Abstract
Background
The aim of our study was to compare the presence of the intI1 gene and its associations with the antibiotic resistance of commensal Escherichia coli strains in children with/without previous antibiotic treatments and elderly hospitalized/healthy individuals.
Methods
One-hundred-and-fifteen intestinal E. coli strains were analyzed: 30 strains from 10 antibiotic-naive infants; 27 from 9 antibiotic-treated outpatient infants; 30 from 9 healthy elderly volunteers; and 28 from 9 hospitalized elderly patients. The MIC values of ampicillin, cefuroxime, cefotaxime, gentamicin, ciprofloxacin, and sulfamethoxazole were measured by E-test and IntI1 was detected by PCR.
Results
Out of the 115 strains, 56 (49%) carried class 1 integron genes. Comparing persons without medical interventions, we found in antibiotic-naive children a significantly higher frequency of integron-bearing strains and MIC values than in healthy elderly persons (53% versus 17%; p < 0.01). Evaluating medical interventions, we found a higher resistance and frequency of integrons in strains from hospitalized elderly persons compared with non-hospitalized ones. Children treated with antibiotics had strains with higher MIC values (when compared with antibiotic-naive ones), but the integron-bearing in strains was similar. In most cases, the differences in resistance between the groups (integron-positive and negative strains separately) were higher than the differences between integron-positive and negative strains within the groups.
Conclusion
The prevalence of integrons in commensal E. coli strains in persons without previous medical intervention depended on age. The resistance of integron-carrying and non-carrying strains is more dependent on influencing factors (hospitalization and antibiotic administration) in particular groups than merely the presence or absence of integrons.
Background
Escherichia coli is one of the most common bacteria in the environment and in the intestinal tract of humans and animals [1-3]. The intensive use of antibiotics in both human and veterinary medicine, as well as in the field of agriculture, is associated with an emerging resistance against therapeutic drugs, followed by the selection of virulence and resistance gene cassettes (such as integrons) carrying E. coli strains in humans, animals, and the environment [4-6].
Integrons are ancient structures that contain determinants of a site-specific recombination system to capture genes encoding antimicrobial resistance [7]. They can locate within transposons or conjugative plasmids and contribute to the gene traffic leading to the acquisition of new genes in bacteria [8-13].
The class 1 integron is prevalent on plasmids. It has been found in both gram-positive and especially in gram-negative bacteria like Enterobacteriaceae, Campylobacter spp., Acinetobacter spp., Pseudomonas spp., Aeromonas spp., and Vibrio spp. [13-16]. The prevalence of the class 1 integron in clinical E. coli strains is 33 to 49% [10,17-20].
Class 1 integrons carrying commensal E. coli strains have been isolated from 11%-42% of healthy adult persons [17,19,21]. The high prevalence of the intI1 gene among the intestinal E. coli suggests that commensals could represent an important reservoir of resistance determinants, where gene cassettes could spread between commensal and pathogenic bacteria [1].
Still, there are few data assessing the association between integron carriage and the antimicrobial resistance patterns of E. coli isolated from the commensal gut microbiota of different age groups like infants and elderly persons [17,19,21,22].
We aimed to compare the presence of the intI1 gene and its association with the antibiotic resistance of commensal E. coli strains in patients of different age groups: children with/without previous antibiotic treatment and elderly hospitalized/healthy individuals.
Methods
Subjects
Four groups of subjects were included into the study: (Group 1) antibiotic-naive infants (n = 10; age 3-12 months; mean 7.5 months); (Group 2) antibiotic-treated outpatient infants (n = 9; age 1 week-12 months; mean 7.4 months); (Group 3) healthy elderly volunteers (n = 9; age 68-80 years; mean 73 years), and; (Group 4) elderly hospitalized patients (n = 9; age 65-77 years; mean 70 years). All children were vaginally born at the Women's Clinic of Tartu University Hospital and were randomly selected from the participants of a prospective study on the bias of data concerning previous antibiotic treatment [23]. The healthy volunteers, randomly selected from a registry of general practitioners in Tartu, were considered to be healthy, had not been following any special dietary routines and had not received antibiotic treatment for at least two months prior to inclusion. The hospitalized elderly, without clinical signs indicating inflammatory disease, were randomly recruited from the Department of Orthopaedics of Tartu University Hospital two to three days before scheduled orthopaedic surgery and had not received antibiotics during the previous two months.
