Abstract
Estrogen Receptor alpha (ERα), plays essential roles in the female reproduction. To investigate the dynamic changes in ERα activity in vivo, we have developed an ER Alpha Activity Indicator (ERAAI) mouse. This ERAAI mouse harbors both a modified ERα Bacterial Artificial Chromosome (BAC) clone and a reporter gene which is regulated specifically by the modified receptor. The ERα modification (Gal4-ERα) consists replacing the DNA binding domain (DBD) of ERα with the DBD of yeast Gal4 transcription factor. This reporter transgene consisting of a humanized renilla Green Fluorescent Protein (hrGFP) sequence controlled by the Upstream Activating Sequences for the Gal4 gene (UASG) was inserted into the modified ERα BAC clone. Expression of Gal4-ERα and hrGFP reliably recapitulates endogenous ERα expression and activity in the estrogen target tissues in response to estrogen stimulation. Therefore, the ERAAI mouse represents a novel animal model to investigate dynamic ERα activity in vivo.
Natural, synthetic and environmental estrogens have numerous effects on the development and physiology of mammals through the binding and modulation of the activity of the Estrogen Receptors (Mueller and Korach, 2001). For example, studies have revealed that both sexes of ER knock-out mice are infertile (Eddy et al., 1996), and that female infertility is due, in part, to the insensitivity of the uterus to the mitogenic and differentiative actions of estrogen (Couse and Korach, 1999; Lubahn et al., 1993). In addition to reproductive functions, estrogen is involved in the prevention of osteoporosis by inhibiting bone resorption (Avioli, 1999). Estrogen has also been reported to affect the cardiovascular system by reducing the incidence of coronary heart disease (Kuller et al., 2000; Vogelvang et al., 2006). In order to investigate this dynamic compound and its cognate receptor activities in vivo, researchers have created a transgenic mouse carrying a genomic estrogen response element (ERE)-luciferase reporter system (Ciana et al., 2001). The ERE-luciferase reporter transgenic mouse clearly reveals dynamic estrogen activity in various estrogen target tissues under specific physiological conditions (Ciana et al., 2006). However, the ERE-luciferase mouse model system presents limited opportunities for the investigation of in vivo ER activity due to ER isoforms, ERα and ERβ (Hewitt and Korach, 2002; Zhao et al., 2008). Since knock-out mice have revealed that the isoforms of ER are distinct regulators of specific cellular processes in vivo (Couse and Korach, 1999), dissection of the activity of the individual ER isoforms is important in the identification and evaluation of novel specific estrogen receptor modulator, SERM, compounds. To achieve this goal, we generated a novel “Estrogen Receptor Alpha Activity Indicator (ERAAI) mouse” model using a modified ERα Bacterial Artificial Chromosome (BAC) clone. The BAC approach is advantageous since it utilizes large fragments of genomic DNA averaging 100–300 kilobase pairs (kb) in size that can be cloned in bacterial vectors and stably propagated in bacteria (Zhao, 2001). The large size of BAC clones allows for faithful tissue-specific expression of heterologous genes in vivo in BAC-transgenic mice when the appropriate regulatory sequences surrounding the gene are retained in the BAC clone (Gong et al., 2003; Heintz, 2000).
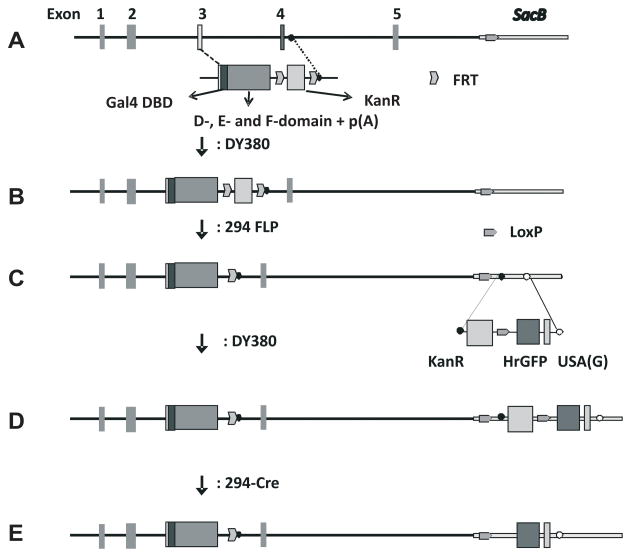
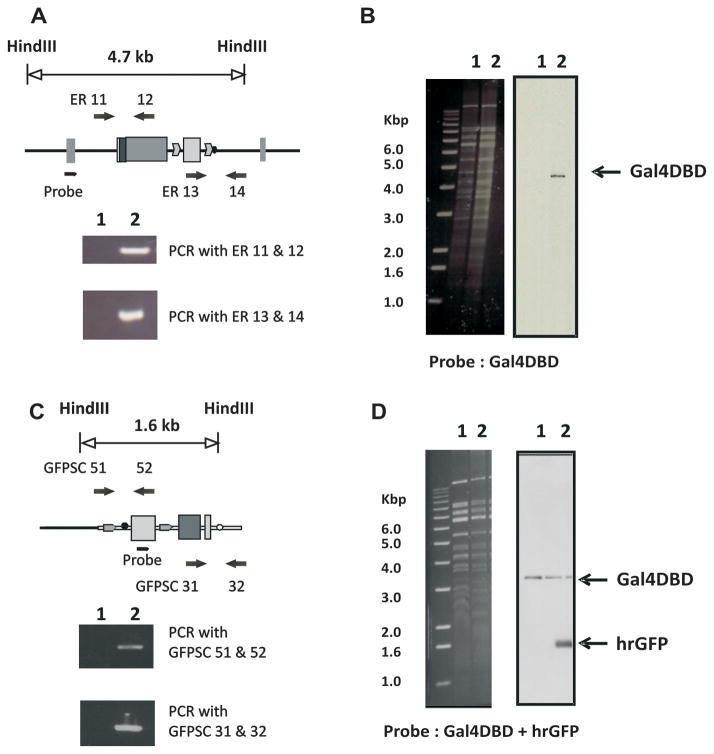
To generate the ERAAI mouse model, several mouse BAC clones containing the mouse ERα gene were selected from the mouse Ensembl Genome Browser (http://www.ensembl.org/Mus_musculus/Info/Index). Among them, the RPCI-23-336H23 mouse ERα BAC clone was selected because this BAC clone maintained its integrity during the recombination process. The RPCI-23-336H23 ERα BAC clone contains ~8 kb of genomic DNA spanning the 5′ end of the ERα BAC clone, and exons 1 to exon 5 of the mouse ERα gene. This clone was modified by exchanging the DNA binding domain (DBD) of the ERα gene, located in exons 3 through 5, with the DBD of the yeast Gal4 transcription factor (Gal4 DBD) using the bacterial recombination system of the DY380 strain (Yu et al., 2000). Because the RPCI-23-336H23 ERα BAC clone did not contain sequences beyond exon 5, sequences from the cDNA that encode the D-, E- and F-domains of mouse ERα were attached to the Gal4-DBD region. For the transcription of the modified ERα gene containing Gal4-DBD to be terminated, a bovine growth hormone (BGH) poly (A) signal sequence was placed just after the F-domain of the modified ERα gene (Fig. 1A and 1B). The kanamycin resistance gene used as the selection marker for the recombination was deleted from modified ERα BAC clones by using the 294-FLP strain as has been previously described (Han et al., 2005) (Fig. 1B and 1C). To confirm the insertion of Gal4-DBD into the modified ERα BAC clone, Southern analysis was performed with a probe against the Gal4 DBD region. As expected, a 4.7 kb hybridization signal was detected from the modified ERα BAC clone with the Gal4-DBD probe (Fig. 2B, line 2), but no signal was observed from the parental ERα BAC clone (Fig. 2B, line 1). The fidelity of 5′ and 3′-recombination events was confirmed by sequencing the PCR products amplified by the ER11 and ER 12 primers or the ER13 and ER 14 primers, respectively (Fig. 2A). This modified ERα, termed Gal4-ERα, will now specifically activate GAL4 reporter genes, and not genes containing the endogenous estrogen responsive elements, EREs.
