Summary
The DAP10 and DAP12 signaling subunits are highly conserved in evolution and associate with a large family of receptors in hematopoietic cells, including dendritic cells, plasmacytoid dendritic cells, neutrophils, basophils, eosinophils, mast cells, monocytes, macrophages, natural killer cells, and some B and T cells. Some receptors are able to associate with either DAP10 or DAP12, which contribute unique intracellular signaling functions. Studies of humans and mice deficient in these signaling subunits have provided surprising insights into the physiological functions of DAP10 and DAP12, demonstrating that they can either activate or inhibit immune responses. DAP10- and DAP12-associated receptors have been shown to recognize both host-encoded ligands and ligands encoded by microbial pathogens, indicating that they play an important role in innate immune responses.
Keywords: DAP10, DAP12, innate immunity, ITAM
Introduction
The innate immune system relies on receptors that recognize molecules present in pathogens, but not in the host (i.e. non-self), and receptors that detect host-encoded molecules that are induced or altered as a consequence of infection. The ligand-binding domains of many of the innate immune receptors have been preserved during evolution to detect conserved structures shared by pathogens, as best exemplified by the Toll-like receptors (TLRs). However, other innate immune receptors, such as the killer immunoglobulin-like receptor (KIR) and Ly49 receptors on natural killer (NK) cells, demonstrate remarkable diversity in their ligand-binding domains and are rapidly evolving, presumably being driven by pathogens (1), as well as by metamorphosis in the host’s major histocompatibility complex (MHC) molecules that are also adapting to microbial pathogens. While changes in the ligand-binding domains of the innate receptors are essential if their pathogen-encoded or host ligands are evolving, the signaling elements of the innate receptors can be conserved and are often shared between families of receptors.
A common strategy used by immune receptors, both of the innate and adaptive immune systems, is to dissociate ligand-binding and signal-transducing elements into separate subunits that are assembled into multi-subunit complexes. This allows the genes encoding the ligand-binding proteins to mutate and diversify as necessary while preserving the signaling functions. Moreover, this allows a conserved signaling subunit to be shared by receptors with different ligand-binding specificities. In the adaptive immune system, the CD3 signaling subunits of the T-cell antigen receptor (TCR) and the CD79 subunits of the B-cell antigen receptor (BCR) are used by the thousands of rearranged TCRs and surface immunoglobulins that comprise the host’s repertoire of antigen-specific receptors. The conserved signaling element in the CD3 and CD79 subunits is the immunoreceptor tyrosine-based activation motif (ITAM), which is defined as (D or E)xxYxx(L or I)X6–8Yxx(L or I) [where x designates any amino acid and X6–8 denotes any 6 to 8 amino acids between the two Yxx(L or I) modules]. TCR or BCR crosslinking results in phosphorylation of the tyrosines in the ITAMs by Src family kinases, which then recruit and activate the ζ-associated protein of 70 kDa (ZAP70) or Syk tyrosine kinases, which are responsible and necessary for TCR and BCR signal transduction, respectively.
DAP12
ITAMs likely evolved before the advent of adaptive immunity, because ITAM sequences are also found in signaling proteins and receptors of cells in the innate immune system. Two ITAM-containing signaling proteins that are abundantly expressed in myeloid cells and NK cells are FcεRIγ (often designated FcRγ) (2) and DAP12 (3). Like CD3ζ (4), FcRγ and DAP12 have a minimal extracellular region, mainly consisting of a cysteine residue that permits the creation of disulfide-bonded homodimers of FcRγ and DAP12, which have no ligand-binding capacity. Intracellularly, FcRγ and DAP12 have no signal-transducing elements other than a single ITAM, which after tyrosine phosphorylation recruits and activates Syk in myeloid cells and Syk and ZAP70 in NK cells (reviewed in 5). Also like the CD3 and CD79 subunits, FcRγ and DAP12 have an acidic amino acid (aspartic acid) embedded within their transmembrane region that allows these subunits to form stable, non-covalent complexes with their associated receptors. The receptors usually have an oppositely charged amino acid (arginine or lysine) in the same relative location in their transmembrane regions for pairing with FcRγ or DAP12. Orthologs of the gene encoding DAP12, designated TYROBP, have been identified in mammals [e.g. humans (3), chimps (GenBank NP_001077092), Rhesus monkey (GenBank NP_001028039), pigs (6), cows (7), water buffalo (GenBank ABO70007), sheep (GenBank EU370549), pigs (GenBank AAF77183), dogs (GenBank XP_533687), mice (3), and rats (GenBank NP_997690)), amphibians (frogs, GenBank EF431894), and fish, i.e. zebrafish (8) and Fugu (9)]. DAP12 has remained remarkably conserved (Fig. 1), demonstrating that it provides essential signaling functions in most vertebrates.
Fig. 1. Conservation of DAP10 and DAP12 during evolution.
Alignment of the protein sequences of DAP12 (A) and DAP10 (B). Sequences were obtained from GenBank: Human DAP12, NP_003323; Mouse DAP12, NP_035792; Cow DAP12, AAI26798; Xenopus DAP12, EF431894.1; Zebrafish DAP12, NP_001093573; Human DAP10, NP_055081; Mouse DAP10, AAH69220; Cow DAP10, AAI42039; Xenopus DAP10, AAP33502; and Zebrafish DAP10 ABO61031. Fugu sequences were from Guselnikov et al. (9). The proteins were aligned starting with the conserved cysteine in the extracellular region. Conserved cysteines in the extracellular region, the aspartic acid in the transmembrane segment, and the conserved signaling motifs in the cytoplasmic domain are in red.
DAP10
Another signaling subunit, DAP10, which is broadly expressed in the innate immune system, was identified by its similarity to DAP12 in the transmembrane region (10). Remarkably, the gene encoding DAP10 (HCST) is adjacent to DAP12 in the genome (the DAP12 and DAP10-encoding genes are separated only by 130 nucleotides in the human genome) but in the opposite transcriptional orientation. DAP10-encoding genes have been identified in mammals (7, 10, 11), amphibians (9), and fish (8, 9) (Fig. 1). The DAP10- and DAP12-encoding genes in zebrafish are adjacent in the genome, indicating that DAP10 and DAP12 have been preserved as a pair for millions of years and that they likely arose by duplication from a common more ancient gene. Like DAP12, DAP10 has a minimal extracellular region, but it also has a conserved cysteine to create a disulfide-bonded homodimer. Despite this similarity, disulfide-bonded heterodimers between DAP10 and DAP12 subunits have not been observed in humans or mice. The distribution of DAP10 and DAP12 in hematopoietic cells is largely overlapping, with both being present in essentially all myeloid cells and NK cells in mice and humans. One significant difference is the expression of DAP10 in CD8+ T cells, which rarely express DAP12. All human CD8+ T cells constitutively express DAP10, and in mice DAP10 is present in all activated CD8+ T cells.
