Abstract
NADPH-cytochrome P450 reductase (CPR), a diflavin reductase, plays a key role in the mammalian P450 mono-oxygenase system. In its crystal structure, the two flavins are close together, positioned for interflavin electron transfer but not for electron transfer to cytochrome P450. A number of lines of evidence suggest that domain motion is important in the action of the enzyme. We report NMR and small-angle x-ray scattering experiments addressing directly the question of domain organization in human CPR. Comparison of the 1H-15N heteronuclear single quantum correlation spectrum of CPR with that of the isolated FMN domain permitted identification of residues in the FMN domain whose environment differs in the two situations. These include several residues that are solvent-exposed in the CPR crystal structure, indicating the existence of a second conformation in which the FMN domain is involved in a different interdomain interface. Small-angle x-ray scattering experiments showed that oxidized and NADPH-reduced CPRs have different overall shapes. The scattering curve of the reduced enzyme can be adequately explained by the crystal structure, whereas analysis of the data for the oxidized enzyme indicates that it exists as a mixture of approximately equal amounts of two conformations, one consistent with the crystal structure and one a more extended structure consistent with that inferred from the NMR data. The correlation between the effects of adenosine 2′,5′-bisphosphate and NADPH on the scattering curve and their effects on the rate of interflavin electron transfer suggests that this conformational equilibrium is physiologically relevant.
Introduction
The cytochrome P450 mono-oxygenase system in the mammalian endoplasmic reticulum is responsible for oxidative metabolism of both endogenous compounds, including fatty acids, steroids, and prostaglandins, and exogenous compounds ranging from therapeutic drugs and environmental toxins to carcinogens (1). Within this enzyme system, NADPH-cytochrome P450 reductase (CPR4; EC 1.6.2.4) plays a central role by catalyzing the transfer of electrons from NADPH, via its two flavin cofactors FMN and FAD, to the cytochromes P450 (2). CPR is also an electron donor to heme oxygenase (3), the fatty acid elongation system (4), and cytochrome b5 (5). More recently, CPR has been found to carry out in vivo reductive activation of anticancer prodrugs in a hypoxia-specific manner, resulting in a markedly cytotoxic effect on tumors (6, 7), and has thus also become a target in anticancer therapy (8, 9). CPR is a member of a small family of diflavin reductases, including the isoforms of nitric-oxide synthase (10), methionine synthase reductase (11), protein NR1 (12), cytochrome P450 BM3 (13), and sulfite reductase (14), each of which catalyzes electron transfer through the pathway NAD(P)H → FAD → FMN → acceptor.
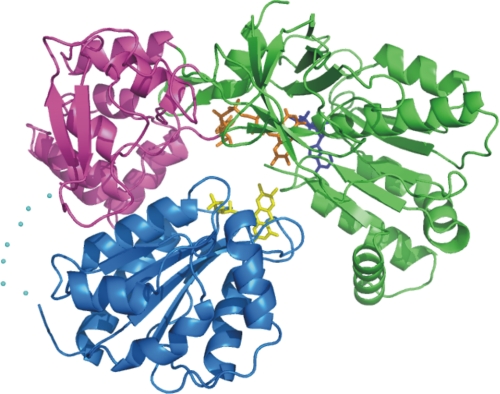
Sequence analysis indicates that CPR comprises three identifiable domains: a hydrophobic N-terminal domain that anchors the enzyme to the membrane, an FMN-binding domain homologous to bacterial flavodoxins, and an (FAD + NADPH)-binding domain homologous to ferredoxin-NADP+ reductases, leading to the proposal that the enzyme evolved as the product of a fusion of two ancestral flavoproteins (15). The latter two putative domains were expressed separately and shown to fold correctly and to bind their respective cofactors (16). The crystal structure of a soluble form of the enzyme lacking the membrane-anchoring N-terminal 70 residues (see Fig. 1) (17) confirmed the existence of these domains and their structural and sequence homology to flavodoxin and ferredoxin-NADP+ reductase, respectively. The structure reveals two additional important features: (i) the existence, as an insert in the FAD-binding domain, of a “linker domain” that may serve to determine the mutual orientation of the FMN- and FAD-binding domains and (ii) the fact that the FMN-binding domain is connected to the rest of the protein through a loop or hinge of 13 residues (positions 232–244) for which little electron density is observed in the crystal structure, implying that it is flexible.
FIGURE 1.
Crystal structure of the truncated soluble form of rat CPR. The FMN-binding domain is shown in blue, the linker domain in magenta, and the FAD-binding domain in green. The loop or hinge connecting the FMN-binding domain to the rest of the molecule, which is not seen in the electron density map, is indicated by blue dots. The cofactors are shown as stick structures: yellow for FMN, orange for FAD, and blue for NADPH. This structure is from Ref. 17 (Protein Data Bank code 1AMO).
The conformation of CPR seen in the crystal structure appears to be ideally suited for electron transfer from FAD to FMN, the two isoalloxazine rings being <4 Å apart (Fig. 1). However, it is more difficult to see how large electron acceptor proteins such as cytochrome P450 could, in this conformation of CPR, approach sufficiently closely to the FMN isoalloxazine ring for electron transfer to occur. As a result, domain movement in CPR is widely anticipated to play an important role in its physiological function (e.g. Refs. 2 and 18–21), and we suggested previously (2) that CPR might exist in an equilibrium between the structure observed in the crystal and one in which the FMN-binding domain could move freely with respect to the rest of the protein, being connected only by the flexible hinge sequence. However, differential scanning calorimetry measurements (22)5 are consistent with a single cooperative unfolding process, suggesting that the FMN-binding domain is closely associated with the FAD-binding domain. Experimental evidence supporting the importance of domain motion in the function of CPR comes from the observations that either the addition of high concentrations of glycerol (23) or the deletion of several amino acids from the flexible loop connecting the FMN-binding domain to the rest of the protein (22, 24) leads to a drastic decrease in the rate of interflavin electron transfer. A wide range of kinetic studies of the isoforms of nitric-oxide synthase, another member of the diflavin reductase family, have also been interpreted in terms of a movement of the FMN-binding domain as an obligatory part of the mechanism (see Refs. 25–27 and references therein).