Faecal samples
From 27 persons, approximately 1 gram of freshly voided stool was collected into sterile plastic containers and frozen at -20°C. Weighed samples of faeces were serially diluted (10-2-10-9) in a pre-reduced phosphate buffer (pH 7.2). A quantitative analysis of the gut bacteria was performed using seeding duplicate samples of 0.1 ml of each dilution on MacConkey (Oxoid, England) agar plates. The colony counts of the different faecal dilutions were recorded, and all colonies of different morphology from the highest dilutions with growth were isolated and identified with the standard methods. From each sample (n = 27), 3-4 fenotypically different dominating E. coli isolates were included. In total, 115 isolates were collected and analyzed.
Antibiotic resistance
Antimicrobial susceptibility testing was performed using the E-test (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar (Oxoid, UK). The values of the minimal inhibitory concentration (MIC) of E. coli isolates to ampicillin, cefuroxime, cefotaxime, gentamicin, ciprofloxacin, and sulfamethoxazole were determined according to the manufacturer's instructions and to Clinical and Laboratory Standard Institute guidelines.
Detection of the class 1 integrons
All E. coli isolates were tested for the presence of class 1 integrons with a polymerase chain reaction (PCR) amplification of a class 1 integrase-specific fragment of the intI1 gene. The total DNA of the E. coli was extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) following the manufacturer's protocol for gram-negative bacteria. The primer sequences used were 5'CS: 5'-GGCATCCAAGCAGCAAG-3' and 3'CS: 5'-AAGCAGACTTGACCTGA-3' [24]. The reactions were carried out in a volume of 50-μl containing 10× PCR buffer, 2.5 mM MgCl2, 2.5 mM dNTPs, 5 pmol/L each primer, 2.5 U Taq polymerase (Fermentas, Lithuania), and 1 μg of template DNA was added. Amplification specifications were as follows: 94°C for 5 min. followed by 35 cycles of 94°C for 30 sec., 55°C for 30 sec., 72°C for 30 sec. and the final extension at 72°C for 7 min. The amplicons were electrophorezed in 1.0% agarose gel and the 1 kb ladder (Fermentas, Lithuania) was used as a molecular size marker.
Statistical analysis
The statistical analyses were performed using the SigmaStat (Jandel Scientific, USA) and Excel (Microsoft Corp.) software programs, employing the Fisher exact test, the chi-square test, and the Mann-Whitney rank sum test. P values less than 0.05 were considered statistically significant.
Approval by ethics committee
The Ethics Committee of Tartu University Medical Faculty approved the study (No. 139/16 20.06.2005 and No. 152/52 18.09.2006).
Results
Prevalence of class 1 integrons in E. coli isolates in different study groups
Out of the 115 dominating commensal E. coli isolates, 56 (49%) carried the intI1 gene. We found a similarly high rate of integrons among dominating E. coli isolates in antibiotic-treated (Group 2; 17/27; 63%) and non-treated (Group 1; 16/30; 53%) infants, as well as in hospitalized elderly patients (Group 4; 18/28; 64%). In elderly healthy volunteers (Group 3), the integron-positive isolates were comparatively low among the dominating E. coli population (5/30; 17%; p < 0.01; Table 1).
Table 1.
Prevalence of class 1 integrons among studied persons and E. coli isolates
| Infants | Elderly | |||
|---|---|---|---|---|
| Group1 | Group2 | Group3 | Group4 | |
| Persons with integrons/total (%) | 7/10 (70%) | 8/9 (89%) | 3/9 (33%) | 8/9 (89%) |
| Isolates with integrons/total (%) | 16/30 (53%) | 17/27 (63%) | 5/30 (17%) | 18/28 (64%) |
P values for comparisons between groups have been shown in the text.
Antibiotic susceptibility of E. coli isolates
Comparing different age groups not influenced by medical intervention, we found that dominating E. coli isolates originating from antibiotic-naive infants (Group1) had higher MIC values to sulfamethoxazole (p < 0.001), cefotaxime (p = 0.003), gentamicin (p < 0.001), and ciprofloxacin (p < 0.001) than those originating from healthy elderly persons (Group3).