Fig. 1. Modification scheme of ERα BAC clones using a bacterial recombination system.
A) Modification scheme of the ERα BAC clone. The unmodified ERα BAC clone (RPCI-23-336H23 ERα BAC) has 5 exons (exons 1 to 5) and 8 kb of 5′-untranslated region (UTR). The DNA binding domain, (DBD) of ERα was replaced by selecting sequences flanking the DBD (exon 3, and exon 4/intron 4 junction regions) as homologous recombination target sequences. In order to express Gal4-ERα from its BAC clone, the targeted DNA fragment consisted of Gal4-DBD, D-, E- and F-domains of mouse ERα and poly (A) additional sequences. B) This is a modified ERα BAC clone containing Gal4-DBD and the D-, E- and F- domains of ERα. C) The KanR gene was excised using FLP recombinase from the modified BAC clone using the 294-FLP strain. D) In order to assay Gal4-ERα activity in vivo, a reporter system that consisted of Gal4-UAS, minimal promoter and hrGFP gene was introduced into the SacB gene region in the BAC clone. E) The KanR gene excised as described above.
Fig. 2. Validation of modified ERα BAC clones.
A) The fidelity of homologous recombination at the 5′-region was validated using by PCR using primer ER11, which binds to the intron 2 region located outside of the targeting region and primer ER12, which binds to the C-terminal region of ERα. The primers ER13, which binds in the KanR gene, and ER14, which binds in the intron 4 region, were used for the PCR to verify homologous recombination at the 3′-region. B) Insertion of the Gal4 DBD target fragment was confirmed using HindIII digestion of parental ERα BAC clone (1) and a modified ERα BAC clone containing Gal4DBD and the D-, E- and F-domains of ERα (2) and Southern analysis with probes specific to the Gal4 DBD. C) The fidelity of homologous recombination at the 3′-region was validated using by PCR was performed using primers designed to generate products verifying the homologous recombination. Both GFPSC51, which binds outside of the 5′-homologous region of SacB gene, and GFPSC52, which binds in the KanR gene, were used to verify homologous recombination in the 5′-region. GFPSC31, which binds in the hrGFP gene, and GFPSC32, which binds outside of the 3′-homologous region of the SacB gene, were used to verify homologous recombination at the 3′-region. D) HindIII-digested modified ERα BAC clone containing Gal4DBD and the D-, E- and F- domains of ERα (1) and a modified ERα BAC clone containing Gal4DBD and the D-, E- and F- domains of ERα and hrGFP reporter (2) DNA was examined by Southern analysis with probes specific to hrGFP and Gal4 DBD.
In order to detect the in vivo activity of Gal4-ERα, a Gal4-responsive humanized renilla GFP (hrGFP) reporter system was inserted into SacB gene in modified ERα BAC clone (Fig. 1C and 1D). The SacB gene that encodes sucrose synthase has been used for the negative selection marker in BAC clones in E. coli (Gong et al., 2002). Therefore, disruption of the SacB gene has no impact on ERα function in BAC clones in vivo. The Southern probe against hrGFP detected 1.6 kb of specific hybridization signal from the ERα BAC: Gal4-DBD: hrGFP clones (Fig. 2D line 2), but not from the ERα BAC: Gal4-DBD clone (Fig. 2D line 1). In addition, the position of the hybridization signal detected by the Gal4-DBD probe did not change before or after the recombination process (Fig. 2D). Therefore, the integrity of the ERα BAC clone was maintained during recombination processes. The fidelity of the 5′ and 3′-recombination events during the integration of the hrGFP reporter system also was confirmed by sequencing PCR products amplified by the GFPSC51 and GFPSC52 primers or the GFPSC31 and GFPSC32 primers, respectively (Fig. 2C). The selection marker, a kanamycin resistance gene, was deleted by using the 294-CRE strain as others have previously described (Han et al., 2005) (Fig. 1D and E). The ERAAI mouse was generated by microinjection of the final modified ERα BAC (Fig. 1E) clone into the fertilized oocytes isolated from the C57BL/6J strain.
