Abstract
We recently demonstrated that Polζ and Rev1 contribute to alleviate the lethal effects of Me-lex, which selectively generates 3-methyladenine, by error prone lesion bypass. In order to determine the role of Polη in the biological fate of Me-lex induced lesions, the RAD30 () gene was deleted in the yIG397 parental strain and in its rev3 (Polζ) derivative, and the strains transformed with plasmid DNA damaged in vitro by Me-lex. While deletion of RAD30 increased the toxicity of Me-lex, the impact on mutagenicity varied depending on the concentration of Me-lex induced DNA damage and the overall TLS capacity of the cells. For the first time the Me-lex induced mutation spectrum in rad30 strain was determined and compared with the spectrum previously determined in WT strain. Overall, the two mutation spectra were not significantly different. The effect on mutation frequency and the features of the Me-lex induced mutation spectra were suggestive of error prone (significant decrease of mutation frequency and significant decrease of AT>TA at a mutation hotspot in rad30 vs RAD30) but also error free (significant increase of AT>GC in rad30 vs RAD30) Polη dependent bypass of lesions. In summary, our previous results with Polζ and Rev1 mutants, the present results with Polη, and the known physical and functional interactions among TLS proteins, lead us to propose that the by-pass of Me-lex induced lesions is a multi-DNA polymerases process that is mostly effective when all three yeast TLS polymerases are present.
Keywords: Me-lex, N3-methyladenine, translesion synthesis, p53, yeast
INTRODUCTION
A number of exogenous {environmental [1] or pharmaceutical [2,3]} compounds, and endogenous methylating agents [4,5] form N3-methyladenine (3-mA) in DNA. Bacteria, yeast and mammals have evolved and maintained specific DNA repair enzymes that are selective for removal of 3-mA from their genomes, a strong indication of the deleterious effect of 3-mA [6]. Using Me-lex, an agent that efficiently and selectively generates 3-mA in A-T rich regions of double-strand DNA due to its minor groove binding conferred by the lexitropsin dipeptide [7,8], we showed that the lesion itself is cytotoxic in E. coli [9], S. cerevisiae and in mammalian cells [10]. In particular, we showed in yeast that the toxicity and mutagenicity of Me-lex, is dependent on the nature of the DNA repair background [8,11]. For example, mag1 and apn1apn2 base excision repair (BER) defective S. cerevisiae strains were both significantly more sensitive to Me-lex toxicity, but only in apn1apn2 cells did Me-lex treatment result in significant mutagenicity. Furthermore, the Me-lex induced mutation spectrum in the human p53 cDNA determined in a yeast-based functional assay and the Me-lex induced methylation pattern determined in vitro in the same p53 target showed minimal overlap. In fact, many strong sites of alkylation were cold spot for base pair substitutions. Those results are consistent with the hypothesis that 3-mA is a strongly cytotoxic but weakly mutagenic DNA lesion [7,8]. The mutation spectra induced by Me-lex were not affected by the status of MAG1 nor by defects in different BER steps. Those observations suggested that 3-mA induced mutagenicity might be associated with the conversion of the primary adduct into an abasic site through the enzymatic activity of Mag1 or spontaneous hydrolysis of the methylated purine. Alternatively, it is possible that the different lesions (3-mA and abasic site) fortuitously afford the same mutation specificity, i.e., the same nucleotide is inserted with similar efficiency opposite both lesions during translesion synthesis (TLS).
The fixation of DNA adducts into mutations depends on the activity of TLS DNA polymerases that can bypass DNA lesions that stall replication forks. In general, TLS polymerases lack 3′→5′ proofreading exonuclease activity, support the bypass of damaged DNA in vitro, and, when copying undamaged DNA, exhibit low fidelity and reduced processivity compared to replicative DNA polymerases. In Saccharomyces. cerevisiae, there are three known TLS polymerases: Polη and Rev1, which belong to the Y family of TLS DNA polymerases, and Polζ, which is a member of the B family [12]. Polζ, composed of, Rev3 and Rev7 subunits, is the TLS polymerase often associated with spontaneous, as well as, damage-induced mutagenesis [12]. Rev3 is the catalytic subunit with DNA polymerase activity while Rev7 has a regulatory function. It was recently shown that Rev7, by forming a protein::protein complex with Rev1 [13], inhibits the binding of Rev1 not only to Rev3 [14] but also to Polη [15]. Polζ is not essential for cell viability in yeast while disruption of REV3 in mice causes embryonic lethality [12]. In vitro, Polζ acts mainly as a mispaired primer extender (frequency 10−1-10−2), although it can also incorporate nucleotides opposite a DNA lesion [12].
The Rev1 protein is required for UV mutagenesis [12,16-19] and mutagenesis resulting from TLS bypass of abasic sites [20] and other damaged bases [21]. Rev1 is specific for the incorporation of a C opposite all template bases and abasic sites [12,20]. but its DNA synthetic activity is not always required for lesion bypass events [20-24], suggesting that Rev1 plays a role in coordinating the assembly of Polζ at the replication fork [12,25-27].
In contrast to Polζ and Rev1, Polη suppresses UV mutagenicity due to its error free TLS activity past UV induced damage [20]. Polη is also able to bypass several bulky lesions with relatively high fidelity. However, DNA lesions that severely impinge upon the minor groove appear to block DNA synthesis by Polη [12]. Zhao et al., (2004) proposed that Polη may also be involved in error prone TLS of some DNA lesions such as abasic sites [28] while others have concluded that this polymerase has no significant impact on the TLS through abasic sites [24,29].
In order to determine which TLS polymerases are involved in the conversion of Me-lex induced lesions into mutations, we previously initiated a systematic genetic approach by deleting the REV3 and REV1 genes in the parental yIG397 yeast strain, and in nucleotide excision repair (NER) defective and base excision repair (BER) defective derivatives [30]. The toxicity and mutagenicity of Me-lex induced lesions were evaluated using a p53 functional assay [11]. Our results were consistent with an involvement of both Polζ and Rev1 in the mutation fixation process of Me-lex induced lesions, however, the involvement of other TLS polymerases could not be excluded.
