Abstract
Uridine diphosphate (UDP)-glucose dehydrogenase (UGDH) catalyzes the oxidation of UDP-glucose to yield UDP-glucuronic acid, a precursor for synthesis of glycosaminoglycans and proteoglycans that promote aggressive prostate cancer (PC) progression. The purpose of this study was to determine if the UGDH expression in normal appearing acini (NAA) from cancerous glands is a candidate biomarker for PC field disease/effect assayed by quantitative fluorescence imaging analysis (QFIA). A polyclonal antibody to UGDH was titrated to saturation binding and fluorescent microscopic images acquired from fixed, paraffin-embedded tissue slices were quantitatively analyzed. Specificity of the assay was confirmed by Western blot analysis and competitive inhibition of tissue labeling with the recombinant UGDH. Reproducibility of the UGDH measurements was high within and across analytical runs. Quantification of UGDH by QFIA and Reverse-Phase Protein Array analysis were strongly correlated (r=0.97), validating the QFIA measurements. Analysis of cancerous acini (CA) and NAA from PC patients vs. normal acini (NA) from non-cancerous controls (32 matched pairs) revealed significant (P<0.01) differences, with CA (increased) vs. NA, NAA (decreased) vs. NA and CA (increased) vs. NAA. Areas under the Receiver Operating Characteristic curves were 0.68 (95% CI: 0.59–0.83) for NAA and 0.71 (95% CI: 0.59–0.83) for CA (both vs. NA). These results support the UGDH content in prostatic acini as a novel candidate biomarker that may complement the development of a multi-biomarker panel for detecting PC within the tumor adjacent field on a histologically normal biopsy specimen.
Keywords: UGDH, biomarker, prostate cancer, tissue proteomics, field disease/effect
INTRODUCTION
One in three men aged >50 years has morphologically identified features of prostate cancer (PC),1 and one in six men will develop clinically significant PC during their lifetime.2 Although diagnosis of prostate cancer (PC) still depends on quantification of the serum prostate-specific antigen (PSA), there is no PSA threshold that establishes the presence of clinically important PC or determines zero risk for PC. Total PSA measurement, free PSA, and PSA velocity, alone and in combination, have not significantly improved the sensitivity and specificity compared to a traditional PSA threshold value of 4 ng/ml, but rather confirmed the need for lowering the threshold for a biopsy3 and/or searching for other markers complementary to PSA testing. Furthermore, prostate biopsy identifies only 26% to 41% of cancer depending on the patient’s age. Re-biopsy successfully establishes the diagnosis in 50%, 62% and 68% of cases following one, two or three re-biopsies, respectively.4 A solution to a personalized rebiopsy is the development of a sensitive and specific biomarker profile for the premalignant field or cancer-induced field effects that is informative in the initial biopsy.5–9 We hypothesize that developing a protein biomarker panel assayed in the initial specimen will provide optimal sensitivity and specificity as a guide to rebiopsy. To obtain an optimal sensitivity and specificity, it is anticipated that functional protein biomarkers will be quantitatively assayed in a multiplex format in histologically normal biopsies from cancer-bearing prostates.
In a previous study,10 quantitative fluorescence imaging analysis (QFIA) was used to quantify β-catenin in prostate biopsies from 42 pairs of PC cases and age-matched controls. Reduced expression in the normal-appearing fields of cancer-bearing glands identified 42% of the cases with 88% specificity. In this study, the expression of uridine diphosphate (UDP)-glucose dehydrogenase (UGDH) was investigated as a potential field disease/effect biomarker. UGDH catalyzes conversion of UDP-glucose to UDP-glucuronic acid, an essential precursor for synthesis of proteoglycans and extracellular matrix glycosaminoglycans (hyaluronan, HA, and other molecules). Increased expression of UGDH has been linked to elevated HA. Profiling HA in PC tissue and non-cancerous prostate tissue revealed that presence of HA accumulation correlated with PC of Gleason sum ≥ 5.11,12 HA, synthesized as a high molecular weight polymer by HA synthases HAS1, HAS2 and HAS3, is cleaved into smaller fragments by hyaluronidases, e.g., HYAL1, a secreted enzyme expressed in epithelial cells but not in stromal cells of PC tissue.11,12 HA-HYAL1 detection has significant