Crystallographic studies of CPR mutants provide evidence for the existence of some mobility of the FMN-binding domain relative to the FAD-binding domain (28), whereas in the structure of the homologous flavoprotein subunit of sulfite reductase, the FMN-binding domain was absent from the electron density maps, implying a substantial degree of mobility (14). The most direct structural evidence for domain mobility has come from recent crystallographic studies of a mutant of rat CPR in which four residues (236TGEE239) were deleted from the hinge (24) and of a chimeric enzyme consisting of the yeast FMN-binding domain and the human linker and FAD-binding domains (29). In both cases, the mutants showed a change in the relative orientation and position of the FMN- and FAD-binding domains such that the FMN was farther from the FAD and more solvent-accessible.
We now report the use of a combination of NMR and small-angle x-ray scattering (SAXS) to address directly the question of domain organization in the soluble form of wild-type human CPR. These experiments provide evidence for the existence in CPR of a two-state equilibrium between a compact conformation (consistent with the crystal structure) and a more extended state that may be the form that transfers electrons to cytochromes P450. The position of this equilibrium is affected by the binding of ligands to the coenzyme-binding site in a way consistent with their effects on interdomain electron transfer.
EXPERIMENTAL PROCEDURES
Materials
NADPH, 2′,5′-ADP, NADH, and horse heart cytochrome c were purchased from Sigma. All other chemicals were of analytical grade.
Protein Expression and Purification
The gene for human fibroblast CPR lacking the N-terminal membrane-anchoring region (a kind gift from Professor C. R. Wolf, University of Dundee) was subcloned into pCS22, a plasmid vector in which expression of the target protein is under the control of the cold shock promoter (30). CPR was expressed in Escherichia coli BL21 Star cells carrying the pCS22 plasmid construct. Cells were grown to mid-log phase in 2YT broth at 42 °C prior to induction by reducing the growth temperature to 18 °C. 2H,15N-Labeled CPR was produced from cells grown in E. coli OD2 DN medium (Silantes). CPR was purified as described previously (31). The protein concentration was calculated using a molar extinction coefficient of ϵ450 nm = 22,000 m−1 cm−1. Cytochrome c reduction assays were carried out in 0.3 m potassium phosphate (pH 7.7) with 50 μm cytochrome c and 50 μm NADPH at 25 °C.
The segment of the CPR gene coding for the FMN domain (residues 61–239) was amplified by PCR, introducing NdeI and SapI restriction sites to enable cloning into the pTWIN2 vector (New England Biolabs) in-frame with the Mth RIR1 intein. This construct allowed expression of the isolated FMN domain with a C-terminal intein/chitin-binding domain affinity tag as a self-cleavable fusion protein. 2H,13C,15N-Labeled FMN domain was produced from cells grown in E. coli OD2 CDN medium (Silantes) at 28 °C and induced at mid-log phase by addition of 0.05 mm isopropyl β-d-thiogalactopyranoside. Harvested cells were resuspended in 20 mm HEPES (pH 7.0) containing 500 mm NaCl and 1 mm EDTA. After sonication and clarification, the extract was applied to a column of 10-ml chitin beads equilibrated in the same buffer. After thorough washing with this buffer, an on-column cleavage reaction was induced by equilibrating the chitin beads in 20 mm HEPES (pH 8.5) containing 500 mm NaCl, 4 mm dithiothreitol, and 1 mm EDTA. The reaction was allowed to continue overnight before elution of the FMN domain in the same buffer. Protein was stored at −20 °C in buffer containing 50% glycerol. The protein concentration was calculated using a molar extinction coefficient of ϵ450 nm = 11,000 m−1 cm−1.
NMR Spectroscopy
NMR samples of 1.5 mm 2H,13C,15N-labeled FMN-binding domain and 0.5 mm 2H,15N-labeled CPR were prepared in 30 mm BES (pH 6.5) containing 10% D2O. All NMR experiments were performed at 25 °C on Bruker AVANCE DRX 600 and AVANCE DRX 800 spectrometers equipped with CryoProbes. Proton chemical shifts were referenced to external 2,2-dimethyl-2-silapentanesulfonic acid. The 15N and 13C chemical shifts were referenced indirectly using recommended gyromagnetic ratios (32). Spectra were processed with TopSpin (Bruker) and analyzed using CCPN Analysis (33). Three-dimensional HNCO, HN(CA)CO, HNCACB, and HN(CO)CACB experiments were used to adjust reported assignments (34) of the isolated FMN-binding domain. Chemical shift differences between the isolated FMN-binding domain and the domain in the full CPR molecule were evaluated from comparison of the 1H,15N heteronuclear single quantum correlation (HSQC) spectra using the minimum chemical shift difference approach. All cross-peaks were picked in the CPR spectrum, and for each cross-peak in the FMN spectrum, the minimum distance to a CPR cross-peak (Δδ(HN,N)) was calculated using the equation Δδ(HN,N) = √[(ΔδHWH)2 + (ΔδNWN)2], where WH and WN are weighting factors for the 1H and 15N amide shifts, respectively (WH = 1, WN = 0.154; BMRB Data Bank; Refs. 35 and 36), and Δδ = δCPR − δFMN. The chemical shifts were compared automatically using a computer script without any attempt to adjust the cross-peak mapping between the two spectra.
Solution X-ray Scattering Data Collection and Analysis
SAXS measurements were carried out at Station 2.1 of the Science and Technology Facilities Council Synchrotron Radiation Source (Daresbury, UK) using a position-sensitive multiwire proportional counter. Scattering profiles were collected at two different sample-to-detector distances, 1 and 4.25 m, using sample concentrations of 15 μm (1 mg/ml), 45 μm (3 mg/ml), 75 μm (5 mg/ml), and 150 μm (10 mg/ml) in 30 mm BES (pH 7.0) at 25 °C. To prepare NADPH-reduced enzyme, solutions of NADPH and CPR were prepared anaerobically in syringes linked by flexible tubing, to give defined concentrations upon mixing, prior to transfer to the observation cell and collection of SAXS data.