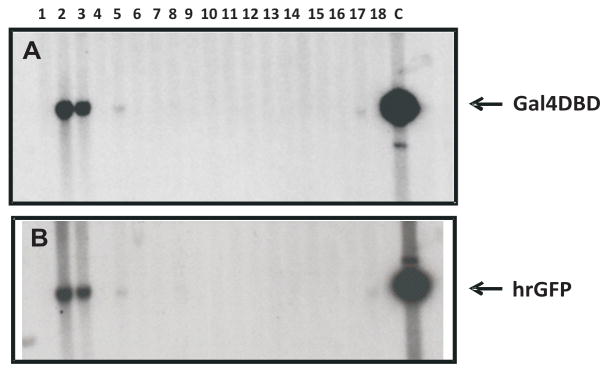
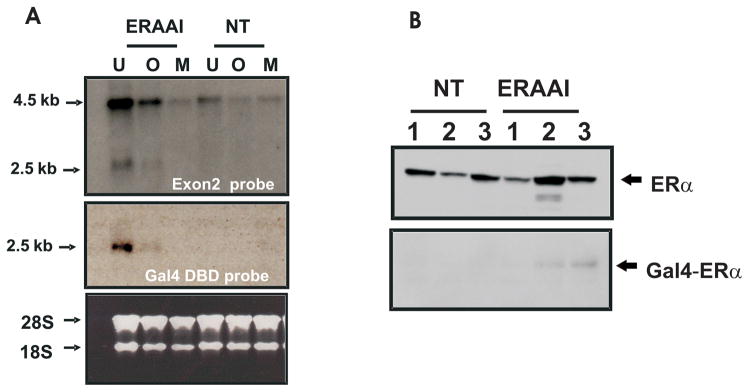
Transgenic ERAAI founder mice were identified by performing Southern analysis with probes against Gal4-DBD and hrGFP. As shown in Fig. 3, 2 of 18 pups were identified as transgenic founders (Fig. 3 line 2 and 3). Unfortunately, one of the transgenic founders (clone number 2 in Fig. 3) died at 32 days after the birth. The surviving transgenic founder (number 3) was bred and germline transmission was achieved. Both male and female ERAAI offspring have normal viability and fertility. Expression of the Gal4-ERα gene in the ERAAI mouse model (clone number 3 in Fig. 3) was assayed by Northern Blot analysis on total RNA isolated from estrogen target tissues (uterus, ovary and mammary gland). A probe corresponding to exon 2 of the mouse ERα gene detected two hybridization signals (one of about 4.5 kb and the other of about 2.5 kb) in these tissues (Fig. 4A, ERAI line in top panel). In contrast, only a 4.5 kb hybridization signal was detected in the estrogen target tissues of non-transgenic mice (Fig. 4A, NT line in top panel). For the identification of the 2.5 kb hybridization signal, a second Northern analysis was performed with a probe against the Gal4-DBD region. This probe specifically detected only a 2.5 kb hybridization signal from the ERAAI mouse tissues, but not from non-transgenic mouse tissues (Fig. 4A, middle panel). Therefore, the 2.5 kb hybridization signal represented the Gal4-ERα RNA transcribed from the modified ERα BAC clone in ERAAI mice. The difference in mRNA size of endogenous ERα versus Gal4-ERα was also investigated. The mouse Ensembl Genome Browser revealed that endogenous ERα has 4.0 kb of 3′-untranslated region (UTR), whereas Gal4-ERα has only 0.2 kb of 3′-UTR. Therefore, a difference in mRNA length from the endogenous ERα versus the Gal4-ERα was likely due to the different lengths of the 3′-untranslated regions. In addition to RNA levels, western analysis with antibody against Gal4-DBD also revealed that Gal4-ERα was expressed in the uteri of ERAAI mice but not in non-transgenic mice (Fig. 4B).
Fig. 3. Screening of transgenic founders of ERAAI mice using Southern blot analysis.
Genomic DNA was isolated from the tail biopsies of 18 pups. Transgenic founders were identified by the presence of an 3.5 kb and 1.6 kb band after Southern analysis of HindIII-digested genomic DNA and hybridization with probes specific to Gal4 DBD (A) and hrGFP (B), respectively. The final modified ERα BAC clone served as a positive control (lane C).
Fig. 4. Expression of Gal4 ERα in ERAAI mouse model.
A) Total RNA was isolated from estrogen target tissues (U: uterus, O: Ovary, M: Mammary gland) in ERAAI mice and non-transgenic mice (NT). Northern analysis was performed with a probe against ERα exon 2 (Top panel) and Gal4-DBD (middle panel). Total ribosomal RNA detected by EtBR staining was used as a loading control (bottom). B) The protein levels of both ERα and Gal4-ERα were determined by Western Blot analysis of protein isolated from the uteri of three non-transgenic mice (NT) or three transgenic mice (ERAAI) (in random, undetermined estrous cycle stages) probed with anti ERα antibody (top) and anti Gal4-DBD antibody (bottom).
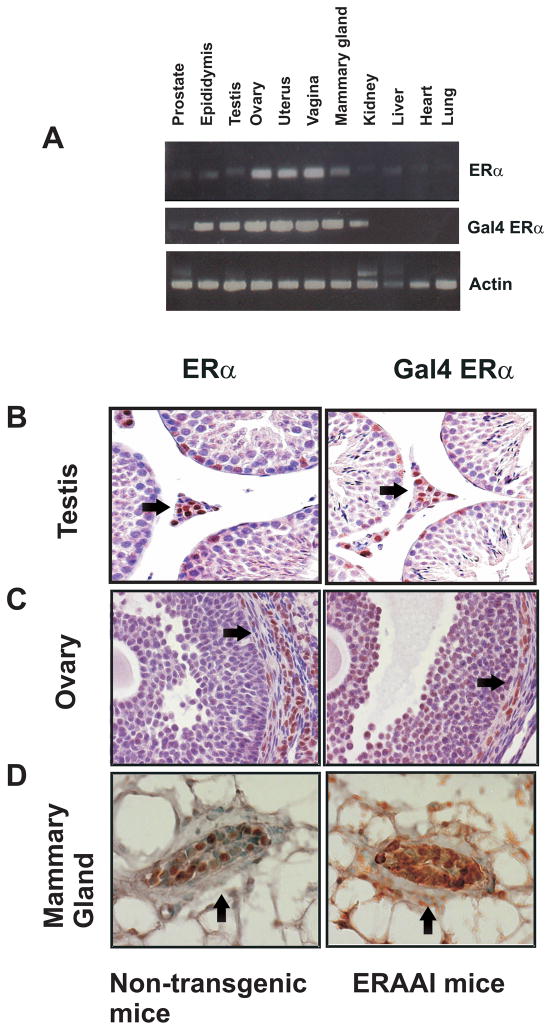
The tissue-specific expression of the Gal4-ERα as compared with the expression of the endogenous ERα, by semi-quantitative RT-PCR analysis performed with total RNA isolated from various tissues of the ERAAI transgenic mice (Fig. 5A). Tissue specific expression patterns of Gal4-ERα were highly correlated with endogenous ERα expression patterns in the tissues of the ERAAI mice (Fig. 5A). Therefore, the Gal4-ERα faithfully recapitulated the tissue-specific expression profile of endogenous ERα in the ERAAI mouse. Since ERα exhibits differential compartment-specific expression in each tissue, we next determined the cellular expression of Gal4-ERα and ERα expression in the testis, ovary and mammary gland of ERAAI mice and non-transgenic mice using immunohistochemistry with anti-Gal4 DBD antibody and anti-ERα antibody, respectively (Edery et al., 1985; Lee et al., 1971; Snijders et al., 1992). ERα expression was detected mainly in Leydig cells in the testis. Consistent with endogenous ERα, Gal4-ERα expression was also detected in this compartment of the testis (Fig. 5B). In female non-transgenic mice (during random, undetermined estrous cycle stages), expression of ERα was detected in theca-interstitial cells of the ovary. Gal4-ERα also displays the same theca-interstitial cells specific expression in the ovaries of ERAAI mice (Fig. 5C). In the mammary glands of female non-transgenic mice (at random, undetermined estrous cycle stages), endogenous ERα was mainly expressed in epithelial compartments as compared with stromal cells. However, expression pattern of Gal4-ERα in mammary glands of ERAAI mice was not clearly detected using Gal4 DBD antibody (Fig. 5D).