The signaling function of DAP10 is distinct from DAP12, in that the only signaling motif in the short cytoplasmic domain of DAP10 is a YINM sequence, which after tyrosine phosphorylation allows the binding of phosphatidylinositol-3 kinase (PI3K) and a Grb2 - Vav1 – son of sevenless 1 (SOS1) complex (reviewed in 5). This YINM motif is similar to the motif in CD28, which provides for costimulatory signaling in conjunction with the ITAM-based TCR/CD3 complex in T cells. Thus, DAP10 might be serving an analogous function to CD28 for DAP12-associated receptors.
Receptors associating with DAP12 and DAP10
The aspartic acid residue centrally located within the transmembranes of the DAP10 and DAP12 proteins provided the initial hint that they might be associated with transmembrane-anchored receptors to provide signaling function (3, 10). Prior biochemical analysis of an activating receptor in the KIR gene family expressed on human NK cells and an activating Ly49 receptor on mouse NK cells had shown co-immunoprecipitation of a small protein designated KARAP (12) or pp16 (13), which subsequently was shown to be encoded by DAP12 (3, 14). Based on its structure, DAP10 was predicted to associate non-covalently with ligand-binding receptors and was confirmed by co-transfection and co-immunoprecipitation to associate with the orphan human NKG2D receptor expressed on NK cells and CD8+ T cells (10, 15). Mutation of the aspartic acid in the transmembrane of DAP10 or DAP12 ablates their ability to pair with the associated receptors, and reciprocally, mutation of the lysine or arginine in the receptors prevents assembly with these signaling subunits. In the decade since discovery of DAP10 and DAP12, numerous receptors have been identified that signal through these subunits, and we are beginning to appreciate their biological relevance, including roles in bone development and neural function as well as innate immunity.
DAP12-associated receptors
Although initially studied in the quest to explain NK cell activation by the KIR and Ly49 receptors, the broad distribution of DAP12 immediately suggested a more general role for this protein in immune responses. Indeed, the original DAP12 cDNA was identified from a human dendritic cell library, and DAP12 transcripts are abundantly present in plasmacytoid dendritic cells, dendritic cells, neutrophils, basophils, eosinophils, monocytes, macrophages, microglial cells, and osteoclasts, as well as NK cells and a subset of NKT cells and γδTCR+ T cells. In addition, low amounts of DAP12 are detectable in B cells (16). Although not expressed in most αβTCR+ CD4+ and CD8+ T cells, subsets of αβTCR+ CD4+ and CD8+ T cells expressing DAP12 have been reported (17-19), in some cases correlated with autoimmune T cells. As summarized in Table 1, numerous receptors have been identified that associate with DAP12 in these different cell lineages. These receptors were discovered either functionally, by expression cloning from cDNA libraries derived from different cells and screened for their ability to express DAP12 on the cell surface in 293T cells (“DAP trapping”) (20-24), or by computational methods to identify candidate receptors with basic amino acids centrally located within their transmembrane regions or by homology with receptors previously shown to associate with DAP12 (3, 14, 25-28).
Table 1.
DAP12-associated receptors
| Gene | Names | Species | Ligand | Expression (predominant cell types) |
Reference |
|---|---|---|---|---|---|
| KIR3DS1 | CD158e2 | human | ? | NK, T subset | (33, 34, 35) |
| KIR2DS5 | CD158g | human | ? | NK, T subset | (36) |
| KIR2DS1 | CD158h | human | HLA-C (weak) | NK, T subset | (33) |
| KIR2DS2 | CD158j | human | ? | NK, T subset | (3) |
| KIR2DS4 | CD158i | human | HLA-C (weak) | NK, T subset | (37) |
| KLRD1/KLRC2 | CD94/NKG2C | human | HLA-E | NK, T subset | (3) |
| KLRD1/KLRC2 | CD94/NKG2C | mouse | Qa-1b | NK | (38) |
| NCR2 | NKp44 | human | ? | NK | (39) |
| Klra4 | Ly49DB6 | mouse | H-2Dd (weak) | NK | (14) |
| Klra8 | Ly49HB6 | mouse | MCMV m157 | NK | (14) |
| Klra12 | Ly49LCBA | mouse | ? | NK | (29) |
| Klra16 | Ly49PMa/My | mouse | H-2Dk + MCMV, H-2Dd (weak) |
NK | (30) |
| Klra | Ly49R129 | Mouse | ? | NK | (31) |
| Klra21 | Ly49UMa/My | mouse | ? | NK | (30) |
| Klra23 | Ly49WNOD | Mouse | H-2Dk (weak); H- 2Dd (weak) |
NK | (32) |
| Klrk1 | NKG2D-short | mouse | Rae-1, H60, Mult1 | NK, T subset | (62,63) |
| SIRPB1 | Sirp-b1 | human | ? | Myeloid | (26,27) |
| Sirpb1 | Sirp-b1 | mouse | ? | Myeloid | (48) |
| PILRB | Pilr-b | human | sialylated O-linked sugars |
Myeloid | (47) |
| Pilrb | Pilr-b | mouse | CD99, sialylated O-linked sugars |
Myeloid, NK | (24) |
| Cd200r3 | CD200R3 (Lb) | mouse | CD200? | Mast cell, myeloid |
(23,49) |
| Cd200r4 | CD200R4 (La) | mouse | CD200? | Myeloid, NK | (23) |
| CLEC5A | MDL-1 | human | Dengue virus | Myeloid (macrophage) |
(20) |
| Clec5a | MDL-1 | mouse | ? | Myeloid (macrophage) |
(20) |
| TREM1 | TREM-1 | human | ? | Myeloid (monocytes, neutrophils) |
(28) |
| Trem1 | Trem-1 | mouse | ? | Myeloid | |
| TREM2 | Trem-2 | human | ? | Myeloid (dendritic cells, microglial cells) |
(57) |
| Trem2 | Trem-2 | mouse | Anionic molecules | Myeloid (microglial cells, macrophage subset) |
(77) |
| Trem3 | Trem-3 | mouse | ? | Myeloid | (58) |
|
CD300LB
(CD300B) |
Trem-5, CLM7 | human | ? | Myeloid | (54,55) |
| CD300lb | LMIR-5 | mouse | ? | Myeloid (mast cells, granulocytes, macrophages, and dendritic cells) |
(55) |
|
CD300LE
(CD300E) |
IREM-2, CLM2 | human | ? | Myeloid | (53) |
| AF251705 | MAIR-II, LMIR-2, DIgR1, CMRF- 35-like molecule 4 |
mouse | ? | Myeloid (macrophage subset), B cell subset (marginal zone) |
(50, 51, 52) |
| Siglech | Siglec-H | mouse | ? | Myeloid (plasmacytoid DC) |
(85) |
| SIGLEC14 | Siglec-14 | human | a2-8-linked oligo Neu5A |
Myeloid | (56) |
| SIGLEC15 | Siglec-15 | human | Neu5Ac2- 6GalNAca |
Myeloid | (60) |
| Siglec15 | Siglec-15 | mouse | Neu5Ac2- 6GalNAca; Neu5Ac(alpha)2– 3Galß1-4Glc |
Myeloid | (60) |
| SIGLEC16 | Siglec-16 | human | ? | Myeloid | (108) |
| pDC-Trem | mouse | ? | Myeloid | (71) |
The DAP12-associated receptors expressed by NK cells include members of the human KIR gene family and mouse Ly49 gene family (3, 14, 29-37), as well as the mouse and human CD94-NKG2C heterodimeric receptors (25, 38) and the human NKp44 receptor (39) present on activated NK cells (Table 1). The KIR and Ly49 receptors are expressed on overlapping subsets of NK cells, providing a diverse repertoire within the NK cell population. Whereas DAP12-associated KIR and CD94-NKG2C receptors have been detected on subsets of human T cells, typically with an effector or memory phenotype, DAP12-associated Ly49 receptors are rarely detected on mouse T cells, even after in vivo activation (40). Wucherpfennig and colleagues (41) have investigated the stoichiometry of these DAP12-associated NK cell receptors and determined that the KIR2DS2 monomer associates with one DAP12 homodimer. Similarly, one DAP12 homodimer assembles with each CD94-NKG2C heterodimer through interactions between the oppositely charged residues in the transmembranes of DAP12 and NKG2C (41). The multimeric nature of these complexes might have evolved to optimize signal transduction by providing two ITAMs for each ligand-binding receptor.
