Abstract
Integrins form mechanical links between the extracellular matrix and the cytoskeleton. Although integrin activation is known to be regulated by an allosteric conformational change, which can be induced from the extracellular or intracellular end of the molecule, little is known regarding the sequence of structural events by which signals propagate between distant sites. Here, we reveal with molecular dynamics simulations of the FnIII10-bound αVβ3 integrin headpiece how the binding pocket and interdomain βA/hybrid domain hinge on the distal end of the βA domain are allosterically linked via a hydrophobic T-junction between the middle of the α1 helix and top of the α7 helix. The key results of this study are: 1) that this T-junction is induced by ligand binding and hinge opening, and thus displays bidirectionality; 2) that formation of this junction can be accelerated by ligand-mediated force; and 3) how formation of this junction is inhibited by Ca2+ in place of Mg2+ at the site adjacent to the metal ion-dependent adhesion site (“ADMIDAS”). Together with recent experimental evidence that integrin complexes can form catch bonds (i.e. become strengthened under force), as well as earlier evidence that Ca2+ at the ADMIDAS results in lower binding affinity, these simulations provide a common structural model for the dynamic process by which integrins become activated.
Introduction
Integrins anchor cells to the extracellular matrix. They are transmembrane heterodimers, composed of non-covalently bound α and β subunits that associate to form the extracellular, ligand-binding head, two multidomain “legs,” two single-pass transmembrane helices, and two short cytoplasmic tails (Fig. 1A). All known integrin heterodimers contain the βA domain (also called I-like or βI domain), located at the extracellular end of the β-subunit. The top of the βA domain contains three metal ion binding sites, termed the “ligand-induced metal binding site” (“LIMBS”),3 the “metal ion-dependent adhesion site” (“MIDAS”), and the “adjacent to the MIDAS” (“ADMIDAS”). The LIMBS has been called the synergistic metal binding site instead, since it was found to contain a metal ion in a crystal structure of the unliganded αIIbβ3 integrin (44).
FIGURE 1.
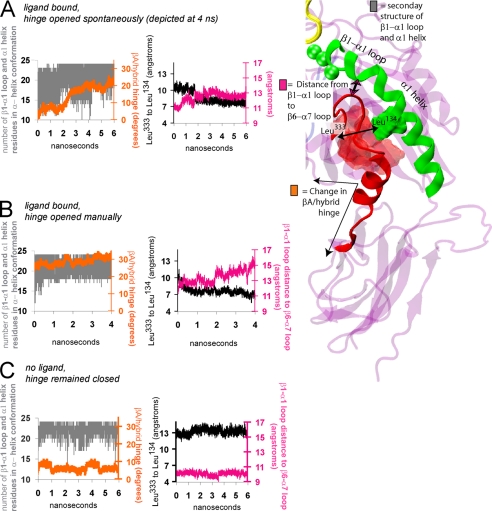
α1/α7 helix T-junction formation and headpiece hinge opening are allosterically linked. A, FnIII10 (15) (yellow) docked to the integrin structure (13). Integrin α- and β-subunits are blue and gray, respectively. Domains not resolved in the crystal structure were added in pink. B, the headpiece complex was simulated with explicit water molecules. C, the region of the α1/α7 helix junction in the integrin headpiece is shown in the box. D, the βA and hybrid domains from the liganded αvβ3 integrin crystal structure (13), in black, are aligned with the same domains from the unliganded αvβ3 integrin crystal structure (11), shown in color (red for the β6-α7 loop and α7 helix, green for the β1-α1 loop and α1 helix, and transparent purple for everything else). E, the βA and hybrid domains from the liganded αvβ3 integrin crystal structure (13), in black, are aligned here with the same domains from the liganded αIIbβ3 integrin structure (12), shown in color. Where the hinge is closed in both structures (D), the greatest structural change is in the binding pocket. Where the difference in the hinge angle is ∼62° (E), the region of greatest change is that of the α1/α7 T-junction. F–H, the three integrin crystal structures are shown separately. MD-derived snapshots (I and J) are depicted beneath the starting structures to which they correspond (F and G, respectively). The β1-α1 loop/α1 helix and α7 helix/hybrid domain regions are shown in green and red, respectively. Residues of the β6-α7 loop and α7 helix shown space-filling in red are Leu343 and Ile344, located at the top of the α7 helix, Val247, Ile307, and Ala309, located on underlying β-strands, and Leu333, located where the β6-strand becomes the β6-α7 loop. Shown in space-filling in green are Leu134 and Leu138 in the α1 helix. Black arrows identify the region where the β1-α1 loop meets the α1 helix, and the region where the α1 helix comes in contact with the β6 strand and α7 helix during T-junction formation.
Integrin-mediated adhesion often occurs under tensile forces such as fluid flow or myosin-mediated contractions that cells exert to sample the rigidity of their surroundings. Thus, to enable mechanosensing, integrins cannot be constitutively active. Rather, integrin activation is regulated by long-range conformational changes that can originate from the cytoplasmic or extracellular end of the integrin molecule (1). For example, ligand binding has been shown to induce the activating conformational change that leads to hinge opening between the βA and hybrid domains in the integrin headpiece (2–4). Vice versa, events originating from the cytoplasmic region of the molecule have been shown to switch the extracellular binding site to the high affinity state (5). This bidirectional reciprocity is typical for allosterically regulated proteins (6).
Although many factors that regulate the allosteric pathway of integrin activation are known, such as ligand binding (7), divalent cations (8), and mechanical force (9, 10), how structural alterations propagate from one end of the molecule to the other remains poorly understood. For instance, x-ray crystallographic structures of the unliganded αvβ3 (11) and liganded αIIbβ3 (12) integrins provide snapshots of the closed and open headpiece hinge conformations, respectively. These differences highlight the structural events that accompany activation. Yet little is known regarding the sequence by which these events occur. In another example, comparison between the liganded αvβ3 (11) and liganded αIIbβ3 (12) integrin crystal structures reveals near identical configurations of the ligand-bound β3 integrin binding pocket, differing only in the backbone hydrogen bonding of the β1-α1 loop (see Fig. 1E) (12, 13). However, these structures display a ∼62° difference in the hinge angle between the βA and hybrid domains, a conformational change that has been shown to govern binding affinity (2–4).