prognostic value for prostate cancer.13,14 HA fragments have potent angiogenic activity, and may promote invasion and metastasis by altering the motility and cell cycle of carcinoma cells, as well as modulating cell adhesion to the extracellular matrix and to endothelial cells.15–17 HA-induced alterations in the adhesion of malignant cells are mediated via activation of their CD44 and CD168 receptors.18–20 Interestingly, prostate stem-like cells and prostate tumor initiating cells involved in the development of prostate cancer21,22 also share CD44 expression.23–25 This provides a hypothetical link between UGDH activity and early tumor invasion and metastases probably mediated by cancer stem-like cells. Interestingly, in PC, both HA and cancer stem-like cells were associated with the progression of androgen-independent disease and metastases.26–28
Alternatively, decreased expression of UGDH, specifically in prostate epithelium, may be linked to increased androgen-dependent cell proliferation and the expression of androgen-sensitive genes, including PSA and vascular endothelial growth factor (VEGF). Consequently, reduced epithelial UGDH may be linked to early pro-carcinogenic effects. UDP-glucuronic acid is required for UDP-glucuronosyltransferase (UGT) catalyzed glucuronidation of other substrates, including dihydrotestosterone (DHT), androstane-3α,17β-diol (3α-diol) and androsterone (ADT).29 UGT2B15 and UGD2B17, UGT enzymes that conjugate glucuronate to DHT and 3α-diol, are localized in prostate luminal and basal cells, respectively.30–32 Inhibition of UGT2B15/B17 by siRNA in LNCaP prostate tumor cells resulted in enhanced proliferation with concomitant increased expression of eight androgen-responsive genes including PSA and VEGF.32
In this study UGDH was quantified in acini/epithelium of archived biopsies from 32 cancer cases and 32 age-matched controls by quantitative fluorescence imaging.10 This study revealed a differential expression of UGDH in the cancer and the premalignant field and identified UGDH as a potential biomarker for PC field/disease effect.
MATERIALS AND METHODS
Study Design
The retrospective analysis of archived tissue blocks was performed using thirty two cancerous and 32 non-cancerous core biopsies fixed in 10% neutral buffered formalin and embedded in paraffin. All blocks were retrieved from the pathology archives of the Veterans Affairs Hospital at Omaha (VAHO). Non-cancerous biopsies were obtained from patients who underwent initial biopsy because of suspect cancer and were biopsy negative. Patients without clinical PC (controls) had no evidence (stable PSA or negative repeat biopsy) of clinical PC for at least 5 years (5–14years) following the initial biopsy. Only patients that were negative for malignancy on a rebiopsy or had a stable PSA level remained in the control cohort. Control prostate core biopsies were random with respect to non-cancerous prostate conditions, i.e., no effort was made to select a particular non-cancerous condition, e.g., prostatitis or benign prostatic hyperplasia (BPH). Control patients were retrospectively matched one-to-one with cancer patients on the basis of age (± 5 years) and year of biopsy (mainly the same year but not more than 3 years), yielding thirty-two matched biopsy pairs. All procedures were in compliance with human studies protocols approved by the Institutional Review Boards (IRB) of the VAHO and the University of Nebraska Medical Center (UNMC).
Slides were prepared with 4-micron sections of each paraffin block containing 3–6 cores and then one slide per block was stained with hematoxylin and eosin (H&E). Two pathologists (S.L. and D.H.) evaluated the H&E-stained slide specimens to determine suitability of each specimen for the study.37 Clinical and demographic data, excluding patient identifiers, were obtained from patient records and associated with each anonymous specimen (Table 1).
Table 1.
Summary of Clinical Data for Prostate Cancer Cases and Controls
| Control cases* | Cancer cases | |
|---|---|---|
| Number of cases/controls | 32 | 32 |
| Mean age at biopsy (years) | 66.5 | 66 |
| Age range at biopsy (years) | 55– 77 | 57 – 77 |
| Year of biopsy (range) | 1992 – 1998 | 1991 – 1997 |
| PSA (ng/ml) at time of biopsy (range) | 0.8 – 13.7 | 1.3 – 70.5 |
| Gleason Score (range) | — | 2 – 9 |
| 2–4 (number) | — | 4 |
| 5–7 (number) | — | 22 |
| 8–10 (number) | — | 6 |
Glandular hyperplasia or no noted pathology defined on a biopsy.