Fig. 5. Tissue-specific expression of Gal4-ERα versus ERα.
A) The tissue specific expression patterns of Gal4-ERα and endogenous ERα were determined by semi-quantitative RT-PCR as described in Methods section. RNA levels of actin were used for the loading control. Immunohistochemical analysis was used to determine the cell specific expression of ERα and Gal4-ERα in the Leydig cells of the testis (arrow head in panel B), theca-interstitial cells of the ovaries (arrow head in panel C) and epithelial cells of the mammary glands (arrow head in panel D) of non-transgenic mice and ERAAI mice, respectively.
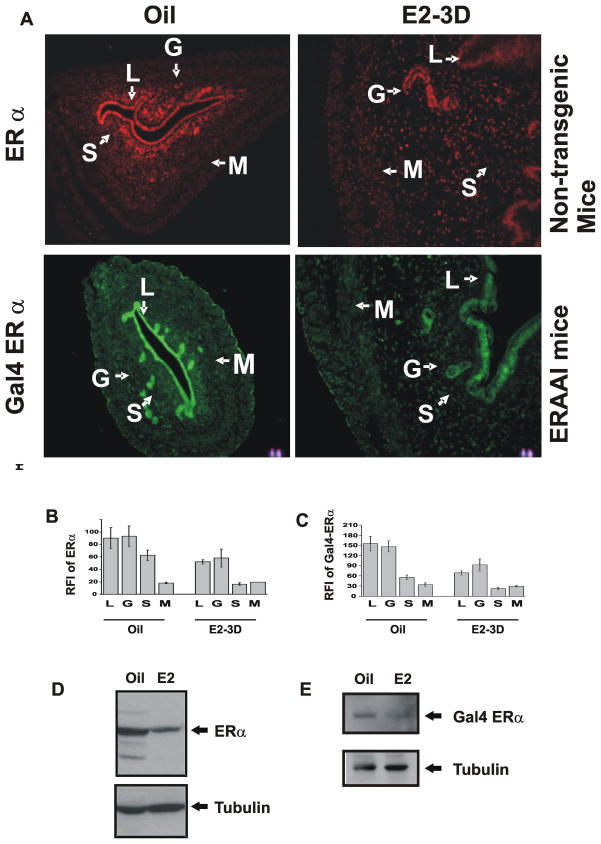
Since ERα levels are dynamically regulated in specific tissues by external hormone stimuli (Buchanan et al., 1999), this is exemplified in the uterus (Tibbetts et al., 1998). The uterine cell specific expression of Gal4-ERα was investigated in response to steroid hormone stimulation. Uterine ERα and Gal4-ERα levels were determined in ovariectomized non- transgenic and ERAAI mice treated with vehicle or estrogen. ERα was expressed in the luminal (LE), glandular epithelial (GE) and stromal (S) compartments of the uteri of ovariectomized non-transgenic mice in the absence of external estrogen stimuli (Fig. 6A and 6B). Chronic estrogen stimulation reduced levels of ERα expression in those compartments when compared to vehicle treatment. Consistent with patterns found with endogenous ERα, expression levels of Gal4-ERα in both the epithelial and stromal compartments in uterus of ERAAI mice were reduced by chronic estrogen treatment as compared with vehicle treatment (Fig. 6A and 6C). Collectively, Gal4-ERα expression generally reflected the expression pattern of endogenous ERα in each uterine cell compartment under the endocrine states tested. Western analysis also confirmed that both ERα and Gal4-ERα levels in the uterus were reduced by estrogen treatment as compared with vehicle treatment (Fig. 6D and 6E).
Fig. 6. Expression of ERα versus Gal4-ERα in the uteri of ERAAI mice.
A) Immunofluorescence analysis of the compartment-specific expression pattern of Gal4-ERα in the uteri of ovariectomized ERAAI mice and endogenous ERα in the uteri of non-transgenic mice treated with oil of estrogen daily once for 3 days (E2-3D). Relative Fluorescence Index (RFI) values for ERα and Gal4-ERα in each compartment of the uterus in response to oil or estrogen (B and C respectively). Western blot analysis of the expression levels of ERα and Gal-4ERα (D and E respectively). Tubulin was used for the loading control. (L – luminal, epithelium, G - Glandular Epithelium, S - Stroma, and M-Myometrium.)