We have now determined the role of in the processing of Me-lex induced lesions by deleting the RAD30 gene in a WT and a rev3 background. While deletion of RAD30 increased Me-lex toxicity, the impact on mutagenicity varied depending on the concentration of Me-lex (i.e. the level of DNA damage) and the overall TLS capacity of the cells. The features of the Me-lex induced mutation spectra were suggestive of both Polη error prone and error free lesion bypass. Based upon the known protein-protein interactions and their functional consequences it is reasonable to suggest that the TLS of Me-lex lesions is a multi-DNA polymerases process that is most effective when all three yeast polymerases are present.
2. MATERIALS AND METHODS
2.1 Hazardous procedures
Me-lex should be considered a toxic compound, and was handled accordingly.
2.2 Compounds
Reagents of the highest purity were purchased from Sigma (St. Louis, MO, USA) or Aldrich (Milwaukee, WI, USA) unless otherwise stated. Me-lex was prepared as previously described [31].
2.3 Vectors, strains and media
The yeast expression vector pTS76 harbours the human wild-type p53 cDNA under the control of an ADH1 constitutive promoter and contains the TRP1 selectable marker. The haploid S. cerevisiae strain yIG397 [32], and its isogenic TLS-deficient derivatives (Table 1), were used as recipients for pTS76. The p53-dependent reporter ADE2 gene allowed the phenotypic selection of p53 mutants as its recombinant promoter, derived from the CYC1 gene, contains three copies of the p53 responsive element RGC [32]. Standard yeast manipulations were performed as previously described [33].
Table 1.
Yeast strains used in this study.
| Name | Genotype | Phenotype | Reference | |
|---|---|---|---|---|
| yIG397 | MATα ade2-1 leu2-3,112 trp1-1 his3-11,15 can1-100 ura3-1 URA3 3xRGC::pCYC1::ADE2 | WT | (Flaman 1995) | |
| yPM6 | same as yIG397 rev3::HYGROR | (rev3) | TLS−(polζ) | (Monti 2008) |
| yPM13 | same as yIG397 rad30::HYGROR | (rad30) | TLS−(polη) | this work |
| yPM14 | same as yPM6 rad30::KANMX4R | (rad30rev3) | TLS−(polζ polη) | this work |
2.4 DNA modification, analysis, and transformation
The in vitro treatment of pTS76 plasmid with Me-lex was performed as previously described [7,11]. Briefly, Me-lex was dissolved in DMSO immediately before the plasmid pTS76 DNA was treated with different concentrations (up to 12 mM) in 10mM Tris–HCl (pH 7.4), 1mM EDTA, 50% EtOH for 1h at 37 C. DNA was purified by EtOH precipitation, washed with 70% EtOH, and resuspended in sterile water. Damaged or undamaged plasmids were then transformed by the LiAc method into the same number of yeast cells (measured by OD600), using the same growth conditions, and transformants were plated on selective synthetic medium plates and incubated for 3 days. The selection for the plasmid marker (TRP1) allowed an indirect determination of the lethal effect of the damaging treatment, calculated as the number of transformants scored in transfections with Me-lex-treated plasmids relative to that obtained with undamaged vector (hereafter indicated as plasmid survival). As transformation plates contained a minimal amount of adenine, adenine auxotrophs produced small red colonies (an Ade− phenotype) due to lack of functional p53. The number of small red colonies with respect to the total number of transformants was used as measurement of the spontaneous or induced MF (sMF or MF, respectively). For each strain and each Me-lex concentration, at least 4 independent experiments were performed. For MF, the cumulative mutation frequencies observed in all experiments are reported [all red colonies scored in all experiments over the total number of colonies (red+white) scored in all experiments]. Plasmid transformation efficiency of undamaged vector was unaffected by deletion of the TLS genes. For each Me-lex concentration tested, the effects on mutagenicity and toxicity were normalized to those observed with the undamaged plasmid in each strain.
2.5 Construction of TLS-deficient yeast strains
The rev3 yeast strain was previously described [30]. The same strategy was used for the construction of rad30 yeast strain. The RAD30 (polη) disruption cassette was obtained by PCR using as a template pCOREUH plasmid [34] containing the selectable marker HYGROr (generous gift of Dr. Francesca Storici). Primers have a sequence homology to the 5′-(or 3′-) end of the gene to be disrupted (underlined) and a sequence (in bold) that is complementary to the 5′-(or 3′-) region of the HYGRO resistance gene present in pCOREUH. The WT strain was transformed with the unpurified PCR product. The deletion of the RAD30 genes was confirmed by phenotypic selection (resistance to hygromycin B) and by yeast colony PCR. The primers used for the construction of the disruption cassette were RAD30-hygro dw: 5′-atg tca aaa ttt act tgg aag gag ttg att cag ctt ggt tcc ccc agt aaa atc tgg gca gat gat gtc g-3′(70mer); RAD30-hygro up: 5′-tca ttt ttt tct tgt aaa aaa tga taa gat gtt ttt gga aga tgt aac ttg cgc gcg ttg gcc gat tca t-3′(70mer). The PCR conditions were 95°C for 40 s, 55°C for 60 s and 72°C for 90 s repeated for 35 cycles. The primers for yeast colony PCR were: RAD30-5′, 5′-ctc taa aaa tgt act aag gat aaa-3; RAD30-3′, 5′-aaa aac ggc tgt atg gat cgg tat gag -3′; and HYGRO-3′, 5′-gtt ttt tta tat tgt agt tgt tc-3′; allowing us to confirm RAD30 deletion at the genomic level. The colony PCR conditions were 94°C for 60s, 50°C for 60 s and 72°C for 120s repeated for 35 cycles; yeast colonies were heated at 94°C for 8 min before starting. The rad30rev3 double mutant was obtained by disrupting the RAD30 gene in the rev3 strain using the specific deletion strain from EUROSCARF (BY4741; MATa; ;; ;YDR419w::kanMX4) as source of the RAD30 disruption cassette (by PCR on yeast colonies). As previously described, the rev3 strain was transformed with unpurified PCR products. The correct replacement of the RAD30 gene with the selectable marker KanMX4 was verified by phenotypic selection (resistance to geneticin, G418) and by the appearance of PCR products of the expected size using primers (A and D) that span the left and right junctions of the deleted region within the genome (primer A: 5′-aaa att aaa cgc tac cta atc ctg c-3′; primer D: 5′-cat cta att gat taa gtc ccc tga a-3′; with wild-type sequence, i.e., in the absence of a deletion, the size of the AD amplicon is 2566 bp, while in the presence of RAD30 deletion the size of the AD amplicon is 2251 bp). The KanB primer (5′-ctg cag cga gga gcc gta at-3′), internal to the KanMX4 module, was also used to confirm the correct targeting of the KanMX4 sequence.