Prostatectomy specimens of consented subjects
In compliance with IRB-approved protocols, tissue slices (200–300 mg) were collected from surgically removed prostate glands of consented patients, at UNMC and the VAHO. Specimens collected from two benign hyperplastic (BH) glands characterized by glandular hyperplasia and three cancerous (CA) glands were processed according to standard procedures established by the research laboratory.10 Specimens were fixed with 4% EM grade formaldehyde (Polysciences, Inc., Warrington, PA) or methacarn fixative, embedded in paraffin, sectioned at 4 microns, and mounted to glass slides. Formaldehyde is conventionally used as a fixative for archiving clinical samples. However, methacarn fixative introduces fewer cross-links in tissue protein, allowing for better extraction and less alteration of protein from tissue specimens,3,24 so it is more compatible with reverse-phase protein array analysis (RPPA). Since our validation of UGDH quantification required use of RPPA, we compared QFIA data from the same tissue fixed with each fixative. A tissue microarray (TMA) prepared with 1 mm cores from formaldehyde-fixed and methacarn-fixed tissue slices included triplicate cores from glandular areas of the two BH glands and normal appearing areas and cancerous areas of the three CA glands. Slide specimens of a formaldehyde-fixed slice from one BH gland were used to validate QFIA measurements of UGDH in slide specimens, by parallel RPPA analyses of proteins extracted from adjacent tissue sections.
PC3 cells
PC3 cells (American Type Culture Collection, Manassas, VA) cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum were harvested at laboratory passage 10–15, fixed in 0.5% EM grade formaldehyde in phosphate-buffered saline (Polysciences, Inc.), and imprinted to glass slides, as previously described.10
Development and Specificity of the Rabbit Anti-UGDH antibodies
Antibodies against human UGDH were developed as previously described.33 Briefly, recombinant UGDH expressed in Escherichia coli was purified from soluble lysates by nickel-NTA chromatography and used to raise polyclonal antibodies in New Zealand white rabbits (performed by Covance Research, Inc.). Sera were ammonium sulfate fractionated before use in experiments. The optimal dilution of the resultant anti-UGDH antibody was determined by titration using standard prostate tissue section slides from a non-cancerous prostate and an established QFIA standard procedure. The dilution (1:500) at which fluorescence yielded a saturation plateau was considered optimal.
Western blot analysis
Cultured 22Rv1 human prostate carcinoma cells (ATCC), cultured in RPMI containing 10% FBS, as recommended by the vendor, were washed with PBS and treated for 30 minutes on ice, with lysis buffer [50 mM Tris, 150 mM NaCl, and 1% (v/v) Nonidet P-40; pH 8.0].
The lysate was centrifuged to remove insoluble material. Ten μL of lysate (10 μg protein) and six-histidine tagged recombinant UGDH antigen (rUGDH; 10 ng) were loaded onto the stacking gel. Proteins were fractionated on a 12% SDS-polyacrylamide gel and blotted to a PVDF membrane (Immobilon™-P; Sigma-Aldrich). The blot was treated for one hour at room temperature with blocking buffer [20 mM Tris, 137 mM NaCl, 5% (wt/vol) dried milk, 0.05% (v/v) Tween 20, and 0.05% (v/v) Nonidet P-40; pH 8.0] and then incubated overnight at 4° C with anti-UGDH antiserum (1/5000 dilution). Rabbit Ab were incubated with peroxidase-conjugated goat anti-rabbit IgG (Amersham Pharamacia Biotech) for one hour and a chemiluminescence signal was generated with the SuperSignal Ultra detection system (Pierce Chemical Co.).
Competitive inhibition of tissue labeling with recombinant UGDH
Increasing concentrations (0, 10−9, 10−8, 10−7, and 10−6 M) of recombinant UGDH were incubated for two hours with an optimum dilution (1/500) of anti-UGDH or pre-immune rabbit serum in an antibody diluent (Invitrogen Corp., Carlsbad, CA 92008). Subsequently, the mixtures were applied to duplicate slide specimens of a BH gland and the slides were incubated overnight at 4° C, washed and treated with Alexa Fluor® conjugated goat anti-rabbit IgG, mounted in ProLong Gold® antifade medium, and sealed with lacquer. Ab-dependent signal in prostate epithelium, corrected for the corresponding Ab-independent signal produced by treatment with pre-immune serum, was quantified by QFIA (below).
Reverse-Phase Protein Array Analysis (RPPA)
Slide specimens to be extracted were stained with Mayer’s hematoxylin and reviewed by light microscopy for structural integrity. The entire section was shaved off the glass slide, with a single-edge razor blade, and deposited into 200 uL of extraction buffer consisting of a 1:1 mixture of 2x Tris-Glycine SDS sample buffer and Tissue Protein Extraction Reagent (TPER; Pierce Biotechnology, Rockford, IL) plus 2.5% β-mercaptoethanol. The sample was heated for two hours at 70° C, sonicated for ten minutes and heated at 95° C for eight minutes. The sample was centrifuged for five minutes at maximum speed and the supernatant was stored at 5° C for a maximum of 48 hours, prior to spotting onto nitrocellulose slides.
