Abstract
Under anaerobic conditions, several species of green algae perform a light-dependent hydrogen production catalyzed by a special group of [FeFe] hydrogenases termed HydA. Although highly interesting for biotechnological applications, the direct connection between photosynthetic electron transport and hydrogenase activity is still a matter of speculation. By establishing an in vitro reconstitution system, we demonstrate that the photosynthetic ferredoxin (PetF) is essential for efficient electron transfer between photosystem I and HydA1. To investigate the electrostatic interaction process and electron transfer between PetF and HydA1, we performed site-directed mutagenesis. Kinetic analyses with several site-directed mutagenesis variants of HydA1 and PetF enabled us to localize the respective contact sites. These experiments in combination with in silico docking analyses indicate that electrostatic interactions between the conserved HydA1 residue Lys396 and the C terminus of PetF as well as between the PetF residue Glu122 and the N-terminal amino group of HydA1 play a major role in complex formation and electron transfer. Mapping of relevant HydA1 and PetF residues constitutes an important basis for manipulating the physiological photosynthetic electron flow in favor of light-driven H2 production.
Introduction
Among all photosynthetic organisms, only green algae can couple light-driven electron transport originating from water splitting with hydrogen production (1). Hydrogen evolution in the unicellular green alga Chlamydomonas reinhardtii is naturally induced upon nutrient deprivation (2). Especially in the absence of sulfur, the photosynthetic oxygen evolution rate drops below the respiratory rate leading to intracellular anaerobiosis. Under anaerobic conditions, the oxygen-sensitive (2, 3) [FeFe] hydrogenase HydA is synthesized and catalyzes light-dependent H2 production, thereby dissipating excess redox equivalents under conditions in which the Calvin cycle is down-regulated (4).
The extraordinarily small monomeric [FeFe] hydrogenases of green algae only consist of the catalytic core unit containing the active site (H-cluster), whereas other [FeFe] hydrogenases possess an additional N-terminal F-domain harboring one to four accessory iron-sulfur clusters (5, 6). Because Chlorophyta-type [FeFe] hydrogenases lack any accessory clusters (7, 8), a direct electron transfer between the native electron donor and the H-cluster has been assumed (9, 10). In C. reinhardtii, HydA1 has been shown to be localized in the chloroplast stroma (11), and first kinetic examinations with purified proteins demonstrated that the plastidic ferredoxin PetF can interact with HydA1. These results and the fact that H2 production in C. reinhardtii is photosystem I (PSI)2-dependent (4) led to the hypothesis that PetF is the native electron donor of the plastidic hydrogenase (12). Derived from in silico analyses, two possible PetF-HydA2 electron transfer complex models were recently suggested (13). However, in contrast to the well studied interaction of PetF with other redox partners like ferredoxin-NADPH oxidoreductase (14, 15), the mechanism of the electron transfer process between PetF and the algal hydrogenase is still an open question (9, 10, 16). Recently, we reported the establishment of an efficient system for the heterologous synthesis of [FeFe] hydrogenases, including HydA1 of C. reinhardtii (17). Using this system, we generated several variants of HydA1 and PetF that were specifically designed on the basis of predicted electrostatic surface distribution and preceding in silico docking analyses. The characterization of the kinetics of electron transfer processes between these protein variants of HydA1 and PetF allowed us to specify the residues that are essential for a proper interaction of the two proteins. To examine the in vivo relevance of these results, we established an in vitro system by reconstituting a part of the photosynthetic electron transport chain consisting of plastocyanin, PSI, PetF, and [FeFe] hydrogenase. This assay verifies the model of PSI-dependent H2 production, and it also allows mechanistic insights into complex formation and electron transfer between HydA1 and PetF. The experimental data demonstrate that especially Lys396 of HydA1, which is particularly conserved among green algal hydrogenases, is crucial for a successful binding and electron transfer between PetF and HydA1.
EXPERIMENTAL PROCEDURES
Generation of Variants of HydA1 and PetF by Site-directed Mutagenesis
Site-directed mutagenesis (SDM) of hydA1 was performed with the host-adapted cDNA that already has been demonstrated to allow efficient heterologous expression in the Clostridium acetobutylicum host strain ATCC 824 (17). SDM variants of cloned cDNA sequences from hydA1 and petF1 were created in a two-step procedure using mismatch primers according to the fusion PCR technique (18). For mutagenesis of C. reinhardtii petF, the cDNA was amplified excluding the sequence encoding the first 33 N-terminal amino acids comprising the transit peptide region (see supplemental material for further details).
Determination of H2 Production Activity
Measurements of in vitro hydrogenase activity were performed as described previously (17). For determining the kinetic parameters, Vmax and Km values via Lineweaver-Burk plots of the H2 production rate using PetF SDM variants as substrate, PetF concentrations up to 80 μm were used. For measuring light-driven H2 production via the in vitro reconstitution system, wild type and mutant forms of HydA1 (1–100 nm) were combined with C. reinhardtii wild type PetF (40 μm) as well as plastocyanin (60 μm) and PSI (20 or 40 μg of chlorophyll a per ml), which were isolated as described previously (19). The 200-μl reaction mix contained 1 mm sodium ascorbate, 100 μm dichloroindophenol, 0.03% dodecyl maltoside, and 10 mm MgCl2 in 20 mm Tricine-KOH buffer (pH 7.4). The assays were prepared under anoxic conditions in 2-ml Eppendorf tubes that were then sealed with Suba-Seal rubber stoppers. After sparging the reaction mixture with argon for 3.5 min, the tubes were light-exposed (1200 μmol photons m−2 s−1) for 30 min under constant shaking at room temperature. H2 production was determined by analyzing 400 μl of the gas phase via gas chromatography (GC-2010, Shimadzu).