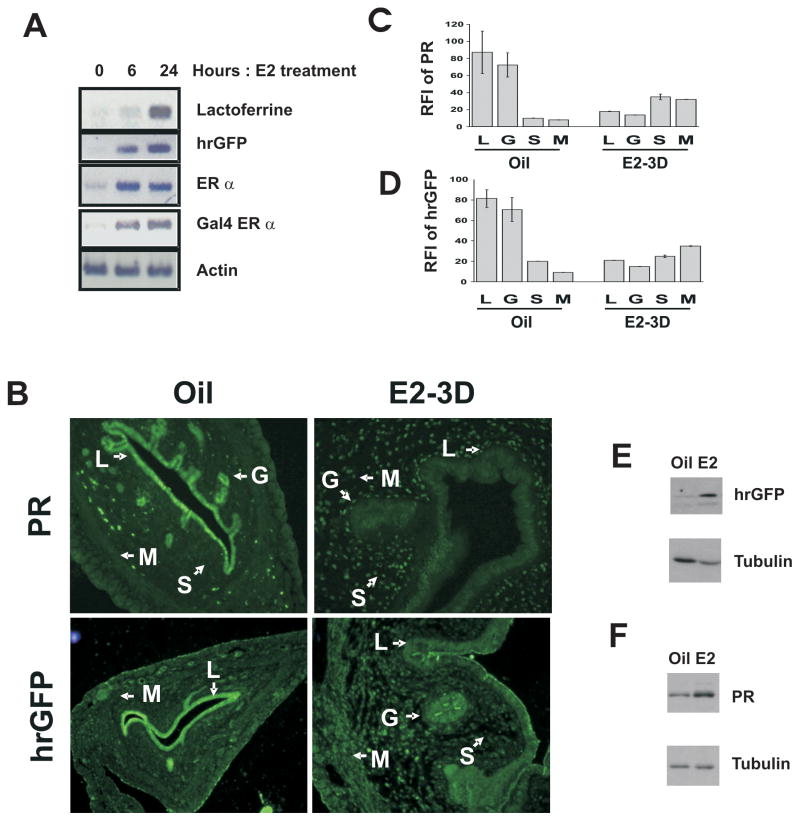
Having determined the fidelity of Gal4-ERα expression to endogenous ERα, we next determined if expression of the hrGFP reporter gene mimics activation of ERα target genes in the uterus in response to estrogen stimulation. Ovariectomized ERAAI transgenic mice were injected with estrogen as described and sacrificed 6 and 24 hours after estrogen injection. Estrogen treatment increased the expression of an endogenous ERα regulated gene, lactoferrin, as compared with vehicle treatment (Fig. 7A). Consistent with the induction of lactoferrin expression, hrGFP expression in the uterus was increased estrogen treatment (Fig. 7A). The short term treatment of estrogen assayed regulation of ERα activity in the epithelial compartment of the uterus. In order to assay the activity in the stroma compartment, mice were treated ovariectomized mice were treated with estrogen once daily for 3 days. ERα activity was assayed by immunohistochemical analysis of the expression of PR and Gal4-ERα activity was assayed by immunohistochemical analysis of hrGFP. As expected, levels of PR expression were reduced in the epithelium, but increased in the uterine stromal and myometrial compartments of mice receiving estrogen treatment (Fig. 7B and 7C). Similarly, hrGFP expression levels in the epithelium were reduced, but as predicted, levels in the stromal and myometrial compartments increased compared to vehicle treatment (Fig. 7B and 7D). Western analysis also revealed that hrGFP expression faithfully recapitulated PR expression in the uterus in response to chronic estrogen treatment (Fig. 7E and 7F).
Fig. 7. Expression of hrGFP mimics ERα activity in the uterus of ERAAI mice.
A) Semiquantitative analysis for the expression of Lactoferrin, hrGFP, ERα, Gal4-ERα and actin in ERAAI mouse uteri 0, 6 and 24 hours after estrogen injection. B) Immunohistochemical analysis of the expression patterns of PR or hrGFP in the uteri of ERAAI mice treated with oil of estrogen once daily for 3 days. The RFI values for PR (C) or hrGFP (D) in each compartment of the uterus in response to oil or estrogen are shown in the graph. Expression levels of hrGFP (E) and PR (F) in the uterus were also determined by Western blot analysis in response to chronic estrogen treatment. Tubulin was used for the loading control. (L - luminal epithelium, G - Glandular Epithelium, S - Stroma, and M – Myometrium).
In summary, we generated a new ERα activity indicator mouse model that can be employed to investigate in vivo ERα activity. For example, the ERAAI mouse model will be quite useful for analysis of in vivo ERα activity in response to the administration of selective estrogen receptor modulator (SERM) drugs or endocrine disruptors of ER activity.
Methods
ERα BAC clone modification
Sequences of all primers that were used for modification of ERα BAC clone are described in Table 1. To replace ERα DBD with Gal4 DBD (147 amino acid) in the ERα BAC clone, a targeting DNA construct was made as follows (Fig. 1A): the part of the exon 3 region that does not contain the DBD region was used for the 5′-homologous recombination region. Gal4 DBD was fused with the D-, E- and F-domains of the ERα and the BGH polyadenylation signal. The intron region located in between exon 4 and exon 5 was used as the 5′-homologous recombination region. For the 5′-homologous recombination region, a part of exon 3 that is not in the ERα DBD was isolated by PCR with primers ER1 and ER2 from the mouse genome. The Gal4-DBD region was isolated by PCR with primers ER3 and ER4 from pGBK7 (BD Bioscience, San Jose, CA). The D-, E- and F-domains of mouse ERα were isolated by PCR with primers ER5 and ER6 from cDNA generated from the total RNA of the mouse uterus. For the transcriptional termination of the modified ERα, the BGH polyadenylation signal was isolated with primers ER7 and ER8 from the pCR3.1 vector. The 3′-homologous recombination target site that is located in between exons 4 and 5 was isolated by PCR with primers ER9 and ER10 from the mouse genome. DY380 cells carrying the ERα BAC clone (RPCI-23-336H23) were electroporated with the Gal4 DBD targeting cassette, as has been previously described (Lee et al., 2001; Yu et al., 2000).
Table 1.
