Abstract
Chlorophyll b is one of the major photosynthetic pigments of plants. The regulation of chlorophyll b biosynthesis is important for plants in order to acclimate to changing environmental conditions. In the chloroplast, chlorophyll b is synthesized from chlorophyll a by chlorophyllide a oxygenase (CAO), a Rieske-type monooxygenase. The activity of this enzyme is regulated at the level of protein stability via a feedback mechanism through chlorophyll b. The Clp protease and the N-terminal domain (designated the A domain) of CAO are essential for the regulatory mechanism. In this study, we aimed to identify the specific amino acid residue or the sequence within the A domain that is essential for this regulation. To accomplish this goal, we randomly introduced base substitutions into the A domain and searched for potentially important residues by analyzing 1,000 transformants of Arabidopsis thaliana. However, none of the single amino acid substitutions significantly stabilized CAO. Therefore, we generated serial deletions in the A domain and expressed these deletions in the background of CAO-deficient Arabidopsis mutant. We found that the amino acid sequence 97QDLLTIMILH106 is essential for the regulation of the protein stability. We furthermore determined that this sequence induces the destabilization of green fluorescent protein. These results suggest that this sequence serves as a degradation signal that is recognized by proteases functioning in the chloroplast.
Introduction
Chlorophyll is a tetrapyrrole molecule that is indispensable for photosynthesis. It harvests light and transfers excitation energy (or electrons) to other components of the photosynthetic electron transport chain. Land plants, green algae, and a few groups of cyanobacteria synthesize both chlorophyll a and chlorophyll b. Chlorophyll b absorbs light at a wavelength slightly different from that of chlorophyll a. Therefore, by utilizing both chlorophyll a and b, these organisms are capable of absorbing a wider range of light. Furthermore, chlorophyll b plays an essential role in regulating the antenna size of both photosystems in response to changing light conditions (1–3).
Chlorophyll b biosynthesis is catalyzed by chlorophyllide a oxygenase (CAO),2 a Rieske-type oxygenase (4). The enzyme incorporates molecular oxygen into the C7-methyl group of the chlorophyll a precursor chlorophyllide a to form chlorophyll b precursor chlorophyllide b (5). In plants, chlorophyllide b is immediately converted to chlorophyll b by the action of chlorophyll synthase (6, 7). It has been suggested that the CAO protein level determines the rate of chlorophyll b synthesis, because the amount of CAO protein correlates well with chlorophyll b levels in various CAO-overexpressing transgenic plants (8, 9). Although transcription of the CAO gene is regulated in response to environmental light intensity (10–13), the level of the CAO protein is mainly regulated by its protein stability rather than transcription (8, 9). Regulation of CAO protein stability is dependent on characteristic functional domains of the CAO protein. CAO consists of three domains (14), designated as the A, B, and C domains. The C-terminal domain (C domain) catalyzes the conversion of chlorophyllide a to chlorophyllide b (14). The B domain seems to function as a linker of the N-terminal domain (A domain) and the C domain (14). The A domain participates in the regulation of CAO protein stability. In previous studies, we examined the role of the A domain in the regulation of CAO stability (8). First, we expressed full-length CAO as well as truncated CAO protein lacking the A domain in Arabidopsis. Transgenic plants expressing CAO without the A domain accumulated a greater amount of the other two domains and exhibited a marked increase in chlorophyll b levels. In contrast, transgenic plants expressing full-length CAO did not accumulate CAO protein, and their chlorophyll b levels were similar to the wild type. These results indicated that the A domain is essential in maintaining the accumulation of CAO at a low level. Subsequently, we expressed a chimeric fusion construct of the A domain and green fluorescent protein (GFP) in Arabidopsis. Interestingly, the fusion protein did not accumulate when it was expressed in wild-type plants, whereas the same fusion protein accumulated to a greater amount when it was expressed in a chlorophyll b-deficient mutant, chlorina1-1 (ch1-1). These experiments demonstrated that the A domain conferred CAO protein instability only when chlorophyll b was synthesized. Furthermore, we also demonstrated that a mutation in the ClpC1 gene, which encodes a subunit of plastid Clp protease, resulted in increased stability of CAO (15). Taken together, we hypothesized that chlorophyll b accumulation induces degradation of CAO by Clp protease, a process that requires the A domain (or a part of it) to trigger degradation.
In order to understand how CAO degradation is regulated, it is important to examine how chlorophyll b conditionally induces destabilization of CAO. We first hypothesized that chlorophyll b may trigger modifications of the A domain by interacting with certain modifying enzymes, including kinases, methylases, or prenylases. Such modifications may then lead to the destabilization of CAO. Alternatively, we postulated the presence of a degradation signal (degron) within the A domain and that proteases are not accessible to the degron of CAO in the absence of chlorophyll b. Chlorophyll b may affect the structure of the A domain so that the degron becomes exposed to the proteases in the presence of chlorophyll b. Interestingly, in bacteria, dozens of proteins have such degrons (16), which are also referred to as protease recognition sequences (17). These sequences are essential in the interaction between the protein substrate and the recognition subunit/domain of the protease. If such a degradation mechanism is present in CAO, it is important to identify a degron in order to better understand how CAO degradation is regulated.
Protein degradation in the chloroplast is not as well understood as in bacteria. Although it has been suggested that there are at least 13 different types of proteases present in the chloroplast (18–20), their protein substrates were only partly identified (20–23). Moreover, since some of these proteases appear to target a large number of proteins (e.g. Clp protease), it is reasonable to assume that dozens of protein substrates remain to be identified. In contrast, over 100 protein substrates were identified in bacteria (16, 24). There are several reasons why this research in bacteria is more advanced than in plants. Generally, bacterial strains are easier to handle and can be grown more rapidly than plants. Furthermore, many degrons were identified in bacteria, which greatly aided in our understanding of the degradation mechanisms mediated by proteases (25–28). Likewise, the identification of plant degron will contribute to the elucidation of the protein degradation mechanisms in plants.
In order to assess the two hypotheses mentioned above, we followed two experimental strategies. The first one is a random mutagenesis approach, which introduces random nucleotide substitutions into the CAO gene in order to identify amino acid residues that are essential for protein function. In this study, we introduced random mutations into the A domain sequence and examined their effects on the accumulation of the CAO protein in transgenic Arabidopsis plants. In the second strategy, we generated serial deletions in the N terminus of CAO and searched for a particular region essential for the regulation of CAO stability. We hoped that this strategy would aid in the identification of the degron. In fact, we successfully identified a sequence of 10 amino acids that was essential for destabilization of CAO using the serial deletion strategy. Subsequently, we found that this particular sequence induces the degradation of GFP when fused to the N terminus of GFP. Therefore, we suggest that this sequence functions as a degron for a chloroplast protease (the CAO degron).