With no experimental techniques currently available to obtain dynamic, atomic level insights into the integrin activation pathway, we used molecular dynamics (MD) simulations and steered molecular dynamics (SMD) simulations to derive these insights computationally. Here, we show how a key structural event at a location halfway between the RGD-binding pocket and the headpiece hinge region directly regulates hinge opening. This event is the formation of a T-junction between hydrophobic residues at the middle of the α1 helix and the top of the β6 strand and C-terminal α7 helix. This hydrophobic contact was originally identified in the description of the first open-hinge crystal structure of an integrin headpiece, that of the αIIbβ3 integrin (12). Here, we show that this event immediately precedes ligand-induced hinge opening. Vice versa, inhibition of this T-junction prevents hinge opening, and we show how Ca2+ at the ADMIDAS conducts this inhibitory role. In addition, we show that simulated mechanical force or “manual” opening of the hinge accelerates formation of this junction. Together with recent experimental data (10), these findings lay the groundwork for a high-resolution model of how the lifetime of the RGD-αvβ3 integrin complex can be lengthened under force, the hallmark of a “catch” bond. Differences between the βA domain model of ADMIDAS-regulated, T-junction formation described here and the pull-spring model for the structurally homologous αA integrin domain proposed elsewhere (14) are discussed.
EXPERIMENTAL PROCEDURES
System Setup
The crystal structures of the αvβ3 integrin in complex with an RGD-containing mimetic ligand (Protein Data Bank entry code 1L5G, 3.2 Å resolution (13)) and FNIII10 from the crystal structure of the FNIII7–10 tetramer (PDB entry code 1FNF, 2.0-Å resolution (15)) were used to build the FNIII10-αvβ3 starting structure with the program VMD (16) as described previously (17). To reduce the integrin complex to a size that can be simulated on a feasible time scale, we used only the βA and hybrid domains of the β3 subunit and the β-propeller domain of the αv subunit. The starting structure was solvated in a 116 Å × 115 Å × 122 Å box of explicit TIP3 (18) water molecules, resulting in 153,570 atoms (Fig. 1B). For comparison, the corresponding domains from the unliganded αvβ3 integrin crystal structure (11) were also solvated in a box of explicit water molecules, resulting in 88,417 atoms. Seven cation binding sites are resolved in the crystal structure of the headpiece, three in the MIDAS, LIMBS, and ADMIDAS binding pocket motifs and four in the solvent-exposed β hairpin loops at the bottom of the α-subunit domain. These were occupied by Mn2+ in the crystal structure and by Mg2+ or Ca2+ in our simulations.
An additional complex was built in which the βA/hybrid domain hinge was opened “manually.” To do this, the FNIII10-αvβ3 integrin complex was aligned after 1 ns of equilibration with the same complex after 7 ns of equilibration, at which point the hinge angle had increased by ∼22°. Alignment was done via the 6 β-strands of the βA domain. Once aligned, the computational coordinates of the (closed hinge) hybrid domain at 1 ns were replaced by those of the (open hinge) hybrid domain at 7 ns. In this fashion, the hinge angle of the FNIII10-αvβ3 integrin complex, after 1 ns of equilibration, was increased manually by ∼22°.
Simulation Procedures and Parameters
All MD simulations were carried out with the program NAMD (19) using the CHARMM27 force fields (20). For a detailed description of the simulation protocol, see Refs. 17 and 21. All complexes remained stable during equilibrations, exhibiting an overall Cα root mean square deviation of less than 2.0 Å in the integrin head. The integrin head is comprised of the β-propeller domain from the αv subunit and the βA domain from the β3 subunit.
To probe the relative kinetics of force-induced hinge angle opening, external forces were applied by SMD protocols at constant forces of 100, 150, 250, 500, 700, 800, and 900 pN (22). To examine the mechanical response of the complex in various pulling geometries, we fixed either one or both of the C-terminal residues of the integrin headpiece (i.e. the Cα atoms of either αV-Arg438 and/or β3-Asp434). In every case, force was applied to the Cα atom at the C terminus of FnIII10 (Thr93). Generally, the direction of the stretching force was along the vector pointing from the fixed atom to the pulling atom. When the C-terminal ends of both the α and β subunits were fixed, the force vector was directed from the midpoint of the two fixed atoms to the pulling atom in the FnIII10 domain.
Simulations were conducted on the Gonzales cluster at ETH Zürich and on the Cray XT3 Palu supercomputer at the Swiss National Computing Centre, Manno, Switzerland. A 1-ns simulation required about 24 h on 64 Dalco nodes with 2.4 GHz AMD Opteron processors, or 10 h on 64 dual core AMD Opteron nodes with 2.6 GHz processors, respectively. All structure alignments were done in VMD (22) via the backbone atoms of the 6 β-strands of the βA domain. Relative hinge angle movement was measured using a script called Hingefind (23) and the frequency of attacks by free water molecules was measured as described previously (21). Customized scripts to measure the secondary structure, the center of mass, or atom-atom distances over specific structural regions were written in Tcl within VMD. Figures were rendered using VMD.
RESULTS
Our MD and SMD approaches take advantage of the existence of the semi-equilibrated, liganded αvβ3 integrin crystal structure, which was obtained by soaking RGD-peptides into preformed, unliganded integrin crystals (13). Previously, our MD equilibrations of this structure revealed that the closed βA/hybrid domain hinge opens on the nanosecond time scale when the constraints of the bent leg domains are lifted and the RGD-ligand is replaced with the 10th type III module of fibronectin (FnIII10, colored yellow in all of our figures) (17).
Here, the FnIII10-αvβ3 integrin headpiece complex was further investigated in 25 separate simulations ranging from 3 to 7 ns each. These simulations consist of 8 MD equilibrations and 17 SMD simulations. Because Mg2+ is known to regulate integrin activation (8) and is present in the integrin under physiological conditions, all integrin metal ion-binding sites were occupied with Mg2+ unless replaced by Ca2+, where specified, for comparison. The studies presented here, totaling 127 ns of simulation time, were then considered together with the 107 ns of simulation time described earlier (17).