PCR primers for the modification of ERα BAC clone
| Primer Name | Sequence |
|---|---|
| ER1 | 5′-AGACCGCGGGGTCTAATTCTGACAATCGACGCCAGA-3′ |
| ER2 | 5′-AGAAGACAGTAGCTTCATGGCAGACTCCATGATCAT-3′ |
| ER3 | 5′-ATGAAGCTACTGTCTTCTATCG-3′ |
| ER4 | 5′-CGATACAGTCAACTGTCTTTGA-3′ |
| ER5 | 5′-AGACAGTTGACTGTATCGGACCGCCGAGGA-3′ |
| ER6 | 5′-CTCGAGAAGGTGGACCTGATCATGGAG-3′ |
| ER7 | 5′-TTCGCCCTATAGTGACTCGTTATTAC-3′ |
| ER8 | 5′-CGCTTACAATTTACGCGTTAAGA-3′ |
| ER9 | 5′-GCAGAAGGATAAAGAATGAATTCGAT-3′ |
| ER10 | 5′-ATGAATATCAACAAGTGCCACAAATC-3′ |
| ER11 | 5′-CCTTCTAGTCACATATAATACTTAGCA-3′ |
| ER12 | 5′-TGTTGCAGGGATTCTCAGAA-3′ |
| ER13 | 5′-CTGCTCTGATGCCGCCGTGTTCCGGCTGTC-3′ |
| ER14 | 5′-CCAGCTCATGGAGCTTTACTCA-3′ |
| kan1 | 5′-CAGTTCATTCAGGGCACCGGACAGGTCGGTC-3′ |
| kan2 | 5′-ACTGTCAGACCAAG-3′ |
| sacb51 | 5′-AGGAGCGACTCAAGCCTTCGCGAA-3′ |
| scab52 | 5′-CGGCCAGCTGTCCCACACATC-3′ |
| sacb31 | 5′-TGTGCTGCAAATGGGTCTTGA-3′ |
| scab32 | 5′-GCCAAGCTTCTTAATGAACATCA-3′ |
| GFPSC51 | 5′-CTGCTCTGATGCCGCCGTGTTCCGGCTGTC-3′ |
| GFPSC52 | 5′-CAGCTGTCCTTGCTCCAGGAT-3′ |
| GFPSC31 | 5′-ACTGTTGGTAAAGCCACCATGGTGAGCAAGCAGATCCTG-3′ |
| GFPSC32 | 5′-GACTGCACTTCTGGCAGGAGG-3′ |
Electroporation was performed using a Bio-Rad Micropulser set at 1.8 kV/cm. After electroporation, cells were incubated in 1 ml of SOC medium for 1 h and spread onto LB/agar plates containing chloramphenicol (12.5 μg/ml), tetracycline (10 μg/ml) and kanamycin (10 μg/ml). Colonies were allowed to form over 24 h at 30°C to isolate those resistant to chloramphenicol/tetracycline/kanamycin. To screen for recombination events, PCR was performed using primers designed to generate products verifying the homologous recombination.
ER11, which binds to the intron 3 region located outside of the targeting region, and ER12, which binds to the C-terminal region of ERα, were used to verify homologous recombination at the 5′-region. The primers for ER13, which bind in the kanamycin resistant (KanR)gene, and ER14, which binds in the intron 4 region (located outside of targeting region), were used to verify homologous recombination at the 3′-region. To confirm the insertion of the Gal4 DBD target fragment, HindIII-digested BAC DNA was examined by Southern analysis with a radio-labeled probe specific to the Gal4 DBD (Fig. 2B). To remove the KanR gene by FLP recombinase, the modified BAC clone was electroporated into 294-FLP in accordance with existing methods (Buchholz et al., 1996; Lee et al., 2001) (Fig. 1B and 1C). After incubating in SOC medium for 1 h, cells were spread onto LB/agar plates containing chloramphenicol and tetracycline. Colonies were allowed to form over 24 h at 30°C to isolate chloramphenicol/tetracycline resistance colonies. PCR was performed using two KanR primers, kan1 and kan2, to verify the excision of the KanR gene.
The hrGFP reporter cassette, which has 5′ homologous region- hrGFP-minimal promoter-5 copies of UAS for the Gal 4 gene [UAS(G)]-loxP- KanR gene-3′ homologous region, was constructed (Fig. 1C). Next, 200 bp of the SacB gene, which is in the BAC plasmid, was isolated by PCR with primers sacb51 and scab52 to generate the 5′-homologous recombination region; the other 200 bp of SacB gene was isolated by PCR with primers sacb31 and scab32 to generate the 3′-homologous recombination region. DY380 cells carrying a modified Gal4-ERα BAC clone were electroporated with the hrGFP reporter targeting cassette (Lee et al., 2001; Yu et al., 2000). After electroporation, cells were incubated in 1 ml of SOC medium for 1 h and spread onto LB/agar plates containing chloramphenicol, tetracycline and kanamycin. Colonies were allowed to form over 24 h at 30°C to isolate those resistant to chloramphenicol/tetracycline/kanamycin. PCR was performed using primers designed to generate products verifying the homologous recombination. Both GFPSC51, which binds in the KanR gene, and GFPSC52, which binds outside of the 5′-homologous region of the SacB gene, were used to verify homologous recombination at the 5′-region. Other primers - GFPSC31, which binds in the hrGFP gene, and GFPSC32, which binds outside of the 3′-homologous region of SacB gene - were used to verify homologous recombination in the 3′-region. To confirm insertion of the hrGFP reporter, HindIII-digested BAC DNA was examined by Southern analysis with radio-labeled probes specific to hrGFP and Gal4 DBD (Fig. 2D). To remove the KanR gene by Cre recombinase, the modified BAC clone was electroporated into a 294-CRE strain as has been previously described (Buchholz et al., 1996; Lee et al., 2001) (Fig. 1D and 1E). After incubating in SOC medium for 1 h, cells were spread onto LB/agar plates containing chloramphenicol and tetracycline. Colonies were allowed to form over 24 h at 30°C to isolate those resistant to chloramphenicol and/or tetracycline. PCR was performed using the two primers, kan1 and kan2, to verify excision of the KanR gene.
Generation of ERAAI transgenic mice
The final modified ERα BAC clone (Fig. 1E) was purified using the ClonTech BAC purification kit (ClonTech, San Jose, CA). After digestion with the PI-SceI restriction enzyme at 37°C overnight, the linearized BAC DNA was dialyzed against a microinjection buffer (10 mM Tris-Cl, pH 7.5, 0.1 mM EDTA and 100 mM NaCl) at 4°C for 4 h. The linearized BAC fragment was then injected at a concentration of 1 ng/μl into fertilized mouse oocytes isolated from the C57BL/6J strain. Oocytes that survived injection were transferred into both oviducts of pseudo-pregnant mothers.
