Abstract
Many bacteria express phosphoenolpyruvate-dependent phosphotransferase systems (PTS). The mannitol-specific PTS catalyze the uptake and phosphorylation of d-mannitol. The uptake system comprises several genes encoded in the single operon. The expression of the mannitol operon is regulated by a proposed transcriptional factor, mannitol operon repressor (MtlR) that was first studied in Escherichia coli. Here we report the first crystal structures of MtlR from Vibrio parahemeolyticus (Vp-MtlR) and its homolog YggD protein from Shigella flexneri (Sf-YggD). MtlR and YggD belong to the same protein family (Pfam05068). Although Vp-MtlR and Sf-YggD share low sequence identity (22%), their overall structures are very similar, representing a novel all α-helical fold, and indicate similar function. However, their lack of any known DNA-binding structural motifs and their unfavorable electrostatic properties imply that MtlR/YggD are unlikely to bind a specific DNA operator directly as proposed earlier. This structural observation is further corroborated by in vitro DNA-binding studies of E. coli MtlR (Ec-MtlR), which detected no interaction of Ec-MtlR with the well characterized mannitol operator/promoter region. Therefore, MtlR/YggD belongs to a new class of transcription factors in bacteria that may regulate gene expression indirectly as a part of a larger transcriptional complex.
Introduction
Phosphoenolpyruvate-dependent phosphotransferase systems (PTS)2 primarily consist of a transmembrane transporter and enzymes responsible for phosphoryl group transfer from phosphoenolpyruvate to a transporter-bound sugar acceptor (1, 2). The PTS are generally substrate-defined, and their components may vary across species (2, 3). d-Mannitol, or 1,2,3,4,5,6-hexanehexol, is a polyol formed by reduction of mannose or fructose. It is one of the hexitols involved in bacteria catabolism pathways, where it serves as a source of fermentable sugar (4, 5). The d-mannitol-specific PTS was first discovered and sequenced in Escherichia coli (6–8). Taking into consideration that the system was regulated like other hexitol PTS systems, an open reading frame was sought and identified within the mannitol operon (9). Experiments indicated that the loss of the gene (mtlR) led to the constitutive expression of the operon, and its gene product MtlR was proposed to be a transcription factor: mannitol operon repressor (MtlR, COG2213) (9). Two putative DNA operator palindromes that might serve as MltR binding sites were identified within the operator-promoter region.
Besides MtlR, a typical mannitol operon also encodes a mannitol-specific PTS system ABC transporter II component (MtlA, COG2213) and a mannitol-1-phosphate 5-dehydrogenase (MtlD, COG0246). The mannitol operon is conserved and has been cloned from many Gram-negative bacterial families, such as Shigella (10), Salmonella (11), Yersinia (12), Klebsiella (13, 14), and Vibrio (15). For example, in the genome sequence of Vibrio parahemeolyticus RIMD 2210633 (15), a typical mannitol operon, including MtlR, MtlA, and MtlD, has been identified on chromosome I. A paralogue gene yggD annotated as putative transcriptional regulator was found clustered with cmtA and cmtB on chromosome II of V. parahemeolyticus. The cmtA and cmtB genes seem to encode the equivalents of MtlA components according to a recent study (16).
In genomes of many other Gram-negative bacteria, a gene named yggD, encoding a sequence homolog of MtlR, has also been identified. The gene yggD is not clustered with mtlA and mtlD as mtlR in a typical mannitol operon. Its gene neighbors vary considerably among organisms, even among strains, and its position provides little insight into its function. In the complete genome sequence of Shigella flexneri serotype 2a strain 2457T (10), for example, the yggD gene is located about 1 megabase pair away from the mannitol operon on the same strand of DNA (10). Its neighboring genes include uncharacterized yggC and yggF. The latter may encode a fructose-1,6-bisphosphatase II-like protein (10). The function of YggD has never been described. However, based on its sequence similarity to MtlR, it has been assigned to the same protein family, Pfam05068 (mannitol operon repressor) (17).
The protein structural information of mannitol operon components has been limited to the IIA domain of MtlA from E. coli (18) and Salmonella typhimurium LT2 (Protein Data Bank code 1XIZ). The enzyme IIA consists of a central β-sheet flanked by α-helices on both sides, which is remarkably different from IIA proteins that belong to the other three EIIs groups of PTS (18). To understand the structure, molecular mechanism, and regulation of the mannitol operon, we have expressed and purified a number of MtlR and YggD from different bacterial species. Several expression vectors were used to improve protein solubility, and reductive methylation was applied to augment crystallization (19).
Here we present the crystal structures of MtlR from V. parahemeolyticus (Vp-MtlR) and YggD from S. flexneri (Sf-YggD), the first structures of the mannitol operon repressor family. The structures of Vp-MtlR and Sf-YggD were determined at 2.75 and 2.50 Å resolution, respectively, and represent a novel all α-helical fold. Although Vp-MtlR and Sf-YggD share low sequence identity, they are structurally very similar and also form similar dimers. The analysis of their structural features and surface electrostatic properties in combination with DNA-binding studies of Ec-MtlR suggests that MtlR/YggD are unlikely to bind DNA directly. We also provide direct biochemical evidence that the MtlR does not bind the regulatory region of the E. coli mannitol operon. Therefore, MtlR seems to belong to a new class of transcription factors in bacteria and may regulate gene expression indirectly, perhaps as a part of a larger transcriptional complex.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
A truncated on C terminus mtlR gene from V. parahemeolyticus RIMD 2210633 was cloned into pMCSG7 vector (20). The gene codes Vp-MtlR without four C-terminal residues, D173SPF. The protocols for the expression in E. coli BL21 (DE3) and purification of the Se-Met-labeled recombinant protein were reported previously (21). The Vp-MtlR expressed well with a final protein yield of about 195 mg/liter of culture.
The full-length yggD gene from S. flexneri 2a 2457T was initially cloned into pMCSG7 vector. The solubility of Sf-YggD was very low, with a soluble protein yield of ∼5 mg/liter of culture. To improve the protein production, pMCSG19 expression vector was utilized (22), which bears a fusion construct with maltose-binding protein followed by the TVMV protease cleavage site, His6-tag, and TEV protease cleavage site fused into the N terminus of the target protein. The expression and purification protocols of Se-Met-labeled recombinant Sf-YggD were the same as for Vp-MtlR (21). With the use of pMCSG19 vector, the Sf-YggD yield increased to 62 mg/liter of culture.