2.6 Molecular characterization of Me-lex induced p53 mutations
In order to analyse the p53 status of red yeast colonies (Me-lex induced) colony PCRs were performed using primers P3 and P4 [32] and conditions described above. The unpurified PCR product (p53 cDNA fragment from nucleotidic position 125 to 1122) and HindIII-StuI linearized pRDI-22 plasmid were co-transformed into yIG397 (gap repair) and transformants were selected on suitable plates. The p53 amplification products that gave approximately 100% of red or pink colonies were sequenced (BMR Genomics, Padua, Italy).
3. RESULTS
3.1 The deletion of either RAD30 or REV3 has no marked effect on spontaneous mutation frequency
In order to determine the role of Polη in the biological fate of Me-lex induced lesions, per se and with respect to Polζ, the RAD30 gene was deleted in the yIG397 parental strain and in the corresponding rev3 derivative (Table 1; see Materials and Methods for details). All strains [yIG397 (WT), yPM6 (rev3); yPM13 (rad30) and yPM14 (rad30rev3)] contain the ADE2 reporter gene under transcriptional control of wild-type human p53.
To determine the spontaneous mutation frequency (sMF), untreated plasmid pTS76, containing the WT p53 cDNA under the control of the constitutive ADH1 promoter, was transformed into the different strains. Transformants were selected on plates lacking tryptophan but containing sufficient adenine for adenine auxotrophs to grow and turn red. Neither the single deletion of RAD30 or REV3, nor the concomitant deletion of both genes caused a significant change in the sMF with respect to WT (Table 2). However, a significantly lower sMF was observed in yPM13 (rad30) relative to yPM6 (rev3).
Table 2.
Effects of REV3, RAD30, single or double, deletion(s) on spontaneous p53 mutant frequencies.
| Strain | genotype | p53 mutant frequency (red colonies/total) ×103 |
statistics |
|---|---|---|---|
| yIG397 | WT |
0.34 (28/82.288)*§# |
*NS; §NS; #NS |
| yPM6 | rev3 |
0.59 (11/18.728)*^† |
^p<0.05; †NS |
| yPM13 | rad30 |
0.29 (62/215.961)§^** |
**NS |
| yPM14 | rad30rev3 |
0.48 (14/28.851)#†** |
NS: not significant
3.2 The deletion of RAD30 or REV3 reduced similarly the survival of Me-lex damaged plasmid
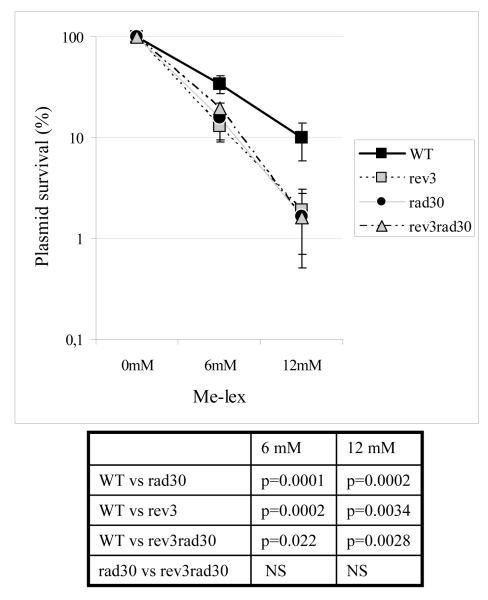
In order to evaluate the influence of Polη on the lethality of Me-lex induced lesions, plasmid pTS76 was damaged in vitro by exposure to increasing Me-lex concentrations and then transformed into the different yeast strains. A general Me-lex concentration-dependent decrease in plasmid survival was observed (Fig. 1). The deletion of the RAD30 or REV3 gene had a similar impact, leading to a significant reduction in plasmid survival as compared to the parental strain. The rev3rad30 double mutant strain was as sensitive as those with the single deletion [yPM6 (rev3) or yPM13 (rad30)]. Together these results indicate that both Rad30 (Polη) and Rev3 (Polζ) provide protection against Me-lex toxicity.
Fig. 1.
Influence of the deletion of the RAD30 gene in WT or rev3 background on the lethality of Me-lex induced lesions (expressed as plasmid survival) with respect to the parental and the rev3 strains. Each point derives from at least 4 independent experiments. Results (p value) of the statistical analyses are reported, NS: not significant.
3.3 The deletion of RAD30 or REV3 has a different impact on Me-lex induced mutagenesis
The impact of a RAD30 or REV3 deletion on Me-lex mutagenicity was examined by exploiting the p53 dependent expression of the ADE2 reporter gene (Table 3). The MF increased proportionally to the Me-lex concentration. Deletion of REV3 led to a decrease in the MF (defined as red colonies over total) with respect to WT strain which was significant at 6 mM Me-lex, (p=0.0004) while at 12 mM Me-lex the decrease did not reach statistical significance. The effect of the deletion of RAD30 on the MF depended on the Me-lex concentration: there was a significant decrease at 6 mM and a significant increase at 12 mM in MF relative to WT.
Table 3.