Reduction and analysis of scattering data were performed as described previously (37). The radius of gyration (Rg), the maximum linear dimension (Dmax), the intraparticle distance distribution function (P(r)), and the forward scattering intensity were calculated from the experimental scattering data using the indirect Fourier transform method with the program GNOM (38). Model-independent molecular shapes were calculated using GASBOR (39). This ab initio procedure represents the protein as an assembly of spherical “dummy residues” and uses simulated annealing to find the three-dimensional arrangement of these dummy residues that gives optimum agreement with the experimental scattering data. Twenty independent GASBOR runs were averaged using DAMAVER (40) and SUPCOMB (41) to obtain a typical molecular shape. Scattering profile simulations from crystal coordinates were carried out using CRYSOL (42). The data were also analyzed using the ensemble optimization method (EOM) (43). In this approach, a pool of 10,000 conformers is calculated by starting from the crystal structure, defining the flexible “hinge” region, and, on this basis, moving the FMN-binding domain relative to the FAD-binding domain. The scattering curve for each of these conformers is calculated, and then a genetic algorithm is used to find the optimum combination of these scattering curves to account for the experimental data. This method is thus able to deal with mixtures of conformers.
RESULTS AND DISCUSSION
NMR Spectroscopy
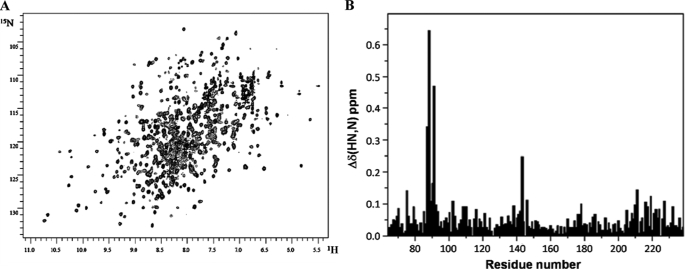
Despite its relatively large size (67 kDa), CPR gives reasonably well resolved NMR spectra (Fig. 2A). This allowed us to compare the 1H,15N HSQC spectra of 2H,15N-labeled CPR with the corresponding spectrum of the isolated FMN-binding domain, which we had previously assigned (34), to compare the environment of the residues in this domain in the two situations (Fig. 2B). The comparison was made using the “minimum chemical shift approach,” in which for each cross-peak in the assigned 1H,15N HSQC spectrum of the isolated FMN domain, we identified the nearest cross-peak in the unassigned spectrum of CPR. This conservative approach may underestimate the shift differences in crowded regions of the spectrum and will clearly not identify all those residues whose environment is different in the two situations, it but has proved to be a reliable indicator of the location of ligand-binding sites in proteins and will similarly provide a valuable indication of which parts of the FMN-binding domain interact with the rest of CPR. It should be noted that the crystal structure of the isolated FMN-binding domain (44) is closely similar to that of the corresponding domain within the crystal structure of CPR (17), with a root mean square deviation of 0.29 Å for the backbone atoms, so any 15N-1H chemical shift differences are very likely to reflect domain-domain interactions rather than structural differences.
FIGURE 2.
NMR spectroscopy of CPR. A, 1H,15N HSQC spectrum of CPR. The sample contained 0.5 mm 2H,15N-labeled CPR in 30 mm BES (pH 6.5) containing 10% D2O. The spectrum was obtained at 800 MHz with a sample temperature of 25 °C. B, chemical shift differences between the isolated FMN-binding domain and the domain in the full CPR molecule evaluated from a comparison of the 1H,15N HSQC spectra using the minimum chemical shift difference approach as described under “Experimental Procedures.”
Fig. 3 depicts those residues in the FMN-binding domain showing minimum NH chemical shift changes of ≥0.075 ppm, mapped onto the crystal structure of CPR. There are no chemical shift changes evident in residues in the “bottom hemisphere” of the FMN-binding domain in the orientation shown in Fig. 3, whereas, as would be expected, residues that are in contact with the FAD-binding domain in the crystal structure, notably those in the loops surrounding the FMN cofactor and in helix 85–93, do display clear chemical shift differences.
FIGURE 3.
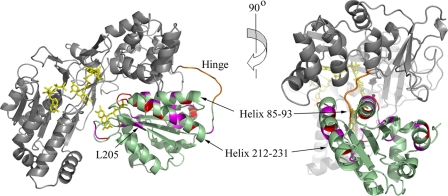
Residues in the FMN-binding domain showing NH chemical shift differences between CPR and the isolated FMN-binding domain mapped onto the crystal structure of CPR. The FMN-binding domain is in pale green, with residues showing chemical shift differences of ≥0.1 ppm highlighted in red and those showing chemical shift differences of between 0.075 and 0.1 ppm highlighted in magenta. The FAD-binding and linker domains are shown in gray, with the hinge (not defined in the crystal structure) in orange and the cofactors as yellow stick structures. Two views, related by a 90° rotation about the vertical axis, are shown.
However, it is notable that residues throughout the “upper hemisphere” of the FMN-binding domain in the orientation in Fig. 3 show differences in chemical shift; these are not limited to residues at the interface with the FAD-binding domain in the crystal structure. Thus, the helix comprising residues 212–231 (helix F (17)) contains many residues showing chemical shift differences, despite lying on a surface of the domain directed away from the interfaces with the linker and FAD-binding domains. Similarly, Leu205, which shows a clear difference in chemical shift, is in a short β-strand lying alongside this helix and is also distant from the interdomain interfaces. There is also a group of residues, including Val70, Lys75, Asn79, Ala102, Arg108, and Met110, at the far end of the FMN-binding domain from the FMN that show clear chemical shift changes; with the sole exception of Ala102, these residues are substantially solvent-exposed in the crystal structure.6
It follows that the FMN-binding domain must spend part of the time in a position that differs both from the crystal structure and from a freely moving “ball on a string” state because, in both these situations, the residues on helix F and at the far end of the domain from the FMN would be solvent-exposed and would thus have chemical shifts very similar to those in the isolated domain. The fact that residues in the FMN-binding domain that are distant from the interdomain interface seen in the crystal structure show chemical shift differences suggests that they may be part of a different interdomain interface.