EXPERIMENTAL PROCEDURES
Plant Materials
The wild ecotype Columbia and the chlorina1-1 (ch1-1) mutant (5) of Arabidopsis thaliana plants were used for the transformation. For propagation, plants were germinated on soil and grown at 23 °C under continuous light conditions (white fluorescent light at 40–60 mmol m−2 s−1). For analysis of transgenic plants, seeds were germinated on plates containing half-strength Murashige-Skoog medium (Wako Pure Chemical Industries, Osaka, Japan), 0.7% (w/v) agar, and 50 mg/liter kanamycin. Plants were grown for 1 week at 23 °C under continuous light conditions.
Plasmid Construction and Plant Transformation
In order to express CAO-GFP fusion proteins, we modified the plasmid vector that was constructed in our previous study (8). The vector contained the cauliflower mosaic virus 35 S promoter, the tobacco mosaic virus ω sequence, the GFP (S65T) sequence (provided by Dr. Niwa, University of Shizuoka) (43), and the nopaline terminator in the backbone of pGreenII-0029 (provided by R. P. Hellens and P. Mullineaux (John Innes Centre)) (29). The GFP (S65T) gene in this plasmid was fused with transgenes (the full-length or truncated CAO cDNA sequences) at the SalI and NotI sites. The plasmids were subsequently transformed into an Agrobacterium tumefaciens (strain GV2260) by electroporation using a cuvette with a 1-mm gap at 25 microfarads and 1.8 kV. Wild-type Arabidopsis and the ch1-1 mutant (5) were transformed by the vacuum infiltration, as described, by Bechtold and Pelletier (44). Primary transformants were selected on agar plates containing 50 mg/ml kanamycin as described above. Lines that showed elevated accumulation of GFP fusion proteins were selected, and their progenies (the third generation) were subjected for further analysis.
Immunoblot Analysis
Ten milligrams of rosette leaves were homogenized with 100 μl of extraction buffer (50 mm Tris, pH 6.8, 2 mm EDTA, 10% (w/v) glycerol, 2% (w/v) SDS, and 6% (v/v) 2-mercaptoethanol). Homogenates were centrifuged at 10,000 × g for 3 min, and supernatants (25 μl) were subjected to 12.5% (w/v) polyacrylamide SDS-PAGE separation. Subsequently, the resolved proteins were electroblotted onto a Hybond-P membrane (GE Healthcare). The CAO-GFP transferred proteins were detected with an anti-GFP rabbit primary antibody (Invitrogen) and an anti-CAO rabbit primary antiserum that was raised against the recombinant Arabidopsis CAO polypeptide (Trp120–Val516) produced in Escherichia coli in our laboratory. Anti-rabbit IgG linked to horseradish peroxidase (GE Healthcare) was used as a secondary antibody. The horseradish peroxidase activity was detected using the ECL Plus Western blotting detection system (GE Healthcare), following the manufacturer's protocol.
Analysis of GFP Expression by Confocal Laser-scanning Microscopy
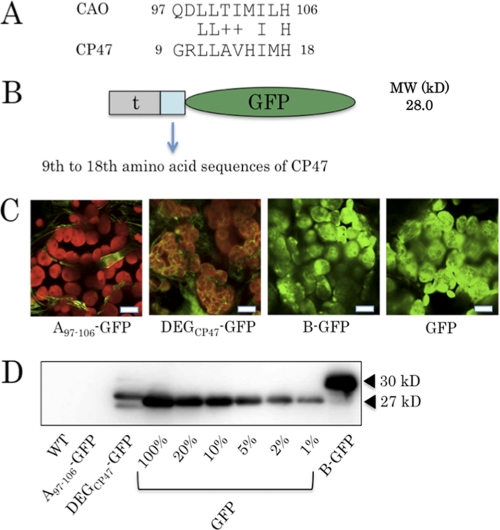
Fluorescence images were recorded at the Nikon Imaging Centre at Hokkaido University on a C1si Spectral Imaging confocal laser-scanning microscopy (CLSM) system with a TE2000-E Inverted microscope (Nikon Corp., Tokyo, Japan). The microscope was equipped with a Nikon CFI60 objective lens series plan apochromat ×100, numerical aperture 1.40 oil immersion type lens (Nikon Corp.). An argon laser (25 milliwatts) was used to generate an excitation source at 488 nm, and GFP and chlorophyll fluorescence were recorded at 500–550 and 600–680 nm, respectively. Images were processed with EZ-C1 Viewer 3.20 (Nikon Corp.). The gain levels of fluorescence intensity for each image were as follows: A107–126-GFP images (Fig. 6), 70; GFP images (Fig. 6), 40; DEGCP47-GFP (Fig. 7), 70; B-GFP and GFP images (Fig. 7), 40; other images, 150.
FIGURE 6.
Characterization of transgenic plants expressing GFP fused to various parts of the A domain. A, schematic presentation of the domain structures of the fusion proteins, containing GFP and various parts of the A domain. A, t, and GFP, the A domain of CAO, the predicted transit peptide sequence, and GFP (S65T), respectively. B, CLSM images of the transgenic plants expressing GFP fused to various parts of the A domain. Cotyledons of these plants were excited with an argon laser at 488 nm, and red chlorophyll and green GFP fluorescence was collected between 500 and 550 nm and between 600 and 680 nm, respectively. Scale bar, 10 μm. C, immunoblot analysis of the transgenic plants described in A. Total protein was extracted from rosette leaves of equal fresh weights (2.5 mg) (see “Experimental Procedures”) and was subjected to SDS-PAGE. The fusion proteins were detected using an anti-GFP antibody. The black arrowheads indicate the predicted molecular sizes of GFP (27 kDa) and the fusion protein (A107–126-GFP, 29 kDa; A-GFP, 40 kDa), respectively.
FIGURE 7.
Characterization of the transgenic plant expressing a fusion of GFP and the hypothetical degron of CP47. A, sequence alignment of the degron of Arabidopsis CAO and the hypothetical degron (DEGCP47) of Arabidopsis CP47. The amino acid letters between the two sequences indicate conserved amino acid residues. The plus sign indicates that two amino acid residues have similar properties. B, schematic presentation of the domain structures of the fusion proteins, containing GFP and DEGCP47. 47, t, and GFP, CP479–18, the predicted transit peptide sequence of CAO, and GFP (S65T), respectively. C, CLSM images of the transgenic plants expressing the CAO degron and GFP (a), the DEGCP47 and GFP (b), the B domain of CAO and GFP (c), and GFP protein (d). Cotyledons of these plants were excited with an argon laser at 488 nm, and red chlorophyll and green GFP fluorescence were collected between 500 and 550 nm and between 600 and 680 nm, respectively. Scale bar, 10 μm. D, immunoblot analysis of the transgenic plants described C. Total protein was extracted from rosette leaves of equal fresh weights (2.5 mg) (see “Experimental Procedures”) and was subjected to SDS-PAGE. Serial dilutions (20, 10, 5, 2, and 1%) of the extract of the GFP-expressing plant were prepared to compare with the DEGCP47-GFP fusion protein. The fusion proteins were detected using an anti-GFP antibody. The black arrowheads indicate the predicted molecular sizes of GFP (27 kDa) and the fusion protein (B-GFP; 30 kDa), respectively. WT, wild type.