Screening of transgenic founder mice by PCR
Transgenic mice were identified by both PCR and Southern analysis with genomic DNA isolated from tail biopsies. To screen transgenic founder mice, the following sets of primers were used: 5′-ATGAAGCTACTGTCTTCTATCG-3′ and 5′-CGATACAGTCAACTGTCTTTGA-3′ (which amplify a 440-bp PCR fragment in the Gal4-DBD region); 5′-AATAGGCTGTCCCCAGTGCAAGT-3′ and 5′-ATGGTCTTCTTCATCACGGGGC-3′ (which amplify a 707 bp PCR fragment in the hrGFP region). For the Southern analysis, genomic DNA was digested with HindIII. Radio-labeled probes specific to hrGFP and Gal4 DBD were hybridized to the Southern blot. ERAAI genotyping was done with genomic PCR using primers that are specific for the Gal4-DBD region described above.
Animals, surgical procedures, and hormone administration
Mice were housed in a pathogen-free animal facility under a standard12-h light/12-h dark cycle and fed standard rodent chow and water. All animal experimentationwas conducted in accordance with accepted standards of humane animal care. In order to mimic the menopausal state in the mice, seven-week-old ERAAI female mice were bilaterally ovariectomized. Two weeks after ovariectomy, a daily dose of oil or estrogen (5 μg/kg/day in sesame oil) was administrated into ERAAI mice (9 weeks old) once daily for 3 days. To investigate the hormonal effects of each drug on ERα activity in the uterus, mice were sacrificed 3 days after the first injection and their tissues were harvested.
Western analysis
Uteri from ERAAI mice in each group were removed and then washed with PBS solution and homogenizedin a buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl,2.5 mM EDTA and 0.125% Nonidet P-40 (vol/vol). Cellular debris was removed by centrifugation at 14,000 rpm for 15 min at 4°C. Protein concentration was determined by Bradford’s method using BSA as a standard. Samples containing 10 μg of total protein were loaded on a 10% SDS-PAGE gel. The separated proteins were then transferred to a polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA). Membranes were blocked overnight with 5% skim milk (wt/vol) in PBS with 0.1% Tween 20 (vol/vol)(Sigma-Aldrich, St. Louis, MO) and probed with an antibody against hrGFP diluted to 1:1000 (240141, Stratagene, La Jolla, CA). The levels of ERα in the uterus were detected by using anti-ERα antibody diluted to1:1000 (SC-542, Santa Cruz Biotechnology, Inc., and Santa Cruz, CA). Immunoreactivity was visualized by incubation with a horseradish peroxidase-linked secondary antibody and treatment with enhanced luminol-based chemiluminescent reagents. For normalization, the membrane was stripped and probed with an anti-tubulin antibody diluted to 1:1000 (T2200, Sigma-Aldrich, St. Louis, MO) and developed again.
RT-PCR
First-strand cDNA was synthesized from total RNA using a SuperScriptII RT kit (Invitrogen, Carlsbad, CA) and 250 ng of random primers according to the manufacturer’s protocol. The PCR primer pairs were as follows: 5′-TTGTCGCAATCATGAAGGGA-3′ and5′-GTGAAGCCAGCATCTTCTCTT-3′ (this primer pair amplified a 199-bp fragment from lactoferrin); 5′-GCT TGG CAT TCC GGT ACT GT-3′ and 5′-GGT TGC CGA ACA GGATGT TG-3′ (this primer pair amplified a 120-bp fragment from hrGFP); 5′-GCATGG GTC GGG ACA AGA AGA-3′ and 5′-CTC CAG CAG GGG GCA CCA CT-3′ (this primer pair amplified a 599-bp fragment from β-actin). To specifically detect endogenous ERα levels, 5′-GGGCGAGGTATACGTGGACAACAG-3′ (ESR1A) and 5′-CTTCGCAGGACCAGACCCCATAAT-3′ (ESR1B) were used because ERS1B was located in the DBD region of ERα. This primer pair amplified a 506-bp fragment. For the Gal4-ERα, 5′-GCCCTCCCGCCTTCTAC -3′ (G4ESR1A) and 5′-TGATGATGTCGCACTTATTCTATG-3′ (G4ESR1B) were used because G4ESR1B recognized the Gal-4 DBD region in the Gal4-ERα genome. This primer pair amplified a 500-bp fragment. Amplifications were performed with a PTC-200 MJ Peltier thermal cycler and PCR temperature profile consisted of denaturation at 94°C for 10 min, followed by 27 cycles at 94°C for 30 s, annealing at 52°C for 30 s and 72°C for 1 min, and a final extension phase at 72°C for 10 min. The PCR reaction was performed with Advantage 2 Polymerase Mix (ClonTech, San Jose, CA) according to the manufacturer’s protocol. The PCR products were analyzed on a 1.5% agarose gel in Tris-acetate-EDTA buffer.
Immunohistochemistry
Mice were anesthetized with Avertin and perfused through the heart with 4% paraformaldehyde in PBS. The uterus was removed and kept in the same fixative for 16 hat 4°C. Samples were dehydrated in an ethanol series, cleared inxylene and embedded in paraffin. Sections were cut at 7 μm. For immunostaining, sections were dewaxed, rehydrated and boiled for 10 min in 10 mM citrate buffer, pH 6.0. To reduce nonspecific binding of antibodies, sections were washed in PBS again and preincubated with 500 μg/ml goat IgG and 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Anti-hrGFP antibody (240141, 1:300; Stratagene, La Jolla, CA), anti-ERα antibody (SC-542, 1:1000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-PR antibody (SC-7208, 1:300; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-Gal4-DBD antibody (SC-577, 1:500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were used to detect expression of each protein in the tissues. For immunofluorescence, sections then were incubated sequentially with biotinylated horse anti-rabbit IgG (1:500) and FITC or Texas Red-conjugated streptavidin (1:1000; Vector Laboratories, Inc., Burlingame, CA). To measure fluorescence intensity in each compartment of the uterus, capture levels were first adjusted to avoid saturation and then kept constant throughout the analysis. For each compartment of the uterus examined, a box was drawn in a random orientation. The pixel intensity value of each box (in arbitrary units) was measured and a mean value was obtained. Values shown in all figures are listed as mean values of intensity (n=3) ± SD. To normalize the fluorescence intensity of each compartment, we used fluorescence intensity from normal serum. Relative Fluorescence Intensity (RFI) is the ratio of specific antibody fluorescence intensity to normal serum fluorescence intensity in each compartment of the uterus. To perform DAB (3, 3′-diaminobenzidine) staining for immunohistochemistry, the VECTASTAIN ABC kit system was applied as described in their protocol (Vector Laboratories, Inc., Burlingame, CA).