Spontaneous Opening of the Hinge Immediately Follows Formation of a Hydrophobic T-junction between the α1 and α7 Helices
The liganded αvβ3 (11) and liganded αIIbβ3 (12) integrin crystal structures display a βA/hybrid domain hinge angle difference of ∼62° (12, 13), a conformational change that has been linked to the switch from the inactive to the active state (2–4). However, comparison between these structures reveals near-identical configurations of the ligand-bound βA domain binding pocket, differing only in the backbone hydrogen bonding of the β1-α1 loop. In contrast, considerable variation is found between the βA domains of these structures in the region surrounding the middle of the α1 helix and the top of the C-terminal α7 helix (Fig. 1, shown overlaid in E and shown separately in G and H). Indeed, a novel hydrophobic packing in this region surrounding the middle of the α1 helix and the top of the C-terminal α7 helix and β6 strand was first identified in the description of the liganded αIIbβ3 integrin crystal structure (12) (Fig. 1H). Here, we report that the transition to this hydrophobic packing arrangement led to hinge opening in the RGD-occupied αvβ3 integrin headpiece. When Leu134, at the middle of the α1 helix, formed a hydrophobic junction with residues surrounding the top of the α7 helix, the hinge increased by more than 10° over a time frame of 1 to 2 ns. This junction is evident by a tight packing between the middle of the α1 helix and the top of the β6-strand, as shown in Fig. 1, H and J (black arrows). Residues of the α1 helix and β6 strand/α7 helix are colored green and red, respectively.
To identify T-junction formation, we measured the decrease in Cβ-atom distance between the middle of the α1 helix (Leu134) and the top of the β6-strand, where it becomes the β6-α7 loop (Leu333). This distance is traced in black in Figs. 2, 4, and 6. Hinge opening is also displayed in supplemental Videos 1 and 2.
FIGURE 2.
Structural characteristics of hinge opening and α1/α7 helix T-junction formation. Traces over the course of MD equilibrations illustrate the opening of the βA/hybrid domain hinge (orange), fluctuations of the β1-α1 loop/α1 helix (gray), the formation of the α1/α7 T-junction (black), and the distance between the β1-α1 loop and β6-α7 loop (pink). More specifically, the orange trace in each panel displays the relative movement of the βA and hybrid domains, the gray trace in each panel displays the number of residues across the β1-α1 loop and α1 helix (residues 120 to 146) that form an α helix over time, the black trace in each panel is the Cβ-atom distance between Leu134 of the α1 helix and Leu333 on the β6 strand, and the pink trace in each panel is the distance between the centers of mass of the β1-α1 loop (residues 122 to 127) and the β6-α7 loop (residues 333 to 340). To obtain the number of residues across the β1-α1 loop and α1 helix that form an α helix over time (gray trace), each of the 26 residues is assigned either a “1” or “0,” corresponding to whether the residue was participating in an α helix (1) or not (0). Then these numbers were summed and traced over time. The maximum value of this measurement was 24, because Leu120 and Ser121, located at the top of the β1-strand, do not ever join the α helix. During spontaneous hinge opening (A, orange trace), the β1-α1 loop and α1 helix fluctuations displayed a wide range (A, gray trace). The α1/α7 T-junction formed when the hinge opened spontaneously (A, black trace) or manually (B, black trace). The α helix fluctuations were relatively less when the hinge was opened manually (B, orange and gray traces) or remained closed (C, orange and gray traces). When the hinge remained closed, the α1/α7 T-junction did not form (C, black trace). A characteristic of the β1-α1 loop that accompanies α1/α7 T-junction formation is an increased separation from the β6-α7 loop (pink trace, A and B versus C). To view repeat simulations with the same analysis, see supplemental Fig. S1.
FIGURE 4.
How Ca2+ in place of Mg2+ at the ADMIDAS inhibits formation of the α1/α7 helix T-junction. A, top, the coordination shell of the Ca2+-occupied ADMIDAS. Water molecules are shown in cyan licorice representation. When Ca2+ replaces Mg2+, the second carboxyl oxygen from Asp126 joins the coordination sphere and a split becomes more prevalent between the β1-α1 loop and α1 helix (see Fig. 3C). Under equilibrium conditions, the hinge does not open on the nanosecond time scale and the α1/α7 T-junction does not form in Ca2+-occupied complexes (compare with Fig. 2A). B, when the charge on the second Asp126 oxygen was turned off, in Ca2+-occupied complexes, creating a Ca2+-specific Asp126-charge off complex, the most prevalent conformation of the β1-α1 loop and α1 helix switched to an uninterrupted helical structure. Also, the α1/α7 T-junction formed (bottom, black trace) and a greater increase is evident in the hinge (middle, orange trace). When force was applied to this Ca2+-specific Asp126-charge off complex, hinge opening was accelerated relative to Ca2+-occupied complexes (supplemental Fig. S3). For further depiction of how the bidentate ADMIDAS coordination governs the β1-α1 loop conformation, see supplemental Fig. S2.
FIGURE 6.
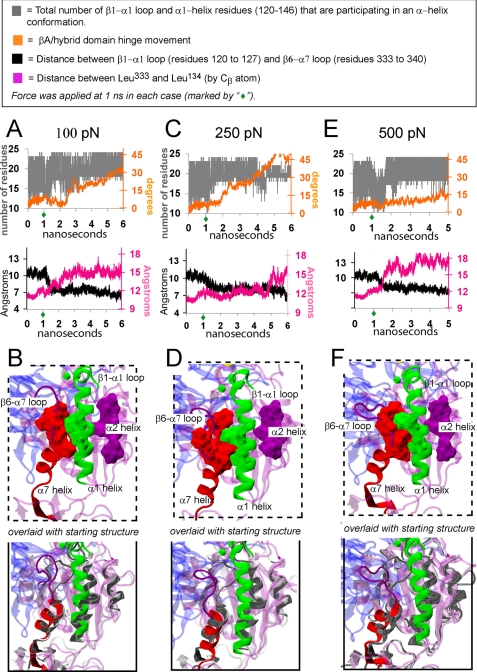
Force-induced α1/α7 helix junction formation. Traces from three separate SMD simulations are shown, with corresponding structural snapshots beneath. In each case, force was applied to FnIII10 after 1 ns of equilibration and was found to induce α1/α7 T-junction formation. Force was applied to the C terminus of the FnIII10 module with the C terminus of the β hybrid domain fixed. When force was applied at 500 pN (E and F), opening of the hinge angle was constrained by also fixing the C terminus of the α subunit β propeller domain. All snapshots are from the 3rd ns under force. In the bottom panels, each snapshot is overlaid with the starting crystal structure (shown in gray).
When the corresponding headpiece domains from the unliganded αVβ3 integrin structure were equilibrated, the hinge remained closed and the α1/α7 helix junction did not form. This is shown in the snapshot in Fig. 1I and the traces in Fig. 2C. These findings were reproduced in repeat simulations of the FnIII10-bound and unliganded integrin headpiece domains (supplemental Fig. S1).