Pigment Analysis
Chlorophyll was extracted from rosette leaves of A. thaliana using acetone. Extracts were centrifuged at 15,000 × g for 10 min at 20 °C. The supernatant was diluted with water to a final acetone concentration of 80% and subjected to high pressure liquid chromatography analysis. Pigments were separated on an octadodecyl column (Shim-pack CLC-ODS column, 6.0 × 150 mm) (Shimadzu, Kyoto, Japan) and eluted with methanol at a flow rate of 1.7 ml/min. Elution profiles were monitored by measuring absorbance at 650 nm. Chlorophyll contents were quantified from the chromatographic peak area.
Random Mutagenesis of the A Domain Sequence
Random mutagenesis was carried out by combining the methods of random PCR (31) and of hydroxylamine treatment (30). The cloned CAO cDNA from Arabidopsis leaves was PCR-amplified using 30 pmol of primers (caoFor2, TCTTGCGTCGACATGAACGCCGCCGTGTTT; caoRev2, ACCGTCCGCGGCCGCTTAGCCGGAGAAAGG), 7 mm MgCl2, 0.5 mm MnCl2, 10 mm Tris-HCl, 50 mm KCl, 0.2 mm dATP, 0.2 mm dGTP, 1.0 mm dCTP, 1.0 mm dTTP, and 5 units/μl Taq DNA polymerase (Sigma). PCR was performed for 35 cycles at 94 °C for 1 min, 45 °C for 1 min, and 72 °C for 1min. PCR products were separated on a 1% agarose gel and visualized by the SyberSafe DNA gel stain (Invitrogen) and extracted using the QIAquick gel extraction kit (Qiagen, Valencia, CA). Extracted DNA (∼1 μg) was treated with 1 m hydroxylamine (adjusted to pH 5 with NaOH) at 70 °C for 8 h. Treated DNA was kept at room temperature for 30 min and was separated on 1% agarose gel and extracted again as described above. DNA was then incorporated into the pGreen-II MH binary vector (8), containing the sequences encoding the B and C domains of CAO and GFP. The constructs were used for Agrobacterium-mediated transformation of ch1-1 as described above.
RESULTS
Experimental Design of the Random Mutagenesis Experiments and the Serial Deletion Approaches
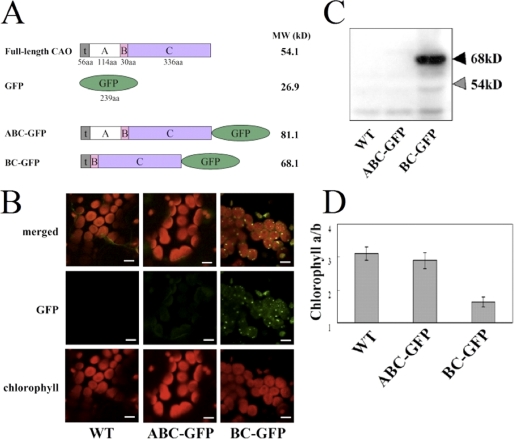
To examine whether a specific amino acid residue or a specific sequence within the A domain is essential for the regulation of CAO stability, we randomly introduced mutations into the A domain sequence and then expressed the mutated CAO sequences in Arabidopsis to assess the effect of the mutations. In the second set of experiments, we introduced serial deletions in the A domain sequence and expressed the truncated CAO sequences in Arabidopsis. For these experiments, we used the pGreenII binary vector, which contains the kanamycin resistance gene for selection of transgenic plants (29). We introduced the following sequences into this vector: the cauliflower mosaic virus 35 S promoter, the ω sequence (for enhancement of translational stability), the full-length sequence for CAO, the GFP coding sequence, and the nopaline synthase terminator. The construct was similar to those used in our previous study (8, 9). However, in contrast to our previous vectors, where the GFP sequence was inserted upstream of the CAO sequence, the GFP sequence was inserted downstream of the C domain of CAO (Fig. 1A). We made this change because we anticipated that genetic manipulations of the A domain sequence might affect the conformation of GFP if the GFP sequence is fused adjacent to the A domain. The vector constructs were then introduced into a CAO-deficient mutant, ch1-1, for expression of the CAO-GFP fusion protein. This mutant was used instead of the wild type in order to eliminate the possibility that the endogenous CAO interferes with the phenotype of the expressors. CLSM and immunoblot analysis showed that the expressor of the full-length CAO-GFP fusion did not accumulate the CAO-GFP protein, whereas the expressor of the B and C domains fused to GFP (BC-GFP) accumulated substantial amounts of protein (Fig. 1, B and C). Considering the same transcriptional levels of transgenes in BC-GFP and CAO-GFP (supplemental Fig. 1), these experiments demonstrated that the CAO-GFP fusion protein was destabilized in the expressor. This was also observed in the GFP-CAO expressors described in our previous study (8). The chlorophyll a to b ratios in these transgenic plants are consistent with the levels of the transgene products. The BC-GFP expressor exhibited a lower chlorophyll a to b ratio than the wild type and the CAO-GFP expressor (Fig. 1D).
FIGURE 1.
Characterization of transgenic plants expressing ABC-GFP and BC-GFP. A, schematic presentation of the domain structures of the transgenic plants expressing CAO, GFP, the full-length CAO-GFP fusion, and the BC-GFP fusion. A, B, C, t, and GFP, the A, B, and C domains of CAO, the predicted transit peptide sequence, and GFP (S65T), respectively. B, CLSM images of the wild type (WT) and transgenic plants expressing full-length (ABC)-GFP and BC-GFP fusions. Cotyledons of these plants were excited with an argon laser at 488 nm, and chlorophyll fluorescence (bottom) and GFP fluorescence (middle) were collected between 500 and 550 nm and between 600 and 680 nm, respectively. Merged images are at the top. Scale bar, 10 μm. C, immunoblot analysis of the CAO-GFP fusion protein in transgenic plants. Total protein was extracted from rosette leaves of equal fresh weight (2.5 mg) (see “Experimental Procedures”) and was subjected to SDS-PAGE. The CAO-GFP fusion or CAO proteins were detected using an anti-CAO antibody. The black arrowheads indicate the predicted molecular sizes of CAO fusion proteins (ABC-GFP, 81 kDa; BC-GFP, 68 kDa). The gray arrowheads indicate the predicted molecular size of the native CAO protein (54 kDa). All signals except that of 68 kDa were nonspecific to CAO. D, chlorophyll a to b ratios in cotyledons of the transgenic plants, n = 10–15. Note that we do not include the chlorophyll a/b ratio of ch1-1, which is the parental strain of our transgenic plants. It is because ch1-1 is unable to produce chlorophyll b, therefore, that its chlorophyll a/b ratio is infinite. Instead, we use here the Columbia WT line as a control, because this line is the background of ch1-1, and it is able to produce chlorophyll b.