The CD94-NKG2C-DAP12 receptor complex (25) recognizes as ligand a non-classical MHC class I protein, human leukocyte antigen-E (HLA-E) in humans (42) and Qa1b in mice (38). Some, but not all, of the DAP12-associated KIR and Ly49 receptors have been shown to recognize HLA-C or H-2 ligands (43), although the interactions appear weak and the physiological relevance has not been established, other than demonstrating that NK cells expressing the Ly49D-DAP12 receptor can mediate rejection of allogeneic bone marrow grafts in certain mouse stains (44, 45).
Physiological ligands for NKp44 have not yet been identified, but presumably self-antigens interacting with NKp44 exist based on the ability of anti-NKp44 mAbs to block NK cell-mediated lysis of certain tumor cell lines and the ability of recombinant NKp44 fusion proteins to bind many different tumor cell lines (39, 46). This putative NKp44 ligand appears to be broadly distributed in many cell types. Mice do not possess an NKp44 ortholog, thus rendering in vivo studies of this receptor in cancer or infectious diseases difficult.
Numerous receptors associating with DAP12 have been identified in mouse and human myeloid cells, including monocytes, macrophages, microglial cells, dendritic cells, plasmacytoid dendritic cells, mast cells, basophils, eosinophils, and neutrophils (Table 1). In some cases, these genes are conserved in mice and humans, and in other cases they are species-specific, implying that evolutionary pressures are shaping the repertoire of these receptors. Like the KIR and Ly49 receptor families, some of these DAP12-associated receptors on myeloid cells are members of a small gene family in which highly homologous genes possess immunoreceptor tyrosine-based inhibitory motifs (ITIMs) [e.g. the paired immunoglobulin-like receptor (PILR) (47), signal regulatory protein (SIRP) (48), CD200 (23, 49), myeloid-associated immunoglobulin-like receptor (MAIR) (50-52), CD300 (53-55), Siglec (56), and triggering receptor expressed by myeloid cells (TREM) (28, 57, 58)] gene families, whereas other receptors are encoded by a single gene without a closely related ITIM-encoding gene (e.g. CLEC5A) (20). In most cases, ligands for these DAP12-associated receptors on myeloid cells have not been identified. However, certain receptors have been shown to recognize carbohydrates as ligands. In particular, the PILRB receptors bind to sialylated O-linked sugars (24, 59), Siglec-14 binds α2-8-linked oligo Neu5A (56), and Siglec-15 recognizes Neu5Ac2-6GalNAca (60). As yet, the physiological significance of carbohydrate binding by these receptors has not been revealed and awaits further experimentation to illuminate their functions.
DAP10-associated receptors
Human NKG2D was the first receptor identified to associate with DAP10 (10), and the receptor complex is a hexamer, composed of one NKG2D homodimer assembled with two DAP10 homodimers (61). Subsequently, it was determined that in mice two alternatively spliced transcripts of NKG2D exist encoding a NKG2D-Long (L) protein that pairs exclusively with DAP10 and a NKG2D-Short (S) protein, lacking 13 amino acids in the cytoplasmic domain compared with NKG2D-L, that promiscuously pairs with either DAP10 or DAP12 (62, 63). Although not yet proven experimentally, based on the prior studies of Wucherpfennig and colleagues, it seems likely that a subset of mouse NKG2D-S receptor complexes might be comprised of one NKG2D-S homodimer paired with one DAP10 homodimer and one DAP12 homodimer. Resting mouse NK cells predominantly (but not necessarily exclusively) transcribe NKG2D-L, whereas activated mouse NK cells transcribe both NKG2D-L and NKG2D-S. At the protein level, resting NK cells in DAP10-deficient (Hcst–/–) mice express little or no detectable NKG2D on the cell surface of NK cells (63, author’s unpublished observations). After activation in vitro or in vivo, NKG2D (presumably NKG2D-S – DAP12) is expressed transiently on the cell surface of DAP10-deficient NK cells but is not expressed on activated DAP10-deficient CD8+ T cells (presumably due to the lack of DAP12 in resting and activated CD8+ T cells)(63, author’s unpublished observations). In contrast, human NKG2D associates only with DAP10 and is unable to pair with DAP12, thus representing a species-specific difference between humans and mice with respect to NKG2D signaling (64).
Several other receptors originally identified by their ability to associate with DAP12 also appear capable of pairing with DAP10. For example, Ly49H and Ly49D were co-immunoprecipitated with DAP10 from mouse NK cells and shown to associate with DAP10 when co-transfected into 293T cells, although the association of Ly49D and Ly49H with DAP10 appeared less efficient than with DAP12 (65). By co-transfection studies, DAP10 has also been shown to associate with human Sirp-b1 in transfected rat RBL-2H3 cells (66). Similarly, human and mouse Siglec-15 (60) and Cd300lb (55) have been shown by co-transfection and co-immunoprecipitation to pair with either DAP10 or DAP12, but apparently with a preference for DAP12. However, these associations with DAP10 have not yet been shown in primary cells expressing Sirp-b1, Cd300lb, or Siglec-15.
The ability of certain receptors to pair with either DAP10 or DAP12 is likely due to the similarity in the transmembrane domains of DAP10 and DAP12 [indeed, DAP10 was originally identified because of the homology between its transmembrane and DAP12’s transmembrane region (10)]. Wucherpfennig and colleagues (67) have reported that the location of the charged residue (lysine or arginine) within the transmembranes of the DAP12- or DAP10-associating receptors is critical for pairing with the aspartic acid residues centrally located within the transmembrane of DAP10 or DAP12. Therefore, a very surprising finding is that the MAIR-II (also named LMIR-2, DIgR1, or CMRF-35-like molecule 4 receptor) apparently associates not only with DAP10 and DAP12 but also with the FcRγ chain (51, 68). The location of the aspartic acid residue in the transmembrane of the FcRγ chain is more proximal to the extracellular region (Fig. 2), and typically the relative position of the charged residues in the transmembrane region of the signaling subunit and the associated receptor is critical to their interaction (69). The lysine is centrally located in the transmembrane of MAIR-II (LMIR-2) so is expected to interact with DAP12 and/or possibly DAP10. However, structurally how this centrally located lysine physically contacts the aspartic acid of the FcRγ chain near the extracellular region is difficult to envision.