The expression constructs for Ec-MtlR, FruR1 (coding for the first 57 amino acids of fructose repressor (FruR)), and FruR2 (the full-length protein) were cloned into the NdeI and XhoI restrictions sites of pET28. The resulting constructs consisting of an N-terminal His6 tag and a thrombin cleavage site were expressed in BL21(DE3) and purified as reported earlier (23). The gene for full-length Ec-MtlR was also cloned into pMCSG7 vector. The protein was expressed and purified as reported previously (21).
Reductive Methylation of Sf-YggD Protein
A reductive methylation of lysine residues of Sf-YggD was performed to improve protein crystallization properties (19). During reductive methylation, the protein is modified through the addition of two methyl groups to Lys residues. The concentration of the protein was first adjusted to about 10 mg/ml. Dimethylamine-borane complex (ABC; 20 μl of 1 m stock) and formaldehyde (40 μl of 1 m stock) were added per 1 ml of the protein solution. The mixture was incubated for 2 h at 4 °C. The procedure was then repeated for the second time. After that, only ABC (10 μl per 1 ml of protein solution) was added, and the mixture was incubated overnight. On the second day, 1 ml of 5 mg/ml glycine and 1 m dithiothreitol to a final concentration of 5 mm were added, and the reaction mixture was incubated on ice for 2 h. The protein was finally dialyzed against crystallization buffer and concentrated.
Limited Proteolysis of Ec-MtlR
Ec-MtlR was first treated with trypsin and chymotrypsin and separated on an SDS-polyacrylamide gel, which shows apparent reduction of molecular mass. The digest was then redone with 100 μg of protein, stopped after 6 h with 1 mm phenylmethylsulfonyl fluoride, and analyzed by mass spectrometry. The mass of 20,359.6 Da corresponds to a fragment of Ec-MtlR truncated of the first 14 N-terminal amino acids (theoretical mass 20,351 Da).
Size Exclusion Chromatography
Size exclusion chromatography was performed on a Superdex-200 10/300 GL column (GE Healthcare) using AKTATM Xpress. The column was pre-equilibrated with crystallization buffer (20 mm HEPES, pH 8.0, 250 mm NaCl, 2 mm dithiothreitol) and calibrated with premixed protein standards, including ribonuclease A (13.7 kDa), chymotrypsinogen A (25 kDa), ovalbumin (43 kDa), albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), and blue dextran (2,000 kDa). For each run, a 5-ml protein sample at a 10-mg/ml concentration was injected into the column. The chromatography was carried out at 4 °C at a flow rate of 2 ml/min. The calibration curve of Kav versus log molecular weight was prepared using the equation, Kav = Ve − Vo/Vt − Vo, where Ve represents the elution volume for the protein, Vo is column void volume, and Vt is total bed volume.
Protein Crystallization
The Se-Met-labeled Vp-MtlR and unmethylated and methylated Sf-YggD proteins were screened for crystallization conditions with the help of the Mosquito robot (TTP Labtech), using the sitting drop vapor-diffusing technique in a 96-well CrystalQuick plate (Greiner). For each condition, 0.4 μl of protein (79 mg/ml for Vp-MtlR and 66 mg/ml for Sf-YggD) and 0.4 μl of crystallization formulation were mixed; the mixture was equilibrated against 135 μl of the reservoir in the well. Crystallization screens used were Index (Hampton Research), Wizard (Emerald Biosystems), and SM4 (Nextal) at both 4 and 18 °C. Diffraction quality crystals of Vp-MtlR appeared under the condition of 0.2 m triammonium citrate, pH 7.0, 20% (w/v) polyethylene glycol 3350. The crystallization condition of methylated Sf-YggD crystals was 0.1 m sodium acetate, 0.1 m MES, pH 6.5, 30% (w/v) polyethylene glycol 2000MME. Prior to data collection, crystals were treated with cryoprotectant complemented from crystallization buffer. For Vp-MtlR, 25% (v/v) glycerol was added. For methylated Sf-YggD, 20% (v/v) glycerol was added, and polyethylene glycol 2000MME was reduced to 20%. No crystal was produced from unmethylated Sf-YggD protein. Extensive screenings of crystallization conditions for Ec-MtlR and its proteolytic fragment were also performed, and no crystal was obtained.
DNA-binding Experiments of Ec-MtlR Electrophoretic Mobility Shift Assay
80 fmol of 5′-32P-labeled DNA-oligonucleotides were incubated with a 4–10-fold excess of protein in TMD-buffer (20 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 1 mm dithiothreitol). Bovine serum albumin was added in a ratio of 100:1 to the protein under investigation to prevent nonspecific binding. The samples were incubated for 30 min and separated on a vertical polyacrylamide gel. An x-ray film was exposed for 16 h at −70 °C. Additional electrophoretic mobility shift assay was carried out on a minigel electrophoresis system (Invitrogen) using a Native PAGE™ 4–16% gradient gel (Invitrogen) and Tris-borate-EDTA buffer (see supplemental material).
Surface Plasmon Resonance
An IASys single channel resonant mirror biosensor (Fisons Applied Sensor Technology, Cambridge, UK) (34, 35) was used to measure the affinity of Ec-MtlR constructs to DNA. The 5′-biotin-labeled oligonucleotides mtlr1_for (5′-CTCGGGCTTCCAGCCTGCG) and mtlr2_for (5′-ACATAAGAAGGGGTGTTTTTATGT) and their complementary strands were obtained from Microsynth, dissolved, and annealed according to the standard procedure. A 100–200-μg excess of streptavidin (Pierce) was coupled to a sensing cuvette manufactured with a biotin layer, and the biotinylated double-stranded DNA was then coupled to the immobilized streptavidin. All binding reactions were performed in phosphate-buffered saline, pH 7.2, 25% glycerol, and a range of additional NaCl concentrations between 100 and 300 mm were tested to optimize the binding.
A SELEX-based Method for Measuring DNA-binding Specificities
A SELEX-based binding site selection assay was developed for the identification of protein-DNA binding specificities. Wells of Nunc immunoplates were coated with 400 μl of 1 mg/ml concentrations of the proteins under test and incubated for between 1 and 8 h. The wells were washed with Tris-HCl buffer (pH 7.4) prior to the binding reaction. 50 μl of binding reaction buffer containing 200 ng of poly(dI-dC)-poly(dI-dC) and 0.2 ng of the random oligonucleotide R76 were added to each test well. The oligonucleotide R76 comprised a random 26-base sequence flanked by two 25-nucleotide PCR primers as follows: 5′-CAGGTCAGTTCAGCGGATCCTGTCG(A/G/C/T)26GAGGCGAATTCAGTGCAACTGCAGC. Plates were incubated for 4 h with shaking. After the binding reaction, wells were washed thoroughly with buffer E, and the bound DNA was eluted with 250 μl of 500 mm NaCl. 2-μl aliquots of the eluate were amplified by PCR. The amplified DNA was then rebound to the protein from which it was eluted, and the whole binding, elution, and amplification procedure was repeated five times to facilitate greater accuracy in the selection of DNA sequences. The final amplification of the selected DNA sequences was performed using Eppendorf Taq polymerase, which produces 3′-A overhangs and facilitates cloning into the pGEMT Easy vector system (Promega). The blue/white screening was used to select clones containing inserts, and the selected DNA was sequenced.