Effects of REV3, RAD30 single or double deletion(s) on Me-lex (6 mM, 12 mM) induced p53 mutant frequencies.
| Strain | genotype | p53 mutant frequency (red colonies/total) ×103 |
statistics |
|---|---|---|---|
| Me-lex induced (6 mM) | |||
| yIG397 | WT |
10.7 (301/28.043)*§# |
*p<0.0004; §p<0.0023; #p<0.0001 |
| yPM6 | rev3 |
2.89 (7/2.421)*^† |
^p<0.0054; †NS |
| yPM13 | rad30 |
8.3 (303/36.319)§^** |
**p<0.0001 |
| yPM14 | rev3rad30 |
2.2 (12/5.459)#†** |
|
| Me-lex induced (12 mM) | |||
| yIG397 | WT |
28.36 (155/5.466)*§# |
*NS; §p<0.0009; #p<0.015 |
| yPM6 | rev3 |
17.87 (6/336)*^† |
^p<0.05; †NS |
| yPM13 | rad30 |
42.6 (129/3.024)§^** |
**p<0.0005 |
| yPM14 | rev3rad30 |
8.25 (4/485)#†** |
NS: not significant
The yPM14 (rev3rad30) strain always showed a significantly lower MF than in the yPM13 (rad30) strain, which made it appear that the REV3 deletion was dominant over the RAD30 deletion in terms of inhibition of mutagenicity. This suggests that Polζ plays a major role in Me-lex mutagenicity. To verify this hypothesis, a representative subset of mutants isolated at 6 mM Me-lex, were isolated and used to determine whether the phenotypically mutant (i.e. red) colonies obtained after transformation of Me-lex treated plasmids in the various TLS mutant strains contained full length mutated p53 cDNA. This analysis was accomplished by yeast colony PCR amplification of the p53 cDNA followed by gap repair with appropriate controls (see Materials and Methods). For the yIG397 (WT) strain 56% (59/105) of the mutant colonies contained mutant p53 cDNA of normal length. However, the percentage decreased to 33% (48/144) for yPM13 (rad30) (WT vs rad30: p<0.0008) and dropped to less than 15% (0/7) for yPM6 (WT vs rev3: p<0.0043). For the yPM14 (rev3rad30) double mutant, the percentage decreased to 9% (1/12) (WT vs rad30rev3: p=0.0017) and very few of the mutant colonies recovered from the rev3 strains yielded an amplification product. This result suggests that defects in TLS capacity could stimulate other redundant or alternative pathways (e.g. recombination repair) that resulted in PCR negative clones. Therefore, the mutant colonies in yPM6 (rev3) and yPM14 (rev3rad30) strains lost the p53 cDNA probably through rearrangements encompassing the whole p53cDNA. Overall, our results suggest that TLS capacity decreases in the different TLS mutant strains, and is virtually absent upon deletion of Polζ in our experimental system.
3.4 Effect of RAD30 deletion on the mutation spectra induced by Me-lex
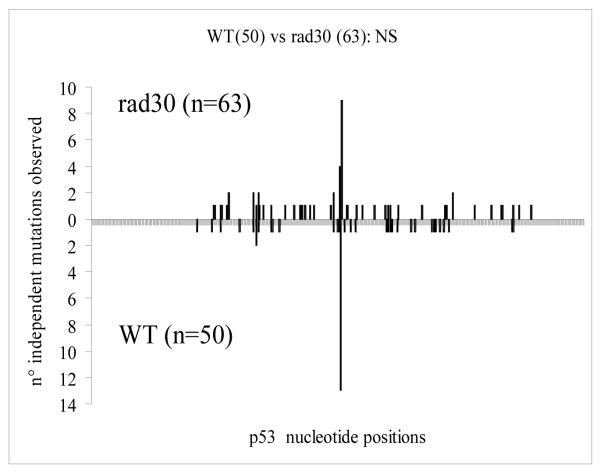
In order to characterize the effects of RAD30 deletion on the mutation spectra, we isolated and characterized 53 Me-lex induced p53 mutants in the yPM13 (rad30) strain for which a normal length p53 cDNA was obtained, with a total of 64 mutations (63 base pair substitution and one +1G insertion) (Table 4). Of the 63 base pair substitutions, 57 (90.5%) were found in 48 mutants isolated at 6mM Me-lex, while the remaining 6 mutations were found in 5 mutants isolated at 12mM concentration. The mutations observed are Me-lex induced based upon several criteria. First, mutants were isolated at least 28-fold above sMF, therefore, less than 4% of the mutations are of a spontaneous origin. Second, by defining a Me-lex binding site as a sequence with at least 3 out of 4 bp being A/T, and considering that the binding can tolerate a single G/C pairing [7], 38/47 (>80%) of AT targeted mutations occur at a Me-lex binding site, while only 5/16 (>31%) GC targeted mutations occur in or in the proximity (within 2bp) of a Me-lex binding site. The mutation spectra obtained in the rad30 and WT strains were compared in terms of type and site of mutations along the p53 cDNA sequence using Cariello’s test [35], which provides a rigorous statistical test of the relatedness of two spectra obtained at the same locus. The two mutation spectra resulted indistinguishable (Fig. 2). We then compared base pair substitutions with those previously obtained in yIG397 (WT) [8] in terms of classes of mutations (Table 5). A decrease in GC-targeted and a concomitant increase in AT-targeted mutations were found in the yPM13 (rad30) strain with respect to WT, although those differences were not statistically significant (Table 5). When the specific types of base pair substitution were considered, a significant increase of AT>GC mutations (p=0.039) and a significant decrease of GC>CG mutations (p=0.043) became apparent. Interestingly, both differences remains statistically significant even when mutations characterized in mutants isolated at 12 mM Me-lex were excluded.
Table 4.