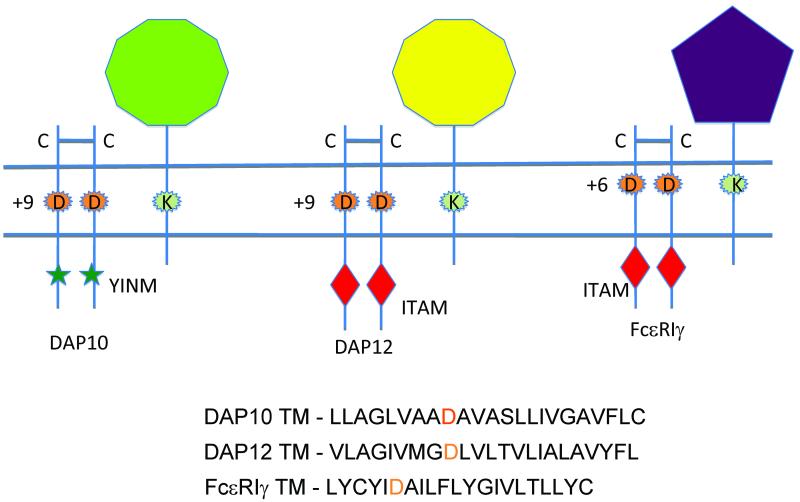
Fig. 2. Charged residues with the transmembranes of DAP10, DAP12, FcRγ, and their associated receptors.
Schematic representation of prototypical receptor complexes with relative location of the charged residues within the transmembrane regions noted. Sequences of the transmembrane regions of human DAP10, DAP12, and FcRγ are shown. The relative position of the aspartic acid (D) in the transmembrane region is indicated, with the number indicating the distance in amino acids below the outer surface of the cell.
It is possible that a MAIR-II–DAP12 (or DAP10) complex might in fact be associating with another unknown receptor in the cell that associates with FcRγ chain. As precedent, the DAP12-associated receptor TREM2 is known to interact with plexin-A1, which in turn interacts with Semaphorin 6D (70). Similarly, DAP12 associates with pDC-Trem, which binds to plexin-A1 bound to Semaphorin 6D (71). Therefore, one might envision a FcRγ-associated receptor that is able to interact with MAIR-II, in turn associating with DAP12 or DAP10 to explain the apparent association between MAIR-II and FcRγ chain.
In the situations where both DAP10 and DAP12 have been shown to associate with a receptor, the stoichiometry of these complexes has not been determined. In addition, if there is competition for binding to the receptor, the abundance of DAP10 versus DAP12 in a cell and the relative affinity of the interactions between these signaling subunits and the receptor are important considerations to understand the physiological relevance of the interactions. Wucherpfennig and colleagues (67) have also noted that the extracellular regions of DAP10 or DAP12 might prevent association with certain receptors due to steric hindrance or render the complexes unstable. Presumably steric hindrance caused by the 13 additional amino acids in the cytoplasmic domain of mouse NKG2D-L prevents its association with DAP12 (62-64).
Recognition of microbial ligands
Many of the DAP10- or DAP12-associated receptors appear to recognize host-encoded molecules, including both carbohydrate and protein ligands. In the case of NKG2D, these ligands [e.g. the MHC class I chain-related gene A (MICA), MICB, and UL16-binding protein (ULBP) and RAET-1 proteins in humans and Rae-1, H60, and murine UL16-binding protein-like transcript-1 (MULT1) proteins in mice] are absent in healthy cells or are sequestered intracellularly until induced by cell stress, such as DNA damage or infection with viruses or bacteria (reviewed in 5, 72, 73). However, other DAP10- or DAP12-associated receptors can directly recognize microbial ligands. The best-characterized example is recognition of the mouse cytomegalovirus (MCMV)-encoded glycoprotein m157 by Ly49H (73, 74). m157 is a GPI-anchored glycoprotein with homology to MHC class I (75), which is displayed on the surface of MCMV-infected cells, resulting in activation of Ly49H+ NK cell-mediated cytotoxicity and cytokine protection that protects the host. Another DAP12-associated receptor, Ly49P, also recognizes MCMV-infected cells. Unlike recognition by Ly49H, Ly49P only recognizes MCMV-infected cells expressing H-2Dk, demonstrating that Ly49P recognition is MHC-restricted, although as yet the viral ligand modifying H-2Dk has not been identified (30).
DAP12-associated receptors on myeloid cells have also been implicated in recognition of microbial ligands. Recently, Clec5 (MDL-1) has been shown to bind to Dengue virus virions (76). Whether Clec5 (a member of the C-type lectin superfamily) interacts with protein or carbohydrate ligands on Dengue virus has not been determined. Much of the pathology caused by Dengue virus infection results from a cytokine storm initiated by binding of Dengue virus to macrophages expressing Clec5, which can be prevented by using a blocking anti-Clec5 monoclonal antibody (76), thereby providing a new therapeutic opportunity for treating this disease.
Microbial ligands have also been suggested for the DAP12-associated Trem2 receptor. Seaman and colleagues have shown that Trem2 binds to a variety of microbial products, including lipopolysaccharide (LPS), a variety of Gram-positive and Gram-negative bacteria, and yeast (77), and can contribute to the immune response against Salmonella in a mouse model (78). In addition to microbial ligands, Trem2 also binds undefined host-encoded ligands on macrophages (79). The binding of Trem2 to such a diverse variety of ligands suggests that these interactions might be charged-based, given the ability of anionic microbial products to block the binding of recombinant Trem2 proteins to both host-encoded and microbial ligands (77). Similarly, a recent study has reported binding of recombinant NKp44 to Mycobacteria and Pseudomonas, suggesting that the DAP12-associated NKp44 receptor, which is expressed on activated human NK cells and a subset of plasmacytoid dendritic cells, might also be a microbial pattern recognition molecule (80). NKp44, which does not exist in mice, is quite similar in structure to mouse Trem2 (39).
Another Trem family member, Trem1, has been implicated in septic shock. Colonna and coworkers (81) demonstrated that septic shock induced by cecal ligation or LPS could be attenuated by administering a Trem1-Fc fusion protein. Presumably, DAP12-mediated signaling through Trem1 augments LPS-induced TLR-dependent activation – contributing to disease pathology. Whether Trem-1 recognizes a microbial ligand or a LPS-induced host-encoded ligand has not been determined. Nonetheless, these studies clearly indicate that DAP12-associated receptors contribute to the innate response against microbes.