X-ray Diffraction Data Collection
Diffraction data were collected at 100 K at the 19ID and 19BM beamlines of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory using the program SBCcollect. A multiple-wavelength anomalous diffraction data set and a single-wavelength anomalous diffraction data set were collected at the wavelengths near selenium absorption peaks from Se-Met-labeled protein crystals of Vp-MtlR and Sf-YggD, respectively. All data were processed and scaled with the HKL3000 suite (24) (Table 1).
TABLE 1.
Crystallographic statistics
| Vp-MtlR | Sf-YggD | |
|---|---|---|
| Data collection | ||
| Space group | P41 | C2 |
| Unit cell | ||
| a (Å) | 63.28 | 71.37 |
| b (Å) | 63.28 | 70.95 |
| c (Å) | 227.1 | 159.0 |
| α (degrees) | 90 | 90 |
| β (degrees) | 90 | 90.43 |
| γ (degrees) | 90 | 90 |
| Wavelength (Å) | 0.97927, 0.97941a | 0.97883 |
| Resolution (Å) | 46-2.75 | 32-2.50 |
| No. of unique reflections | 23,045b | 26,126b |
| Redundancy | 4.8 | 4.0 |
| Completeness (%) | 99.2 (91.6)c | 94.2 (78.9)c |
| Rmerge (%) | 10.0 (64.2)c | 7.4 (51.9)c |
| I/σ(I) | 17.5 (1.5)c | 30.8 (1.9)c |
| Phasing | ||
| RCullis (anomalous) (%) | 89, 94a | 63 |
| Figure of merit (%) | 23.8 | 27.6 |
| Refinement | ||
| Resolution | 46-2.75 | 32-2.50 |
| Reflections (work/test) | 21,778/1443 | 24,064/2020 |
| Rcrystal/Rfree | 20.7/27.3 | 20.7/27.0 |
| Bond length (Å)/angle (degrees) r.m.s. deviation from ideal geometry | 0.018/1.93 | 0.014/1.72 |
| Protein atom average B value (Å2), main chain/side chain | 51.1/52.8 | 63.2/63.8 |
a Peak and inflection.
b Including Bijvoet pairs.
c Last resolution bin.
Structure Determination and Refinement
The Vp-MtlR and Sf-YggD crystal structures were determined using multiple-wavelength anomalous diffraction and single-wavelength anomalous diffraction methods, respectively. For both structures, selenium sites were first located using the program SHELXD (25), and they were used for initial phasing with the program MLPHARE (26). After density modification (26), partial models (Vp-MtlR, 378 residues with 26 side chains placed; Sf-YggD, 392 residues with 17 side chains placed) were obtained from automatic model-building trials using the program RESOLVE (27). All of the above programs are integrated within the program suite HKL3000 (24). After cycles of model buildings using the program COOT (28), the models were refined using the program REFMAC (29) (Table 1). After final refinements, electron density calculated at 1.0 σ is well connected for main chains of three Vp-MtlR monomers. There were density breaks between Met-127 and Asp-138 for the fourth Vp-MtlR monomer. In case of Sf-YggD, electron density breaks were observed only at the loop region from residue Phe-137 to residue Lys-142 in three of four monomers. Electron densities were observed for methyl groups of 22 of 32 lysine residues in the methylated Sf-YggD, and these methyl groups were built into the structural model.
RESULTS
Expression, Purification, Crystallization, and Structure Determination
The recombinant proteins Vp-MtlR and Sf-YggD were overexpressed in the E. coli BL21(DE3) strain and purified to homogeneity using His6 tag-specific immobilized metal affinity chromatography columns as described previously (21). Sf-YggD was further subjected to reductive methylation of lysine residues to improve crystallization (19). The x-ray diffraction data of Vp-MtlR (2.75 Å) and Sf-YggD (2.50 Å) were both collected near the selenium absorption K-edge of their Se-Met-labeled protein crystals (Table 1) with their structures solved by multiple-wavelength anomalous diffraction and single-wavelength anomalous diffraction techniques, respectively (Table 1). The Ec-MtlR and its proteolytic fragment failed to crystallize.
Structures of Vp-MtlR and Sf-YggD Monomers
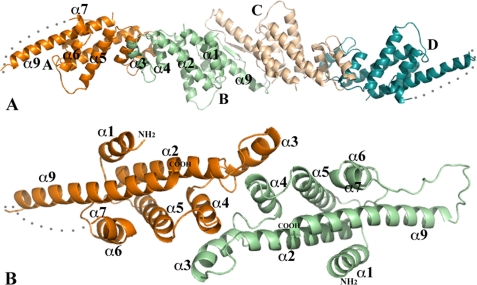
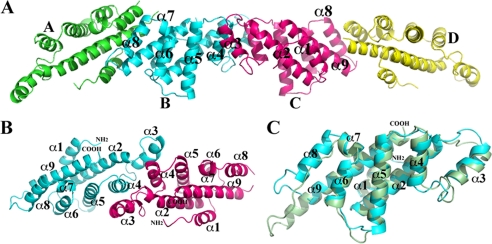
There are four monomers in one asymmetric unit of both Vp-MtlR and Sf-YggD crystals (Figs. 1A and 2A). Although different in their molecular packing, the structures of the MtlR and the YggD monomers are highly similar. Each monomer assumes an all α-helical fold rather than the predicted α/β structure of E. coli MtlR (9). The Sf-YggD monomer has nine α-helices arranged in three layers (Fig. 2). In Vp-MtlR the α8 helix is replaced by a flexible loop (Figs. 1B and 2C). Except for the short α7 helix within the loop between α6 and α8, all helices that are adjacent in the amino acid sequence are antiparallel. The first layer includes α4, α5, α6, and α8 as well as the short helix α7. The four helices α4, α5, α6, and α8 are antiparallel to each other with a relative rotation of about 20° to each other. The second layer or the central helical sheet includes α2, α3, and α9. The α2 and α9 helices are parallel to each other. They are the longest and the most prominent helices of the structure. The second layer has a rotation of about 50° relative to the first layer. The packing between two adjacent individual α-helices within a helical layer or between two helical layers is largely representative of two typical helix-helix packing modes, with the ridges of one helix fitting into grooves of the other and vice versa (30). A single N-terminal helix (α1) is stacked onto the second layer, interacting with α2 and α9. The interactions between these three helices are similar to what was observed in a coiled-coil helical structure, although there are only two layers of hydrophobic interactions in both Vp-MtlR and Sf-YggD. Furthermore, the majority of interhelical interactions in both monomers are predominantly hydrophobic in nature, suggesting the high stability of this all α-helical fold.