To evaluate the influence of medical interventions on dominating E. coli populations, we compared the MIC values of isolates of antibiotic-naive versus treated infants and healthy elderly persons versus hospitalized. The isolates from antibiotic-treated infants (Group2) were more resistant to different antibiotics (range 0-5, median 2 versus range 0-3, median 1; p = 0.025) and had higher MIC values to sulfamethoxazole (p = 0.048), cefuroxime (p < 0.001), cefotaxime (p = 0.013), gentamicin (p = 0.003), and ciprofloxacin (p = 0.002), compared to antibiotic-naive infants (Group1). In isolates from hospitalized patients (Group 4), the resistance to tested antibiotics (range 0-6, median 1 versus range 0-2 median 0; p = 0.01) and MIC values were higher to sulfamethoxazole (p < 0.001), ampicillin (p < 0.001), cefuroxime (p < 0.001), cefotaxime (p < 0.001), and ciprofloxacin (p < 0.001), when compared to their outpatient counterparts (Group 3).
Relation between the presence of the intI1 gene and antibiotic susceptibility
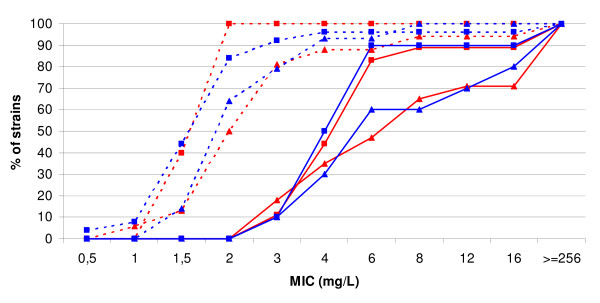
In comparing strains from all the groups together, the MIC-s of intI1-positive isolates were significantly higher than integron-negative ones in the case of several antibiotics (sulfamethoxazole, p = 0.001; ampicillin, p < 0.001; cefuroxime, p = 0.003; cefotaxime, p < 0.001; ciprofloxacin, p = 0.038). However, in comparing the MIC values of intI1-positive and negative isolates in particular groups, we found a significant difference only in non-treated infants (Group 1) in the case of cefotaxime (p = 0.023). Comparing only intI1-negative isolates, we found significantly higher MIC values in groups with medical interventions (i.e. antibiotic-treated infants and hospitalized patients versus non-treated infants and non-hospitalized patients). Similar trends were also evident if only intI1-positive isolates were compared (Table 2). Thus, differences in MIC values were related to age in groups without medical interventions (infants versus elderly) and with medical interventions (antibiotic treatment and hospitalization), but not to the carriage or absence of integrons within a particular group. Figure 1 shows the cumulative MIC values of different groups in the case of cefuroxime.
Table 2.
MIC50 values of intestinal dominant E. coli population in studied groups
| Antibiotics | MIC50 (mg/L) of all isolates MIC50 of integron-negative/integron-positive isolates |
Statistical significance* | |||
|---|---|---|---|---|---|
| Children | Elderly | ||||
| Group 1 | Group 2 | Group 3 | Group 4 | ||
| Ampicillin | 5 31/7 |
6 101/6 |
2 22/256 |
6 62/36 |
1 p = 0.05 2 p = 0.001 |
| Cefuroxime | 2 21/2.52 |
6 61/82 |
2 23/24 |
6 53/64 |
1 p < 0.001 2 p < 0.001 3 p < 0.001 4 p < 0.001 |
| Cefotaxime | 0.064 0.0321/0.094 |
0.125 0.1251/0.125 |
0.047 0.0472/0.0473 |
0.125 0.1252/0.1253 |
1 p = 0.028 2 p < 0.001 3 p = 0.001 |
| Gentamicin | 0.38 0.38/0.441 |
1.5 1.0/1.51 |
0.5 0.5/0.38 |
0.38 0.38/0.38 |
1 p = 0.011 |
| Ciprofloxacin | 0.012 0.012/0.0121 |
0.023 0.023/0.0231 |
0.008 0.0082/0.0123 |
0.016 0.0162/0.0163 |
1 p = 0.004 2 p < 0.001 3 p = 0.048 |
| Sulfamethoxazole | 512 20/512 |
512 512/512 |
24 241/512 |
64 2281/56 |
1 p = 0.03 |
*Statistical significance (by the Mann-Whitney test) has been shown separately for integron-positive and negative strains comparing groups with medical intervention to groups without intervention (i.e. antibiotic-naive children versus treated ones; hospitalized elderly versus non-hospitalized). P values for the comparison of the other groups have been shown in the text.