Features of Me-lex induced p53 mutations in rad30 (polη) yeast strain
| N° mutation | sequenze context 5′->3′a |
base pair changes |
codon | Aminoacid substitution |
|---|---|---|---|---|
| #1 | GGA AAT TTG | AT>GC | 200 | Asn>Asp |
| #2 | TAC AAG CAG | AT>TA | 164 | Lys>Stop |
| #3 | GGA AAT TTG | AT>GC | 200 | Asn>Asp |
| #4 | GGT TTC CGT | AT>GC | 109 | Phe>Ser |
| #5 | CAG CAT CTT | GC>TA | 193 | His>Asn |
| #6 | AAT TTG CGT | AT>TA | 201 | Leu>Stop |
| #7 | AAT TTG CGT | AT>TA | 201 | Leu>Stop |
| #8 | GAG AAT CTC | AT>TA | 288 | Asn>Tyr |
| #9 b | AAT TTG CGT | AT>TA | 201 | Leu>Stop |
| #10 | CAG GGC AGC | GC>TA | 105 | Gly>Cys |
| #11 | GAG CGC TTC | GC>AT | 337 | Arg>Cys |
| #12 | AAT TTG CGT | AT>TA | 201 | Leu>Stop |
| #13 | GAG AAT CTC | AT>TA | 288 | Asn>Tyr |
| #14 | CAG CAC ATG | AT>GC | 168 | His>Arg |
| #15 | GGA AAT TTG | AT>GC | 200 | Asn>Asp |
| #16 | CAG CTG TGG | AT>TA | 145 | Leu>Gln |
| #17 | TGT AAC AGT | AT>GC | 239 | Asn>Asp |
| #18 | TAT TTC ACC | AT>TA | 328 | Phe>Tyr |
| #19 | TTT TGC CAA | AT>GC | 135 | Cys>Arg |
| #20 b | CAC CAT GAG | AT>TA | 179 | His>Gln |
| #21 | CTT ATC CGA | AT>GC | 195 | Ile>Thr |
| #22 | Cga Gtg>Cg(+1g)A Gtg | +1G | insertion | |
| #23 | AAG ATG TTT | AT>CG | 133 | Met>Arg |
| #24 | GTG GTG CCC | AT>CG | 218 | Val>Gly |
| #25 | AAT TTG CGT | AT>CG | 201 | Leu>Trp |
| #26 | AAC TAC ATG | AT>CG | 236 | Tyr>Ser |
| #27 | GCC AAG ACC | AT>CG | 138 | Lys>KQ |
| #28 | ATG TTT TGC | AT>CG | 134 | Phe>Leu |
| #29 b | CTT ATC CGA | AT>GC | 195 | Ile>Thr |
| #30 | GGC ATG AAC | AT>GC | 246 | Met> Thr |
| #31 | CGC TGC CCC | AT>GC | 176 | Arg>Cys |
| #32 | ATC ATC ACA | AT>GC | 255 | Ile>Thr |
| #33 | AAG AAG AAA | AT>GC | 320 | Lys>Glu |
| #34 | GAG TAT TTG | AT>GC | 205 | Cys>Tyr |
| #35 | ATG TTC CGA | AT>GC | 341 | Phe>Ser |
| #36 | GAA GGA AAT | AT>GC | 199 | Cys>Cys silent |
| #37 | TTT TGC CAA | AT>GC | 135 | Cys>Arg |
| #38 | TAT TTC ACC | AT>GC | 328 | Phe>Leu |
| #39 | AAT TTG CGT | AT>TA | 201 | Leu>STOP |
| #40 | CTC AAG GAT | AT>TA | 351 | Lys>STOP |
| #41 | CTC AAC AAG | AT>TA | 131 | Asn>Tyr |
| #42 | AAT TTG CGT | AT>TA | 201 | Leu>STOP |
| #43 | CGT CTG GGC | AT>TA | 111 | Leu>Gln |
| #44 | AAT TTG CGT | AT>TA | 201 | Leu>STOP |
| #45 | GGA AAT TTG | AT>TA | 200 | Asn>Tyr |
| #46 | AAT TTG CGT | AT>TA | 201 | Leu>STOP |
| #47 | CTC AAC AAG | AT>TA | 131 | Asn>Tyr |
| #48 | AAT TTG CGT | AT>TA | 201 | Leu>STOP |
| #49 b | TAT TTG GAT | AT>TA | 206 | Leu>STOP |
| #50 b | CGT CTG GGC | AT>TA | 111 | Leu>Gln |
| #51 | GAC CGG CGC | GC>AT | 282 | Arg> Gln |
| #52 | GGC TCT GAC | GC>AT | 227 | Ser>Phe |
| #53 | TTT CGA CAT | GC>AT | 213 | Arg>STOP |
| #54 | CCT TCC CAG | GC>AT | 99 | Ser>Ser silent |
| #55 | AAC AGT TCC | GC>AT | 240 | Ser>Asn |
| #56 | AAG CGA GCA | GC>AT | 306 | Arg>Gln |
| #57 | CGC GTC CGC | GC>AT | 157 | Val>Ile |
| #58 | GAG GTT GTG | AT>TA | 172 | Val>Asp |
| #59 | CGC ACA GAG | GC>AT | 284 | Thr>Ile |
| #60 | CCT TCC CAG | GC>AT | 99 | Ser>Phe |
| #61 b | ATG ACG GAG | GC>AT | 170 | Thr>Met |
| #62 | TTT CGA CAT | GC>CG | 213 | Arg>Pro |
| #63 | TTT TGC CAA | GC>TA | 135 | Cys>STOP |
| #64 | CAG GGC AGC | GC>TA | 105 | Gly>Val |
in bold: nucleotide position; underlined: codon triplet where the mutation causes the indicated aminoacid change.
mutations found in mutant isolated at 12mM Me-lex, all the other 57 mutations were found in mutants isolated at 6mM Me-lex.
Fig. 2.
Sequence distribution along the p53 nucleotide positions of independent p53 base pair substitutions induced by Me-lex and isolated in WT (50 mutations) [8] and in rad30 (63 mutations). Mutation spectra were compared using the Cariello’s program [35]. NS: not significant.
Table 5.