FIGURE 1.
Crystal packing of Vp-MtlR monomers and Vp-MtlR dimer. A, the four Vp-MtlR monomers in one asymmetric unit. The monomers A and B form a dimer, and monomers C and D form a dimer. Both dimers are of 2-fold pseudosymmetry. For clarity, only the front helices of the AB-dimer are labeled. The AB-dimer and CD-dimer form extensive interactions across their interface and potentially form a tetramer, as discussed under “MtlR and YggD Dimers and Tetramers.” B, a ribbon drawing of a Vp-MtlR dimer (AB-dimer). The orientation of the dimer is related to its position in A by a rotation about 90° around the horizontal axis. All helices of the novel layered α-helical fold are labeled. The α8 helix is missing in Vp-MtlR in comparison with the structure of Sf-YggD shown in Fig. 2. This figure and other ribbon diagram figures were prepared using the program PyMOL (available on the World Wide Web).
FIGURE 2.
Crystal packing of Sf-YggD monomers and Sf-YggD dimer. A, the four Sf-YggD monomers in one asymmetric unit. The monomers B and C form a pseudo-2-fold dimer, whereas monomers A and D form dimers with their 2-fold symmetry-related monomers, respectively. For clarity, only front helices of the BC-dimer are labeled. The interaction between monomers A and B is different from the interaction between monomers C and D, by skipping one turn of the helix. They represent two types of primary dimer-dimer contacts found in the structure. B, a ribbon drawing of a Sf-YggD dimer (BC-dimer). The orientation of the dimer is related to its position in A by a rotation about 90° around the horizontal axis. The α8 helix is unique to the YggD structure. C, a structural alignment of Vp-MtlR (dark green) and Sf-YggD (cyan) monomers.
There is no significant conformational variation between four MtlR monomers in Vp-MtlR. A pairwise superposition of four MtlR monomers results in root mean square (r.m.s.) deviation values within a range of 0.66–1.20 Å. The primary variable region is the loop between α7 and α9, which has poorly defined electron density in three of four monomers. The loop also influences conformation changes in the N-terminal region of the α9 helix. A similar pairwise superposition of four YggD monomers results in much smaller r.m.s. deviation values (0.40–0.61 Å). The only variation was found in the disordered or partially disordered loop region between α8 and α9. Multiple sequence alignment (Fig. 3) suggests that the region between α7 and α9 is the most variable part in MtlR and YggD. A typical superimposition of MtlR and YggD monomers gives an r.m.s. deviation of 1.60 Å with 140 residues from each monomer aligned (Fig. 2C). The primary difference between the two structures is the replacement of the α8 helix in Sf-YggD by a flexible loop in Vp-MtlR as discussed earlier. In addition, the α3 helix shows a small conformational variation; α3 is an edge helix and is involved in dimerization of MtlR and YggD.
FIGURE 3.
Multiple sequence alignment of MtlR and YggD. The identical residues are highlighted in red, and the similar residues are highlighted in cyan. *, residues forming the hydrophobic core of MtlR and YggD; ⋀, residues contributing to their dimerization. The helices of Vp-MtlR and Sf-YggD structures are underlined with blue coils. The first 14 N-terminal amino acids of Ec-MtlR were removed with limited proteolysis. The proteins used in the alignment include Ec-MtlR, Vp-MtlR I (from chromosome I), Sf-YggD, E. coli YggD (Ec-YggD), and Vp-MtlR II (from chromosome II). The S. flexneri MtlR (Sf-MtlR) and S. sonnei MtlR I (Ss-MtlR) are identical to Ec-MtlR in primary sequences. The S. dysenteriae MtlR (Sd-YggD) is identical to Sf-YggD.
MtlR and YggD Dimers and Tetramers
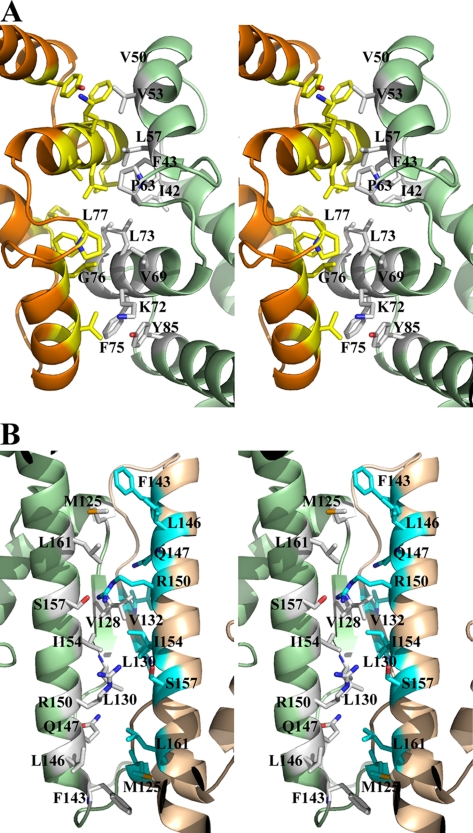
Analysis of crystal packing and interactions across monomer/monomer interfaces suggests that the most likely biological unit of Vp-MtlR and Sf-YggD is a dimer. In Vp-MtlR, monomers A and B and monomers C and D form two dimers (AB-dimer and CD-dimer) related through pseudo-2-fold axis with total buried surface areas of 1585 and 1554 Å2, respectively (Fig. 1). In Sf-YggD, monomers B and C form a similar BC-dimer with a total buried surface area of 1735 Å2 (Fig. 2, A and B), whereas monomer A and monomer D form dimers with their symmetry-related mates, respectively. The resulting buried surface areas of these two symmetric dimers are 1764 and 1783 Å2, respectively. These values are consistent with the “standard size” protein interfaces in which the total area buried by the components in the protein/protein interface is 1600 ± 400 Å2 (31). Both MtlR and YggD dimers are formed by the contacts mainly between the α2, α3, and α4 helices of two monomers (Figs. 1B and 2B), with α3 and α4 providing the most important hydrophobic interactions. As a result of the dimerization, an extended five-layered helical structure forms, with the central layer contributed primarily by six (MtlR) or eight (YggD) α-helices (Figs. 1B and 2B). The dimerization interface is highly hydrophobic, and the residues involved in the interactions across the interface include Ile-42(MtlR)/Leu-41(YggD), Phe-43/Phe-42, Val-44/Val-43, Ala-49/Ile-49, Val-53/Ala-53, Val-54/Val-54, Leu-57/Leu-57, Phe-63/Phe-63, Val-69/Val-69, Lys-72/Arg-72, Leu-73/Leu-73, Phe-75/Tyr-75, Gly-76/Ala-76, Leu-77/Leu-77, and Tyr-85/Tyr-85 (Fig. 4A). A total of 8 aromatic residues from two monomers are involved in the interaction across the interface in each case. It is remarkable that these residues contributing to the dimer interface are either identical (69%) or highly conserved (31%) in the family, although the sequence identity between Vp-MtlR and Sf-YggD is only 22% (Fig. 3). Multiple-sequence alignment shows the region including the α3 and α4 helices as being the most conserved across different species. Additionally, neither a specific hydrogen bond nor a salt bridge is observed across the dimer interface.