Homology Modeling and in Silico Docking Analyses
For structure modeling of HydA1 and PetF, the automated protein-modeling server Swiss-Model was used. The coordinates for the structure models are available as supplemental material. Calculations of the electrostatic surface potentials were done using Swiss-PdbViewer 3.7. In silico docking was carried out with the BiGGER module of the structure analysis software Chemera (20). Distances between FeS-clusters as well as potential salt bridge interactions were determined using WebLab-Viewer 3.2.
RESULTS
Electron Transfer Complex Formation between PetF and HydA1 Depends on Electrostatics
As already known from interactions with other electron transfer partners (21–23), PetF tends to form complexes on the basis of electrostatic charge attraction. Protein interactions depending on intermolecular electrostatic contacts are weakened at increasing ionic strength as demonstrated between ferredoxin and ferredoxin-NADPH oxidoreductase (24). Using methyl viologen (MV) as an artificial electron donor, the specific activity of HydA1 remained unaffected by increasing the ionic strength up to 600 mm NaCl, demonstrating that the redox reaction with MV is independent from intermolecular charge attraction. The same experiment using PetF as the electron donor for HydA1 demonstrated a decreasing specific hydrogenase activity under elevated salt concentrations, finally reaching a residual activity of only 33% (supplemental Fig. S1). Thus, electrostatic interactions seem to be crucial for the interaction between PetF and HydA1.
Michaelis-Menten Kinetics of HydA1 SDM Variants with MV and PetF
For analyzing protein-protein interaction between heterologously produced PetF and hydrogenase HydA1 (25, 26), we performed site-directed mutagenesis on both proteins and tested the respective exchange effects in kinetic assays. Because electrostatic contacts seemed to play a major role in complex generation, the surface charge distribution on HydA1 was calculated. The model revealed a distinct area of positive net charge caused by several basic amino acid residues that originate from the edge of a structural invagination that directly leads to the hydrophobic niche harboring the H-cluster (Fig. 1A). From this region, eight basic amino acids were chosen for site-directed mutagenesis on HydA1.
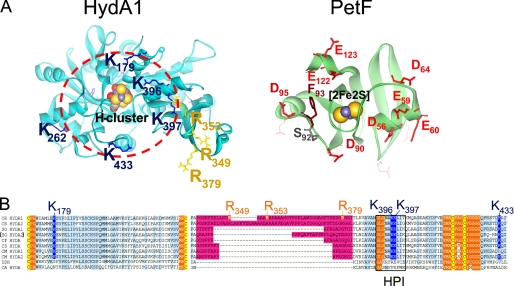
FIGURE 1.
Localization of exchange positions in structure models of HydA1 and PetF. Structures of HydA1 and PetF from C. reinhardtii (solid ribbon style; FeS-clusters in CPK style) were modeled using the crystal structures of CpI from C. pasteurianum (6) and PetF from Chlorella fusca (40) as archetypes. A, left side, detailed view on the assumed PetF-docking site, resolving the positively charged surface area of HydA1 with basic amino acid residues protruding from the brim of a surface recess leading to the [4Fe4S] subcluster of the active center. The depicted dark blue and yellow residues have been chosen for site-directed mutagenesis. The right side sketches the putative interaction surface on PetF. Residues that have been exchanged in this study are presented as red (acidic) or brown (aromatic) stick models. B, multiple sequence alignment of two parts of [FeFe] hydrogenase domains. A light blue background indicates a high level of sequence conservation. A pink background marks the large green alga-specific insertion. Cluster integrating cysteines are presented with a yellow background. Hydrophobic residues enclosing the H-cluster are indicated by an orange background. A black frame marks the sequence of the HPI. CR, C. reinhardtii (13, 16); SO, Scenedesmus obliquus (9); CF, C. fusca (10); CS, Chlorococcum submarinum (8); CM, Chlamydomonas moewusii (8); DDH, Desulfovibrio desulfuricans (41); CA, C. acetobutylicum (17).
Among these, three arginine residues (Arg349, Arg353, and Arg379) originate from a polypeptide insertion (including 45 amino acids in the case of C. reinhardtii HydA1) restricted to [FeFe] hydrogenases of green algae (Fig. 1, A and B) (8–10, 16). So far, the function of this alga-specific peptide insertion is unknown and has been speculated to be involved in PetF interaction (16). From the main area of positive net charge, which encircles the putative docking site (see red circle in Fig. 1A), another five residues (Lys179, Lys262, Lys396, Lys397, and Lys433) were chosen for single or double exchanges. Amino acid alignments of different green algal hydrogenases as well as prokaryotic sequences demonstrated that most of the chosen residues are especially conserved among green algal hydrogenases (see Fig. 1B). According to the structure model of HydA1, residues Arg96 and Asp129 have an inter-residue distance of ∼2.9 Å and presumably form a structure-stabilizing intramolecular salt bridge, which is also conserved in the crystal structure of CpI from C. pasteurianum (Arg234–Asp267). The exchange variant R96Q was generated to serve as a control for detecting structure-destabilizing side effects that might result from the performed amino acid exchanges.