Solution X-ray Scattering Studies
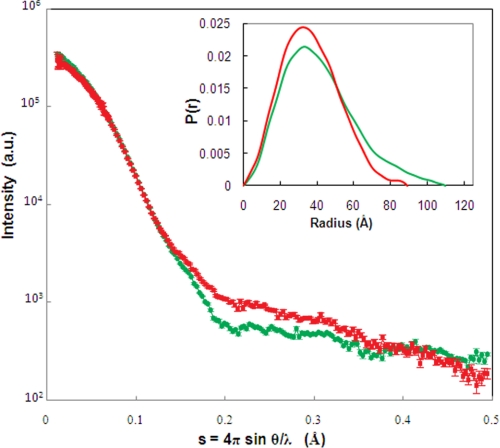
The evidence from NMR thus suggests that CPR exists in an equilibrium between (at least) two conformations that differ in the orientation of the FMN-binding domain. We investigated this further by SAXS studies of CPR under a number of different conditions. Fig. 4 compares the experimental scattering profiles of oxidized CPR (CPRox) (22) and 4-electron reduced CPR (CPRhq; prepared under anaerobic conditions with a 10-fold excess of NADPH).7 It is clear that these curves are not superimposable, demonstrating that CPR undergoes significant structural changes upon reduction by NADPH. This is further illustrated by the values for Dmax and Rg (distribution of scattering mass) calculated from the scattering data, which are presented in Table 1. The clear differences in these two parameters between CPRox and the fully reduced, coenzyme-bound enzyme show that CPR is able to adopt at least two distinct conformations in solution. The differences between the two conformations are even more apparent when comparing the P(r) functions, directly calculated from the scattering curves. This function provides a histogram of the interatomic distances within the molecule, giving information on shape and maximum size. The comparison of the P(r) functions for CPRox and CPRhq (Fig. 4, inset) clearly shows that CPR adopts different conformations in these two states, with CPRox having a more extended shape with a broader P(r) (in agreement with its larger Dmax and Rg values).
FIGURE 4.
Small-angle x-ray scattering of CPR. The experimental scattering profiles of CPRox (green) and CPRhq (prepared under anaerobic conditions with a 10-fold excess of NADPH; red) are compared. The inset compares the P(r) functions for CPRox and CPRhq, calculated using the indirect Fourier transform method with the program GNOM (38). a.u., arbitrary units.
TABLE 1.
Dmax and Rg values for oxidized and reduced CPRs in the presence and absence of coenzymes
Values were derived from SAXS data for the enzyme using the indirect Fourier transform method with the program GNOM (38). Estimated precision is ±2 Å for Dmax and ±0.2 Å for Rg.
| Sample | Dmax | Rg |
|---|---|---|
| Å | Å | |
| Oxidized | 110 | 32.3 |
| Oxidized + 2′,5′-ADP | 100 | 30.1 |
| 4e−-reduced (by NADH, no coenzyme remaining bound) | 109 | 32.4 |
| 2e−-reduced (by NADPH, with NADP+ remaining bound) | 99 | 30.5 |
| 4e−-reduced (by NADPH, with NADP+ remaining bound) | 89 | 28.1 |
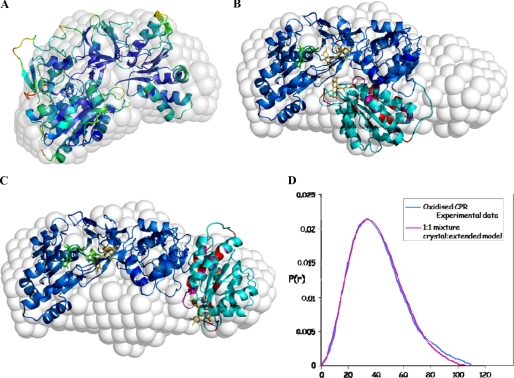
Shape reconstructions from the experimental scattering profiles of CPRox and CPRhq were performed using the established ab initio dummy residue procedure of the program GASBOR (39) and averaged by DAMAVER (40). Fig. 5A shows the low resolution molecular envelope obtained for CPRhq, the 4-electron reduced state. As implied by the Dmax and P(r) data reported above, the calculated envelope indicates a compact, more or less spherical shape for this state of the enzyme, which corresponds reasonably well to the crystal structure of rat CPR (17). The few parts of the crystal structure that extend somewhat outside the calculated envelope tend to be those with higher B-factors.
FIGURE 5.
Shape reconstructions from the experimental SAXS profiles of CPRox and CPRhq. Reconstructions were performed using the ab initio dummy residue procedure of the program GASBOR (39) as described under “Experimental Procedures.” A, low resolution molecular envelope obtained for CPRhq with the crystal structure superimposed. The crystal structure is colored by B-factor, from blue (low) to red (high). B and C, the molecular envelope obtained for CPRox. In B, the crystal structure is superimposed, and in C, a model of the extended structure (constructed as described under “Results and Discussion”) is superimposed. In B and C, the FAD-binding and linker domains are dark blue, and the FMN-binding domain is light blue, with the residues identified by NMR highlighted as described in the legend to Fig. 3. D, comparison of the experimental P(r) curve with that calculated for a 50:50 mixture of the compact (crystal) and extended (modeled) structures.
By contrast, the molecular envelope derived from the SAXS data for CPRox, the oxidized form of the enzyme without bound coenzyme, is significantly more elongated (Fig. 5B). In this case, the crystal structure is a very poor fit to the envelope, leaving one end of the calculated volume empty while the FMN-binding domain protrudes outside the envelope. This observation demonstrates clearly that the crystal structure is not the only conformation accessible to wild-type CPR in solution and supports the idea of domain reorganization within CPR upon changes in redox state and/or coenzyme binding.
Model for an Extended Conformer of CPR
Because the SAXS data show that in the oxidized state CPR can adopt a conformation that is more extended than that seen in the crystal, we sought to construct a model that accounts both for the SAXS data and for the information from NMR implicating a group of residues in and near helix F of the FMN-binding domain. We postulated that the FMN-binding domain might rotate across the surface of the linker domain to make a new interface involving these latter residues. This simple model of an extended CPR is shown in Fig. 5C. When superimposed on the molecular envelope for CPRox, it is apparent that this extended model does help to account for the extended molecular envelope, occupying space that is left empty by the crystal structure. However, it also leaves some unoccupied space midway along the envelope, which is occupied by the FMN-binding domain of the crystal structure in Fig. 5B.