FIGURE 4.
The dimer interface and potential tetramer interface of Vp-MtlR. A, stereo ribbon drawing of the dimeric interface of the AB-dimer as shown in Fig. 1. The predominant hydrophobic residues involved in the dimer formation are drawn in sticks and colored differently according to monomers. Since the dimer is of a pseudo-2-fold symmetry, only those residues from one monomer are labeled. B, stereo ribbon drawing of the potential tetrameric interface, which corresponds to the interface between the monomers B and C and represents the interaction between the AB-dimer and the CD-dimer as shown in Fig. 1A. The residues from two dimers involved in the dimer-dimer interactions are drawn in sticks and colored in different codes. The two flexible loops between α7 and α9 helices form a pair of antiparallel strands with five main chain-main chain hydrogen bonds.
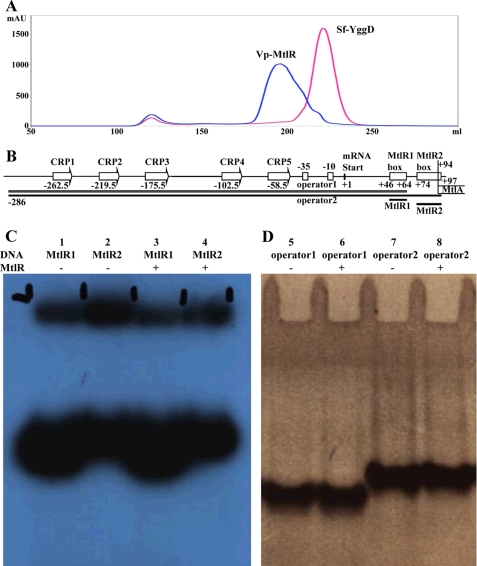
In the Sf-YggD crystal, all contacts between dimers seem insignificant, suggesting that no higher oligomers can be formed. The observation agrees with the results of size exclusion chromatography. The elution profiles of Sf-YggD showed a single major peak (Fig. 5A) with an apparent molecular mass of 47.8 kDa (data not shown). The calculated molecular mass of the Sf-YggD monomer from its amino acid sequence is 19.5 kDa (including 3 N-terminal vector-derived residues, SNA), and estimated molecular masses for the dimer and trimer are 39.0 and 58.5 kDa, respectively. Therefore, it appears that Sf-YggD is a dimer in the solution. Its slightly larger apparent molecular mass obtained from size-exclusion chromatography might be a result of its elongated shape.
FIGURE 5.
Size exclusion chromatography and DNA-binding studies. A, size exclusion chromatographies of Vp-MtlR and Sf-YggD. The elution peak of Sf-YggD is sharp, and its apparent molecular size was estimated to be 47.8 kDa, slightly larger than a Sf-YggD dimer. The Vp-MtlR peak is broad, and the eluted Vp-MtlR was estimated to be 90.4 kDa, roughly a tetramer size. A possible mixture of oligomers is proposed, as discussed under “DNA-binding Studies of Ec-MtlR.” B, a schematic drawing of the mannitol operator/promoter region and the four constructs used in electrophoretic mobility shift assay experiments (operator 1, operator 2, MtlR1, and MtlR2). Shown is an electrophoretic mobility shift assay with DNA fragments corresponding to the two palindromic boxes MtlR1-box and MtlR2-box (C) and the whole operator region (D).
The calculated molecular mass of the Vp-MtlR monomer is 19.7 kDa and is very similar to that of Sf-YggD. However, its elution volume from size exclusion chromatography was significantly different from that of Sf-YggD, and its peak profile was much broader (Fig. 5A). The Vp-MtlR molecular mass was estimated to be 90.4 kDa, more than 4 times that of the Vp-MtlR monomer (78.8 kDa). Therefore, it appears that Vp-MtlR can form a tetramer, possibly through the interaction between the AB-dimer and the CD-dimer, as shown in our structure (Fig. 1A). In fact, the two Vp-MtlR dimers interact much more extensively than in the case of Sf-YggD, resulting in a total buried interface of area as high as 2135 Å2. The dimer-dimer contact is also the primary contact between any two dimers in the crystal structure of Vp-MtlR. Across the dimer-dimer interface, the flexible loops between α7 and α9 form two short antiparallel strands with five main chain-main chain hydrogen bonds (Fig. 4B). In addition to these contacts, there are also hydrogen bonds contributed by side chains across the interface and a number of hydrophobic interactions (Fig. 4B). However, the tetramer cannot be very stable because a tetramer with the AB-dimer/CD-dimer interactions would lead to polymerization of Vp-MtlR dimers as is observed in the crystal. In solution, at high protein concentration, the tetramer is likely to exist as a well as other oligomeric forms of Vp-MtlR in an equilibrium with a dimer. This may explain the broadening of the elution peak of Vp-MtlR and its shift toward high molecular mass (Fig. 5A).
Similar results were obtained for the Ec-MtlR.3 The results from size exclusion chromatography, native PAGE, dynamic light scattering, and analytical ultracentrifugation showed a polydisperse state of MtlR with multimers formed in a concentration-dependent manner. This polydispersity may be the primary reason why Ec-MtlR failed in our crystallization trials. Our data show that the MtlR (as well as YggD) dimer is a basic biological unit, but proteins in different species and under different conditions can form higher level aggregates.