Class of base pair substitutions of Me-lex induced p53 mutations in different genetic backgrounds
| WTa | rad30 | WT vs rad30b | |
|---|---|---|---|
| GC-targeted | 19(38%) | 16 (25%) | NS |
| GC>AT | 7 (14%) | 11 (17%) | NS |
| GC>TA | 6 (12%) | 4 (6%) | NS |
| GC>CG | 6 (12%) | 1 (2%) | p=0.043 |
| AT-targeted | 31 (62%) | 47 (75%) | NS |
| AT>TA | 21 (42%) | 23 (37%) | NS |
| AT>GC | 6 (12%) | 18 (29%) | p=0.039 |
| AT>CG | 4 (8%) | 6 (9%) | NS |
| 50 (100%) | 63 (100%) |
data from (8)
Fisher’s exact test: events in a specific class vs total
Of the 63 base pair substitutions reported, 57 were found in mutants isolated at 6 mM Me-lex, while the remaining 6 mutations were found in mutants isolated at 12mM.
Note that the statistically significant differences remain even if the six mutations found at 12mM are excluded (p=0.048; p=0.034 respectively).
4. DISCUSSION
The fixation of promutagenic DNA damage into mutations depends predominantly on the activity of TLS DNA polymerases that allow bypass of DNA lesions that stall replication forks. Accordingly, we have demonstrated in Saccharomyces cerevisiae, that disruption of Polζ activity (through deletion of REV3 that codes for the catalytic subunit of Polζ) or Rev1, increased Me-lex lethality and decreased Me-lex mutagenicity in different repair backgrounds [30]. Thus Polζ and Rev1 contribute to a decrease in the lethal effects of Me-lex-induced lesions, with an associated increase in mutagenic events. In the present study, we have found that in a WT background the deletion of RAD30 (Polη) resulted in a significant increase in Me-lex lethality, similarly to those observed in rev1 or rev3 strains. However, the mutagenicity results are more complicated in that there is a concentration-dependent effect: the MF decreased at 6 mM; but increased at 12 mM. In the rev3 background, deletion of RAD30 does not further increase Me-lex toxicity (Fig. 1) and it also does not reduce the antimutator effect due to the REV3 deletion alone (Table 3). A possible explanation of this concentration-dependent difference is that level of lesions (i.e., lesions/plasmids) at 6 mM and at 12 mM is both quantitatively and qualitatively different. The different biological effects observed in a rad30 background suggest that repair of Me-lex-induced DNA damage can indeed be a modulating factor for the MF. From a qualitative point of view, it is known that 3-mA can generate AP sites via non-enzymatic hydrolysis. While at 6 mM the contribution of this phenomenon may be of no consequences, the level and the biological importance of AP sites might become quantitatively not negligible at the 12 mM concentration [11].,
In order to verify at the molecular level the influence of Polη in the fixation of mutations, base pair substitutions induced by Me-lex [mainly (57/63) at the 6mM concentration] in the yPM13 (rad30) strain were compared with those previously obtained in a WT background [8]. The two mutation spectra, compared using the Cariello’s test, appear indistinguishable (Fig. 2). Even excluding the common hot spot present in both spectra, (position 602, codon 201), the spectra do not differ (not shown). However, the fact that MF is slightly but significantly reduced in yPM13 (rad30) with respect to yIG397 at 6mM suggests a role of Polη in some error prone processes. Furthermore, when only AT targeted base pair substitutions are considered, AT>TA mutations at hot spot position 602 appear to be significantly less frequent in yPM13 (rad30) than in the WT strain (9/47 vs 13/31, p=0.04, Fisher’s exact test). Consistent with our result, Plosky et al. found that human Polη can incorporate A, albeit inefficiently, opposite 3-methyl-3-deaza-Adenine [36]. The deletion of RAD30 gene was also associated with a significant decrease in GC>CG transversions. On the other hand, a role of Polη in an error free process related to AT-targeted mutations is supported by the fact that a small yet significant increase in AT>GC mutations (p=0.039) in yPM13 (rad30) with respect to WT strain was observed. One possible interpretation is that in the absence of Polη, Me-lex induced DNA damage at certain AT sites is processed by other error prone TLS polymerases (i.e., Polζ and/or Rev1). Taken collectively, these results suggest that Polη participates in TLS processing of Me-lex induced lesions in an error prone way, albeit with some limited error free bypass.
Two recent papers describe the yeast TLS DNA polymerases involved in the replication bypass of methyl methanesulfonate (MMS)-induced DNA damage in vivo [29,36]. They report that WT and the rad30 derivative have a similar sensitivity towards MMS. This is in contrast with previous work showing that the rad30 strain is somewhat more sensitive to MMS with respect to the parental strains [28,37,38]. In our work reported here, the rad30 strain was more sensitive than WT towards the Me-lex induced lesionsThe discrepancies observed between the different reports can be attributed to differences in the experimental design (in vivo treatment versus in vitro treatment of plasmid followed by transfection) and/or, to the chemical properties of the methylating agents used. MMS generates 3-mA as less than 15% of the total DNA adducts [39] and without any sequence selectivity. In contrast, Me-lex efficiently yields almost exclusively 3-mA (~ 95%) and with a marked sequence selectivity for A/T-rich sequences. Despite some differences, it was generally observed that in a mag1 background, where the level of 3-mA will be increased due to the BER deficiency, even the single deletion of RAD30 increased the sensitivity to MMS [29,36]. Using Me-lex, which produces >1000-fold more 3-mA than MMS based upon an equimolar concentration [9], we observe a similar enhanced sensitivity of the rad30 mutant. . In summary, these data are consistent with 3-mA being a cytotoxic lesion in the absence of Polη and might also suggest that MMS would behave differently than Me-lex in our experimental system. Indeed, in WT yeast the MMS mutation spectrum (unpublished) differs significantly from that induced by Me-lex; in the latter the mutation spectrum is associated with Me-lex equilibrium binding sites.