The difference in the α1/α7 helix junction region between the liganded αvβ3 and liganded αIIbβ3 structures displayed in Fig. 1E contrasts the close alignment of this region between the liganded and unliganded αvβ3 integrin structures displayed in Fig. 1D. In both the liganded and unliganded αvβ3 integrin structures, the headpiece hinge is closed and the α1/α7 helix junction is not formed (Fig. 1, shown overlaid in D and shown separately in F and G) (11, 13). Thus, the link between α1/α7 T-junction formation and hinge opening is consistent with crystallographic data.
Characteristic Structural Alterations That Accompany Formation of the α1/α7 Helix Junction
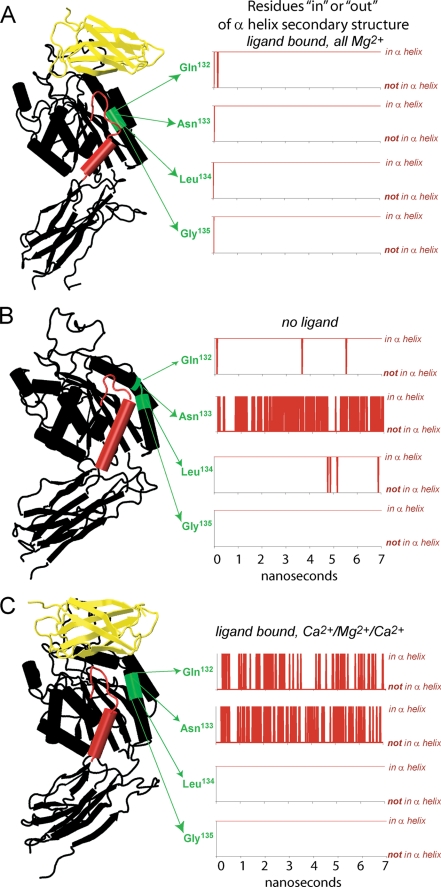
In the absence of the α1/α7 T-junction, a break exists in the α1 helix. During equilibrations of the unliganded headpiece domains, this break in the α1 helix was maintained at Asn133 (Fig. 3B). In contrast, with the junction in place, the β1-α1 loop and α1 helix tended to join into one continuous helix structure as shown in Fig. 1, H and J, and traced over time for residues at the center of the helix (132 to 135) in Fig. 3A. To quantify the fluctuations of the α1-helix conformation, we added the number of residues in the β1-α1 loop and α1 helix that join in the α-helix conformation over time (residues 120 to 146). This tally is traced in gray in Figs. 2, 4, and 6. Another feature of α1/α7 T-junction formation is an increase in the distance between the β1-α1 loop and the β6-α7 loop, as shown in the pink traces in Figs. 2, 4, and 6.
FIGURE 3.
The α1 helix split. The helical conformation of the middle of the α1 helix (residues 132 to 135) was assessed by residues over time using the “ssrecalc” command in VMD. A, when the hinge opened spontaneously, all of these residues were maintained in the α helix conformation. B, in the unliganded structure, Asn133 fluctuated in and out of the α helix conformation, resulting in a split helix. C, when Ca2+ replaced Mg2+ at the LIMBS and ADMIDAS, Gln132 and Asn133 fluctuated in and out of the α helix conformation, again resulting in a split helix. In all of our simulations, we found the prevalence of the split α1 helix to be a characteristic feature of complexes in which the hinge did not open on the nanosecond time scale (e.g. unliganded or Ca2+-occupied complexes).
Bidirectionality of the α1/α7 T-Junction/Hinge Opening Pathway
Experiments have shown that structural factors distal to the binding site can transition the integrin headpiece to the high affinity state by opening the βA/hybrid domain hinge (2, 4, 24). Thus, we asked if opening the hinge manually would induce the α1/α7 T-junction to form. The hinge was opened manually by replacing only the hybrid domain coordinates after 1 ns of equilibration (at which time the hinge had not yet opened), with the hybrid domain coordinates after a hinge increase of ∼22°. In this fashion, a starting structure was obtained in which the α1/α7 helix junction was not in place and the hinge was increased by ∼22°. When this complex was equilibrated in three separate simulations of 4 ns each, the α1/α7 T-junction formed within the first 40 ps in each case. As expected for a bidirectional allosteric process junction formation was accompanied by a decrease in the average Leu333–Leu134 Cβ-atom distance (Fig. 2B, black trace), an increase in the separation of the β1-α1 and β6-α7 loops (Fig. 2B, pink trace) and reduced fluctuations in the α1 helix conformation (Fig. 2B, gray line).
How Ca2+ in Place of Mg2+ at the ADMIDAS Inhibits Formation of the α1/α7 T-Junction
Previously, we described 4 separate FnIII10-αvβ3 integrin equilibrations in which small structural perturbations near the binding pocket prevented hinge opening on the nanosecond time frame (17). Here, we report that in each of these cases, hinge opening was deterred by a common mechanism: inhibition of α1/α7 T-junction formation. In one case, the structural perturbation was the replacement of Mg2+ with Ca2+ at the LIMBS and ADMIDAS in the binding pocket, which was found to promote an outwards break between the β1-α1 loop and α1-helix (17). Now, we report that in two separate repeat equilibrations of the Ca2+-occupied complex, this outward break consistently prevailed (Fig. 3C) and the T-junction between the α1 and α7 helices did not form. Because Ca2+ has long been known to inhibit integrin activation (8), we asked how Ca2+ in place of Mg2+ promotes the split between the β1-α1 loop and α1-helix and thus abrogates T-junction formation.