Michaelis-Menten kinetic measurements were done with the artificial donor MV and reduced wild type PetF. Three enzyme variants (R96Q, R349Q, and R379Q) showed a diminished maximal activity (Vmax) with MV. At least for R96Q and R379Q, a similarly decreased activity can be measured in assays with PetF. The exchange R353Q had no significant influence on enzyme activity (Table 1). Thus, a direct participation of the green alga-specific peptide insertion in the electrostatic interaction between HydA1 and PetF seems questionable. The in vitro activity of R96Q was also significantly affected under an elevated ambient temperature. Although at 67 °C the wild type protein still exhibited 61% of the activity measured at the optimum temperature of 37 °C, the mutant protein showed a residual activity of only 27% under the same conditions. A similar effect could be observed analyzing the HydA1 variant R379Q, which had a residual activity of 26% at 67 °C (supplemental Fig. S2). It is therefore conceivable that at least Arg379 is involved in the structural integration of the alga-specific peptide loop rather than specifically forming transient salt bridges with PetF during the electron transfer process.
TABLE 1.
In vitro H2 production activity with wild type and SDM variants of HydA1 and PetF
Each value represents the mean of at least three independent assays. Underlined values indicate a significantly negative influence on HydA1 activity. Vmax and H2 production (H2-prod.) is given as μmol of H2 mg−1 min−1. The Km values refer to micromoles of ferredoxin. WT, wild type.
| HydA1 variant | Vmax (MV) | H2 prod. (40 μm PetF) | PetF variant | Vmax | Km |
|---|---|---|---|---|---|
| WT | 730 ± 54 | 278 ± 19 | WT | 435 ± 21 | 21 ± 2 |
| R96Q | 277 ± 22 | 147 ± 15 | D56N | 243 ± 10 | 39 ± 2.0 |
| R349Q | 476 ± 31 | 245 ± 7 | E59Q | 345 ± 26 | 27 ± 3.1 |
| R353Q | 667 ± 25 | 244 ± 12 | E60Q | 571 ± 38 | 36 ± 6.2 |
| R379Q | 370 ± 17 | 205 ± 10 | D64N | 352 ± 31 | 45 ± 7.1 |
| K179Q | 588 ± 26 | 182 ± 5 | D90N | 463 ± 22 | 50 ± 2.2 |
| K262Q | 667 ± 38 | 214 ± 10 | F93L | 138 ± 39 | 86 ± 12 |
| K433Q | 712 ± 12 | 165 ± 15 | S92A | 435 ± 41 | 30 ± 2.9 |
| K396Q | 714 ± 22 | 51 ± 17 | D95N | 406 ± 35 | 53 ± 11 |
| K397E | 770 ± 34 | 17 ± 15 | E122Q | 115 ± 29 | 519 ± 87 |
| K396Q/K397Q | 715 ± 25 | 23 ± 9 | E123Q | 500 ± 28 | 41 ± 3.3 |
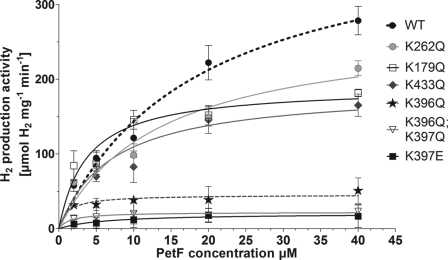
The remaining seven mutant proteins (K179Q, K262Q, R353Q, K396Q, K397E, K396Q/K397Q, and K433Q) exhibited kinetic profiles similar to that of the wild type protein. However, HydA1 variants K262Q and K179Q only reached 75 and 63%, respectively, of the wild type activity when assayed with PetF. The specific activity of K433Q in PetF assays was even more diminished to about 59% of the wild type level (Table 1 and Fig. 2). Strong effects on the specific activity of HydA1 using PetF as electron donor were observed in variants having exchanges of the adjacent residues Lys396 and Lys397. Although neutralizing Lys397 (K397Q) still allowed a residual enzyme activity of 68%, the replacement of this residue by an oppositely charged amino acid (K397E) resulted in a HydA1 variant that exhibited only 6% of the wild type activity while showing normal activity in MV assays. Even more drastic effects were noticed when Lys396 was changed. The mutant protein K396Q and the double variant K396Q/K397Q exhibited only 18 and 8%, respectively, of the wild type activity and therefore had the lowest PetF-specific activities among all charge compensating alterations (Table 1 and Fig. 2).
FIGURE 2.
PetF-dependent in vitro hydrogen production activity of HydA1 wild type and mutant proteins. In vitro tests were performed with 50–100 ng of HydA1 wild type (WT) or mutant protein as described earlier (17). Each value represents the mean of at least three independent assays.
Hydrogen Production Activity of HydA1 with Wild Type PetF and Mutant Proteins
Photosynthetic ferredoxins share a high degree of homology, even between cyanobacterial PetF proteins and those of higher plants such as spinach or maize (27). Interactions between PetF and various redox partners usually involve three polypeptide motifs (28) that correspond to Asp56–Asp64, Asp90–Asp96, and Glu122–Glu123 in PetF of C. reinhardtii. These three motifs contain highly conserved acidic residues that direct the first stages of complex generation dominated by electrostatic attraction and intermolecular salt bridge formation (22, 29, 30). Out of all three motifs, eight acidic residues were chosen for site-directed mutagenesis (Fig. 1B) of C. reinhardtii PetF. Furthermore, the aromatic residue Phe93 was changed to leucine to examine the possibility of its contribution to complex stabilization via hydrophobic interactions with PetF.