Hence, neither of these two conformations alone is able to account for the molecular envelope of CPRox calculated from the SAXS data; rather, a good fit appears to require a mixture of the two. This is indicated by the P(r) plots in Fig. 5D, which show that a 50:50 mixture of the two does fit the experimental data.
Effects of Reduction and Ligand Binding
To further probe the effect of reduction and coenzyme binding on the shape of CPR, we collected SAXS data for CPR reduced to the 2-electron level by stoichiometric amounts of NADPH (CPR semiquinone; CPRsq), and CPR reduced to the 4-electron level by NADH. NADP+ binds tightly to the enzyme, so in the two samples reduced by NADPH, the reduced CPR will have NADP+ bound, whereas after reduction by NADH, the weakly binding NAD+ will not remain bound at the concentrations used (46). The Dmax and Rg values for these samples are given in Table 1, and the P(r) curves are in the supplemental figure. After addition of stoichiometric amounts of NADPH, CPRsq exists as a mixture of the FAD•/FMN• and FAD/FMNH2 species (23, 47) with NADP+ bound. In this case, as shown in Table 1, Dmax and Rg are decreased (corresponding to an increase in the proportion of the compact form) but not to the same extent as by the binding of NADPH and reduction to the CPRhq state. By contrast, reduction to the CPRhq state by NADH has essentially no effect on the shape of the enzyme as judged by the Dmax and Rg values, indicating that reduction alone is not sufficient to lead to the change in the position of the conformational equilibrium produced by addition of NADPH.
We also further analyzed the data on the 2′,5′-ADP complex reported previously (22). As shown in Table 1, the Dmax and Rg values for this sample are intermediate between the extremes represented by unliganded CPRox and NADP+-bound CPRhq and are essentially identical to the values for NADP+-bound CPRsq. Importantly, this shows that coenzyme binding alone, without any change in the redox state of the enzyme, is sufficient to produce a significant change in the position of the conformational equilibrium, although reduction to the CPRhq state, as well as coenzyme binding, appears to be required to see the full effect.
In a calorimetric comparison of the binding of 2′,5′-ADP to CPR and to the isolated FAD-binding domain (31), although the free energy changes upon 2′,5′-ADP binding were very similar, the changes in heat capacity and entropy were larger (more negative) for the FAD-binding domain than for intact CPR. There is thus a thermodynamic interaction between the coenzyme-binding site and the FMN-binding domain, and the effect of deletion of the flexible hinge region (22) indicates that this thermodynamic influence is related to domain movements. In our model of the extended conformation of human CPR, the solvent-accessible surface area is ∼900 Å2 greater than in a model in which the human sequence is built into the crystal structure of the rat enzyme, and the change in relative population of the two states will contribute to the thermodynamics of 2′,5′-ADP binding. An accurate estimate of this contribution must, however, await a better structural model of the extended conformation and a detailed SAXS study of the temperature dependence of the equilibrium.
We have shown that interflavin electron transfer in human CPR is kinetically gated by domain motion. In human CPR reduced to the 2-electron level with dithionite, the interflavin electron transfer rate is 11 s−1, but this is increased to 35 s−1 in the presence of 2′,5′-ADP and to 55 s−1 when the reduction is carried out by NADPH (i.e. when NADP+ is bound) (23, 48), correlating well with the effects of these ligands on the conformational equilibrium.
Analysis of the SAXS Data Using the EOM
Having shown that the scattering data could be accounted for by a mixture of conformers, we analyzed the data using the recently described EOM (43), which allows an objective approach to the analysis of scattering data from flexible proteins. This program uses a genetic algorithm to select an ensemble of conformers (from a pool of 10,000 possibilities), which account optimally for the experimental scattering data.
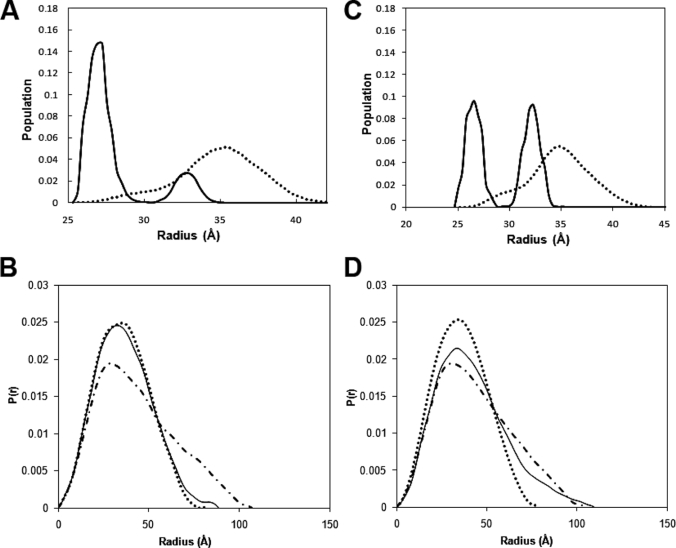
Fig. 6A shows the frequency distribution of conformers as a function of the Rg obtained by this analysis for CPRhq, with the P(r) curves shown in Fig. 6B. This indicates that the scattering curve is best described by an ∼85:15 mixture of two families of conformations with Rg values of ∼27 and ∼33 Å, respectively. This is in broad agreement with the conclusion drawn above, that the reduced enzyme with coenzyme bound exists in solution predominantly8 in a compact conformation similar to the crystal structure (hydrated Rg value calculated from the crystal structure = 26.6 Å), and indeed the fit to the experimental scattering curve is very much the same for the mixture derived from the EOM analysis as for the crystal structure. Representative structures for the compact form within the EOM ensemble clearly resemble the crystal structure (Fig. 7). Fig. 6 (C and D) shows the corresponding results of the EOM analysis for CPRox. Although the fit to the experimental data is less good than for the reduced enzyme (χ2 value of ∼12 versus ∼3 for the reduced enzyme), two families of conformations are again selected by the EOM analysis, with equivalent Rg values of ∼27 and ∼32 Å and similar widths, and these are structurally very similar to those obtained from the analysis of the data for the reduced enzyme (Fig. 7). However, in the case of the oxidized enzyme with no ligand bound, the proportions of the two conformational families reveal that equal amounts of the compact and extended forms give the best fit to the data8; in Fig. 6, the proportions are 50.3% compact and 49.7% extended conformation, in agreement with the basic analysis in Fig. 5.