Figure 1.
Cumulative MIC values of cefuroxime. red lines/marks - integron-positive strains; blue lines/marks - integron-negative strains; triangles - strains of children; squares - strains of adults; dotted line - strains of uninfluenced persons (antibiotic-naive children or healthy elderly); continuous line - strains of persons with medical interventions (antibiotic-treated children or hospitalized elderly).
Discussion
We found a significantly higher frequency of the intI1 gene in the dominating intestinal E. coli populations in healthy antibiotic-naive children (Group 1; 53%) compared to healthy elderly persons (Group3; 17%). The MIC values for several antibiotics for dominant E. coli isolates were lower in elderly persons than in children. Although some previous studies have shown variations of integron prevalence in E. coli isolates from the intestinal tracts of healthy persons, no significant differences in different age groups (not influenced by antibiotics and hospitalization) in the same region has been described yet [5,21,22,25].
Our finding is somewhat surprising, since one can assume a higher rate of resistant and integron-containing bacteria in older people due to the selective pressure of antibiotics consumed during a lifespan. The possible explanation is that in young children (<1 years), the gastrointestinal micro-ecosystem is not yet fully completed and colonization resistance (supported by anaerobes, lactobacilli, and other endogenous commensal bacteria) against exogenous bacteria is weaker; hence, they may be colonized by high numbers of E. coli strains from the environment, containing different virulence or resistance genes, e.g. integrons. Furthermore, during the development of commensal intestinal microflora, the strains without resistance probably have advantages in microbial competition in the intestinal micro-ecosystem, and due to the fitness cost the resistant population would be suppressed (in the case of the absence of an antibiotic pressure) [26].
Comparing the dominating E. coli populations of antibiotic-naive and antibiotic-influenced infants, we found a similar high rate in the presence of class 1 integrons. However, the resistance of E. coli was significantly higher in antibiotic-treated infants. Our results indicate that the higher resistance in antibiotic-treated infants is probably mediated by integron-independent mechanisms, or the expression of resistance genes in integrons is due to antibiotic consumption.
We found that the dominating E. coli population in the intestinal tracts of hospitalized elderly patients was more resistant and more frequently harbored integrons than did those in healthy persons. However, this resistance was not mainly integron-mediated, since similar differences in MIC values were found when separately comparing integron-positive and negative isolates. The published data concerning the prevalence of integrons in E. coli strains isolated from a hospital and community are contradictory. Some investigators have found that higher numbers of integron-positive isolates originated from patients of intensive care units, when compared to non-intensive care unit ones and outpatients (82% versus 10% and 8%) [25]. To the contrary, Lee and co-workers indicated that the prevalence of class 1 integrons among commensal E. coli isolates was similar to that of clinical E. coli isolates from hospital-acquired infections [27].
Increasing resistance in the dominating populations of intestinal bacteria could be caused by the acquisition of resistant and/or integron-positive strains from the hospital environment or by the overgrowth of normally suppressed resistant populations. In both cases, the disruption of commensal microflora responsible for the maintenance of colonization resistance is a conducive factor. This can happen due to a recent exposure to antibiotics, a different diet, increased hygiene habits, and a stressful situation, in essence describing a hospital environment [23,26,28]. It has been previously indicated that exposure to stress results in a significant alteration of intestinal microflora: the counts of beneficial bacteria decrease and enterobacteria like E. coli increase [29,30].