In our initial random mutagenesis screens, we only analyzed one seedling per line. However, for lines that showed higher GFP signals in the first screening, we analyzed several T2 progenies per line to confirm their phenotypes. In the serial deletion experiments, we analyzed several transgenic lines for each construct to circumvent the positional effects of transgene insertion into the genome. Because we always observed similar phenotypes among the lines that contained the same transgene construct, we present here the results from one representative line for each construct. Note that most CAO expressors exhibited stable phenotypes among transgenic lines expressing the same construct. This suggests that the modification of CAO protein structure gave a more pronounced effect on protein stability than the positional effects of transgene insertion into the genome on the CAO transcript levels. This observation may reflect the characteristics of the regulatory mechanism of CAO, in which the CAO protein level is primarily determined by protein stability (8).
Random Mutagenesis of the A Domain
We searched for specific amino acids that are essential for the regulation of CAO in the A domain by employing a random mutagenesis approach. We introduced mutations into the DNA sequence encoding the A domain in vitro by the combination of two methods: hydroxylamine treatment (30) and random PCR mutagenesis (31). The former method induces C-to-T or G-to-A substitution. In contrast, the other method induces A-to-T/G or T-to-C substitution. By combining both methods, it is theoretically possible to cause all types of base substitutions in double-stranded DNA. Mutagenized sequences were then cloned into the pGreenII vector for subsequent expression of the CAO-GFP fusion protein in Arabidopsis (see supplemental Fig. 2).
To estimate the frequency of base substitution, we determined the nucleotide sequences for the A domain from 100 independent and randomly selected expressors. The average number of nucleotide mutations per sequence was 2.36, causing an average of 1.52 amino acid substitutions/sequence. Among the 100 expressors we sequenced, 86 lines had at least a single mutation (see supplemental Table 1). This mutation frequency was comparable with those reported previously (31). Among the 342 bases of the sequence encoding the A domain, there are 77 guanine, 102 adenine, 54 cytosine, and 109 thymine bases. Thymine was the most frequently substituted (see supplemental Fig. 3A); 51.3% of the T bases were substituted to any other bases in at least one expressor. In contrast, the lowest frequency of substitution was 25.9% for G bases (supplemental Fig. 3A). Using the results from this analysis, we estimated, using the recurrence equation, how many expressors we should analyze to find at least one substitution in any base in the A domain sequence (supplemental Fig. 3B). We calculated that more than 99% of the total bases should be mutated at least once among a population of 1,000 expressors. Therefore, by screening 1,000 expressors, we would expect at least one mutation in any base in the A domain sequence with a probability of 99%.
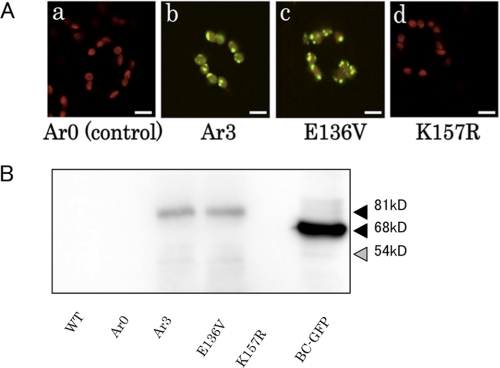
Accordingly, we screened 1,000 expressors for increased accumulation of the CAO-GFP protein by CLSM. We found 35 expressors exhibiting GFP fluorescence above the background level, although the fluorescence intensities of these expressors were much lower than those of the BC-GFP expressor (Fig. 2A). We found that accumulation of the CAO-GFP protein was most intense in guard cells compared with other leaf cells. This could be explained by the fact that guard cells produce less chlorophyll than mesophyll cells, which may result in lower degradation activity for CAO (32). Fig. 2A shows a CLSM image of a representative single line, which we designated Ar3 (for A-domain randomly mutagenized mechanism, line 3). Using immunoblot analysis and a GFP-specific antibody, we found that CAO-GFP protein accumulated in Ar3, although that protein level was much lower than that of BC-GFP. In contrast, CAO-GFP protein could not be detected in the control line, Ar0, which expressed the intact A domain sequence fused with GFP (Fig. 2B). Sequence analysis showed that Ar3 had two amino acid substitutions (E136V and K157R) in the A domain. In order to confirm whether one of these substitutions contributed to the phenotype of Ar3, we generated two site-directed mutants, containing either E136V or K157R substitutions. In the E136V site-directed mutant, the CAO(E136V)-GFP protein accumulated to the same level as in Ar3. In contrast, the CAO(K157R)-GFP protein was not detected. These results demonstrate that the E136V substitution is responsible for the increased stability of CAO in the Ar3 expressor.
FIGURE 2.
Characterization of a selected random mutagenesis mutant and site-directed mutants. A, CLSM images of transgenic leaves expressing the intact A domain sequence (a), of a selected random mutagenized mutant, Ar3 (b), and of the site-directed mutants, E136V and K157R (c and d, respectively). Cotyledons of these plants were excited with an argon laser at 488 nm, and red chlorophyll and green GFP fluorescence were collected between 500 and 550 nm and between 600 and 680 nm, respectively. Guard cells of the plants are shown in the center of each image. These cells tend to show the highest GFP fluorescence, if any. Scale bar, 10 μm. B, immunoblot analysis of the CAO-GFP fusion protein in transgenic plants. Total protein was extracted from rosette leaves of equal fresh weights (2.5 mg) (see “Experimental Procedures”) and was subjected to SDS-PAGE. The CAO-GFP fusion or CAO proteins were detected with an anti-CAO antibody. The black and gray arrowheads indicate the predicted molecular sizes of the CAO fusion proteins (81 kDa) and the native CAO protein (54 kDa), respectively. WT, wild type.