Analysis of specimen extracts was implemented as previously described. Serial two-fold dilutions of each extract and human recombinant UGDH starting from (10 ng/100 μL) were transferred to a 384-well polypropylene plate. Samples were arrayed to nitrocellulose glass slides (LI-COR Biosciences, Lincoln, NE) with an 8-pin arrayer (VP478) according to the instructions of the manufacturer (V&P Scientific, Inc., San Diego, CA). Arrayed nitrocellulose slides were placed in a light-tight box that contained desiccant and stored at −20° C for no more than five days prior to protein quantification.
For protein labeling, arrays were treated with Re-Blot™ antibody stripping solution (Chemicon, Temecula, CA) for 15 min at room temperature, washed in PBS and then incubated overnight in blocking solution (1.2 g I-Block™ [Tropix, Bedford, MA] and 600 μL Tween® 20 [Sigma, St. Louis, MO] dissolved in 600 mL Dulbecco’s PBS) at 4° C. Duplicate arrays were treated with anti-UGDH or pre-immune control serum (both at 1:500) for 2 hours at room temperature and then washed twice for 10 min with TBS-T solution. Subsequently, the slides were treated for 30 min (in the dark, at room temperature) with IRDye™ 800 CW-conjugated goat anti-rabbit IgG (LI-COR Biosciences, Lincoln, NE) at a dilution of 1:2500 and then washed with TBS-T and air-dried.
Slides were scanned with the Odyssey infrared imaging system (LI-COR) at a resolution of 84 μm, a sensitivity of 7.0 and a background setting of ‘medium’. The images were analyzed with the Odyssey software. Integral fluorescence intensity, corrected for background, was determined for each spot and the mean of each set of triplicate spots was determined.
Quantitative Fluorescence Imaging Analysis (QFIA)
Fluorescence labeling of slide-mounted tissues and imprinted PC3 cells
Slide specimens of paraffin-embedded tissue slices and core biopsies were deparaffinized with EZ-AR Common™ (BioGenex Laboratories, San Ramon, CA) and treated with EZ-AR1™ (BioGenex Laboratories) for epitope recovery, according to manufacturer specifications. Fluorescence labeling was implemented with a computer-controlled autostainer (BioGenex i6000™) as previously described.10 Briefly, moist slides were soaked in Super Sensitive Wash Buffer™ (BioGenex, San Ramon, CA) for 15 min, blocked with 10% goat serum (Zymed Laboratories, Inc., San Francisco, CA) for 20 min, treated overnight at 4°C with an epitope-saturating dilution of anti-UGDH (1:500) and then labeled at room temperature for one hour with Alexa Fluor® 568 conjugated goat anti-rabbit IgG (200 μg/mL; Molecular Probes, Inc., Eugene, OR). Controls were treated with pre-immune serum. Labeled specimens were mounted with ProLong Gold® anti-fade medium (Molecular Probes/Invitrogen, Eugene, OR), sealed with clear lacquer and stored at −20°C. PC3 cells were labeled with the same procedures, except that epitope recovery was not required.
Since it was not possible to label all core biopsy specimens of the 32 matched pairs of cases and controls in one run, we implemented a design to minimize run-to-run variations and their effects on measurement of UGDH expression.10 The first run included duplicate slides of seven cases and matched controls, six slide specimens each of the tissue standard and PC3 cells. One slide of each duplicate was treated with anti-UGDH serum and the second with pre-immune serum. The second run was a replicate of the first, yielding two slides each of antibody-treated and control slides for each cell and tissue specimen. This cycle was repeated with some variation in the number of matched cases and controls, until all core biopsies were labeled.
Fluorescence imaging and image capture
Fluorescence images were captured with a Leica epifluorescence microscope as previously described.10 The Leica microscope (DMRXA2; North Central Instruments, Plymouth, MN) was equipped with a mercury/xenon excitation lamp (Model E7536; Hamamatsu Corp., Bridgewater, NJ) and digital b/w CCD camera (Model ORCA-C4742-95-12ER; Hamamatsu Corp.). We captured twelve-bit grayscale images with Image-Pro® Plus software (MediaCybernetics, Inc., Bethesda, MD), using a 10x objective and a Texas Red filter set (Chroma 41004). Each image capture session was initiated with a protocol to confirm consistent system performance across analytical runs.10 Briefly, three slides were prepared with suspensions of InSpeck™ fluorescent microspheres. Grayscale images of 200–400 microspheres per slide were captured and the mean pixel intensity (MPI) of each was determined with Image-Pro® Plus software. Mean and standard deviation of the MPIs for spheres from each suspension were computed and compared to the values of previous image capture sessions.