In our study, all integron-positive isolates together had higher MIC values to antibiotics than integron-negative isolates, but susceptibility was similar in studied particular groups. In previous studies, resistant phenotypes usually correlated with the presence of integrons [25,31,32]. Several studies have described the association between the presence of class 1 integrons and non-susceptibility to sulfamethoxazole, β-lactams (ampicillin, piperacillin, cefuroxime), aminoglycosides (gentamicin, tobramycin), and fluoroquinolone (ciprofloxacin) in E. coli strains [20,24]. It has been supposed that the screening of integrons could assist in guiding treatment regimens and could complement existing antibiotic resistance surveillance programs by providing information about both aspects: resistance and its dissemination between commensal and infectious strains [32]. On the one hand, the absence of significant differences in resistance between integron-positive and negative isolates within particular groups in our study may be explained with the small number of strains/persons. However, on the other hand, significant differences in resistance between these groups (separately comparing integron-positive and negative isolates) indicate that the presence of integrons is not important for the predisposition or correlation of the general antimicrobial resistance. Thus, other factors are more important in determining the resistance of the predominant population of E. coli and probably other facultative pathogens as well. For a more complete understanding of these results, further studies detecting particular resistance genes and the presence of empty integrons are needed.
Conclusion
In our study, the prevalence of the intl1 gene in commensal E. coli isolates differs between age groups and depends on hospitalization. Resistance (to the antibiotics selected in our study) of intl1-carrying and non-carrying isolates is more dependent on the influencing factors (such as hospitalization or antibiotic administration) on particular groups than merely the presence or absence of integrons.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ES - study design, drafting the manuscript; PN - statistics and data interpretation, revising the manuscript; JS - molecular studies; SK - antimicrobial resistance testing, drafting the manuscript; KL - collection of the samples and clinical data, interpretation of data; MM - study design, revision of the manuscript; KT - microbiological studies, final correction of the manuscript. All authors have read and approved the final version of manuscript.
Contributor Information
Epp Sepp, Email: epp.sepp@ut.ee.
Jelena Stsepetova, Email: jelena.stsepetova@ut.ee.
Krista Lõivukene, Email: krista.loivukene@kliinikum.ee.
Kai Truusalu, Email: kai.truusalu@ut.ee.
Siiri Kõljalg, Email: siiri.koljalg@ut.ee.
Paul Naaber, Email: paul.naaber@kliinikum.ee.
Marika Mikelsaar, Email: marika.mikelsaar@ut.ee.
Acknowledgements
This work was supported by the Estonian Science Foundation (Grants no. 7497 and no. 6782) and the Estonian Ministry of Education and Research (Grant no. 0132).
References
- Roe MT, Vega E, Pillai SD. Antimicrobial resistance markers of class 1 and class 2 integron-bearing Escherichia coli from irrigation water and sediments. Emerg Infect Dis. 2003;9:822–826. doi: 10.3201/eid0907.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijkeren E, Box ATA, Schellen P, Houwers DJ, Fluit ADC. Class 1 integrons in Enterobacteriaceae isolated from clinical infections of horses and dogs in The Netherlands. Microb Drug Resist. 2005;11:383–386. doi: 10.1089/mdr.2005.11.383. [DOI] [PubMed] [Google Scholar]
- Ozgumus OB, Celik-Sevim E, Alpay-Karaoglu S, Sandalli C, Sevim A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J Microbiol. 2007;45:379–387. [PubMed] [Google Scholar]
- Asai T, Kojima A, Harada K, Ishihara K, Takahashi T, Tamura Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Japan J Infect Dis. 2005;58:369–372. [PubMed] [Google Scholar]
- Kang HY, Jeong YS, Oh JY, Jeong JH, Seol SY, Cho DT, Kim J, Lee YC. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli from humans and animals in Korea. J Antimicrob Chemother. 2005;55:639–644. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- Cocchi S, Grasselli E, Gutacer M, Benagli C, Convert M, Piffaretti JC. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol. 2007;50:126–132. doi: 10.1111/j.1574-695X.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- Johansson C. PhD thesis. Uppsala University, Department of Medical Biochemistry and Microbiology; 2007. Mechanisms and DNA specificity in site-specific recombination of integron cassettes. [Google Scholar]