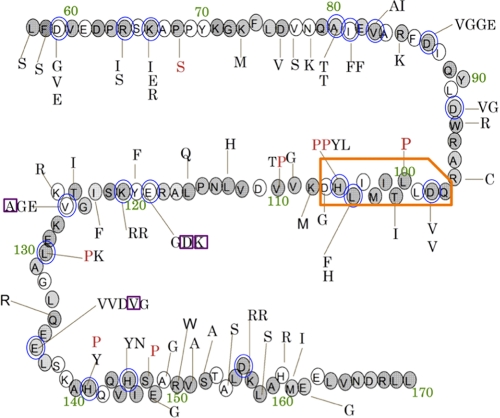
We found that among the 35 expressors, six lines contained the same mutations. This may be explained by the fact that we amplified the plasmid constructs after mutagenesis, which were then used for plant transformation. We therefore conclude that the 35 expressors were transformants of 29 independent mutant plasmids. In Fig. 3, we summarized the identified substitutions of amino acids within the 29 expressors. Redundant mutations from the same clones were omitted from this summary. The average number of amino acid substitutions in the 29 expressors was 2.8. When multiple substitutions were present in one clone, it is possible that only one of them contributed to the increase in CAO stability. Therefore, it is possible that a substantial portion of these substitutions did not contribute to the increase in CAO stability. When a clone contained a single substitution, it is most likely that this mutation contributed to increased CAO stability. We found three such mutations in three independent clones (purple boxes in Fig. 3). Alternatively, when the substitutions in the same amino acid were found in different clones, it is possible that the amino acid contributed to increased CAO stability. For 19 amino acids, we found multiple substitutions in different clones (blue circles in Fig. 3). These amino acids, whose mutations probably contributed to increased CAO stability in the expressors, were distributed throughout the A domain sequence (Fig. 3). Taking into account our observation that none of the 35 expressors showed a level of GFP fluorescence equivalent to that of the BC-GFP expressor, we concluded that there is no unique amino acid in the A domain sequence vital in the regulation of CAO stability. It is likely that multiple amino acid substitutions at one time may be required to eliminate the function of the A domain. We therefore speculate that modification of a single specific amino acid is not involved in the regulation.
FIGURE 3.
Summary of amino acid substitutions within the 29 selected transformants in the random mutagenesis experiments. Shown are amino acid substitutions in 29 transformants that exhibited GFP fluorescence. Original residues are shown in circles. Replacing residues are indicated with lines pointing to the original residues. The red letters indicate the substitution by proline, which is known to disrupt protein structure. The blue circles indicate the amino acids that have multiple substitutions in different clones. The shaded residues indicate conserved amino acids within the CAO sequences of five higher plant species. (A. thaliana, Oryza sativa, Lycoperiscon esculentum, Poplus trichocarpa, and Hordeum vulgare). The green characters indicate the position numbers of the amino acid residues of CAO. The orange box indicates the degron of CAO (the CAO degron was determined in a subsequent set of experiments). The purple box indicates mutations of single mutation lines (see supplemental Table 2).
Identification of the Degradation Signal Sequence in the A Domain
In a second set of experiments, we generated serial deletions of the A domain sequence by consecutively deleting 10 amino acids (30 nucleotides) using the plasmid construct that contains the A domain fused to B and C domains of CAO and GFP (Fig. 4A). Consequently, we obtained ten plasmid constructs, containing different lengths of the A domain (Fig. 4A). These constructs were then introduced into the ch1-1 mutant. Deletions of the A domain sequence up to amino acid Gln-97 had no effect on GFP fluorescence in the expressors (Fig. 4B). In contrast, deletions up to amino acid His-106 or further deletions resulted in significant accumulation of the CAO-GFP fusion proteins in the respective expressors (Fig. 4B). The intensities of GFP fluorescence in the deletion mutants up to or beyond His-106 were equivalent to that in the expressor lacking the entire A domain (the BC-GFP expressor). The same consequences of protein level of transgene products were detected by immunoblot analysis (Fig. 4C). Deletions up to or beyond His-106 resulted in a drastic increase in CAO-GFP protein levels in the expressors (Fig. 4C). Chlorophyll a to b ratios were ∼2.8 in the expressors with deletions up to Arg-96, whereas ratios decreased to less than 2.0 in the expressors with deletions beyond Gln-97 (Fig. 4D). Collectively, these results indicate that the amino acid sequence from Gln-97 to His-106 is essential in the regulation of the CAO protein level.
FIGURE 4.
Effects of serial deletions of the A domain sequence. A, schematic presentation of the consecutively deleted A domain sequences, which were fused to the transit peptide (t), the B and C domains, and GFP. The predicted molecular masses for the fusion proteins are indicated. B, CLSM observations of transgenic plants expressing serially deleted A domain sequences. Cotyledons of these plants were excited with an argon laser at 488 nm, and red chlorophyll and green GFP fluorescence was detected between 500 and 550 nm and between 600 and 680 nm, respectively. C, immunoblot analysis of the CAO-GFP fusion protein in transgenic plants. Total protein from rosette leaves of equal fresh weight (2.5 mg) was extracted (see “Experimental Procedures”) and was subjected to SDS-PAGE. The CAO-GFP fusion or CAO proteins were detected using an anti-CAO antibody. The black arrowheads indicate predicted molecular sizes of CAO-fusion proteins (ABC-GFP, 81 kDa; BC-GFP, 68 kDa), respectively. The gray arrowheads indicate the predicted molecular size of the native CAO protein (54 kDa), which was not detected. D, the chlorophyll a to b ratios in cotyledons of the transgenic plants (n = 5–8). WT, wild type.
In order to assess whether the sequence from Gln-97 to His-106 was sufficient to induce destabilization of CAO, this sequence was fused to BC-GFP and expressed in the ch1-1 mutants (A97–106-BC-GFP; Fig. 5A). As controls, the other parts of the A domain were fused to BC-GFP and expressed in the same mutant (A57–96-BC-GFP, A107–126-BC-GFP, A127–146-BC-GFP, and A147–170-BC-GFP). In the A97–106-BC-GFP expressor, GFP fluorescence was not detected, whereas the other expressors showed significant GFP fluorescence (Fig. 5B). Consistent with these results is the fact that the A97–106-BC-GFP expressor did not have detectable amounts of the fusion protein, whereas the expressors for the other parts of the A domain accumulated significant amounts of the fusion proteins (Fig. 5C). Accordingly, the chlorophyll a to b ratio was similar to that of wild type in the A97–106-BC-GFP expressor. In contrast, the chlorophyll a to b ratios decreased to ∼2.0 in the other expressors, which did not contain the sequence from Gln-97 to His-106. Taken together, we conclude that the 10-amino acid sequence QDLLTIIMILH in the A domain is the degradation signal sequence. We designated it the CAO degron.
FIGURE 5.