FIGURE 6.
Analysis of SAXS data for CPR by the EOM. A and B, CPRhq; C and D, CPRox. A and C show the frequency distribution of CPR conformers as a function of Rg obtained by this analysis (solid lines) and the distribution of the initial pool of structures generated by the EOM (dotted lines). B and D compare the P(r) curves of the compact (dotted lines) and extended (dotted-dashed lines) conformations generated by the EOM analysis with the experimental P(r) curve (solid lines).
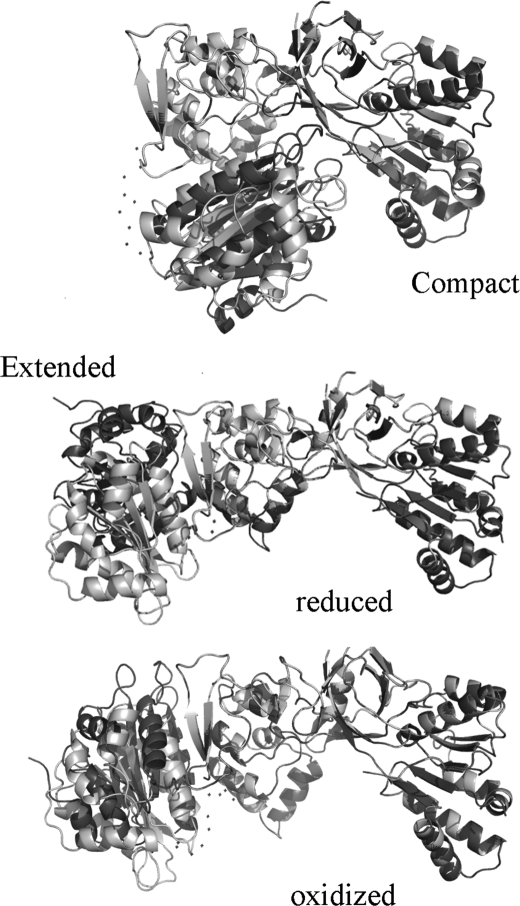
FIGURE 7.
Typical structures of CPR obtained from the EOM analysis. Structures are shown for the extended and compact forms, and in the case of the extended form, structures derived from the analysis of data from both CPRox and CPRhq are shown. In each case, several structures are shown superimposed on the FAD-binding domain to give an indication of the range of positions for the FMN-binding domain obtained from this analysis.
It is particularly notable that the EOM analysis provides independent evidence that CPR exists in a two-state equilibrium. The widths of each of the two peaks in Fig. 6 (A and C) are very similar to that of the single peak obtained by starting with the crystal structure, generating a theoretical scattering curve using the program CRYSOL, and analyzing this using the EOM. This suggests that each of the two forms is a reasonably well defined conformation. The two conformations are, within the limitations of the low resolution data, very similar for both the oxidized and reduced enzymes (Fig. 7); the compact conformation is similar to the crystal structure. The results of the EOM analysis are thus in agreement with the conclusions from the analysis with GASBOR, using our simple model for the extended form of the enzyme, that CPR reduced by NADPH exists essentially wholly in a compact conformation consistent with the crystal structure, whereas in the oxidized state the enzyme exists approximately half the time in an extended form.
The data for all the complexes listed in Table 1 were analyzed by the EOM. In each case, the experimental data were satisfactorily accounted for by a mixture of the same two families of conformations, with Rg values of ∼27 and ∼33 Å. The proportion of the extended conformation in the 2′,5′-ADP complex was estimated to be 33%, intermediate between the values estimated for the NADPH-reduced and oxidized enzymes; this is consistent with the discussion above on the basis of the Dmax and Rg values.
Relation to the Structures of Mutant CPRs
Hamdane et al. (24) have recently reported studies of a rat CPR mutant, ΔTGEE, in which four residues were deleted from the hinge region; this resulted in a marked decrease in the ability of the enzyme to catalyze reduction of cytochrome c or cytochrome P450 2B4 due to a defect in interflavin electron transfer. The crystal structure of the NADP+ complex of the ΔTGEE mutant shows a marked reorientation of the FMN-binding domain relative to the FAD-binding domain in each of the three molecules in the asymmetric unit (24). Molecule A has a relatively compact structure, but with the FMN-binding domain rotated relative to the FAD-binding domain so as to make the FMN more solvent-accessible; however, residues 211–222 and the residues at the far end of the domain from the FMN remain solvent-exposed, so molecule A cannot account for the NMR results reported here. Molecules B and C have more extended conformations, and the electron density for the FMN-binding domain is ill defined in both cases, suggesting a significant degree of mobility. Complete coordinates are not available for these less well ordered molecules, but modeling (on the basis of the incomplete coordinates and Fig. 1 in Ref. 24) suggests that both are broadly similar to the extended conformations shown in Fig. 7. Aigrain et al. (29) have reported the structure of a chimeric enzyme consisting of the FMN-binding domain from yeast CPR with the hinge and the linker and FAD-binding domains from human CPR. In this structure, the enzyme adopts a very extended conformation, with the FMN some 86 Å away from the FAD. This structure extends beyond the molecular envelope derived from the SAXS data for human CPRox and yields a calculated Rg value of 35.2 Å, which is significantly greater than the 32 Å obtained for the extended conformation of human CPR from the EOM analysis (see above). This structure is therefore not consistent with the SAXS data reported here for the human enzyme and in addition cannot account for our NMR data because the residues identified as showing chemical shift differences between the isolated FMN domain and intact CPR are solvent-exposed in the structure of the yeast-human chimeric CPR. Thus, although the exact domain positions in these two mutant CPR crystal structures cannot account for the NMR and SAXS data for human wild-type CPR presented here, they do lend clear support to the idea that CPR can exist in both compact and extended forms.