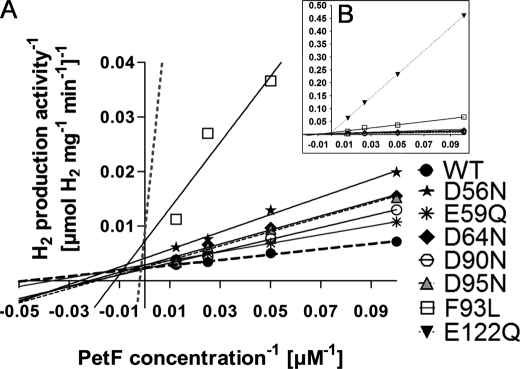
Although in vitro HydA1 activity was not negatively affected when the PetF mutant proteins E60Q, S92A, and E123Q served as electron donors, other changes in the PetF protein led to decreased in vitro H2 evolution rates (Table 1 and Fig. 3). The exchanges D90N, D64N, and D95N resulted in higher Km values. Although the reaction of HydA1 with wild type PetF exhibited a Km of 21 μm, values were 2.5-fold (D90N), 3.3-fold (D64N), or 4.1-fold (D95N) increased in the reactions with these mutated forms of PetF. On the other hand, the exchanges E59Q and D56N affected Vmax. Although E59Q was only slightly influenced (84% residual activity), the in vitro H2 production rate of HydA1 reached only half of the maximum obtained in combination with wild type PetF when the D56N variant was used as electron donor (Table 1 and Fig. 3A).
FIGURE 3.
Lineweaver-Burk plot of the HydA1 in vitro hydrogen production activity under increasing concentrations of PetF wild type or PetF mutant proteins. A, in vitro tests were anaerobically prepared as described in Fig. 2. B, larger scale to depict the strongly deviating graphs of E122Q and F93L. Each value represents the mean of at least three independent assays. For the sake of clarity, error bars are not presented in this figure. WT, wild type.
The strongest effects on the kinetics of H2 production were observed when analyzing PetF variants E122Q and F93L. These exchanges strongly affected both Vmax and Km. The Vmax of H2 production via HydA1 with either of these two ferredoxin variants as electron donors reached only 33% of the maximum rate obtained with wild type PetF, and the substrate affinity was reduced to values of 25% (F93L) and 3% (E122Q), respectively (Table 1 and Fig. 3B).
In Vitro Reconstitution of the Light-driven HydA-dependent Hydrogen Production
Light-dependent H2 production by C. reinhardtii cells has been attributed to electron transfer from PSI via PetF to the hydrogenase (9, 10, 16). Although a direct interaction of PetF and HydA1 has proven to be possible in vitro (see above), the coupling of HydA1 to the photosynthetic electron transport chain via PetF has not yet been analyzed on the biochemical level (11, 12, 31).
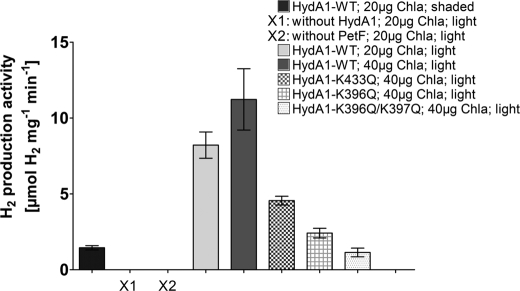
In this study, we reconstituted a part of the photosynthetic electron transfer chain to simulate the in vivo situation. Purified C. reinhardtii plastocyanin, PSI, PetF, and HydA1 were combined in a reaction mixture, supplied with sodium ascorbate and dichloroindophenol, and illuminated to allow light-dependent electron transport from ascorbate via plastocyanin and PSI to PetF. H2 evolution could only be achieved when both HydA1 and PetF were included in the assays (Fig. 4). Furthermore, incubating the complete reaction mixture, including PSI in the dark, resulted only in a low H2 production rate. However, when the complete reaction mixture was illuminated, H2 evolution rates of 8.2 μmol of H2 per mg of HydA−1 min−1 were obtained in the presence of PSI amounts equivalent to 20 μg of chlorophyll a per ml.
FIGURE 4.
Light-dependent hydrogen production activity of HydA1 wild type and mutant proteins. The samples were prepared as described above. X1 and X2 indicate inactive control samples that lack either HydA1 or PetF. After incubating the reaction mixtures for 30 min in the light (1200 μmol photons × m−2 s−1) or, as a control, in the dark (shaded), the H2 content of the gas phase was determined by gas chromatography. Chla, chlorophyll a; WT, wild type.
Doubling the amount of PSI to an equivalent of 40 μg of chlorophyll a per ml resulted in a further increase of the H2 production rate to 11.2 μmol of H2 mg of HydA1−1 min−1. On the other hand, an increase of the amount of hydrogenase enzyme did not enable higher H2 production (supplemental Fig. S3).
Using several mutated hydrogenase variants for analyzing light-driven PSI-dependent H2 production, the observed rates were comparable with those observed in in vitro assays containing PetF reduced by sodium dithionite (Fig. 2). The light-dependent H2 production rates obtained with these HydA1 variants reached 43% (K433Q), 21% (K396Q), and 10% (K396Q/K397Q), respectively, of the wild type activity (Fig. 4), which corresponds to the respective residual activities measured with artificially reduced PetF.