Characterization of transgenic plants expressing different parts of the A domain sequence. A, schematic presentation of the domain structures of the CAO-GFP fusion proteins that expressed various parts of the A domain. A, B, C, t, and GFP, the A, B, and C domains of CAO, the predicted transit peptide sequence, and GFP (S65T), respectively. B, CLSM images of the transgenic plants shown in A. Cotyledons of these plants were excited with an argon laser at 488 nm, and red chlorophyll and green GFP fluorescence was detected between 500 and 550 nm and between 600 and 680 nm, respectively. Scale bar, 10 μm. C, immunoblot analysis of the CAO-GFP fusion protein in transgenic plants. Total protein was extracted from rosette leaves of equal fresh weights (2.5 mg) (see “Experimental Procedures”) and was subjected to SDS-PAGE. The expressed proteins were detected using an anti-CAO antibody. The black arrowheads indicate the predicted molecular sizes of ABC-GFP (81 kDa) and BC-GFP (68 kDa). The gray arrowheads indicate the predicted molecular size of the native CAO protein (54 kDa). D, the chlorophyll a to b ratios in the cotyledons of the transgenic plants, expressing various parts of the A domain sequence (n = 5–8). WT, wild type.
The CAO Degron Targets the Attached Protein for Degradation
Subsequently, we tested whether the CAO degron regulates degradation of proteins other than CAO. We fused the CAO degron to GFP and expressed the protein (A97–106-GFP) in wild type and ch1-1 (Fig. 6A). As a control, we expressed the fusion of the sequence from Asp-107 to Val-126 with GFP (A107–126-GFP) in wild type and ch1-1 (Fig. 6A). Expression of GFP alone in the chloroplast resulted in strong GFP fluorescence in entire chloroplasts (Fig. 6B, d and h) of the wild type and ch1-1 backgrounds. The A107–126-GFP expressor also showed significant GFP fluorescence (Fig. 6B, b and f). In contrast, when the CAO degron was fused to GFP, the GFP signal was detected neither in the wild type nor ch1-1 (Fig. 6B, a and e). There was a notable difference in GFP fluorescence of the A97–106-GFP and A-GFP constructs when expressed in ch1-1: GFP fluorescence was detected in the A-GFP expressor but not in the A97–106-GFP expressor (Fig. 6, e and g). The results of the CLSM analysis were consistent with the immunoblot analysis. The A-GFP expressor accumulated a substantial amount of the A-GFP protein, whereas the A97–106-GFP expressor did not accumulate detectable protein levels (Fig. 6C). These results demonstrate that the CAO degron functions as a degradation signal sequence for proteins other than CAO. Our data also suggest that the other part of the A domain is necessary to confer chlorophyll b dependence of the degradation process.
Arabidopsis CP47 Has CAO Degron-like Sequences
Next, we examined whether the other sequences similar to the CAO degron participated in the regulation of protein stability in the chloroplast. In order to find such sequences, we first searched the Arabidopsis protein data base for sequences similar to the CAO degron, using the Patmatch program at the web site of the Arabidopsis Information Resource. We did not find any sequences that matched more than 7 residues of the 10 CAO degron residues. There were six sequences that matched the CAO degron with 7 residues. However, these proteins are all predicted to localize outside of the chloroplast. Therefore, these sequences may not function as degrons. These results are not surprising because bacterial degrons are not strictly conserved, as described above (16). When we allowed various patterns of mismatches in our sequence search, we obtained a greater number of sequences using the Arabidopsis protein data base. For example, if we searched the data base with “QXLLTJMJLX” as a query (where X denotes any residue, and J denotes any hydrophobic residue) and if we allowed 2 mismatches for the search, we found that 1,343 sequences matched these conditions. In these 1,343 sequences, we found that CP47, a core subunit of photosystem II (33), contains a sequence similar to the CAO degron. This sequence shares four identical amino acids and common features with the CAO degron, containing a charged residue followed by a stretch of hydrophobic residues and ending with a histidine residue (Fig. 7A). We tentatively named this sequence the CP47 degron-like sequence (DEGCP47). The sequence locates near the N terminus of CP47. To examine whether the amino acid sequence functions as a degron, we made a transgenic plant that expressed GFP fused with DEGCP47 (DEGCP47-GFP) (Fig. 7B).
In order to determine the level of DEGCP47-GFP, we carried out confocal microscopy and immunoblot analyses. The GFP fluorescent signal in the DEGCP47 expressor was substantially lower than those of B-GFP or GFP expressors (Fig. 7C), but it was higher than that in A97–106-GFP expressor. By immunoblotting analysis, we estimated that the level of the GFP fusion protein in the DEGCP47 expressor was about 5% of the GFP expressor (Fig. 7D). These results indicate that DEGCP47 functions as a weak degron in the chloroplast.
DISCUSSION
Random Mutagenesis of the A Domain
The random mutagenesis experiments of the A domain showed that substitutions of many amino acid residues enhanced the stability of the CAO protein (Fig. 3). These results suggest that there is no specific residue in the A domain that induces destabilization of the CAO protein. Therefore, we have to refute our first hypothesis, which predicted that the modification of a specific residue is involved in the regulation of CAO stability. Instead, it is more likely that the mutations shown in Fig. 3 modified the correct folding of the A domain, which interferes with the destabilization mechanism of the CAO protein. This idea is consistent with our observation that about 10% of the substitutions affecting CAO stability were changes from or to proline residues (Fig. 3). Since proline is less flexible in its conformation than the other amino acids, it tends to disrupt the secondary structure of a protein in the middle of regular structural elements, such as an α-helix, whereas it tightens these elements when it locates their edges (34). Therefore, it is possible that substitutions from or to proline had a greater effect on the structure of the CAO protein than the other substitutions. We speculate that the mutations shown in Fig. 3 result in tighter folding of the CAO protein, thereby impairing access of proteases to the A domain. This hypothesis may explain why the amino acid substitutions shown in Fig. 4 stabilized the CAO protein, whereas the deletions up to Arg-96 (Fig. 5) did not affect CAO stability. It is plausible that deletions shown in Fig. 4 never contributed to strengthen the folding of the CAO protein; thus, the A domain may not be protected from attack by proteases.
It is noteworthy that the levels of CAO protein in any random mutagenesis mutant were much lower compared with mutants in which the A domain was completely removed (BC-GFP). GFP fluorescence in the selected random mutagenesis mutants was only marginally increased in the mesophyll cells, whereas it was significantly increased in the guard cells (Fig. 2A). These observations suggest that even if the structure of the A domain affected the regulation of CAO as we discussed above, the structural changes might not be sufficient to prevent access of proteases to the CAO protein.