Bidentate coordination is known to be a characteristic feature of Ca2+ ions, relative to Mg2+ (25), and this distinction was consistently displayed in all of our MD and SMD simulations (17). While the preferred number of atoms coordinating Mn2+ or Mg2+ in integrin crystal structures is six, the preferred coordination number of the Ca2+ ion occupying the ADMIDAS is seven (11, 12). Three of the four residues that directly coordinate the ion occupying the ADMIDAS are located along the β1-α1 loop. These are Ser123, Asp126, and Asp127. When Ca2+ occupies the ADMIDAS instead of Mn2+ or Mg2+ the seventh atom that joins the ADMIDAS coordination sphere in β3-integrin crystal structures is the second carboxylate oxygen from Asp126, located where the bottom of the β1-α1 loop joins the top of the α1-helix (Fig. 4A) (12, 13). To investigate the influence of this Ca2+-specific, bidentate ADMIDAS coordination, we turned off the charge on the second Asp126 oxygen. In this fashion, we produced a complex in which the ADMIDAS displayed an “Mg2+ or Mn2+ mimetic” configuration while occupied by Ca2+. This configuration is characterized by a single ADMIDAS-coordinating oxygen from each of the Asp126 and Asp127 β1-α1 loop residues (Fig. 4B), as is the case when Mg2+ or Mn2+ occupy the ADMIDAS. Hereafter, we shall refer to this as the “Ca2+-specific Asp126-charge off” complex.
In three of four separate equilibrations of the Ca2+-specific Asp126-charge off complex (2 ns each), the 6-fold coordination of the ADMIDAS ion was maintained. In two of these simulations, the α1/α7 T-junction formed. In both of these cases, the most prevalent conformation of the β1-α1 loop and α1-helix switched from that of the split conformation that is typical of Ca2+ occupation (Fig. 4A) to the continuous helix conformation that we otherwise found in Mg2+-occupied complexes (Fig. 4B). In the third simulation, a hydrogen bond between the top of the α1-helix (at Trp129) and the β6-α7 loop (at Ser338) remained formed, which caused the split between the β1-α1 loop and α1-helix to remain in place. In the fourth simulation, 7-fold coordination of the Ca2+-occupied ADMIDAS was restored when the second carboxylate oxygen from Asp127, instead of Asp126, joined the Ca2+ coordination sphere. Strikingly, in this case the split between the β1-α1 loop and α1-helix became exaggerated, relative to the split configuration that is typical of “wild-type” Ca2+-occupied complexes (supplemental Fig. S2).
Together with experimental evidence linking Ca2+ at the ADMIDAS with integrin activation inhibition (26) and mutations of the ADMIDAS with changes in the headpiece hinge angle (27), these simulations suggest that the structural basis for activation inhibition by Ca2+ is as follows: Ca2+ at the ADMIDAS promotes bidentate coordination by a β1-α1 loop Asp, which in turn favors the outwards split between the β1-α1 loop and α1-helix, which in turn hinders α1/α7 helix junction formation, which obstructs hinge opening (Fig. 5).
FIGURE 5.
Model of the structural pathway across the βA domain by which Ca2+ in place of Mg2+ at the ADMIDAS in the ligand-bound headpiece inhibits βA/hybrid domain hinge angle opening. The RGD loop from FnIII10 is visible in yellow and the α-subunit domain is visible in blue. A, with Mg2+ occupying the ADMIDAS of the ligand-bound headpiece, monodentate coordination by the β1-α1 loop aspartates promotes the uninterrupted conformation of the α1 helix and thus α1/α7 T-junction formation and hinge angle opening. B, with Ca2+ occupying the ADMIDAS in the ligand-bound headpiece, bidentate coordination by a β1-α1 loop aspartate promotes the split conformation of the α1 helix, which hinders α1/α7 T-junction formation and hinge angle opening.
Force Accelerates Formation of the α1/α7 Helix T-Junction
Previously, we used SMD simulations to show that tensile force accelerates the opening of the integrin headpiece hinge (17). Here, we asked if force also accelerates the formation of the T-junction between the α1 and α7 helices. In nine separate SMD simulations, force was applied to the Mg2+-occupied integrin complex after 1 ns of equilibration, at which point the α1/α7 T-junction was not yet formed and the hinge was still closed. Forces of 100 to 900 pN were applied via the C terminus of the FnIII10 module. Forces in the hundreds of piconewtons were chosen to observe structural changes over a computationally feasible time frame. To vary the direction of strain, force was applied with the C terminus of only the hybrid domain fixed (at 100, 150, and 250 pN) or with the C termini of both the α subunit β-propeller domain and the hybrid domain fixed (at 500, 700, 800, and 900 pN).
We found that force induced the α1/α7 T-junction to form within the 5-ns time frame in seven (of nine) simulations. These include two in which the hinge was free to open under force and five in which the closed hinge was constrained. Even in the latter case, force varied the constrained hinge angle by as much as 15°. For representative depictions, see Fig. 6 and supplemental Videos 3 and 4.
We found that force could induce the T-junction to form prior to inducing a significant change in the hinge angle. This is illustrated in Fig. 6 by the decrease in the Leu333–Leu134 Cβ-atom distance (black trace) that occurs prior to the corresponding hinge increase (orange trace). When the α1/α7 T-junction did not form under force (two of nine times) the split between the β1-α1 loop and the α1 helix displayed greater prevalence and the β1-α1 loop and β6-α7 loop remained in close proximity (supplemental Video 5, in contrast to supplemental Videos 3 and 4).
Once the α1/α7 T-junction was formed in a mechanically strained integrin complex, it remained stable (seven simulations). In three additional simulations, forces of 500, 800, and 900 pN were applied for several nanoseconds after the junction was formed. Even at these high pulling forces, the junction was not disrupted.
Next, we asked if force could induce formation of the α1/α7 T-junction in Ca2+-occupied complexes, because we did not observe this event under equilibrium conditions (three of three Ca2+-occupied equilibrations). When the hinge of Ca2+-occupied complexes was opened under forces of 100, 150, and 250 pN, according to the same protocol described above for Mg2+-occupied complexes, formation of the α1/α7 T-junction accompanied hinge opening two of three times (at 100 and 250 pN).
Hinge opening in Ca2+-occupied complexes occurred with a delay relative to Mg2+-occupied complexes. Thus, we asked if the same structural feature regulates hinge opening and α1/α7 T-junction formation under force that we found to regulate this pathway under equilibrium conditions: namely, the β1-α1 loop/ADMIDAS coordination. We found that when the same constant forces were applied to the Ca2+-specific Asp126-charge off complex, force-induced hinge opening was accelerated toward the time scale of Mg2+-occupied complexes (supplemental Fig. S3). In other words, like ligand-induced hinge opening, force-accelerated hinge opening was found to be regulated by the influence of the bidentate β1-α1 loop ADMIDAS coordination.