Signal transduction: evidence for positive and negative regulation
The only signaling element in the cytoplasmic domain of DAP12 is an ITAM, which has a well-defined downstream signaling cascade that is also used by the ITAM-signaling subunits of the BCR, TCR, and FcR (reviewed in 5). Mutation of either tyrosine in the ITAM of DAP12 abrogates its biological activity, as does abrogation of Syk or ZAP70, the tyrosine kinases binding to phosphorylated DAP12. Ligation of receptors signaling through ITAM-bearing subunits typically initiates cellular activation, resulting in the production of cytokines and triggers degranulation in cytolytic effector cells. A surprising finding was that in some cases DAP12 could suppress the activation of macrophages or dendritic cells stimulated through TLRs, inhibiting the production of pro-inflammatory cytokines such as interleukin-6 (IL-6) and IL-12 (82, 83). Trem2 mediates the DAP12-dependent suppression of macrophages (79, 84). The DAP12-associated receptors Siglec-H in mice (85) and NKp44 in humans (86) can also suppress the production of type I interferon by TLR-activated plasmacytoid dendritic cells. By contrast, blocking the DAP12-associated NKp44 receptor in NK cells partially inhibits killing of certain tumor cell lines expressing putative NKp44 ligands (39). As yet, the biochemical basis for the ability of DAP12 to either suppress or activate an immune response, depending upon which receptor or cell type is involved, has not been determined. The outcome might be influenced by the affinity or avidity of the interaction between the DAP12-associated receptor and its ligands (reviewed in 87). For example, Monteiro and colleagues (88) have shown that binding of monomeric IgA to the FcRγ-associated FcαRI receptor recruits the tyrosine phosphatase SHP-1, which suppresses immune responses, whereas aggregated, multimeric immunoglobulin A complexes cause cellular activation. Therefore, it is possible that the nature of the ligand, as well as the receptor involved and the cell type, will determine whether DAP12 activates or inhibits.
Signal transduction by DAP10 is mediated by its cytoplasmic YINM motif, which after tyrosine phosphorylation recruits PI3K or a Grb2 – Vav1 – SOS1 signaling complex (reviewed in 5). Horng et al. (89) have proposed that DAP10 is coupled with the IL-15 receptor and that Janus kinase 3 (Jak3), recruited and activated by IL-15, is responsible for phosphorylation of DAP10. The biological roles of DAP10 and DAP12 are distinct, in that activation through DAP12-associated receptors potently induces the production of cytokines, whereas DAP10 is much less efficient at this activity. DAP10 is generally considered important for augmentation (‘costimulation’) of signals initiated through other receptors. Although DAP10 has been clearly shown to mediate costimulation, for example through NKG2D in activated CD8+ T cells (90, 91), this is dependent on the activation status of the T cells (92). The ability of DAP10 to activate the PI3K – Akt pathway suggests a role in cell survival, although this has not been evaluated.
DAP10 and DAP12 deficiency
The initial clues about the physiological relevance of DAP12 emerged from the discovery of humans with a recessive genetic disorder caused by loss-of-function of the TYROBP gene, designated Nasu-Hakola disease or polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (93). These patients present at adolescence with problems in bone development and later develop dementia due to plaque formation in the central nervous system (CNS). These same clinical symptoms are present in patients with a loss-of-function in the TREM2 gene, demonstrating that loss of Trem2 function is responsible for the disease (94). The bone phenotype is likely caused by the defective function of osteoclasts lacking Trem2 or DAP12 (95, 96), whereas the brain pathology probably stems from dysfunctional microglial cells, which are myeloid cells resident in the CNS that are the predominant cell type expressing DAP12 and Trem2 (97). In DAP12-deficient mice, osteoclast development and function are impaired (98-101). Takai and colleagues (98) have reported abnormalities in the brains of their DAP12-null mice; however, we have not observed brain pathology in our Tyrobp–/– mice (author’s unpublished observation).
Given the number of receptors that use DAP12 for signal transduction, it is quite surprising that humans lacking DAP12 do not have problems resolving infections with viruses or bacteria. Mice lacking DAP12 are more susceptible to MCMV infection because of a poorly functioning Ly49H receptor (102); however, the plasmacytoid dendritic cells in DAP12-deficient mice are hyper-responsive in mice infected with MCMV (103). Similarly, we observed that macrophages from DAP12-deficient mice hyper-respond, producing higher amounts of tumor necrosis factor (TNF), IL-12, and IL-6, when stimulated in vitro by various TLR ligands. DAP12-deficient mice control infection with Listeria monocytogenes, Mycobacterium bovis, and Mycobacterium tuberculosis better than wildtype mice (82, 104, author’s unpublished observations). NK cells in DAP12-deficient mice are also hyper-responsive (103, 105); however, this might be predominately due to hyperactive dendritic cells and macrophages secreting elevated amounts of type I interferons and stimulatory cytokines such as IL-12. Thus, the increased production of cytokines in response to TLR signaling in the absence of DAP12 might compensate for the loss-of-function of the DAP12-associated receptors in dealing with many infections. Future studies using mice with conditionally deleted DAP12-encoding genes in myeloid cells or NK cells would be informative to address this issue.
In humans lacking DAP12, DAP10 is expressed and functional (64, 93). Therefore, the receptors able to pair with either DAP12 or DAP10 would retain partial function in the absence of DAP12 and perhaps contribute to immune responses against pathogens involving these receptors. In mice lacking DAP10 (Hcst–/–), NKG2D-dependent immune functions are impaired, including the ability to reject NKG2D-ligand bearing tumors and the ability to reject bone marrow grafts, although the ability of mouse NKG2D to associate with DAP12 provides partial retention of NKG2D-mediated functions (63, 106).
Interestingly, a hyper-response phenotype has been reported in DAP10-deficient mice, reminiscent of the hyper-responsiveness in DAP12-deficient mice. Phillips and colleagues (107) have shown that rejection of B16 melanomas is improved in DAP10-deficient mice, attributed to the hyper-responsiveness of NKT cells because of defective CD4+ T-regulatory cells in these animals. It is possible that in the absence of DAP10, receptors able to pair with either DAP10 or DAP12 might preferentially pair with DAP12 and mediate stronger activation. Further studies are warranted to define the receptors involved and the basis for the phenotype in the DAP10-deficient mice.
Concluding remarks
More than 20 immune receptors in humans or mice have been shown to associate with DAP10 or DAP12. Recent findings suggested that many of these receptors are promiscuous, able to associate with either DAP10 or DAP12, at least in overexpression systems in vitro. This raises intriguing questions about the relative contributions of these signaling proteins in vivo in immune responses induced by the different receptors and in different cell types. Will the relative abundance of DAP10 or DAP12 proteins dictate their association with these receptors? Do DAP10 and DAP12 have different affinities for the different receptors and will they compete for association? In animals lacking DAP10 or DAP12 will the remaining signaling subunit provide compensation or alter the cell’s development? Finally, how do the downstream signaling pathways initiated by DAP10 and DAP12 intersect, and do they provide any feedback circuitry to positively or negatively affect signaling by their receptors or other independent receptor signaling pathways? Since DAP10 and DAP12 are highly conserved during evolution and have been maintained in the genome as closely linked genes for millions of years, it seems likely they have been preserved to provide important functions for the innate and adaptive immune systems.