Northern analysis
Total RNA was prepared from mouse tissues by guanidinium isothiocyanate extraction as others have previously described (Chomczynski and Sacchi, 1987). It was analyzedin 1% formaldehyde agarose gels by standard methods. RNA was transferred to nitrocellulose membranes in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Prehybridization and hybridization were performed in 6× SSC, 5× Denhardt’s solution,1% sodium dodecyl sulfate and 100 μg of single-stranded DNA per ml at 65°C. Final washes were carried out at 65°C in 0.2× SSC and 0.1% sodium dodecylsulfate.
Acknowledgments
This research was supported by the Eunice Kennedy Shriver NICHD/NIH through cooperative agreement U54 HD007495-34 (Bert W. O’Malley) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and HD07857 (Bert W. O’Malley).
References
- Avioli LV. SERM Drugs for the Prevention of Osteoporosis. Trends Endocrinol Metab. 1999;10:317–319. doi: 10.1016/s1043-2760(99)00176-9. [DOI] [PubMed] [Google Scholar]
- Buchanan DL, Setiawan T, Lubahn DB, Taylor JA, Kurita T, Cunha GR, Cooke PS. Tissue Compartment-Specific Estrogen Receptor-{alpha} Participation in the Mouse Uterine Epithelial Secretory Response. Endocrinology. 1999;140:484–491. doi: 10.1210/endo.140.1.6448. [DOI] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Ciana P, Di Luccio G, Belcredito S, Pollio G, Vegeto E, Tatangelo L, Tiveron C, Maggi A. Engineering of a mouse for the in vivo profiling of estrogen receptor activity. Mol Endocrinol. 2001;15:1104–1113. doi: 10.1210/mend.15.7.0658. [DOI] [PubMed] [Google Scholar]
- Ciana P, Scarlatti F, Biserni A, Ottobrini L, Brena A, Lana A, Zagari F, Lucignani G, Maggi A. The dynamics of estrogen receptor activity. Maturitas. 2006;54:315–320. doi: 10.1016/j.maturitas.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Couse JF, Korach KS. Estrogen Receptor Null Mice: What Have We Learned and Where Will They Lead Us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- Eddy EM, Washburn TF, Bunch DO, Goulding EH, Gladen BC, Lubahn DB, Korach KS. Targeted disruption of the estrogen receptor gene in male mice causes alteration of spermatogenesis and infertility. Endocrinology. 1996;137:4796–4805. doi: 10.1210/endo.137.11.8895349. [DOI] [PubMed] [Google Scholar]
- Edery M, Imagawa W, Larson L, Nandi S. Regulation of estrogen and progesterone receptor levels in mouse mammary epithelial cells grown in serum-free collagen gel cultures. Endocrinology. 1985;116:105–112. doi: 10.1210/endo-116-1-105. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Han SJ, Jeong J, DeMayo FJ, Xu J, Tsai SY, Tsai M-J, O’Malley BW. Dynamic Cell Type Specificity of SRC-1 Coactivator in Modulating Uterine Progesterone Receptor Function in Mice. Mol Cell Biol. 2005;25:8150–8165. doi: 10.1128/MCB.25.18.8150-8165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Analysis of mammalian central nervous system gene expression and function using bacterial artificial chromosome-mediated transgenesis. Hum Mol Genet. 2000;9:937–943. doi: 10.1093/hmg/9.6.937. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Korach KS. Estrogen receptors: structure, mechanisms and function. Rev Endocr Metab Disord. 2002;3:193–200. doi: 10.1023/a:1020068224909. [DOI] [PubMed] [Google Scholar]
- Kuller LH, Matthews KA, Meilahn EN. Estrogens and women’s health: interrelation of coronary heart disease, breast cancer and osteoporosis. J Steroid Biochem Mol Biol. 2000;74:297–309. doi: 10.1016/s0960-0760(00)00106-0. [DOI] [PubMed] [Google Scholar]
- Kurita T, Lee KJ, Cooke PS, Taylor JA, Lubahn DB, Cunha GR. Paracrine regulation of epithelial progesterone receptor by estrogen in the mouse female reproductive tract. Biol Reprod. 2000;62:821–830. doi: 10.1093/biolreprod/62.4.821. [DOI] [PubMed] [Google Scholar]
- Lee C, Keyes PL, Jacobson HI. Estrogen receptor in the rabbit corpus luteum. Science. 1971;173:1032–1033. doi: 10.1126/science.173.4001.1032. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of Reproductive Function but Not Prenatal Sexual Development After Insertional Disruption of the Mouse Estrogen Receptor Gene. Proceedings of the National Academy of Sciences. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SO, Korach KS. Estrogen receptors and endocrine diseases: lessons from estrogen receptor knockout mice. Curr Opin Pharmacol. 2001;1:613–619. doi: 10.1016/s1471-4892(01)00105-9. [DOI] [PubMed] [Google Scholar]
- Snijders MP, de Goeij AF, Debets-Te Baerts MJ, Rousch MJ, Koudstaal J, Bosman FT. Immunocytochemical analysis of oestrogen receptors and progesterone receptors in the human uterus throughout the menstrual cycle and after the menopause. J Reprod Fertil. 1992;94:363–371. doi: 10.1530/jrf.0.0940363. [DOI] [PubMed] [Google Scholar]
- Tibbetts TA, Mendoza-Meneses M, O’Malley BW, Conneely OM. Mutual and intercompartmental regulation of estrogen receptor and progesterone receptor expression in the mouse uterus. Biol Reprod. 1998;59:1143–1152. doi: 10.1095/biolreprod59.5.1143. [DOI] [PubMed] [Google Scholar]
- Vogelvang TE, van der Mooren MJ, Mijatovic V, Kenemans P. Emerging selective estrogen receptor modulators: special focus on effects on coronary heart disease in postmenopausal women. Drugs. 2006;66:191–221. doi: 10.2165/00003495-200666020-00005. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K, Gustafsson JA. Estrogen receptor beta: an overview and update. Nucl Recept Signal. 2008;6:e003. doi: 10.1621/nrs.06003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. A comprehensive BAC resource. Nucleic Acids Res. 2001;29:141–143. doi: 10.1093/nar/29.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]