In silico docking between both protein models was accomplished using the structure analysis software Chemera 3.0 in the soft docking mode with HydA1 as target and PetF as probe model. Out of 5000 preselected docking orientations, one complex solution was selected, using the electrostatic complementarity, the cluster to cluster distance, and the results of the site-directed mutagenesis experiments as filtering parameters (Fig. 6) (for further details see supplemental material).
FIGURE 6.
Working model for complex formation and electron transfer between PetF and HydA1. Electrostatic contacts of HydA1 (white arrows) with possible influence on the hydrogen bond network (green lines) that stabilizes the redox state of PetF. The electron exchanging [FeS] cluster are presented as CPK model.
DISCUSSION
The objective of this work was to characterize the interaction of photosynthetic electron transport and hydrogen production, which is uniquely found in several green algal species (7–10, 16). Earlier physiological studies have demonstrated that a significant H2 production in green algae is dependent on photosystem I activity (31). It was further shown that the C. reinhardtii [FeFe] hydrogenase HydA1 can accept electrons from artificially reduced ferredoxin PetF in vitro (12). However, so far neither the molecular mechanism of the HydA1-PetF interaction nor the electron transport chain from reduced PSI to HydA1 has been analyzed.
Reconstitution of Light-driven Hydrogen Production
In this study, the in vivo situation was successfully reconstituted by combining isolated elements of the photosynthetic electron transport chain beginning with artificially reduced plastocyanin. The light-dependent assay confirmed that PetF works as an efficient electron donor for HydA1 and demonstrated that the interaction between both proteins could indeed constitute a functional interface between photosynthetic electron transport and hydrogen production in living C. reinhardtii cells. A direct electron transfer between HydA1 and plastocyanin or PSI can be excluded. Interestingly, increasing the concentration of HydA1 above 10 nm had no influence on H2 production in this system, demonstrating that the amount of HydA1 is not the rate-limiting factor. However, H2 production could be enhanced by increasing the PSI concentration, indicating that the electron supply rather than HydA1 catalytic activity is the rate-limiting factor of in vivo H2 production in C. reinhardtii.
HydA1 Residues That Are Important for the Reaction with PetF
The mechanism of the HydA1-PetF interaction was further analyzed on the molecular level performing site-directed mutagenesis. Alterations of conserved lysine residues to noncharged amino acids led to a specific decrease of PetF-dependent hydrogenase activity, although the MV-dependent H2 production rate was left unaffected. These residues therefore seem to be specifically important for the electron transfer between HydA1 and PetF. Because their exchange exhibited only a moderate decrease in PetF-dependent activity, it can be assumed that Lys262, Lys179, Lys397, and Lys433 at least take part in the guidance of the two electron transfer partners into a final stable intermolecular electron transfer complex. The low residual activity of only 18% resulting from exchanging Lys396 of HydA1 by glutamine indicates a strong influence of this lysine residue on forming and stabilizing the electron transfer complex between PetF and HydA1.
As most of the examined lysine residues are specifically conserved among hydrogenases of green algae (Fig. 1B), it seems as if these positions have not gained their importance until the differentiation of the chlorophyta type [FeFe] hydrogenases (8–10, 16), which can be distinguished from other [FeFe] hydrogenases by lacking any accessory FeS clusters (7, 8). Therefore, a close interaction between HydA1 and PetF is necessary to allow electron transfer between the active site redox clusters.
Residues of PetF Important for Binding and Electron Transfer to HydA1
The docking complex between HydA1 and PetF was further examined by analyzing mutant variants of PetF. Although exchanging the amino acids PetF-Glu60 and PetF-Glu123 had no significant influence on PetF-dependent HydA1 activity, charge neutralization of positions Asp90, Asp64, and Asp95 resulted in a decreased affinity of HydA1 for these PetF variants, indicating their importance for intermolecular attraction and orientation in the first stages of complex formation. On the other hand, the exchange of Asp56 to asparagine mainly influenced the Vmax value. Obviously, Asp56 has a more important role in stabilizing the final complex orientation.
Residues Glu122 and Phe93 constitute a third group, because neutralizing exchanges of these resulted in strong effects on both the Vmax and the Km values of PetF-dependent hydrogenase activity. Because the in vitro assays were conducted using sodium dithionite in excess to completely reduce the respective PetF variants (32), a dominating influence of a possible change in the PetF redox potential is unlikely (33). Exchanging the residues Glu94 and Phe65 in PetF of Anabaena (corresponding to Glu122 and Phe93 in PetF of C. reinhardtii) had a similar impact on electron transfer between PetF and ferredoxin-NADPH oxidoreductase as described here for the PetF-HydA1 complex (33, 34). Accordingly, in the crystal structure of the PetF-ferredoxin-NADPH oxidoreductase complex from Anabaena, residue Glu94 has been located as part of an important intermolecular salt bridge, whereas Phe65 turned out to be involved in hydrophobic interactions that stabilize the final complex orientation (14).
Although a comparison might be difficult as two different isoenzymes are examined, the suggested patterns of electrostatic interactions derived from the published in silico models of the PetF-HydA2 complex do not match the experimental results for the PetF-HydA1 interaction presented here (13). In the favored PetF-HydA2 complex (solution 16) residues Glu122 and Asp56 of PetF would be of no importance, although Glu60, which has been experimentally demonstrated to have no influence on complex formation with HydA1 in this study, was assumed to participate in electrostatic interaction. In both models that were suggested to be the most likely ones (13), position Lys407 of HydA2 has no significant influence on PetF-HydA2 complex stability, whereas residue Lys396 of HydA1, which corresponds to Lys407 of HydA2, was proven in this study to be the most important residue for complex formation between PetF and HydA1.