Alterations of the β1-α1 Loop Shield the Force-bearing RGD-Integrin Bond from Water Access
Having established a link between β1-α1 loop contacts and the α1/α7 T-junction, we next asked if the β1-α1 loop could regulate the lifetime of the principal RGD-αvβ3 integrin bond. Previous SMD studies have shown that force-bearing bonds break when free water molecules compete with the bonding partners to form hydrogen bonds (21, 28). This indicates that the lifetime of a force-bearing bond can be increased by small structural perturbations that lessen access of free water to the bond. In the αvβ3 integrin complex, the principal force-bearing bond occurs between a carboxyl oxygen on AspRGD and the MIDAS ion (21). Thus, we investigated whether the β1-α1 loop can regulate access of free water molecules to this bond.
The top of the β1-α1 loop packs flush against the force-bearing AspRGD-MIDAS ion bond via direct contacts with both the MIDAS ion and AspRGD (Fig. 7A). The contact with AspRGD involves the “second” carboxyl oxygen (i.e. the AspRGD side chain oxygen that is not bound to the MIDAS ion) and Tyr122. In our simulations, we found that this contact was sometimes lost in Mg2+-occupied complexes when the second AspRGD side chain oxygen joined the first in coordinating the MIDAS ion (Fig. 7D). Although formation of the bidentate MIDAS ion coordination by AspRGD may not be physiologically relevant, as it is not currently found in crystal structures, we used this structural event to compare the influence of the AspRGD–Tyr122 bond on the access of water molecules to the force-bearing AspRGD–MIDAS ion bond.
FIGURE 7.
The β1-α1 loop regulates the stability of the force-bearing AspRGD-MIDAS bond. A, the force-bearing bond between Oδ-2 of AspRGD (black) and the MIDAS ion (red) occurs at the top of the β1-α1 loop, shown in green, schematic, and transparent surface representations. When the AspRGD and Tyr122 hydrogen bond is formed (dashed line), the β1-α1 loop packs in tightly with the AspRGD side chain. Water molecules within 5 Å of the Oδ-2 atom are shown in blue van der Waals representation. B, when the bond between AspRGD–Tyr122 was formed under force, as evident from the trace of the heavy atom distance (right side, traced in black), shielding of force-bearing Oδ-2 atom from attack by free water molecules was increased (left side, traced in blue). Water contact events to the Oδ-2 atom were counted for each time step (picosecond intervals) by identifying the number of water molecules within 3 Å of the Oδ-2 atom. C, when the AspRGD–Tyr122 bond was already formed under force, this bond remained formed (right side, black trace) and the Oδ-2 atom remained shielded from attack by water molecules (left side, blue trace). When instead both of the Oδ-1 and Oδ-2 atoms coordinated the MIDAS ion (D) and the AspRGD–Tyr122 bond did not form (E, right side), the AspRGD atom that forms the force-bearing bond with the MIDAS ion, Oδ-2, was subject to a relatively higher level of water molecule attacks.
Analysis of the number of collision events between free water molecules and the MIDAS ion-coordinating AspRGD oxygen (Oδ-2) reveals a considerable shielding when the AspRGD–Tyr122 bond is in place (Fig. 7, C versus E). During SMD simulations begun with the bidentate AspRGD-MIDAS ion coordination in place, it either remained formed (six of nine) or switched to monodentate AspRGD-MIDAS ion coordination together with AspRGD–Tyr122 bond formation (three of nine). In the latter cases, we found that the frequency of water attacks on the ion-coordinating AspRGD oxygen decreased when the AspRGD–Tyr122 bond was formed, as illustrated in Fig. 7B. These findings suggest a critical role for the AspRGD–Tyr122 bond, and thus of the β1-α1 loop, in regulating the lifetime of the principal RGD–αvβ3 integrin bond.
DISCUSSION
Although experiments have linked the switch to high affinity binding in the integrin headpiece with the opening of the βA/hybrid domain hinge (2–4), the structural basis for this link has remained unclear. On the one hand, the first crystal structure of the ligand-bound integrin headpiece (13) was formed under conformational constraints (17) and displays a closed βA/hybrid domain hinge. On the other hand, this crystal structure displays a conformation of the ligand-bound binding pocket that is nearly identical to that of the open-hinge β3 integrin headpiece structure, which was solved subsequently by co-crystallization with ligand (12). Thus, these crystallographic snapshots beg the question of how the conformation of the βA/hybrid domain hinge governs structural events in the vicinity of the βA domain binding pocket.
The simulations presented here were based on a semi-equilibrated crystal structure, in which activating structural changes local to the binding pocket had already occurred as a result of ligand binding but long-range propagation of the activation signal had been arrested by domain-domain contacts in the pre-existing crystal lattice. Previously, we showed with molecular dynamics simulations based on this structure that a small inwards (versus outwards) movement of the α1 helix promotes (versus impedes) opening of the βA/hybrid domain hinge (17). Here, we have shown how a contact of the α1 helix that occurs approximately halfway between the RGD-binding pocket and the βA/hybrid domain hinge links the signal propagation between these two sites. This contact is an α1/α7 T-junction between hydrophobic amino acid side chains in the middle of the α1-helix, the top of the β6-strand, and the top of the C-terminal α7-helix. We found that formation of this junction results from RGD occupation of the binding pocket and, as expected for allosterically regulated proteins, can also be induced by βA/hybrid domain hinge opening. A characteristic of this α1/α7 T-junction is the fusion of the α1-helix and β1-α1 loop, resulting in one continuous helix structure flanking the side of the βA domain (Figs. 1, H and J, and 3A). A change in ADMIDAS ion coordination, resulting from Ca2+ in place of Mg2+, inhibits formation of the α1/α7 T-junction by stabilizing a break between the β1-α1 loop and the α1-helix (Figs. 3C and 4A). Mechanical force was found to accelerate formation of the α1/α7 T-junction (Fig. 6). Taken in consideration with mutational, monoclonal antibody, electron micrographic and tomographic experiments that link the βA/hybrid domain hinge opening with integrin activation (2–4), together with recent evidence of force-accelerated integrin activation (9, 10), formation of the α1/α7 T-junction appears to be a key event along the integrin activation pathway.