MtlR/YggD Represent a Unique Protein Fold
A structural similarity search using the DALI server (available on the World Wide Web) (32) resulted in no significant hits. The four top structural homologues from a search by using Sf-YggD as a search template, for example, generally represent a four-helix bundle with one additional short helical turn and one three-turn α-helix with Z score of 5.3–6.7, r.m.s. deviation of 3.4–4.3, number of aligned residues 96–103, and sequence identity of 5–11%. They include proteins of unknown function from bacteria and the C-terminal domain of kanamycin nucleotidyltransferase from Haemophilus influenzae (Protein Data Bank code 1KNY). A part of the C-terminal domain of kanamycin nucleotidyltransferase involves nucleotide binding (33). Although part of the MtlR structure can be aligned with the C-terminal domain of kanamycin nucleotidyltransferase-like proteins, the eight- or nine-helix layered MtlR and YggD monomers represent a novel protein fold.
DNA-binding Studies of Ec-MtlR
The mannitol operon has been best described in E. coli, and the effect of Ec-MtlR on the transcription of the operon has been studied (Fig. 5B) (9). However, the in vitro binding of MtlR to the operator/promoter region has not been established so far. Similar to the results described by Figge et al. (9), we were able to show an inhibitory effect of MtlR on the uptake of mannitol into E. coli by plating on MacConkey agar plates. In vitro studies using several different techniques were undertaken to identify the DNA binding site of MtlR within the mannitol operon. In experiments with an optical biosensor (IAsys) (34, 35), we tried to prove the hypothesis that MtlR may bind to two palindromic sequence stretches identified on the mtlR promoter (Fig. 5B).
Two different DNA sequences from the mannitol operator site were investigated in electrophoretic mobility shift assay studies with Ec-MtlR. They were named MtlR1 and MtlR2, for operator 1 and operator 2, respectively (Fig. 5B). MtlR1 consists of sequence 5′-CTCGGGCTTCCAGCCTGCG, and MtlR2 consists of sequence 5′-ACATAAGAAGGGGTGTTTTTATGT; operator 1 and 2 were created by PCR with mtlR_pro1f (5′-TATGACGAAGGCATAACATGC) as forward primer and the reverse oligonucleotides of the MtlR1-box (mtlR1_rev) and the mtlR2-box (mtlR2_rev) as reverse primer. No interaction could be detected between the MtlR protein and the two palindromic sequence stretches on the promoter (MtlR1 and MtlR2) (Fig. 5, C and D). In order to exclude the possibility of another binding site on the operator-promoter region, the whole region from the first cyclic AMP receptor protein (CRP) binding site to the two palindromic sequence sites MtlR1 and MtlR2 was investigated in gel shift experiments. Again, no binding of MtlR to the DNA could be detected. Different buffer conditions, in the presence and absence of mannitol and various protein concentrations, were tested without success. The binding of the global regulatory protein, FruR, to the mtlR operon was used as a positive control in these experiments (36).
We have also investigated the DNA-binding properties of MtlR using a SELEX-based approach. This allows the identification of the DNA sequence, which binds to the protein of interest. The proteins tested were the well characterized sequence-specific FruR, the structure-specific HMG-box protein Cmb1, MtlR, and a bovine serum albumin control. Binding to the random sequence oligonucleotide R76 as well as to specific binding sequences could be detected for FruR and Cmb1. However, no binding was observed for MtlR and bovine serum albumin.
DISCUSSION
The MtlR has been experimentally studied and proposed to be a transcriptional regulator (9). Its gene, mtlR, is one of the conserved components of the mannitol operon across many bacterial species. The presence of two putative palindromes within the operator-promoter region implied possible MltR binding sites in the early studies of the E. coli mannitol operon (9). Many bacterial genomes code for the MtlR sequence homolog named YggD. However, the function of YggD has never been experimentally determined, and the yggD gene location in chromosomes provides little clue about its function. In this study, we have established that Ec-MtlR does not bind regulatory regions of mannitol operon, and we have determined the crystal structures of both MtlR and YggD. It is apparent that MtlR and YggD resemble each other in both their monomer and dimer structures, implying potentially similar functions.
Although assigned to the same protein sequence family (Pfam05068), MtlR and YggD are largely different in their molecular sizes. Generally, MtlRs are longer, having about 195 residues, whereas YggDs are shorter, with about 170 amino acids. The primary difference between MtlR and YggD seems to be the presence of an additional 20 plus N-terminal residues in MtlR. For example, MtlRs of Shigella and phylogenetically indistinguishable E. coli are 26 residues longer than their YggDs (Fig. 3). However, this rule is not observed by all MtlRs. In most Vibrio species, MtlRs have 176 residues with exceptions like V. cholerae 623-39, which has an MtlR of 193 residues (17). Therefore, these extra N-terminal residues in many MtlRs are unlikely to be determinants of the mannitol operon repressor's function. Rather, it could be a species-specific factor. Structurally, the extra N-terminal sequence could lead to an elongated α1 helix (Fig. 2B) and a loop linked to a possible short helix at the very N terminus, based on secondary structure prediction (available on the Rost laboratory's web site) (37). The prediction seems to be consistent with the results of limited proteolysis of Ec-MtlR, which showed the 14 N-terminal residues removed when Ec-MtlR was treated with trypsin or chymotrypsin (see “Experimental Procedures”).
Another line of evidence suggesting that MtlR and YggD may function similarly is that MtlR and YggD do not necessarily coexist in one species, whereas we believe that the mannitol-specific PTS is a common and vital molecular system in all bacterial species. For example, only the yggD gene was detected in Shigella dysenteriae Sd197, whereas only the mtlR gene was reported in Shigella sonnei Ss046 (38). In S. dysenteriae Sd197, the yggD gene is clustered with cmtA and cmtB, the two genes that are the frequent neighbors to yggD. They are generally annotated as cryptic mannitol-specific PTS enzyme:IIB/IIC components and IIA component, respectively, which sequentially corresponds to the components of MtlA. A recent solution structure study of E. coli K12 CmtB revealed that the overall structure of CmtB is highly similar to that of the IIA domain of MtlA (16). Although the exact phosphoryl transfer mechanism of CmtB could be different due to the position change of the catalytic arginine residue, the function of CmtB may be similar to that of the IIA domain of MtlA. This information further suggests that YggD could play a role similar to that of MtlR in the regulation of mannitol-specific PTS.
With the structures of Vp-MtlR and Sf-YggD determined, we analyzed the presence of potential DNA-binding motifs in these proteins. It is especially intriguing that they both form homodimers, which is expected for prokaryotic transcription factors binding to palindromic operator sequences within the promoter region (9). However, no known DNA-binding motif is found in both structures that could correspond to the DNA double helix spacing in major or minor grooves (39, 40). Although MtlR/YggD are all α-helical, there is no helix-turn-helix motif or other related structural motif typically found in transcriptional regulators (40). The surface of MtlR/YggD dimer is rather flat without the protruding structural element that could specifically recognize bases of DNA in the major or minor groove. The lack of an apparent DNA-binding motif also seems to be consistent with the results from the DALI structural homolog search mentioned previously. Even low score hits did not include DNA-binding proteins.