Modeling the Final and Productive Electron Transfer Complex between HydA1 and PetF
On the basis of the results obtained by site-directed mutagenesis, a single complex model was filtered out of 5000 pre-designed in silico docking complexes. With about 670 Å2, the contact interface of the complex lies at the lower range of typical protein contacts (800 ± 200 Å2) and thus corresponds to values of transient interactions (35) characteristic for electron transfer complexes.
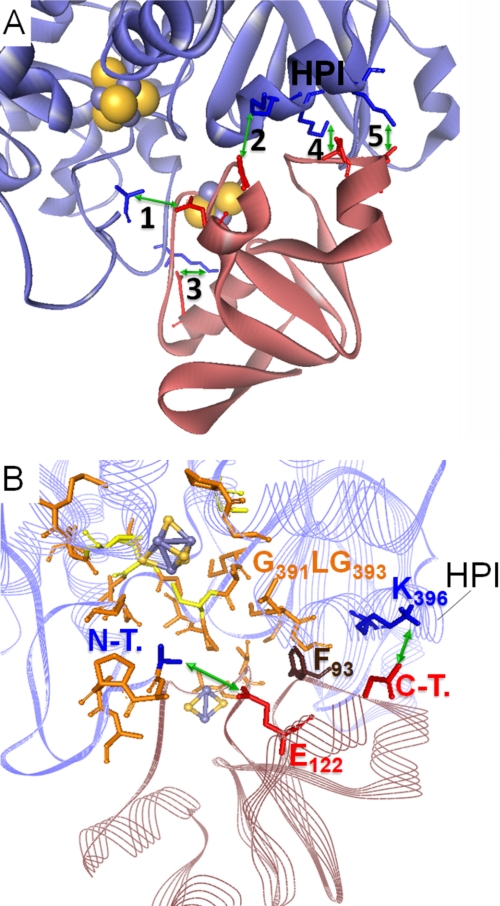
The salt bridge contacts include some of the residues examined in this study. Interestingly, for the respective participants of pairings 2 and 3 (see Fig. 5A), the experimentally determined level of influence on the electron transfer between PetF and HydA1 corresponds to each other. Three of the HydA1 residues (marked with an asterisk in the legend of Fig. 5) originate from a single α-helix (helix of PetF interaction, HPI), which has a high sequence conservation level among green algal [FeFe] hydrogenase enzymes (see Fig. 1B). Furthermore, according to this model complex, Phe93 of PetF would be located close to the hydrophobic N-terminal part of the HPI (from Gly391 to Gly393) and thus might further stabilize the contact of the HPI with PetF on the level of hydrophobic interactions.
FIGURE 5.
Model of the PetF-HydA1 electron transfer complex derived from the SDM effect-based screening of possible in silico docking complexes. A, zoomed view on the model complex interface with five possible salt bridge contacts (green arrows) between conserved residues of PetF (light red ribbon model) and HydA1 (blue ribbon model). 1, N-terminal amino group of HydA1-Ala57⇔PetF-Glu122; 2, HydA1-Glu396*⇔C-terminal carboxyl group of PetF-Tyr126; 3, HydA-Lys433⇔PetF-Asp56; 4, HydA1-Lys397*⇔PetF-Asp95; 5, HydA1-Lys501*⇔PetF-Asp96. Residues marked with * are localized within a single α-helix that is especially conserved in green algal hydrogenases and termed HPI in this study. B, integration of PetF into the putative binding side on HydA1 near the active center that is enclosed by hydrophobic residues. Hydrophobic residues probably participating in complex stability are depicted as orange stick models. N-T., N-terminal amino group of HydA1; C-T., C-terminal carboxyl group of PetF.
As indicated by the electron transfer complex model, electrostatic contacts of HydA1 residue Lys396 with the C terminus of PetF (Tyr126) on the one hand and between Glu122 of PetF and the N terminus of HydA1 (Ala57) of the other hand could play a central role in complex formation, which corresponds to the experimental results obtained with the variants HydA1-K396Q and PetF-E122Q, respectively. The interaction of Glu122 with the amino group of the hydrophobic HydA1 N terminus might even open up a “hydrophobic lid” covering the passage to the H-cluster generated by the first five amino acids of the processed HydA1 molecule (A1APAA5) (Fig. 5B).
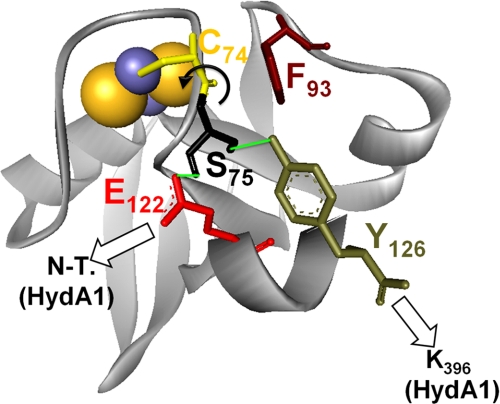
Furthermore, the homologous positions of Glu122 and the C-terminal tyrosine group (Tyr126) in PetF of Anabaena have been described to take part in a hydrogen bond network originating from the loop that surrounds the 2Fe2S cluster (33, 36). Comparative examinations on the crystal structures of reduced and oxidized PetF indicate that this network stabilizes the redox state of PetF by fixing the position of the peptide bond between Ser75 and Cys74, which has been demonstrated to perform a flipping movement upon the switch of the redox center from the reduced to the oxidized state (36). Considering the importance of the Ser75–Cys74 bond in C. reinhardtii PetF, residue Phe93 could exert its effect by influencing this bond, because it seems to be very close to the Ser75–Cys74 pair. Additionally, the interaction of Phe93 and Tyr126 might play an important role for the stabilization of PetF, because it has been discussed that the homologous residues fulfill this function in Anabaena PetF (36). Therefore interactions of the C. reinhardtii PetF amino acids Glu122, Tyr126, and Phe93 with residues of HydA1 might even be involved in the change of the redox state that initiates the electron transfer (Fig. 6).
