S., Johnson L. N. (2008) EMBO J. 27, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anand K., Schulte A., Vogel-Bachmayr K., Scheffzek K., Geyer M. (2008) Nat. Struct. Mol. Biol. 15, 1287–1292 [DOI] [PubMed] [Google Scholar]
- 43.Barboric M., Yik J. H., Czudnochowski N., Yang Z., Chen R., Contreras X., Geyer M., Peterlin B. M., Zhou Q. (2007) Nucleic Acids Res. 35, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dames S. A., Schönichen A., Schulte A., Barboric M., Peterlin B. M., Grzesiek S., Geyer M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14312–14317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. (2009) Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 46.Hargreaves D. C., Horng T., Medzhitov R. (2009) Cell 138, 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VanDemark A. P., Kasten M. M., Ferris E., Heroux A., Hill C. P., Cairns B. R. (2007) Mol. Cell 27, 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mujtaba S., He Y., Zeng L., Yan S., Plotnikova O., Sachchidanand, Sanchez R., Zeleznik-Le N. J., Ronai Z., Zhou M. M. (2004) Mol. Cell 13, 251–263 [DOI] [PubMed] [Google Scholar]
- 49.Mujtaba S., Zeng L., Zhou M. M. (2007) Oncogene 26, 5521–5527 [DOI] [PubMed] [Google Scholar]
- 50.Liu Y., Wang X., Zhang J., Huang H., Ding B., Wu J., Shi Y. (2008) Biochemistry 47, 6403–6417 [DOI] [PubMed] [Google Scholar]
- 51.Huang B., Yang X. D., Zhou M. M., Ozato K., Chen L. F. (2009) Mol. Cell Biol. 29, 1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]