Recently, five activating point mutations have been identified in the β-subunit headpiece domain (29). A preliminary analysis of the solvent accessible surface areas of these residues in our simulations indicates changes before and after α1/α7 T-junction formation that are consistent with the model proposed here. These include decreased fluctuations in residues located close to junction formation on the α1 and α2 helices and increased fluctuations in residues located at the βA/hybrid domain interface.4
Residues of the α1/α7 Helix T- junction Are Highly Conserved
The hydrophobic α1/α7 T-junction residues Leu138, Leu333, Leu343, Val247, Ile307, and Ala309 are completely conserved across the 8 integrin β-subunits, whereas Leu134 and Ile344 are conserved across 6 β-subunits. These residues are shown in green and red surface representations in Figs. 1 and 6. In the integrin β8A domain, both Leu134 and Ile344 are replaced by the similarly hydrophobic Val, whereas in the β4A domain, they are replaced by hydrophobic residues Met and Leu, respectively.
In accord with the premise of the model described here for the αvβ3 integrin, experiments have shown that mutations of hydrophobic residues on the α1 and α7 helices influence activation of the α5β1 integrin as well (30). Although the key residues identified experimentally differ from the α1/α7 T-junction residues identified here, there is a large degree of hydrophobic redundancy across highly conserved residues of the α1- and α7-helices (supplemental Fig. S4). For example in a preliminary study, we found that when Leu131, located above Leu134 in the α1 helix, is computationally “mutated” to Ala that a different kind of α1/α7 T-junction formed. In this case, a hydrophobic residue from the top of the α7 helix, Ile344, became inserted into a hydrophobic pocket in the α1 helix between Leu138 and Met142 and the headpiece hinge angle displayed an increase of ∼20° after 5 ns of equilibration (data not shown). Experimentally mutating the equivalent Leu residue in the α5β1 integrin was shown to promote activation (30). Thus, we suggest that a variety of putative hydrophobic interactions between the α1 and α7 helices could influence the dynamics of the β1-α1 loop and thereby influence binding affinity of the RGD-binding pocket.
A Unique Role for the ADMIDAS in βA Domain Allostery
Although all 24 known integrin heterodimers include the three headpiece domains investigated here, 9 types of integrins additionally contain the αA domain (also called the I domain) inserted into the extracellular end of the α-subunit. In those integrins that contain it, the αA domain mediates ligand binding directly. Although the αA and βA domains share considerable functional and structural homology, our simulations indicate that their mechanisms of activation differ. Although αA domain activation has been shown to proceed via the ratchet-like movement of the β6-α7 loop through an underlying pocket of the hydrophobic protein core (31), βA domain activation involves a distinctive, key role for the α1 helix and the ADMIDAS, a metal ion binding site that is not present in the αA domain (17, 26). Our simulations indicate that the change in bidentate ADMIDAS coordination that accompanies the change in occupation from Mg2+ or Mn2+ to Ca2+ regulates βA domain activation via an influence on α1/α7 T-junction formation.
Recent mutational studies of the β1-α1 loop ADMIDAS coordinating residues have shown disparate effects on integrin binding affinity. In one case, mutations equivalent to Asp126 induced firm adhesion in flow chamber experiments of α4β7 integrins (32). In the other case, the corresponding mutation induced weakened adhesion in solid phase binding assays of α5β1 (33) and α2β1 integrins (27). In this case, mutationally induced activation inhibition was partially rescued by monoclonal antibodies 12G10 or TS2/16, which bind at or near the α1 helix (2, 26). In all cases, ADMIDAS was found to play a critical role in relaying allostery from the ligand binding pocket to the rest of the integrin molecule. Notably, cell rolling assays probe integrin binding under tensile force conditions imposed by fluid flow, whereas solid phase binding assays probe integrin binding interactions in the absence of tensile forces. Thus, these seemingly disparate findings are also consistent with the model described here, whereby we predict that force applied to Ca2+-occupied complexes could override the α1 helix split that otherwise occurs, allowing the α1/α7 T-junction to form. In other words, this is how force could reverse the inhibitory influence of the Ca2+-occupied ADMIDAS. It should also be pointed out that ADMIDAS residues are completely conserved across all integrin β subunits except the β8 subunit, where the two β1-α1 loop/α1 helix aspartates are replaced by similarly acidic asparagines.
Force-accelerated Allostery and the Integrin Catch Bond
Several receptor-ligand complexes have been shown to increase their binding affinity under force, the hallmark of a catch bond. These include the bacterial adhesin FimH with mannose (34), the myosin-actin motor protein complex (35), the cellular adhesins P- and L-selectin with their respective ligands (36, 37) and, most recently, the α5β1 integrin with FnIII7–10 (10). In each case, a structural change has been implicated that is first induced or accelerated by force and then extends the lifetime of the receptor-ligand bond (for reviews, see Refs. 38–40). Like integrins, the FimH ligand binding site is allosterically coupled to a distal hinge region (40). Also like integrin catch bond complexes, activation of the FimH and selectin catch bond complexes can be stabilized by point mutations or monoclonal antibodies that promote the opening of the headpiece hinge, even if their epitopes are located distal to the ligand-binding site. Yet, for each of the receptor-ligand catch bond complexes currently known, the structural changes in the binding pocket that enable the extension of the bond lifetime remain experimentally undetermined (36, 41).
Here, we have shown that in the case of the RGD-αvβ3 integrin complex, formation of a bond between the β1-α1 loop (Tyr122) and AspRGD will extend the lifetime of the principal RGD-integrin bond by shielding it from frequent attacks by free water molecules (Fig. 6). This finding is consistent with available pharmacophoric refinement data, which identified the same bond as critical for RGD-integrin binding stability (42). It is also consistent with a general mechanism for allosteric proteins, whereby the dual character of regions of low and high structural stability in the binding site leads to the propagation of structural change via the region of low structural stability (43). In accord with this general mechanism, the β1-α1 loop both connects directly to the α1/α7 T-junction and, as shown in comparison across crystal structures of various presumed affinities (Fig. 1, D–H), is the only loop in the immediate vicinity of the AspRGD-MIDAS ion bond that displays significant structural change. This model is also consistent with the recent finding that monoclonal antibodies that activate integrins by their influence on the α1 helix also shift the integrin catch bond to a lower force regime (10).