It was also reported that Polζ and Polη act synergistically [29,36] since the rev3rad30 strain was more sensitive to MMS with respect to the rev3 strain. Our results do not confirm this observation. Indeed, the sensitivities of the yPM6 (rev3) [or yPM13 (rad30] or the yPM14 (rev3rad30) strains to Me-lex are similar (Fig. 1). It has to be noted that at ~1% survival levels, our results are consistent with those reported by Plosky et al., (see Fig. 6 in [36]) not only in the WT background, but also in the mag1 background. In fact, a synergistic effect between rad30 and rev3 deletions was visible only in the mag1 background [36] at survivals levels 4-orders of magnitude lower (~10−6) then those reached in the present study. An alternative explanation is that since plasmids were damaged randomly, and assuming a Poisson’s distribution, even with 4-5 lesions /plasmids (compatible with our treatment), there will be still a fraction (approximately 0.5-1%) of plasmids without lesions. Therefore, this fraction of undamaged plasmid after transformation in yeast will generate a 0.5-1% background level of colonies. This background makes it impossible to reach the lower survival level that could mask a potential synergistic effect in our experimental system.
Overall, the results in the various TLS deficient strains reveal that the absence of a single TLS polymerase (Rev3or Rev1 [30]; Rad30, in the present work) causes a similar increase in the lethality of Me-lex induced lesions. Based upon the known protein-protein interactions of Polη, Rev1, Polζ and the functional consequences of these interactions [15]; Rev1 interacts with Polζ and enhances Polζ proficiency of extension of primer termini opposite DNA lesions [14]), it is reasonable to suggest that the TLS of Me-lex lesions is a multi-DNA polymerases process that is most effective when all three polymerases are present. The absence of even one of the three yeast polymerases has a similar consequence in terms of blocking DNA replication (similar lethality). This implies that all three TLS Pols (Polη, Rev1, Polζ) participate in the lesion bypass process.
It was reported using MMS as the methylating agent that Polη and Polζ provide alternate means by which replication through the 3-mA could be accomplished in a predominantly error free way [29,36]. Yeast Polη shows some in vitro capacity of being an error free TLS; indeed it prefers to insert T opposite 3-methyl-3-deaza-A with an efficiency that is ~3% of that observed opposite an A. Polη is also able to extend from the resulting 3-methyl-3-deaza-A:T terminus with an extension efficiency that is ~13% of that observed from an A:T base pair [36]. In our experiments, we see events suggestive of error free bypass that are dependent on Polη, e.g. a significant increase of AT>GC transitions in yPM13 (rad30) vs yIG397 (WT). Despite this indication of a potentially error-free bypass, the deletion of RAD30 results in a slight, yet significant, decrease in MF. This suggests an error prone bypass dependent on Polη, which is consistent also with the observation that position 602 is a significantly less hotspot for mutations in yPM13 (rad30) vs yIG397 (WT). These contrary effects, can explain why a significant increase in the induced MF in the absence of Polη, an expected outcome if Polη was a pure error free TLS, was not observed (at least at 6mM).
Which Me-lex-induced lesion(s) is (are) bypassed by Polη? The two candidates are 3-mA, which is directly formed from Me-lex, and AP sites, which are derived from glycosylase mediated excision or non-enzymatic hydrolysis of 3-mA. It is important to recognize that the hydrolytic stability of 3-mA in DNA is significantly reduced in single-strand vs. double-strand DNA [40], which may be the environment at a stalled replication fork. The 3-mA is highly cytotoxic and poorly mutagenic, while AP sites are both cytotoxic and highly mutagenic. Our previous results suggested that both lesions are substrates for TLS bypass [30], an hypothesis that could be tested in vivo or in vitro using DNA modified with a single lesions introduced at a specific location (work in progress). However, it is important to note, that AP sites can produce secondary lesions (such as 3′-blocked single strand breaks with 3′-dRP termini), spontaneously (via hydrolysis) or through the action of different enzymes. The potential contribution of these secondary lesions to the end point analyzed in the present work is unknown. Thus, we cannot unequivocally pinpoint the composition of lesions present at the replication fork.
The specificity of the mutations that significantly increased in yPM13 (rad30) with respect to yIG397 (WT) (AT>GC) might suggest a role for Rev1, which specifically incorporates C opposite all template bases and abasic sites [12,20]. From a mechanistic point of view this hypothesis is appealing. The N-terminal portion of Rev1 approaches the template–primer duplex from the minor groove side and forces the template base to flip out of the double helix, and, in its place, it supplies an arginine side chain to hydrogen bond with the incoming dCTP [41]. Since 3-methyl-3-deaza-A (and very likely 3-mA) appears to have the propensity to unstack (Gold et al., unpublished), it is reasonable to imagine how the activity of Rev1 might be facilitated. Rev1 polymerase activity is dispensable for the fixation of mutation involving other lesions [20-24], and it has been reported to promote replication through abasic sites and N2-adducted-Gs [42] that, similar to 3-mA, impinge upon the minor groove [12].
In summary, we have demonstrated that Polη is involved in the processing of Me-lex induced lesion(s) and that this involvement, being mainly error prone, is dose-dependent and influenced by the sequence context.
ACKNOWLEDGEMENTS
This work was supported by NIH grant RO1 CA29088 (BG) and partially by AIRC (GF, AI).