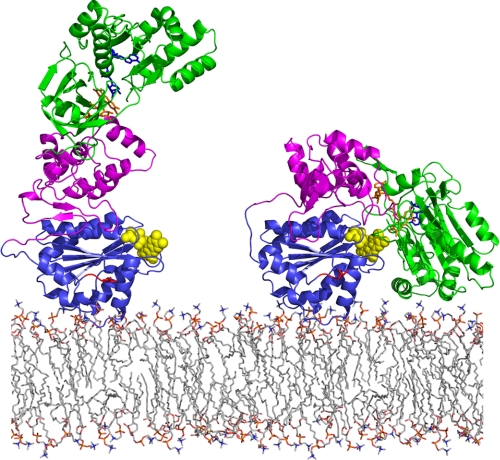
In conclusion, the combination of NMR and SAXS data presented here provides clear evidence for a model in which CPR in solution can be described by a two-state conformational equilibrium. The SAXS data indicate that one conformation of the enzyme is compact and that the other is more extended. Although analysis of the SAXS data can of course provide only low resolution pictures of these two conformations, the data are entirely consistent with the idea that the compact state, which predominates in the reduced, coenzyme-bound state, corresponds to the crystal structure. The extended conformation is as yet structurally less well defined, and experiments aimed at obtaining more detailed structural information are in progress. However, the NMR data indicate that a group of residues on and near the last helix of the FMN-binding domain must, in some state of the protein, interact with the rest of the protein. A model of the extended conformation was generated based on the NMR data, and a mixture of approximately equal amounts of this and the compact state is consistent with the SAXS data for the oxidized enzyme. If the models of the extended conformation in Figs. 5C and 7 are reasonably accurate, it is possible to envision a mechanism of domain reorganization wherein the FMN-binding domain slides or rolls across the surface of the linker domain to form a new interface involving the surface of the FMN-binding domain, including helices B (residues 86–102) and F (residues 212–231); residues on the hinge itself; and surface charges such as Glu388, Arg403, and Asp442 on the linker domain. This is shown in Fig. 8, which represents CPR on the membrane as it would be when attached by its N-terminal hydrophobic sequence. The extended conformation appears to be appropriate for electron transfer from FMN to electron acceptors such as cytochrome P450 because the FMN is readily accessible.
FIGURE 8.
Model of the conformational equilibrium in CPR. CPR is represented on the membrane as it would be when attached by its N-terminal hydrophobic sequence. Right, the compact form, represented by the crystal structure, appropriate for interflavin electron transfer. Left, the extended form, modeled as described under “Results and Discussion,” with the FMN exposed, appropriate for electron transfer to cytochrome P450. The FMN-binding domain is shown in blue, the linker domain in magenta, and the FAD-binding domain in green. The cofactors are shown: FMN, yellow space-filling representation; FAD, orange; and NADPH, blue.
The observation that the effects of ligand binding on the equilibrium between the extended and compact forms of the enzyme correlate well with the effects on the rate of interflavin electron transfer (22, 23, 48) strongly suggests that the extended form is physiologically relevant, and the observations with the ΔTGEE mutant (24) indicate that the same is true for the physiological electron transfer to cytochromes P450. Kinetic studies (23, 48) suggest that domain movement is rate-limiting for interflavin electron transfer, and there is evidence that a similar domain movement is rate-limiting in nitric-oxide synthase, another member of the diflavin reductase family (Refs. 25–27 and references therein). This indicates the crucial importance of this major structural change for the function of this enzyme.
Acknowledgments
We gratefully acknowledge the Science and Technology Facilities Council for awarding x-ray scattering beam time at the Daresbury Synchrotron Radiation Source. We are grateful to Professor C. R. Wolf for the gift of the gene for human fibroblast CPR lacking the N-terminal membrane-anchoring region.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council and the Wellcome Trust.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
A. Gutierrez, unpublished data.
In the crystal structure, the ϵ-amino group of Lys75 is within hydrogen-bonding distance of the carboxylate of Glu354, a residue on a loop of the linker domain; however, these residues are essentially completely solvent-exposed, and this interaction may not be significant in solution.
The 4-electron reduced species may not be physiologically relevant (45), but it represents a stable reduced state of the enzyme that can conveniently be studied under the conditions of the SAXS experiment.
The accuracy of the EOM is determined by the extent to which the large pool of structures it generates provides an adequate portrayal of all the potential conformers and by the procedure of the filtering process (43). Looking at the pool of conformations generated for CPR by the EOM (indicated by the dotted line in Fig. 6A, which represents the Rg distribution derived from the pool of structures), some bias toward extended structures is apparent. It is therefore possible that the family of structures characterized by the larger Rg value may be over-represented.
- CPR
- NADPH-cytochrome P450 reductase
- CPRox
- oxidized CPR
- CPRhq
- 4-electron reduced CPR
- CPRsq
- 2-electron reduced CPR (CPR semiquinone)
- SAXS
- small-angle x-ray scattering
- BES
- N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid
- HSQC
- heteronuclear single quantum correlation
- EOM
- ensemble optimization method.