Besides PETF, the nuclear genome of C. reinhardtii contains at least five additional genes encoding proteins with significant homology to plant-type ferredoxins (FDX2-6) (37, 38). Compared with PetF, Fdx2 exhibits the highest level of sequence homology (65% identity), although for the other ferredoxins the similarity lies between 47% (Fdx5) and 29% (Fdx6) sequence identity. Indeed, several of the PetF residues that contribute to complex formation with HydA1 are conserved in Fdx2, Fdx3, and Fdx5 (supplemental Fig. S4).
Recent examinations demonstrated the tendency of Fdx2 for NADPH oxidation via ferredoxin-NADPH oxidoreductase. Furthermore, reduced Fdx2 obviously has a substrate specificity for nitrite reductase. This behavior suggests an equivalent role of Fdx2 and root-type ferredoxin of higher plants, which operates independently from the photosynthetic electron transport (38).
Due to the fact that Glu122, one of the most important residues for complex formation with HydA1, is substituted by a serine, it further seems unlikely that Fdx3 would effectively interact with HydA1. Even though the transcript of FDX5 strongly accumulates under anaerobic conditions, corresponding to the expression pattern of HYDA1 (16), recent experimental evidence suggests that Fdx5 is not able to efficiently donate electrons to HydA1 (26).
In the end, it would be surprising if one of the other ferredoxins could replace PetF as an efficient electron mediator for HydA1, as Phe93, another important PetF position, is not conserved in any of the other ferredoxins. Even the fact that the C-terminal end of PetF is unique among the ferredoxins of C. reinhardtii could potentially exclude the other ferredoxins as interaction partners (compare Fig. 6 and supplemental Fig. S4).
The effects on PetF-dependent hydrogenase activity observed in this study indicate that most of the residues that are involved in PetF-ferredoxin-NADPH oxidoreductase interaction (21, 30, 33) also participate in complex formation between PetF and HydA1. Nevertheless, there are some significant exceptions. Mutagenesis at positions Glu59 and Glu60 have only minor or no effect on interaction with HydA1, although their importance for the PetF- ferredoxin-NADPH oxidoreductase complex was demonstrated for different plants such as spinach and maize (21, 30). Under competitive conditions, PetF mainly delivers electrons to the ferredoxin-NADPH oxidoreductase instead of sustaining H2 production (39). Indeed, the availability of electrons has been pointed out to be a critical aspect in light-driven hydrogen production (see above). Mapping of PetF residues relevant for the interaction with each PetF redox partner will allow us to select positions for site-directed mutagenesis to favor and direct photosynthetic electron flow to the hydrogenase and thus for enhancing light-driven H2 production.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants HI 739/4-1 and SFB 480 (to T. H. and M. W., respectively) and the European Union/Energy Network SolarH2 FP7 Contract 212508 (to T. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Tables S1 and S2, and additional references.
- PSI
- photosystem I
- SDM
- site-directed mutagenesis
- Tricine
- N-tris(hydroxymethyl)methylglycine
- MV
- methyl viologen
- HPI
- helix of PetF interaction.
REFERENCES
- 1.Melis A., Happe T. (2004) Photosynth. Res. 80, 401–409 [DOI] [PubMed] [Google Scholar]
- 2.Melis A., Zhang L., Forestier M., Ghirardi M. L., Seibert M. (2000) Plant Physiol. 122, 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wykoff D. D., Davies J. P., Melis A., Grossman A. R. (1998) Plant Physiol. 117, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemschemeier A., Fouchard S., Cournac L., Peltier G., Happe T. (2008) Planta 227, 397–407 [DOI] [PubMed] [Google Scholar]
- 5.Peters J. W. (1999) Curr. Opin. Struct. Biol. 9, 670–676 [DOI] [PubMed] [Google Scholar]
- 6.Peters J. W., Lanzilotta W. N., Lemon B. J., Seefeldt L. C. (1998) Science 282, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 7.Happe T., Hemschemeier A., Winkler M., Kaminski A. (2002) Trends Plant Sci. 7, 246–250 [DOI] [PubMed] [Google Scholar]
- 8.Kamp C., Silakov A., Winkler M., Reijerse E. J., Lubitz W., Happe T. (2008) Biochim. Biophys. Acta 1777, 410–416 [DOI] [PubMed] [Google Scholar]
- 9.Florin L., Tsokoglou A., Happe T. (2001) J. Biol. Chem. 276, 6125–6132 [DOI] [PubMed] [Google Scholar]