As evidenced in crystallographic data (12, 13), formation of the α1/α7 T-junction occurs only once the βA domain is free to re-equilibrate after a binding event. Equilibration of the βA domain might be hindered by domain-domain contacts such as those formed in the bent-knee conformation of inactivated integrins. Although the two available liganded crystal structures display near identical configurations of the liganded β3 integrin binding pocket, differing only in backbone hydrogen bonding of the β1-α1 loop, the α1/α7 T-junction is absent where the ligand was soaked into preformed integrin crystals (13) that where constrained in the closed-hinge conformation by the pre-existing crystal lattice (17). In contrast, the junction is formed where the ligand was co-crystallized with the integrin headpiece in the absence of the legs domains (12). Thus, we propose that the physiological role of the bent-knee conformation is to tune, by domain-domain contact, the height of the energy barrier necessary to transition the headpiece to the high affinity state via hinge opening and α1/α7 T-junction formation. This may be the mechanism by which force accelerates the integrin activation process.
Acknowledgments
We gratefully acknowledge Drs. M. Gao, M. Smith, and V. HytÖnen and Profs. K. Schulten and M. P. Sheetz for discussions.
This work was supported by ETH Zurich, the Swiss National Supercomputing Center (CSCS, ALPS Project), and National Institutes of Health Roadmap for Medical Research Grant PN2 EY016586 to the Nanotechnology Center for Mechanics in Regenerative Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Videos 1–5.
E. Puklin-Faucher and V. Vogel, unpublished data.
- LIMBS
- ligand-induced metal binding site
- MIDAS
- metal ion-dependent adhesion site
- ADMIDAS
- adjacent to the metal ion-dependent adhesion site
- MD
- molecular dynamics
- SMD
- steered molecular dynamics
- N
- newton.
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Mould A. P., Barton S. J., Askari J. A., McEwan P. A., Buckley P. A., Craig S. E., Humphries M. J. (2003) J. Biol. Chem. 278, 17028–17035 [DOI] [PubMed] [Google Scholar]
- 3.Luo B. H., Strokovich K., Walz T., Springer T. A., Takagi J. (2004) J. Biol. Chem. 279, 27466–27471 [DOI] [PubMed] [Google Scholar]
- 4.Luo B. H., Springer T. A., Takagi J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. (2007) Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 6.Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 7.Takagi J., Strokovich K., Springer T. A., Walz T. (2003) EMBO J. 22, 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mould A. P., Akiyama S. K., Humphries M. J. (1995) J. Biol. Chem. 270, 26270–26277 [DOI] [PubMed] [Google Scholar]
- 9.Friedland J. C., Lee M. H., Boettiger D. (2009) Science 323, 642–644 [DOI] [PubMed] [Google Scholar]
- 10.Kong F., García A. J., Mould A. P., Humphries M. J., Zhu C. (2009) J. Cell Biol. 185, 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 14.Jin M., Andricioaei I., Springer T. A. (2004) Structure 12, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 15.Leahy D. J., Aukhil I., Erickson H. P. (1996) Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 16.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 17.Puklin-Faucher E., Gao M., Schulten K., Vogel V. (2006) J. Cell Biol. 175, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 19.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., Shulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 21.Craig D., Gao M., Schulten K., Vogel V. (2004) Structure 12, 2049–2058 [DOI] [PubMed] [Google Scholar]
- 22.Isralewitz B., Baudry J., Gullingsrud J., Kosztin D., Schulten K. (2001) J. Mol. Graph. Model. 19, 13–25 [DOI] [PubMed] [Google Scholar]
- 23.Wriggers W., Schulten K. (1997) Proteins 29, 1–14 [PubMed] [Google Scholar]
- 24.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 25.Katz A. K., Glusker J. P., Beebe S. A., Bock C. W. (1996) J. Am. Chem. Soc. 118, 5752–5763 [Google Scholar]
- 26.Mould A. P., Askari J. A., Barton S., Kline A. D., McEwan P. A., Craig S. E., Humphries M. J. (2002) J. Biol. Chem. 277, 19800–19805 [DOI] [PubMed] [Google Scholar]
- 27.Valdramidou D., Humphries M. J., Mould A. P. (2008) J. Biol. Chem. 283, 32704–32714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu H., Schulten K. (2000) Biophys. J. 79, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo B. H., Karanicolas J., Harmacek L. D., Baker D., Springer T. A. (2009) J. Biol. Chem. 284, 3917–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barton S. J., Travis M. A., Askari J. A., Buckley P. A., Craig S. E., Humphries M. J., Mould A. P. (2004) Biochem. J. 380, 401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimaoka M., Xiao T., Liu J. H., Yang Y., Dong Y., Jun C. D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J. H., Springer T. A. (2003) Cell 112, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Salas A., Springer T. A. (2003) Nat. Struct. Biol. 10, 995–1001 [DOI] [PubMed] [Google Scholar]
- 33.Mould A. P., Barton S. J., Askari J. A., Craig S. E., Humphries M. J. (2003) J. Biol. Chem. 278, 51622–51629 [DOI] [PubMed] [Google Scholar]
- 34.Thomas W. E., Trintchina E., Forero M., Vogel V., Sokurenko E. V. (2002) Cell 109, 913–923 [DOI] [PubMed] [Google Scholar]
- 35.Guo B., Guilford W. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9844–9849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. (2003) Nature 423, 190–193 [DOI] [PubMed] [Google Scholar]
- 37.Sarangapani K. K., Yago T., Klopocki A. G., Lawrence M. B., Fieger C. B., Rosen S. D., McEver R. P., Zhu C. (2004) J. Biol. Chem. 279, 2291–2298 [DOI] [PubMed] [Google Scholar]
- 38.Zhu C., Lou J., McEver R. P. (2005) Biorheology 42, 443–462 [PubMed] [Google Scholar]
- 39.Thomas W. E., Vogel V., Sokurenko E. (2008) Annu. Rev. Biophys. 37, 399–416 [DOI] [PubMed] [Google Scholar]
- 40.Tchesnokova V., Aprikian P., Yakovenko O., Larock C., Kidd B., Vogel V., Thomas W., Sokurenko E. (2008) J. Biol. Chem. 283, 7823–7833 [DOI] [PubMed] [Google Scholar]
- 41.Yakovenko O., Sharma S., Forero M., Tchesnokova V., Aprikian P., Kidd B., Mach A., Vogel V., Sokurenko E., Thomas W. E. (2008) J. Biol. Chem. 283, 11596–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marinelli L., Lavecchia A., Gottschalk K. E., Novellino E., Kessler H. (2003) J. Med. Chem. 46, 4393–4404 [DOI] [PubMed] [Google Scholar]
- 43.Luque I., Freire E. (2000) Proteins 4, (suppl.) 63–71 [DOI] [PubMed] [Google Scholar]
- 44.Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., Springer T. A. (2008) Mol. Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]