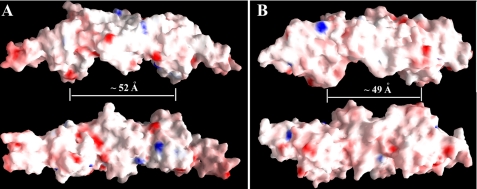
Moreover, both Vp-MtlR and Sf-YggD are very acidic proteins with calculated pI values of 4.46 and 4.33, respectively. They are both highly negatively charged with the same net charges of −28e per dimer. The net charges per amino acid for two molecules are −0.083 and −0.080, respectively. The surface electrostatic potential shows the overwhelming negative potentials of the surfaces of both MtlR and YggD dimers; there are no major positive charge patches present on the surface (Fig. 6). The electronic dipole moments of Vp-MtlR and Sf-YggD dimers are calculated to be 90 e·Å and 71.6 e·Å, respectively. The calculation was performed by using the Web Server to Calculate Dipole Moments of Proteins (41, 42). The electrostatic properties, including net charge and dipole moment, are believed to be important to a protein's ability to bind DNA (43, 44). It was found that these bulk electrostatic properties of DNA-binding proteins are significantly different from those of non-binding proteins (44). Based on these calculations, both Vp-MtlR and Sf-YggD are not favored as DNA-binding proteins due to the net negative charges and small dipole moments.
FIGURE 6.
The electrostatic potential surface representations of Vp-MtlR and Sf-YggD. A, the Vp-YggD dimer at the top is in an orientation similar to the orientation of the AB-dimer in the left panel of Fig. 1A. The dimer at the bottom is rotated about 90° along the horizontal axis. B, the Sf-YggD dimer at the top is in an orientation similar to the orientation of the BC-dimer in the top panel of Fig. 1B. The dimer at the bottom is rotated about 90° along the horizontal axis. The approximate distances between two ridges from both Vp-MtlR and Sf-YggD dimers are about 50 Å as marked. This figure was prepared with the program GRASP (51).
It has been reported that the mannitol operon of E. coli is subject to activation by the CRP (45). There are five unusual CRP binding sequences in the promoter region, and all of them have been proved to bind CRP in vitro in DNA band mobility shift experiments (Fig. 5B) (45). Additionally, the fructose repressor, FruR, also binds to the regulatory region of the mannitol operon as one common regulatory element of almost every major carbon metabolism pathway (36). The FruR recognition element was indicated to be between the −35 and −10 promoter region of the mannitol operon (Fig. 5B). The mannitol repressor MtlR negatively regulates the expression of the mannitol operon, and it was proposed to block expression through direct interaction with DNA at the two palindromic sequences located within the mtlR promoter (9). However, thus far, there is no corroborating experimental evidence that MtlR binds directly to the operator regulatory region (9). Our in vitro DNA-binding experiments clearly showed there was no binding of MtlR to the promoter region, even at high protein concentrations. A search for a different DNA regulatory sequence has also been unsuccessful. Considering the multiple binding sites for transcriptional factors in the mannitol operon, five for CRP, one for FurR, two for RNA polymerase, and potentially one or two for MtlR (or other regulatory protein), the operon control was regarded as one of the most complex found in bacteria (45). However, with the structures of Vp-MtlR and Sf-YggD determined and their properties tested, it is doubtful whether MtlR and/or YggD can bind DNA directly and regulate transcription in chorus or in competition with other DNA-binding regulators.
It is noticed that the mannitol uptake PTS of many Gram-positive bacteria, such as Bacillus (3, 46), Streptococcus (47), and Clostridium (48), have quite different mannitol operon repressors. They are similar to antiterminators and assigned to a different protein family (Pfam00874). For example, B. stearothermophilus MtlR is composed of 697 amino acids and contains a helix-turn-helix DNA-binding motif and two antiterminator-like PTS regulatory domains (49). This MtlR can bind to the promoter region and probably control the expression of the mannitol operon by blocking RNA polymerase binding to the promoter (3). Moreover, the activity of MltR itself can be regulated through phosphorylation by PTS components (3, 50).
It is possible that there is yet another transcriptional factor (TFX) that recognizes mtlR operator 1 and 2 sequences and links expression from the mtlR operon to other cellular function or external signal. We propose that the mannitol repressor MtlR and possibly YggD are a part of a transcriptional complex that controls the mannitol operon expression of E. coli, Shigella, Vibrio, and other Gram-negative bacteria. Instead of directly binding to DNA, they may interact directly or indirectly with other DNA-binding protein(s), resulting in the repression of mannitol operon transcription. Therefore, the true function of MtlR may be a competition with RNA polymerase in the initiation complex involving several proteins, including CPR, FurR, and TFX. This could represent a transcription initiation complex comparable with simple eukaryotic initiation complexes but functioning in bacteria.
Acknowledgments
We thank members of the Structural Biology Center at Argonne National Laboratory for help with data collection at the 19ID beamline, and Youngchang Kim of the Midwest Center for Structural Genomics for assistance in protein expression and characterization, Lindsey Butler for assistance in the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM074942. This work was also supported by the United States Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357. This work was created by UChicago Argonne, LLC, Operator of Argonne National Laboratory.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3BRJ and 3C8G) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
K. Tan, S. Clancy, M. Borovilos, M. Zhou, S. Hörer, J. Sassoon, U. Baumann, and A. Joachimiak, unpublished data.
- PTS
- phosphotransferase system(s)
- Se-Met
- selenomethionine
- MES
- 4-morpholineethanesulfonic acid
- r.m.s.
- root mean square.