Supplementary Material
Acknowledgements
L.L.L. is an American Cancer Society Research Professor and supported by grants from the National Institutes of Health.
References
- 1.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank U, Ra C, White K, Metzger H, Kinet J-P. Complete structure and expression in transfected cells of high affinity IgE receptor. Nature. 1989;337:187–189. doi: 10.1038/337187a0. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 4.Weissman AM, Samelson LE, Klausner RD. A new subunit of the human T-cell antigen receptor complex. Nature. 1986;324:480–482. doi: 10.1038/324480a0. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yim D, Jie HB, Lanier LL, Kim YB. Molecular cloning, gene structure, and expression pattern of pig immunoreceptor DAP12. Immunogenetics. 2000;51:436–442. doi: 10.1007/s002510050642. [DOI] [PubMed] [Google Scholar]
- 7.Fikri Y, Nyabenda J, Content J, Huygen K. Cloning, sequencing, and cell surface expression pattern of bovine immunoreceptor NKG2D and adaptor molecules DAP10 and DAP12. Immunogenetics. 2007;59:653–659. doi: 10.1007/s00251-007-0226-6. [DOI] [PubMed] [Google Scholar]
- 8.Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–164. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guselnikov SV, Najakshin AM, Taranin AV. Fugu rubripes possesses genes for the entire set of the ITAM-bearing transmembrane signal subunits. Immunogenetics. 2003;55:472–479. doi: 10.1007/s00251-003-0599-0. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, et al. An activating receptor complex on natural killer and T cells formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 11.Yim D, et al. Molecular cloning and characterization of pig immunoreceptor DAP10 and NKG2D. Immunogenetics. 2001;53:243–249. doi: 10.1007/s002510100321. [DOI] [PubMed] [Google Scholar]
- 12.Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E. Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by Natural Killer cells. J Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- 13.Mason LH, et al. Characterization of an associated 16 kDA tyrosine phosphoprotein required for Ly-49D signal transduction. J Immunol. 1998;160:4148–4152. [PubMed] [Google Scholar]
- 14.Smith KM, Wu J, Bakker ABH, Phillips JH, Lanier LL. Cutting edge: Ly49D and Ly49H associate with mouse DAP12 and form activating receptors. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 15.Bauer S, et al. Activation of natural killer cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–730. [PubMed] [Google Scholar]
- 16.Yotsumoto K, et al. Paired activating and inhibitory immunoglobulin-like receptors, MAIR-I and MAIR-II, regulate mast cell and macrophage activation. J Exp Med. 2003;198:223–233. doi: 10.1084/jem.20021825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namekawa T, et al. Killer cell activating receptors function as costimulatory molecules on CD4+CD28null T cells clonally expanded in rheumatoid arthritis. J Immunol. 2000;165:1138–1145. doi: 10.4049/jimmunol.165.2.1138. [DOI] [PubMed] [Google Scholar]
- 18.Snyder MR, Nakajima T, Leibson PJ, Weyand CM, Goronzy JJ. Stimulatory killer Ig-like receptors modulate T cell activation through DAP12-dependent and DAP12-independent mechanisms. J Immunol. 2004;173:3725–3731. doi: 10.4049/jimmunol.173.6.3725. [DOI] [PubMed] [Google Scholar]
- 19.Dhanji S, Teh SJ, Oble D, Priatel JJ, Teh HS. Self-reactive memory-phenotype CD8 T cells exhibit both MHC-restricted and non-MHC-restricted cytotoxicity: a role for the T-cell receptor and natural killer cell receptors. Blood. 2004;104:2116–2123. doi: 10.1182/blood-2004-01-0150. [DOI] [PubMed] [Google Scholar]
- 20.Bakker AB, Baker E, Sutherland GR, Phillips JH, Lanier LL. Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc Natl Acad Sci USA. 1999;96:9792–9796. doi: 10.1073/pnas.96.17.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daws MR, Lanier LL, Seaman WE, Ryan JC. Cloning and characterization of a novel mouse myeloid DAP12-associated receptor family. Eur J Immunol. 2001;31:783–791. doi: 10.1002/1521-4141(200103)31:3<783::aid-immu783>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 22.Naper C, et al. Ly-49s3 Is a Promiscuous Activating Rat NK Cell Receptor for Nonclassical MHC Class I-Encoded Target Ligands. J Immunol. 2002;169:22–30. doi: 10.4049/jimmunol.169.1.22. [DOI] [PubMed] [Google Scholar]
- 23.Wright GJ, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol. 2003;171:3034–3046. doi: 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- 24.Shiratori I, Ogasawara K, Saito T, Lanier LL, Arase H. Activation of Natural Killer Cells and Dendritic Cells upon Recognition of a Novel CD99-like Ligand by Paired Immunoglobulin-like Type 2 Receptor. J Exp Med. 2004;199:525–533. doi: 10.1084/jem.20031885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 26.Tomasello E, et al. Association of signal-regulatory proteins beta with KARAP/DAP-12. Eur J Immunol. 2000;30:2147–2156. doi: 10.1002/1521-4141(2000)30:8<2147::AID-IMMU2147>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Dietrich J, Cella M, Seiffert M, Buhring HJ, Colonna M. Cutting edge: signal-regulatory protein beta1 is a DAP12-associated activating receptor expressed in myeloid cells. J Immunol. 2000;164:9–12. doi: 10.4049/jimmunol.164.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 29.Makrigiannis AP, Etzler J, Winkler-Pickett R, Mason A, Ortaldo JR, Anderson SK. Identification of the Ly49L protein: evidence for activating counterparts to inhibitory Ly49 proteins. J Leukoc Biol. 2000;68:765–771. [PubMed] [Google Scholar]
- 30.Desrosiers MP, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McVicar DW, et al. Aberrant DAP12 signaling in the 129 strain of mice: implications for the analysis of gene-targeted mice. J Immunol. 2002;169:1721–1728. doi: 10.4049/jimmunol.169.4.1721. [DOI] [PubMed] [Google Scholar]
- 32.Silver ET, Gong D, Hazes B, Kane KP. Ly-49W, an activating receptor of nonobese diabetic mice with close homology to the inhibitory receptor Ly-49G, recognizes H-2D(k) and H-2D(d) J Immunol. 2001;166:2333–2341. doi: 10.4049/jimmunol.166.4.2333. [DOI] [PubMed] [Google Scholar]
- 33.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting edge: KIR3DS1, a Gene Implicated in Resistance to Progression to AIDS, Encodes a DAP12-Associated Receptor Expressed on NK Cells That Triggers NK Cell Activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pascal V, et al. Detection of KIR3DS1 on the Cell Surface of Peripheral Blood NK Cells Facilitates Identification of a Novel Null Allele and Assessment of KIR3DS1 Expression during HIV-1 Infection. J Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 35.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 36.Chiesa MD, et al. Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur J Immunol. 2008 doi: 10.1002/eji.200838434. in press. [DOI] [PubMed] [Google Scholar]