Protease Recognition Sequence of CAO
Our experiments, in which the CAO degron was removed from the A domain, clearly demonstrated that this region is essential in the destabilization of CAO (Figs. 4 and 5). In addition, the CAO degron was shown to induce destabilization of GFP (Fig. 6). Thus, the CAO degron fulfills the criteria by which bacterial degrons were identified (16). Therefore, we suggest that the CAO degron is a plant degron. A degron usually interacts with the substrate recognition domain (or subunit) of a protease (16). Such an interaction is essential in determining the substrate specificity of a protease. Degrons are most extensively studied in E. coli. Several types of recognition signal sequences have been identified for bacterial proteases (16, 35–37). In contrast, to the best of our knowledge, the CAO degron is the first such recognition sequence identified for chloroplast proteins. In bacterial protease studies, determination of protease recognition signals helped in the identification of interactions between the substrate and the substrate recognition domain (or subunit) of proteases (24, 38, 39). Likewise, the identification of the CAO degron will result in further experiments that will contribute to our understanding of how overall protein degradation is controlled in the chloroplast.
In bacteria, some degrons share common properties. For example, one type of Clp degron located at the N terminus of the substrate protein has a consensus sequence consisting of polar-T/hydrophobic-hydrophobic-basic-hydrophobic residues (16).
One intriguing point is whether or not the CAO degron-like sequences in chloroplast function as degrons. In order to answer this question, we searched the public Arabidopsis protein data base, and we found that CP47 protein contains a sequence similar to the CAO degron. We demonstrated that the CP47 degron compromised the accumulation of GFP protein (Fig. 7, C and D), implying that this sequence functions as a degron (CP47 degron) in the chloroplast. It is possible that some other sequences among the 1343 sequences that we identified also function as degrons. Our study may prompt researchers to test the function of these possible degrons with their own proteins of interest. Collection of the results of such functional studies may lead to identification of the consensus sequence, if any, of the chloroplast sequences.
The CAO degron is well conserved in CAO sequences from various plant species (supplemental Fig. 4). 8 of the 10 CAO degron residues are perfectly conserved, and the other two residues are replaced by similar amino acids. In addition, the sequences surrounding the CAO degron are also well conserved. Although there is no information on the function of these sequences, one might speculate that these sequences are involved in the correct folding of the A domain and/or in the chlorophyll b dependence of protein destabilization.
Conditional Destabilization of CAO in the Presence of Chlorophyll b
The majority of bacterial degrons determines the turnover rates of substrate proteins (16, 17, 40). The CAO degron is distinct from most other degrons identified so far, because the CAO degron conditionally accelerates the degradation of CAO in the presence of chlorophyll b. The CAO degron therefore resembles the degron of the Salmonella glutamyl-tRNA reductase (41, 42); this protein undergoes a rapid turnover in the presence of heme. In addition, a truncated glutamyl-tRNA reductase lacking the degron becomes stable with or without heme (42). Since the CAO degron and the degron of the glutamyl-tRNA reductase share hydrophobic residues (supplemental Fig. 5), one may speculate that the hydrophobic properties of these residues are essential in the function of these proteins. Indeed, Wang et al. (41) demonstrated that substitution of two leucine residues into 2 lysine residues eliminated the heme-dependent destabilization of glutamyl-tRNA reductase. Because hydrophobic residues tend to reside in the interior of a soluble protein, it is plausible that these sequences are structurally embedded in the CAO protein or glutamyl-tRNA reductase. The interior location of degrons may prevent access of proteases (Fig. 8). This hypothesis may explain why these proteins are resistant to degradation in certain conditions, such as in the absence of chlorophyll b or heme. Furthermore, this hypothesis also suggests that a structural change of the A domain may occur in the presence of chlorophyll b. Such a structural change is predicted to be necessary to expose the CAO degron to the exterior of the A domain, which in turn is thought to enable the interaction between proteases and the CAO degron (Fig. 8).
FIGURE 8.
Tentative model for the mechanism of CAO degradation. a, stable form. In the absence of chlorophyll b, the CAO degron lies within the hydrophobic (interior) region of the protein and is protected by the A domain structure. The thick gray lines designate the A domain. The red box indicates the CAO degron. b, degradation form. When chlorophyll b is sufficiently synthesized by CAO, chlorophyll b and a yet unidentified factor or factors, depicted as X, may interact with the A domain. As a result, the A domain unfolds and thus exposes the CAO degron to the exterior of the protein. c, degradation by protease. The ClpC subunit of the ClpCP protease and the recognition subunit/domain of other chloroplast proteases recognize the CAO degron of the A domain and pull the CAO protein to the proteolytic subunit/domain of the proteases. Finally, the proteolytic subunit/domain of the proteases degrades the entire CAO protein. ClpCP protease is depicted as blue and red spheres.
Two hypotheses may explain how chlorophyll b induces destabilization of CAO. First, chlorophyll b modifies the structure of the A domain to increase the susceptibility of the A domain to proteolysis. Second, chlorophyll b activates the protease. We believe that the second possibility is unlikely, because if the activation of the protease is required for degradation of the CAO degron, the A97–106-GFP fusion protein should have been stabilized in the absence of chlorophyll b. Our experiments demonstrated that A97–106-GFP was degraded despite the absence of chlorophyll b (see Fig. 6). Therefore, we suggest that the first hypothesis is more likely; chlorophyll b may modulate the structure of the A domain either by direct interaction with the A domain or by other factors. If we assume that chlorophyll b directly interacts with the A domain and if only a single or a few residues are involved in the interaction, it would be likely that substitution of a single specific amino acid residue greatly decreases the effects of chlorophyll b. However, we did not find any mutations that led to elimination of the destabilization effect of chlorophyll b (Fig. 2; see “Random Mutagenesis of the A Domain”). Thus, we postulate that chlorophyll b modulates the structure of the A domain by other yet unidentified factors (Fig. 8). However, we cannot exclude the possibility that multiple and direct binding of chlorophyll b to the A domain modulates its structure. Taken together, in our current model, chlorophyll b modulates the structure of the A domain by the aid of some unknown factors, which may lead to the exposure of the CAO degron to the exterior of the protein. Then the recognition subunit (or domain) of Clp and/or other proteases recognize the CAO degron, drawing the whole protein into the proteolytic subunit (or domain) of the proteases. If this model is correct, it will be important to identify the unknown factors that are involved in the regulation of CAO in order to understand the entire regulatory mechanism. We are in the process of identifying such factors using a genetic approach (32). The conditional destabilization mechanism of CAO may be used as a model system to elucidate how protein degradation is controlled within chloroplast.
Acknowledgments
We thank Dr. Makio Yokono for the construction of the recurrence equation. We also thank Dr. Kobayashi (Nikon Imaging Center, Hokkaido University) for technical assistance with the use of the confocal laser-scanning microscope.