- 10.Winkler M., Heil B., Heil B., Happe T. (2002) Biochim. Biophys. Acta 1576, 330–334 [DOI] [PubMed] [Google Scholar]
- 11.Happe T., Mosler B., Naber J. D. (1994) Eur. J. Biochem. 222, 769–774 [DOI] [PubMed] [Google Scholar]
- 12.Happe T., Naber J. D. (1993) Eur. J. Biochem. 214, 475–481 [DOI] [PubMed] [Google Scholar]
- 13.Chang C. H., King P. W., Ghirardi M. L., Kim K. (2007) Biophys. J. 93, 3034–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales R., Charon M. H., Kachalova G., Serre L., Medina M., Gómez-Moreno C., Frey M. (2000) EMBO Rep. 1, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palma P. N., Lagoutte B., Krippahl L., Moura J. J., Guerlesquin F. (2005) FEBS Lett. 579, 4585–4590 [DOI] [PubMed] [Google Scholar]
- 16.Happe T., Kaminski A. (2002) Eur. J. Biochem. 269, 1022–1032 [DOI] [PubMed] [Google Scholar]
- 17.von Abendroth G., Stripp S., Silakov A., Croux C., Soucaille P., Girbal L., Happe T. (2008) Int. J. Hydrogen Energy. 33, 6076–6081 [Google Scholar]
- 18.Yon J., Fried M. (1989) Nucleic Acids Res. 17, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hippler M., Drepper F., Farah J., Rochaix J. D. (1997) Biochemistry 36, 6343–6349 [DOI] [PubMed] [Google Scholar]
- 20.Krippahl L., Moura J. J., Palma P. N. (2003) Proteins 52, 19–23 [DOI] [PubMed] [Google Scholar]
- 21.Kurisu G., Kusunoki M., Katoh E., Yamazaki T., Teshima K., Onda Y., Kimata-Ariga Y., Hase T. (2001) Nat. Struct. Biol. 8, 117–121 [DOI] [PubMed] [Google Scholar]
- 22.Saitoh T., Ikegami T., Nakayama M., Teshima K., Akutsu H., Hase T. (2006) J. Biol. Chem. 281, 10482–10488 [DOI] [PubMed] [Google Scholar]
- 23.Fischer N., Hippler M., Sétif P., Jacquot J. P., Rochaix J. D. (1998) EMBO J. 17, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colvert K. K., Davis D. J. (1983) Arch. Biochem. Biophys. 225, 936–943 [DOI] [PubMed] [Google Scholar]
- 25.Girbal L., von Abendroth G., Winkler M., Benton P. M., Meynial-Salles I., Croux C., Peters J. W., Happe T., Soucaille P. (2005) Appl. Environ. Microbiol. 71, 2777–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009) FEBS Lett. 583, 325–329 [DOI] [PubMed] [Google Scholar]
- 27.Fukuyama K. (2004) Photosynth. Res. 81, 289–301 [DOI] [PubMed] [Google Scholar]
- 28.Xu X., Kim S. K., Schürmann P., Hirasawa M., Tripathy J. N., Smith J., Knaff D. B., Ubbink M. (2006) FEBS Lett. 580, 6714–6720 [DOI] [PubMed] [Google Scholar]
- 29.Jacquot J. P., Stein M., Suzuki A., Liottet S., Sandoz G., Miginiac-Maslow M. (1997) FEBS Lett. 400, 293–296 [DOI] [PubMed] [Google Scholar]
- 30.De Pascalis A. R., Jelesarov I., Ackermann F., Koppenol W. H., Hirasawa M., Knaff D. B., Bosshard H. R. (1993) Protein Sci. 2, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melis A., Happe T. (2001) Plant Physiol. 127, 740–748 [PMC free article] [PubMed] [Google Scholar]
- 32.Mayhew S. G., Petering D., Palmer G., Foust G. P. (1969) J. Biol. Chem. 244, 2830–2834 [PubMed] [Google Scholar]
- 33.Hurley J. K., Weber-Main A. M., Stankovich M. T., Benning M. M., Thoden J. B., Vanhooke J. L., Holden H. M., Chae Y. K., Xia B., Cheng H., Markley J. L., Martinez-Júlvez M., Gómez-Moreno C., Schmeits J. L., Tollin G. (1997) Biochemistry 36, 11100–11117 [DOI] [PubMed] [Google Scholar]
- 34.Mayoral T., Martínez-Júlvez M., Pérez-Dorado I., Sanz-Aparicio J., Gómez-Moreno C., Medina M., Hermoso J. A. (2005) Proteins 59, 592–602 [DOI] [PubMed] [Google Scholar]
- 35.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 36.Morales R., Charon M. H., Hudry-Clergeon G., Pétillot Y., Norager S., Medina M., Frey M. (1999) Biochemistry 38, 15764–15773 [DOI] [PubMed] [Google Scholar]
- 37.Merchant S. S., Allen M. D., Kropat J., Moseley J. L., Long J. C., Tottey S., Terauchi A. M. (2006) Biochim. Biophys. Acta 1763, 578–594 [DOI] [PubMed] [Google Scholar]
- 38.Terauchi A. M., Lu S. F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J. N., Knaff D. B., Farmer P. J., Lemaire S. D., Hase T., Merchant S. S. (2009) J. Biol. Chem. 284, 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cinco R., Macinnis J., Greenbaum E. (1993) Photosynth. Res. 38, 27–33 [DOI] [PubMed] [Google Scholar]
- 40.Bes M. T., Parisini E., Inda L. A., Saraiva L. M., Peleato M. L., Sheldrick G. M. (1999) Structure 7, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 41.Nicolet Y., Piras C., Legrand P., Hatchikian C. E., Fontecilla-Camps J. C. (1999) Structure 7, 13–23 [DOI] [PubMed] [Google Scholar]