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;289:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- Martinez-Freijo P, Fluit AC, Schmitz FJ, Grek VSC, Verhoef J, Jones ME. Class 1 integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- Roy PH. Horizontal transfer of genes in bacteria. Microbiol Today. 1999;26:168–170. [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Mazel D. Integron: agent of bacterial evolution. Nat Rev Microbiol. 2006;4:608–720. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- Nandi S, Maurer J, Hofacre C, Summers AO. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. PNAS. 2004;101:7118–7122. doi: 10.1073/pnas.0306466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevskaja NP. Mobile gene cassettes and integrons. Mol Biol. 2002;36:261–267. [PubMed] [Google Scholar]
- Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- Machado E, Ferreira J, Novais A, Peixe L, Canton R, Baquero F, Coque TM. Preservation of integron types among Enterobacteriacae producing extended-spectrum β-Lactamase in a Spanish hospital over a 15-year period (1988 to 2003) Antimicrob Agents Chemother. 2007;51:2201–2204. doi: 10.1128/AAC.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Larivière S, Harel J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol. 2004;42:5444–5452. doi: 10.1128/JCM.42.12.5444-5452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heir E, Lindstedt BA, Leegaard TM, Gjernes E, Kapperud G. Prevalence and characterization of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class 1 integron-located lincosamide resistance gene. Ann Clin Microbiol Antimicrob. 2004;3:12. doi: 10.1186/1476-0711-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AN, Barlow M, Clark LA, Boring JR, Tenover FC, McGowan JE. Class 1 integrons in resistant Escherichia coli and Klebsiella spp., US Hospitals. Emerg Infect Dis. 2006;12:1011–1014. doi: 10.3201/eid1206.051596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik D, Menac'h AL, Zurakowski D, Mazel D, Courvalin P, Denamur E, Andremont A, Ruimy R. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotics exposure. Antimicrob Agents Chemother. 2005;49:3062–3065. doi: 10.1128/AAC.49.7.3062-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante B, Grape M, Larsson M, Kristiansson C, Pallecchi L, Rossolini GM, Kronvall G. Acquired sulphonamide resistance genes in faecal Escherichia coli from healthy children in Bolivia and Peru. Int J Antimicrob Agents. 2005;25:308–312. doi: 10.1016/j.ijantimicag.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Sepp E, Naaber P, Voor T, Mikelsaar M, Björksten B. Development of intestinal microflora during the first month of life in Estonian and Swedish infants. Micr Ecol Health Dis. 2000;12:22–26. doi: 10.1080/089106000435554. [DOI] [Google Scholar]
- Leverstein-van Hall MA, Paauw A, Box ATA, Blok HFM, Verhoef J, Fluit AC. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J Clin Microbiol. 2002;40:3038–3040. doi: 10.1128/JCM.40.8.3038-3040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grape M, Farra A, Kronvall G, Sundström L. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin Microbiol Infect. 2005;11:185–192. doi: 10.1111/j.1469-0691.2004.01059.x. [DOI] [PubMed] [Google Scholar]
- Nahid A. PhD thesis. Göteburg University, Department of Clinical Bacteriology; 2007. Antibiotic resistance and fitness of Escherichia coli in the infantile commensal microbiota. [Google Scholar]
- Lee JC, Kang HY, Oh JY, Jeong JH, Seol SY, Cho DT, Kim J, Lee YC. Antimicrobial resistance and integrons found in commensal Escherichia coli isolates from healthy humans. J Bacteriol Virol. 2006;36:133–139. [Google Scholar]
- Nys S. PhD thesis. Maastricht University, Department of Medical Microbiology; 2005. Antibiotic resistance and the commensal flora. [Google Scholar]
- Lizko NN. Stress and intestinal microflora. Nahrung. 1987;31:443–447. doi: 10.1002/food.19870310530. [DOI] [PubMed] [Google Scholar]
- Lyte M, Bailey MT. Neuroendocrine-bacterial interaction in a neurotoxin-induced model of trauma. J Surg Res. 1997;70:195–201. doi: 10.1006/jsre.1997.5130. [DOI] [PubMed] [Google Scholar]
- Chang C-Y, Chang L-L, Chang Y-H, Lee T-M, Chang S-F. Characterisation of drug resistance gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J Med Microbiol. 2000;49:1097–1102. doi: 10.1099/0022-1317-49-12-1097. [DOI] [PubMed] [Google Scholar]
- Jones LA, Mclver CJ, Rawlinson WD, White PA. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intell. 2003;27(Suppl):103–110. [PubMed] [Google Scholar]