REFERENCES
- 1.Ortiz de Montellano P. R. (ed) (2005) Cytochrome P450: Structure, Mechanism and Biochemistry, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 2.Paine M. J. I., Scrutton N. S., Munro A. W., Gutierrez A., Roberts G. C. K., Wolf C. R. (2005) in Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano P. R. ed) 3rd Ed., pp. 115–148, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 3.Schacter B. A., Nelson E. B., Marver H. S., Masters B. S. (1972) J. Biol. Chem. 247, 3601–3607 [PubMed] [Google Scholar]
- 4.Ilan Z., Ilan R., Cinti D. L. (1981) J. Biol. Chem. 256, 10066–10072 [PubMed] [Google Scholar]
- 5.Enoch H. G., Strittmatter P. (1979) J. Biol. Chem. 254, 8976–8981 [PubMed] [Google Scholar]
- 6.Bailey S. M., Lewis A. D., Patterson L. H., Fisher G. R., Knox R. J., Workman P. (2001) Biochem. Pharmacol. 62, 461–468 [DOI] [PubMed] [Google Scholar]
- 7.Saunders M. P., Jaffar M., Patterson A. V., Nolan J., Naylor M. A., Phillips R. M., Harris A. L., Stratford I. J. (2000) Biochem. Pharmacol. 59, 993–996 [DOI] [PubMed] [Google Scholar]
- 8.Jaffar M., Williams K. J., Stratford I. J. (2001) Adv. Drug Delivery Rev. 53, 217–228 [DOI] [PubMed] [Google Scholar]
- 9.Rooseboom M., Commandeur J. N., Vermeulen N. P. (2004) Pharmacol. Rev. 56, 53–102 [DOI] [PubMed] [Google Scholar]
- 10.Griffith O. W., Stuehr D. J. (1995) Annu. Rev. Physiol. 57, 707–736 [DOI] [PubMed] [Google Scholar]
- 11.Leclerc D., Wilson A., Dumas R., Gafuik C., Song D., Watkins D., Heng H. H., Rommens J. M., Scherer S. W., Rosenblatt D. S., Gravel R. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paine M. J., Garner A. P., Powell D., Sibbald J., Sales M., Pratt N., Smith T., Tew D. G., Wolf C. R. (2000) J. Biol. Chem. 275, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 13.Munro A. W., Leys D. G., McLean K. J., Marshall K. R., Ost T. W., Daff S., Miles C. S., Chapman S. K., Lysek D. A., Moser C. C., Page C. C., Dutton P. L. (2002) Trends Biochem. Sci. 27, 250–257 [DOI] [PubMed] [Google Scholar]
- 14.Gruez A., Pignol D., Zeghouf M., Covès J., Fontecave M., Ferrer J. L., Fontecilla-Camps J. C. (2000) J. Mol. Biol. 299, 199–212 [DOI] [PubMed] [Google Scholar]
- 15.Porter T. D., Kasper C. B. (1986) Biochemistry 25, 1682–1687 [DOI] [PubMed] [Google Scholar]
- 16.Smith G. C., Tew D. G., Wolf C. R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8710–8714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M., Roberts D. L., Paschke R., Shea T. M., Masters B. S., Kim J. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sevrioukova I. F., Li H., Zhang H., Peterson J. A., Poulos T. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall D. A., Vander Kooi C. W., Stasik C. N., Stevens S. Y., Zuiderweg E. R., Matthews R. G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9521–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayburt T. H., Sligar S. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6725–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyanagi T. (2005) Biochem. Biophys. Res. Commun. 338, 520–528 [DOI] [PubMed] [Google Scholar]
- 22.Grunau A., Geraki K., Grossmann J. G., Gutierrez A. (2007) Biochemistry 46, 8244–8255 [DOI] [PubMed] [Google Scholar]
- 23.Gutierrez A., Paine M., Wolf C. R., Scrutton N. S., Roberts G. C. (2002) Biochemistry 41, 4626–4637 [DOI] [PubMed] [Google Scholar]
- 24.Hamdane D., Xia C., Im S. C., Zhang H., Kim J. J., Waskell L. (2009) J. Biol. Chem. 284, 11374–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcin E. D., Bruns C. M., Lloyd S. J., Hosfield D. J., Tiso M., Gachhui R., Stuehr D. J., Tainer J. A., Getzoff E. D. (2004) J. Biol. Chem. 279, 37918–37927 [DOI] [PubMed] [Google Scholar]
- 26.Ilagan R. P., Tiso M., Konas D. W., Hemann C., Durra D., Hille R., Stuehr D. J. (2008) J. Biol. Chem. 283, 19603–19615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welland A., Garnaud P. E., Kitamura M., Miles C. S., Daff S. (2008) Biochemistry 47, 9771–9780 [DOI] [PubMed] [Google Scholar]
- 28.Hubbard P. A., Shen A. L., Paschke R., Kasper C. B., Kim J. J. (2001) J. Biol. Chem. 276, 29163–29170 [DOI] [PubMed] [Google Scholar]
- 29.Aigrain L., Pompon D., Moréra S., Truan G. (2009) EMBO Rep. 10, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mujacic M., Cooper K. W., Baneyx F. (1999) Gene 238, 325–332 [DOI] [PubMed] [Google Scholar]
- 31.Grunau A., Paine M. J., Ladbury J. E., Gutierrez A. (2006) Biochemistry 45, 1421–1434 [DOI] [PubMed] [Google Scholar]
- 32.Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 33.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 34.Barsukov I., Modi S., Lian L. Y., Sze K. H., Paine M. J., Wolf C. R., Roberts G. C. (1997) J. Biomol. NMR 10, 63–75 [DOI] [PubMed] [Google Scholar]
- 35.Ayed A., Mulder F. A., Yi G. S., Lu Y., Kay L. E., Arrowsmith C. H. (2001) Nat. Struct. Biol. 8, 756–760 [DOI] [PubMed] [Google Scholar]
- 36.Seavey B. R., Farr E. A., Westler W. M., Markley J. L. (1991) J. Biomol. NMR 1, 217–236 [DOI] [PubMed] [Google Scholar]
- 37.Grossmann J. G., Crawley J. B., Strange R. W., Patel K. J., Murphy L. M., Neu M., Evans R. W., Hasnain S. S. (1998) J. Mol. Biol. 279, 461–472 [DOI] [PubMed] [Google Scholar]
- 38.Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 39.Petoukhov M. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 540–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 41.Kozin M. B., Svergun D. I. (2001) J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 42.Svergun D. I., Barberato C., Koch M. H. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 43.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I. (2007) J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 44.Zhao Q., Modi S., Smith G., Paine M., McDonagh P. D., Wolf C. R., Tew D., Lian L. Y., Roberts G. C., Driessen H. P. (1999) Protein Sci. 8, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murataliev M. B., Feyereisen R., Walker F. A. (2004) Biochim. Biophys. Acta 1698, 1–26 [DOI] [PubMed] [Google Scholar]
- 46.Sem D. S., Kasper C. B. (1993) Biochemistry 32, 11548–11558 [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez A., Lian L. Y., Wolf C. R., Scrutton N. S., Roberts G. C. (2001) Biochemistry 40, 1964–1975 [DOI] [PubMed] [Google Scholar]
- 48.Gutierrez A., Munro A. W., Grunau A., Wolf C. R., Scrutton N. S., Roberts G. C. (2003) Eur. J. Biochem. 270, 2612–2621 [DOI] [PubMed] [Google Scholar]