This work was supported by Ministry of Education, Culture, Sports, Science and Technology, Japan, Grant-in-aid for Scientific Research 17770027 (to R. T.) and Grant-in-Aid for Creative Scientific Research 17GS0314 (to A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1–5.
- CAO
- chlorophyllide a oxygenase
- ch1-1
- chlorina1-1
- CP47
- chlorophyll-binding protein 47
- GFP
- green fluorescent protein
- CLSM
- confocal laser-scanning microscopy
- BC-GFP
- chimeric protein consisting of the B and C domains of CAO and GFP
- DEG
- degron.
REFERENCES
- 1.Bellemare G., Bartlett S. G., Chua N. H. (1982) J. Biol. Chem. 257, 7762–7767 [PubMed] [Google Scholar]
- 2.Kuttkat A., Edhofer I., Eichacker L. A., Paulsen H. (1997) J. Biol. Chem. 272, 20451–20455 [DOI] [PubMed] [Google Scholar]
- 3.Melis A. (1991) Biochim. Biophys. Acta Bioenergetics 1058, 87–106 [Google Scholar]
- 4.Tanaka A., Ito H., Tanaka R., Tanaka N. K., Yoshida K., Okada K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12719–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oster U., Tanaka R., Tanaka A., Rudiger W. (2000) Plant J. 21, 305–310 [DOI] [PubMed] [Google Scholar]
- 6.Oster U., Bauer C. E., Rüdiger W. (1997) J. Biol. Chem. 272, 9671–9676 [DOI] [PubMed] [Google Scholar]
- 7.Gaubier P., Wu H. J., Laudié M., Delseny M., Grellet F. (1995) Mol. Gen. Genet. 249, 58–64 [DOI] [PubMed] [Google Scholar]
- 8.Yamasato A., Nagata N., Tanaka R., Tanaka A. (2005) Plant Cell 17, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakuraba Y., Yamasato A., Tanaka R., Tanaka A. (2007) Plant Physiol. Biochem. 45, 740–749 [DOI] [PubMed] [Google Scholar]
- 10.Masuda T., Tanaka A., Melis A. (2003) Plant Mol. Biol. 51, 757–771 [DOI] [PubMed] [Google Scholar]
- 11.Harper A., von Gesjen S., Linford A., Peterson M., Faircloth R., Thissen M., Brusslan J. (2004) Photosynth. Res. 79, 149–159 [DOI] [PubMed] [Google Scholar]
- 12.Pattanayak G., Biswal A., Reddy V., Tripathy B. (2005) Biochem. Biophys. Res. Commun. 326, 466–471 [DOI] [PubMed] [Google Scholar]
- 13.Tanaka R., Tanaka A. (2005) Photosynth. Res. 85, 327–340 [DOI] [PubMed] [Google Scholar]
- 14.Nagata N., Satoh S., Tanaka R., Tanaka A. (2004) Planta 218, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 15.Nakagawara E., Sakuraba Y., Yamasato A., Tanaka R., Tanaka A. (2007) Plant J. 49, 800–809 [DOI] [PubMed] [Google Scholar]
- 16.Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. (2003) Mol. Cell. 11, 671–683 [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S. (2003) Annu. Rev. Cell. Dev. Biol. 19, 565–587 [DOI] [PubMed] [Google Scholar]
- 18.Adam Z. (2000) Biochimie 82, 647–654 [DOI] [PubMed] [Google Scholar]
- 19.Adam Z., Clarke A. K. (2002) Trends Plant Sci. 7, 451–456 [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto W. (2006) Annu. Rev. Plant Biol. 57, 599–621 [DOI] [PubMed] [Google Scholar]
- 21.Majeran W., Wollman F. A., Vallon O. (2000) Plant Cell 12, 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sjögren L. L., Stanne T. M., Zheng B., Sutinen S., Clarke A. K. (2006) Plant Cell 18, 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y., Murakami S., Yamamoto Y., Chatani H., Kondo Y., Nakano T., Yokota A., Sato F. (2004) Planta 220, 97–104 [DOI] [PubMed] [Google Scholar]
- 24.Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoskins J. R., Kim S. Y., Wickner S. (2000) J. Biol. Chem. 275, 35361–35367 [DOI] [PubMed] [Google Scholar]
- 26.Hoskins J. R., Yanagihara K., Mizuuchi K., Wickner S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11037–11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien P., Perchuk B. S., Laub M. T., Sauer R. T., Baker T. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6590–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogk A., Haslberger T., Tessarz P., Bukau B. (2008) Biochem. Soc. Trans. 36, 120–125 [DOI] [PubMed] [Google Scholar]
- 29.Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000) Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 30.Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. (1976) Mol. Gen. Genet. 145, 101–108 [DOI] [PubMed] [Google Scholar]
- 31.Cadwell R. C., Joyce G. F. (1992) PCR Methods Appl. 2, 28–33 [DOI] [PubMed] [Google Scholar]
- 32.Kanematsu S., Sakuraba Y., Tanaka A., Tanaka R. (2008) Photochem. Photobiol. Sci. 7, 1196–1205 [DOI] [PubMed] [Google Scholar]
- 33.Bricker T. M., Frankel L. K. (2002) Photosynth. Res. 72, 131–146 [DOI] [PubMed] [Google Scholar]
- 34.Boguski M. S., Freeman M., Elshourbagy N. A., Taylor J. M., Gordon J. I. (1986) J. Lipid Res. 27, 1011–1034 [PubMed] [Google Scholar]
- 35.Levchenko I., Luo L., Baker T. A. (1995) Genes Dev. 9, 2399–2408 [DOI] [PubMed] [Google Scholar]
- 36.Levchenko I., Yamauchi M., Baker T. A. (1997) Genes Dev. 11, 1561–1572 [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez M., Frank E. G., McDonald J. P., Levine A. S., Woodgate R. (1998) Acta Biochim. Pol. 45, 163–172 [PubMed] [Google Scholar]
- 38.Hoskins J. R., Wickner S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D. A., Bukau B. (2006) Nature 439, 753–756 [DOI] [PubMed] [Google Scholar]
- 40.Komenda J., Tichy M., Prásil O., Knoppová J., Kuviková S., de Vries R., Nixon P. J. (2007) Plant Cell 19, 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Wilson S., Elliott T. (1999) J. Bacteriol. 181, 6033–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Elliott M., Elliott T. (1999) J. Bacteriol. 181, 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999) Plant J. 18, 455–463 [DOI] [PubMed] [Google Scholar]
- 44.Bechtold N., Pelletier G. (1998) Methods Mol. Biol. 82, 259–266 [DOI] [PubMed] [Google Scholar]