Abbreviations
- 3-mA
N3-methyladenine
- Me-lex
{1-methyl-4-[1-methyl-4-(3-methoxysulfonylpropanamido)pyrrole-2-carboxamido]pyrrole-2-carboxamido}propane
- MF
mutation frequency
- BER
base excision repair
- NER
nucleotide excision repair
- TLS
translesion synthesis
- WT
wild-type
- MMS
methyl methanesulfonate
Footnotes
Conflict of Interest. None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- [2].Bennett RA, Pegg AE. Alkylation of DNA in rat tissues following administration of Streptozotocin. Cancer Res. 1981;41:2786–2790. [PubMed] [Google Scholar]
- [3].Tentori L, Graziani G. Pharmacological strategies to increase the antitumor activity of methylating agents. Curr Med Chem. 2002;9:1285–1301. doi: 10.2174/0929867023369916. [DOI] [PubMed] [Google Scholar]
- [4].Rydberg B, Lindahl T. Nonenzymatic methylation of DNA bythe intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barrows LR, Magee PN. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3:349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- [6].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. AMS Press; Washington, DC: 2006. [Google Scholar]
- [7].Kelly JD, Inga A, Chen FX, Dande P, Shah D, Monti P, Aprile A, Burns PA, Scott G, Abbondandolo A, Gold B, Fronza G. Relationship between DNA methylation and mutational patterns induced by a sequence selective minor groove methylating agent. J Biol Chem. 1999;274:18327–18334. doi: 10.1074/jbc.274.26.18327. [In Process Citation] [DOI] [PubMed] [Google Scholar]
- [8].Monti P, Iannone R, Campomenosi P, Ciribilli Y, Varadarajan S, Shah D, Menichini P, Gold B, Fronza G. Nucleotide excision repair defect influences lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent in the absence of base excision repair. Biochemistry. 2004;43:5592–5599. doi: 10.1021/bi035968x. [DOI] [PubMed] [Google Scholar]
- [9].Shah D, Kelly J, Zhang Y, Dande P, Martinez J, Ortiz G, Fronza G, Tran H, Soto AM, Marky L, Gold B. Evidence in Escherichia coli that N3-methyladenine lesions induced by a minor groove binding methyl sulfonate ester can be processed by both base and nucleotide excision repair. Biochemistry. 2001;40:1796–1803. doi: 10.1021/bi0024658. [DOI] [PubMed] [Google Scholar]
- [10].Bobola MS, Varadarajan S, Smith NW, Goff RD, Kolstoe DD, Blank A, Gold B, Silber JR. Human glioma cell sensitivity to the sequence-specific alkylating agent methyl-lexitropsin. Clin Cancer Res. 2007;13:612–620. doi: 10.1158/1078-0432.CCR-06-1127. [DOI] [PubMed] [Google Scholar]
- [11].Monti P, Campomenosi P, Ciribilli Y, Iannone R, Inga A, Shah D, Scott G, Burns PA, Menichini P, Abbondandolo A, Gold B, Fronza G. Influences of base excision repair defects on the lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent. J Biol Chem. 2002;277:28663–28668. doi: 10.1074/jbc.M203384200. [DOI] [PubMed] [Google Scholar]
- [12].Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- [13].Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–9563. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase eta. Mol Cell Biol. 2007;27:8401–8408. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics. 1979;92:397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. I. rev1 Mutant strains. J Mol Biol. 1978;122:1–21. doi: 10.1016/0022-2836(78)90104-3. [DOI] [PubMed] [Google Scholar]
- [18].Lawrence CW, Nisson PE, Christensen RB. UV and chemical mutagenesis in rev7 mutants of yeast. Mol Gen Genet. 1985;200:86–91. doi: 10.1007/BF00383317. [DOI] [PubMed] [Google Scholar]
- [19].Lawrence CW, O’Brien T, Bond J. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants ofyeast. Mol Gen Genet. 1984;195:487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- [20].Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Baynton K, Bresson-Roy A, Fuchs RP. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomycescerevisiae. Mol Microbiol. 1999;34:124–133. doi: 10.1046/j.1365-2958.1999.01583.x. [DOI] [PubMed] [Google Scholar]
- [22].Gibbs PE, Borden A, Lawrence CW. The T-T pyrimidine (6-4) pyrimidinone UV photoproduct is much less mutagenic in yeast than in Escherichia coli. Nucleic Acids Res. 1995;23:1919–1922. doi: 10.1093/nar/23.11.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gibbs PE, Kilbey BJ, Banerjee SK, Lawrence CW. The frequency and accuracy of replication past a thymine-thymine cyclobutane dimer are very different in Saccharomyces cerevisiae and Escherichia coli. J Bacteriol. 1993;175:2607–2612. doi: 10.1128/jb.175.9.2607-2612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- [27].Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- [28].Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Monti P, Ciribilli Y, Russo D, Bisio A, Perfumo C, Andreotti V, Menichini P, Inga A, Huang X, Gold B, Fronza G. Rev1 and Polzeta influence toxicity and mutagenicity of Me-lex, a sequence selective N3-adenine methylating agent. DNA Repair (Amst) 2008;7:431–438. doi: 10.1016/j.dnarep.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Y, Chen FX, Mehta P, Gold B. Groove- and sequence-selective alkylation of DNA by sulfonate esters tethered to lexitropsins. Biochemistry. 1993;32:7954–7965. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- [32].Flaman JM, Frebourg T, Moreau V, Charbonnier F, Martin C, Chappuis P, Sappino AP, Limacher IM, Bron L, Benhattar J, et al. A simple p53 functional assay for screening cell lines, blood,and tumors. Proc Natl Acad Sci U S A. 1995;92:3963–3967. doi: 10.1073/pnas.92.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guthrie C, Fink GR. Academic press; San Diego: 1991. [Google Scholar]
- [34].Storici F, Resnick MA. Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet Eng (N Y) 2003;25:189–207. [PubMed] [Google Scholar]
- [35].Cariello NF, Piegorsch WW, Adams WT, Skopek TR. Computer program for the analysis of mutational spectra: application to p53 mutations. Carcinogenesis. 1994;15:2281–2285. doi: 10.1093/carcin/15.10.2281. [DOI] [PubMed] [Google Scholar]
- [36].Plosky BS, Frank EG, Berry DA, Vennall GP, McDonald JP, Woodgate R. Eukaryotic Y-family polymerases bypass a 3-methyl-2′-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic Acids Res. 2008;36:2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- [38].Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- [39].Varadarajan S, Shah D, Dande P, Settles S, Chen FX, Fronza G, Gold B. DNA damage and cytotoxicity induced by minor groove binding methyl sulfonate esters. Biochemistry. 2003;42:14318–14327. doi: 10.1021/bi0353272. [DOI] [PubMed] [Google Scholar]
- [40].Fujii T, Saito T, Nakasaka T. Purines. XXXIV. 3-Methyladenosine and 3-methyl-2′-deoxyadenosine: their synthesis, glycosidic hydrolysis, and ring fission. Chem. Pharm. Bull. 1989;37:2601–2609. [Google Scholar]
- [41].Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- [42].Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Mol Cell Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]