- 37.Augugliaro R, et al. Selective cross-talk among natural cytotoxicity receptors in human natural killer cells. Eur J Immunol. 2003;33:1235–1241. doi: 10.1002/eji.200323896. [DOI] [PubMed] [Google Scholar]
- 38.Vance RE, Jamieson AM, Raulet DH. Recognition of the Class Ib molecule Qa-1b by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitale M, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith HR, Chuang HH, Wang LL, Salcedo M, Heusel JW, Yokoyama WM. Nonstochastic Coexpression of Activation Receptors on Murine Natural Killer Cells. J Exp Med. 2000;191:1341–1354. doi: 10.1084/jem.191.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng J, Garrity D, Call ME, Moffett H, Wucherpfennig KW. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B, and C. Nature. 1998;391:795–798. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 43.Mason LH, Willette-Brown J, Mason AT, McVicar D, Ortaldo JR. Interaction of Ly-49D+ NK Cells with H-2Dd Target Cells Leads to Dap-12 Phosphorylation and IFN-gamma Secretion. J Immunol. 2000;164:603–611. doi: 10.4049/jimmunol.164.2.603. [DOI] [PubMed] [Google Scholar]
- 44.Raziuddin A, Longo DL, Mason L, Ortaldo JR, Bennett M, Murphy WJ. Differential effects of the rejection of bone marrow allografts by the depletion of activating versus inhibiting Ly-49 natural killer cell subsets. J Immunol. 1998;160:87–94. [PubMed] [Google Scholar]
- 45.George TC, Mason LH, Ortaldo JR, Kumar V, Bennett M. Positive Recognition of MHC Class I Molecules by the Ly49D Receptor of Murine NK Cells. J Immunol. 1999;162:2035–2043. [PubMed] [Google Scholar]
- 46.Byrd A, Hoffmann SC, Jarahian M, Momburg F, Watzl C. Expression Analysis of the Ligands for the Natural Killer Cell Receptors NKp30 and NKp44. PLoS ONE. 2007;2:e1339. doi: 10.1371/journal.pone.0001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mousseau DD, Banville D, L’Abbe D, Bouchard P, Shen S-H. PILRα, a novel immunoreceptor tyrosine-based inhibitory motif-bearing protein, recruits SHP-1 upon tyrosine phosphorylation and is paired with the truncated counterpart PILRβ. J Biol Chem. 2000;275:4467–4474. doi: 10.1074/jbc.275.6.4467. [DOI] [PubMed] [Google Scholar]
- 48.Hayashi A, et al. Positive regulation of phagocytosis by SIRPbeta and its signaling mechanism in macrophages. J Biol Chem. 2004;279:29450–29460. doi: 10.1074/jbc.M400950200. [DOI] [PubMed] [Google Scholar]
- 49.Voehringer D, Rosen DB, Lanier LL, Locksley RM. CD200 receptor family members represent novel DAP12-associated activating receptors on basophils and mast cells. J Biol Chem. 2004;279:54117–54123. doi: 10.1074/jbc.M406997200. [DOI] [PubMed] [Google Scholar]
- 50.Chung DH, Humphrey MB, Nakamura MC, Ginzinger DG, Seaman WE, Daws MR. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. J Immunol. 2003;171:6541–6548. doi: 10.4049/jimmunol.171.12.6541. [DOI] [PubMed] [Google Scholar]
- 51.Kumagai H, et al. Identification and characterization of a new pair of immunoglobulin-like receptors LMIR1 and 2 derived from murine bone marrow-derived mast cells. Biochem Biophys Res Commun. 2003;307:719–729. doi: 10.1016/s0006-291x(03)01245-2. [DOI] [PubMed] [Google Scholar]
- 52.Luo K, et al. DIgR1, a novel membrane receptor of the immunoglobulin gene superfamily, is preferentially expressed by antigen-presenting cells. Biochem Biophys Res Commun. 2001;287:35–41. doi: 10.1006/bbrc.2001.5539. [DOI] [PubMed] [Google Scholar]
- 53.Aguilar H, Alvarez-Errico D, Garcia-Montero AC, Orfao A, Sayos J, Lopez-Botet M. Molecular characterization of a novel immune receptor restricted to the monocytic lineage. J Immunol. 2004;173:6703–6711. doi: 10.4049/jimmunol.173.11.6703. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Barriocanal A, Sayos J. Molecular and Functional Characterization of CD300b, a New Activating Immunoglobulin Receptor Able to Transduce Signals through Two Different Pathways. J Immunol. 2006;177:2819–2830. doi: 10.4049/jimmunol.177.5.2819. [DOI] [PubMed] [Google Scholar]
- 55.Yamanishi Y, et al. Analysis of mouse LMIR5/CLM-7 as an activating receptor: differential regulation of LMIR5/CLM-7 in mouse versus human cells. Blood. 2008;111:688–698. doi: 10.1182/blood-2007-04-085787. [DOI] [PubMed] [Google Scholar]
- 56.Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20:1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- 57.Bouchon A, Hernandez-Munain C, Cella M, Colonna M. A dap12-mediated pathway regulates expression of cc chemokine receptor 7 and maturation of human dendritic cells. J Exp Med. 2001;194:1111–1122. doi: 10.1084/jem.194.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung DH, Seaman WE, Daws MR. Characterization of TREM-3, an activating receptor on mouse macrophages: definition of a family of single Ig domain receptors on mouse chromosome 17. Eur J Immunol. 2002;32:59–66. doi: 10.1002/1521-4141(200201)32:1<59::AID-IMMU59>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Tabata S, et al. Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J Biol Chem. 2008;283:8893–8901. doi: 10.1074/jbc.M709793200. [DOI] [PubMed] [Google Scholar]
- 60.Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17:838–846. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- 61.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci USA. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diefenbach A, et al. Selective associations with signaling proteins determine stimulatory versus costimulatory activity of NKG2D. Nat Immunol. 2002;3:1142–1149. doi: 10.1038/ni858. [DOI] [PubMed] [Google Scholar]
- 63.Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3:1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
- 64.Rosen DB, Araki M, Hamerman JA, Chen T, Yamamura T, Lanier LL. A Structural Basis for the Association of DAP12 with Mouse, but Not Human, NKG2D. J Immunol. 2004;173:2470–2478. doi: 10.4049/jimmunol.173.4.2470. [DOI] [PubMed] [Google Scholar]
- 65.Coudert JD, Scarpellino L, Gros F, Vivier E, Held W. Sustained NKG2D engagement induces cross-tolerance of multiple distinct NK cell activation pathways. Blood. 2008;111:3571–3578. doi: 10.1182/blood-2007-07-100057. [DOI] [PubMed] [Google Scholar]
- 66.Anfossi N, et al. Contrasting roles of DAP10 and KARAP/DAP12 signaling adaptors in activation of the RBL-2H3 leukemic mast cell line. Eur J Immunol. 2003;33:3514–3522. doi: 10.1002/eji.200324573. [DOI] [PubMed] [Google Scholar]
- 67.Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4:e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakahashi C, et al. Dual assemblies of an activating immune receptor, MAIR-II, with ITAM-bearing adapters DAP12 and FcRgamma chain on peritoneal macrophages. J Immunol. 2007;178:765–770. doi: 10.4049/jimmunol.178.2.765. [DOI] [PubMed] [Google Scholar]