REFERENCES
- 1.Saier M. H., Jr., Reizer J. (1994) Mol. Microbiol. 13, 755–764 [DOI] [PubMed] [Google Scholar]
- 2.Barabote R. D., Saier M. H., Jr. (2005) Microbiol. Mol. Biol. Rev. 69, 608–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henstra S. A., Tuinhof M., Duurkens R. H., Robillard G. T. (1999) J. Biol. Chem. 274, 4754–4763 [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz D. (1971) J. Bacteriol. 105, 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lengeler J. (1975) J. Bacteriol. 124, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C. A., Saier M. H., Jr. (1983) J. Biol. Chem. 258, 10761–10767 [PubMed] [Google Scholar]
- 7.Davis T., Yamada M., Elgort M., Saier M. H., Jr. (1988) Mol. Microbiol. 2, 405–412 [DOI] [PubMed] [Google Scholar]
- 8.Jiang W., Wu L. F., Tomich J., Saier M. H., Jr., Niehaus W. G. (1990) Mol. Microbiol. 4, 2003–2006 [DOI] [PubMed] [Google Scholar]
- 9.Figge R. M., Ramseier T. M., Saier M. H., Jr. (1994) J. Bacteriol. 176, 840–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei J., Goldberg M. B., Burland V., Venkatesan M. M., Deng W., Fournier G., Mayhew G. F., Plunkett G., 3rd, Rose D. J., Darling A., Mau B., Perna N. T., Payne S. M., Runyen-Janecky L. J., Zhou S., Schwartz D. C., Blattner F. R. (2003) Infect. Immun. 71, 2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu C. H., Tang P., Chu C., Hu S., Bao Q., Yu J., Chou Y. Y., Wang H. S., Lee Y. S. (2005) Nucleic Acids Res. 33, 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Y., Tong Z., Wang J., Wang L., Guo Z., Han Y., Zhang J., Pei D., Zhou D., Qin H., Pang X., Han Y., Zhai J., Li M., Cui B., Qi Z., Jin L., Dai R., Chen F., Li S., Ye C., Du Z., Lin W., Wang J., Yu J., Yang H., Wang J., Huang P., Yang R. (2004) DNA Res. 11, 179–197 [DOI] [PubMed] [Google Scholar]
- 13.Otte S., Lengeler J. W. (2001) FEMS Microbiol. Lett. 194, 221–227 [DOI] [PubMed] [Google Scholar]
- 14.Otte S., Scholle A., Turgut S., Lengeler J. W. (2003) J. Bacteriol. 185, 2267–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., Iijima Y., Najima M., Nakano M., Yamashita A., Kubota Y., Kimura S., Yasunaga T., Honda T., Shinagawa H., Hattori M., Iida T. (2003) Lancet 361, 743–749 [DOI] [PubMed] [Google Scholar]
- 16.Yu C., Li Y., Xia B., Jin C. (2007) Biochem. Biophys. Res. Commun. 362, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 17.Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. L., Studholme D. J., Yeats C., Eddy S. R. (2004) Nucleic Acids Res. 32, D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Montfort R. L., Pijning T., Kalk K. H., Hangyi I., Kouwijzer M. L., Robillard G. T., Dijkstra B. W. (1998) Structure 6, 377–388 [DOI] [PubMed] [Google Scholar]
- 19.Kim Y., Quartey P., Li H., Volkart L., Hatzos C., Chang C., Nocek B., Cuff M., Osipiuk J., Tan K., Fan Y., Bigelow L., Maltseva N., Wu R., Borovilos M., Duggan E., Zhou M., Binkowski T. A., Zhang R. G., Joachimiak A. (2008) Nat. Methods 5, 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 21.Kim Y., Dementieva I., Zhou M., Wu R., Lezondra L., Quartey P., Joachimiak G., Korolev O., Li H., Joachimiak A. (2004) J. Struct. Funct. Genomics 5, 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donnelly M. I., Zhou M., Millard C. S., Clancy S., Stols L., Eschenfeldt W. H., Collart F. R., Joachimiak A. (2006) Protein Expr. Purif. 47, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassoon J., Hörer S., Stoop J., Mooibroek H., Baumann U. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 711–713 [DOI] [PubMed] [Google Scholar]
- 24.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 25.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger T. C. (2003) Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30.Branden C., Tooze J. (1991) Introduction to Protein Structure, pp. 36–37, Garland Publishing, Inc., New York [Google Scholar]
- 31.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 32.Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 33.Pedersen L. C., Benning M. M., Holden H. M. (1995) Biochemistry 34, 13305–13311 [DOI] [PubMed] [Google Scholar]
- 34.George A. J., French R. R., Glennie M. J. (1995) J. Immunol. Methods 183, 51–63 [DOI] [PubMed] [Google Scholar]
- 35.Rubio I., Buckle P., Trutnau H., Wetzker R. (1997) BioTechniques 22, 269–271 [DOI] [PubMed] [Google Scholar]
- 36.Ramseier T. M., Bledig S., Michotey V., Feghali R., Saier M. H., Jr. (1995) Mol. Microbiol. 16, 1157–1169 [DOI] [PubMed] [Google Scholar]
- 37.Rost B., Liu J. (2003) Nucleic Acids Res. 31, 3300–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F., Yang J., Zhang X., Chen L., Jiang Y., Yan Y., Tang X., Wang J., Xiong Z., Dong J., Xue Y., Zhu Y., Xu X., Sun L., Chen S., Nie H., Peng J., Xu J., Wang Y., Yuan Z., Wen Y., Yao Z., Shen Y., Qiang B., Hou Y., Yu J., Jin Q. (2005) Nucleic Acids Res. 33, 6445–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. (1981) Nature 290, 754–758 [DOI] [PubMed] [Google Scholar]
- 40.Luscombe N. M., Austin S. E., Berman H. M., Thornton J. M. (2000) Genome Biol. 1, REVIEWS001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felder C. E., Botti S. A., Lifson S., Silman I., Sussman J. L. (1997) J. Mol. Graphics Model. 15, 318–327 [DOI] [PubMed] [Google Scholar]
- 42.Botti S. A., Felder C. E., Sussman J. L., Silman I. (1998) Protein Eng. 11, 415–420 [DOI] [PubMed] [Google Scholar]
- 43.Stawiski E. W., Gregoret L. M., Mandel-Gutfreund Y. (2003) J. Mol. Biol. 326, 1065–1079 [DOI] [PubMed] [Google Scholar]
- 44.Ahmad S., Sarai A. (2004) J. Mol. Biol. 341, 65–71 [DOI] [PubMed] [Google Scholar]
- 45.Ramseier T. M., Saier M. H., Jr. (1995) Microbiology 141, 1901–1907 [DOI] [PubMed] [Google Scholar]
- 46.Watanabe S., Hamano M., Kakeshita H., Bunai K., Tojo S., Yamaguchi H., Fujita Y., Wong S. L., Yamane K. (2003) J. Bacteriol. 185, 4816–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honeyman A. L., Curtiss R., 3rd (2000) Microbiology 146, 1565–1572 [DOI] [PubMed] [Google Scholar]
- 48.Behrens S., Mitchell W., Bahl H. (2001) Microbiology 147, 75–86 [DOI] [PubMed] [Google Scholar]
- 49.Henstra S. A., Tolner B., ten Hoeve Duurkens R. H., Konings W. N., Robillard G. T. (1996) J. Bacteriol. 178, 5586–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henstra S. A., Duurkens R. H., Robillard G. T. (2000) J. Biol. Chem. 275, 7037–7044 [DOI] [PubMed] [Google Scholar]
- 51.Nicholls A., Sharp K. A., Honig B. (1991) Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]