- 69.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 70.Takegahara N, et al. Plexin-A1 and its interaction with DAP12 in immune responses and bone homeostasis. Nat Cell Biol. 2006;8:615–622. doi: 10.1038/ncb1416. [DOI] [PubMed] [Google Scholar]
- 71.Watarai H, Sekine E, Inoue S, Nakagawa R, Kaisho T, Taniguchi M. PDC-TREM, a plasmacytoid dendritic cell-specific receptor, is responsible for augmented production of type I interferon. Proc Natl Acad Sci USA. 2008;105:2993–2998. doi: 10.1073/pnas.0710351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–2274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 73.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams EJ, et al. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci USA. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen ST, et al. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453:672–676. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- 77.Daws MR, Sullam PM, Niemi EC, Chen TT, Tchao NK, Seaman WE. Pattern recognition by TREM-2: binding of anionic ligands. J Immunol. 2003;171:594–599. doi: 10.4049/jimmunol.171.2.594. [DOI] [PubMed] [Google Scholar]
- 78.Charles JF, et al. The innate immune response to Salmonella enterica serovar Typhimurium by macrophages is dependent on TREM2-DAP12. Infect Immun. 2008;76:2439–2447. doi: 10.1128/IAI.00115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting Edge: Inhibition of TLR and FcR Responses in Macrophages by Triggering Receptor Expressed on Myeloid Cells (TREM)-2 and DAP12. J Immunol. 2006;177:2051–2055. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 80.Esin S, et al. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surface of mycobacteria and other bacteria. Infect Immun. 2008;76:1719–1727. doi: 10.1128/IAI.00870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 82.Hamerman JA, Tchao NK, Lowell CA, Lanier LL. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat Immunol. 2005;6:579–586. doi: 10.1038/ni1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chu CL, Yu YL, Shen KY, Lowell CA, Lanier LL, Hamerman JA. Increased TLR responses in dendritic cells lacking the ITAM-containing adapters DAP12 and FcRgamma. Eur J Immunol. 2008;38:166–173. doi: 10.1002/eji.200737600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Turnbull IR, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 85.Blasius AL, Cella M, Maldonado J, Takai T, Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107:2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fuchs A, Cella M, Kondo T, Colonna M. Paradoxical inhibition of human natural interferon-producing cells by the activating receptor NKp44. Blood. 2005;106:2076–2082. doi: 10.1182/blood-2004-12-4802. [DOI] [PubMed] [Google Scholar]
- 87.Hamerman JA, Lanier LL. Inhibition of immune responses by ITAM-bearing receptors. Sci STKE. 2006;2006:re1. doi: 10.1126/stke.3202006re1. [DOI] [PubMed] [Google Scholar]
- 88.Pasquier B, et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation; dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 89.Horng T, Bezbradica JS, Medzhitov R. NKG2D signaling is coupled to the interleukin 15 receptor signaling pathway. Nat Immunol. 2007;8:1345–1352. doi: 10.1038/ni1524. [DOI] [PubMed] [Google Scholar]
- 90.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 91.Markiewicz MA, et al. Costimulation through NKG2D enhances murine CD8+ CTL function: similarities and differences between NKG2D and CD28 costimulation. J Immunol. 2005;175:2825–2833. doi: 10.4049/jimmunol.175.5.2825. [DOI] [PubMed] [Google Scholar]
- 92.Ehrlich LIR, et al. Engagement of NKG2D by cognate ligand or antibody alone is insufficient to mediate costimulation of human and mouse CD8+ T cells. J Immunol. 2005;174:1922–1931. doi: 10.4049/jimmunol.174.4.1922. [DOI] [PubMed] [Google Scholar]
- 93.Paloneva J, et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet. 2000;25:357–361. doi: 10.1038/77153. [DOI] [PubMed] [Google Scholar]
- 94.Paloneva J, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cella M, Buonsanti C, Strader C, Kondo T, Salmaggi A, Colonna M. Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J Exp Med. 2003;198:645–651. doi: 10.1084/jem.20022220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paloneva J, et al. DAP12/TREM2 Deficiency Results in Impaired Osteoclast Differentiation and Osteoporotic Features. J Exp Med. 2003;198:669–675. doi: 10.1084/jem.20030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sessa G, et al. Distribution and signaling of TREM2/DAP12, the receptor system mutated in human polycystic lipomembraneous osteodysplasia with sclerosing leukoencephalopathy dementia. Eur J Neurosci. 2004;20:2617–2628. doi: 10.1111/j.1460-9568.2004.03729.x. [DOI] [PubMed] [Google Scholar]
- 98.Kaifu T, et al. Osteopetrosis and thalamic hypomyelinosis with synaptic degeneration in DAP12-deficient mice. J Clin Invest. 2003;111:323–332. doi: 10.1172/JCI16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Humphrey MB, et al. The signaling adapter protein DAP12 regulates multinucleation during osteoclast development. J Bone Miner Res. 2004;19:224–234. doi: 10.1359/JBMR.0301234. [DOI] [PubMed] [Google Scholar]
- 100.Koga T, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 101.Mocsai A, et al. The immunomodulatory adapter proteins DAP12 and Fc receptor {gamma}-chain (FcR{gamma}) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci USA. 2004;101:6158–6163. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sjolin H, et al. Pivotal Role of KARAP/DAP12 Adaptor Molecule in the Natural Killer Cell-mediated Resistance to Murine Cytomegalovirus Infection. J Exp Med. 2002;195:825–834. doi: 10.1084/jem.20011427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sjolin H, et al. DAP12 signaling regulates plasmacytoid dendritic cell homeostasis and down-modulates their function during viral infection. J Immunol. 2006;177:2908–2916. doi: 10.4049/jimmunol.177.5.2908. [DOI] [PubMed] [Google Scholar]
- 104.Divangahi M, et al. Critical negative regulation of type 1 T cell immunity and immunopathology by signaling adaptor DAP12 during intracellular infection. J Immunol. 2007;179:4015–4026. doi: 10.4049/jimmunol.179.6.4015. [DOI] [PubMed] [Google Scholar]
- 105.Takaki R, Watson SR, Lanier LL. DAP12: an adapter protein with dual functionality. Immunol Rev. 2006;214:118–129. doi: 10.1111/j.1600-065X.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 106.Ogasawara K, Benjamin J, Takaki R, Phillips JH, Lanier LL. Function of NKG2D in natural killer cell-mediated rejection of mouse bone marrow grafts. Nat Immunol. 2005;6:938–945. doi: 10.1038/ni1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hyka-Nouspikel N, Lucian L, Murphy E, McClanahan T, Phillips JH. DAP10 deficiency breaks the immune tolerance against transplantable syngeneic melanoma. J Immunol. 2007;179:3763–3771. doi: 10.4049/jimmunol.179.6.3763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.