Digital images of labeled tissue specimens and the corresponding isotype controls were captured no more than three days after the slides were processed with the BioGenex autostainer. Camera frames were selected to maximize the number of events (single cells, acini or epithelial strips) captured. In addition, frames of cancerous cores were selected to capture both cancerous acini/epithelium (CA) and normal appearing acini (NAA). Categories of acini/epithelium included CA, NAA, and non-cancerous acini (NA) of control cores and the tissue standard. Typically, 30–300 acini per slide were captured in each category and 200–400 single cells were captured from each slide preparation of the PC3 cells. Images were stored as 12-bit grayscale files. Labeled specimens matched to those specimens analyzed by RPPA were treated differently. Contiguous, non-overlapping images of antibody-labeled specimens and isotype controls were captured with the 4x objective to encompass the entire tissue section.
Quantification of fluorescence signal in captured images
Images were partitioned into relevant events (e.g.; prostatic acini or single cells) and fluorescence signal was quantified with the Image-Pro® Plus software.10 Partitioning of prostate epithelium was achieved by setting a lower threshold just above background emission, typically ca. 230–250 grayscale units (gsu), and an upper threshold at the upper limit of the 12-bit grayscale, i.e., 4095 gsu. Image-Pro® Plus generated an array of measurements that included event number, area, mean pixel intensity (MPI), sum of pixel intensities, and maximum and minimum pixel intensity. The algorithm for event MPI excluded the background pixels of each partitioned event.
Quantification of the UGDH signal in slide specimens matched to those analyzed by RPPA did not entail partitioning individual acini. The sum of pixel intensities corrected for background was determined for the entire tissue section for comparison to total UGDH content of the paired tissue section as determined by RPPA.
Data Analysis
UGDH expression under different analytical conditions
Concordance of UGDH expression in methacarn-fixed tissues analyzed by RPPA and QFIA, and in methacarn-fixed and formaldehyde-fixed tissues analyzed by QFIA was evaluated by linear regression analysis. The correlation coefficient (r value) was calculated as an indicator of the strength of the linear relationship.
UGDH expression in archived core biopsies and PC3 cells
MPIs of labeled acini/epithelium and single cells were corrected for non-specific immunoglobulin labeling and background emission by subtracting the average MPI (AMPI) of the corresponding isotype control (typically 250–300 gsu). Arithmetic average, standard deviation, and standard error were computed for each type of event (single cell, acinus or epithelial strip). Differences between the AMPIs of acinar categories (NA, NAA, CA) were evaluated by a paired t-test (PlotIT® software, Scientific Programming Enterprises, Haslett, MI) for matched pairs and unpaired t-test (GraphPadInstat, GraphPad Software, Inc., San Diego CA) for unmatched cohorts.
Receiver operating characteristic (ROC) curves were generated by GraphPad Prism, version 4 (GraphPad Software) for case-associated AMPIs of CA or NAA in relation to control-associated AMPIs of NA, plotted as a function of increasing threshold AMPI. Fractional area-under-curve (AUC) along with 95% confidence intervals were determined for each plot. An AUC ≥ 0.67 indicated significant discrimination of cases (NAA or CA) and controls (NA). Sensitivities and specificities and their 95% confidence intervals were determined at selected threshold values. Bootstrapping was used to estimate confidence intervals for the AUC and for the sensitivity and specificity at selected threshold values, taking into consideration the repeated measures design of the tests (two slides per person). Two-hundred bootstrapped samples were used to estimate the confidence intervals, re-sampling at the person level within a group (NA, NAA, and CA) to preserve the correlation structure from two slides within a person.
RESULTS
Specificity and Reproducibility of the Analysis of UGDH in Single Cells and in Prostate Tissue Sections, by QFIA
The specificity of the fluorescence signal produced by the rabbit antibody (Ab) against UGDH was determined by both Western analysis of a cell lysate and competitive blockade of the Ab-dependent signal produced in tissue sections. The Ab recognized a single band in whole-cell lysate prepared from a human prostate adenocarcinoma cell line (22Rv1) (Figure 1a). The molecular mass was the same as that of purified recombinant UGDH (rUGDH) adjusted for its epitope tag. In cancerous prostate tissue sections, the Ab produced a bright signal that was progressively inhibited with increasing concentrations of rUGDH (Figure 1b). Inhibition was nearly complete by the addition of 10−7 M rUGDH to the Ab solution.
Figure 1. Specificity of rabbit antiserum prepared against UGDH.
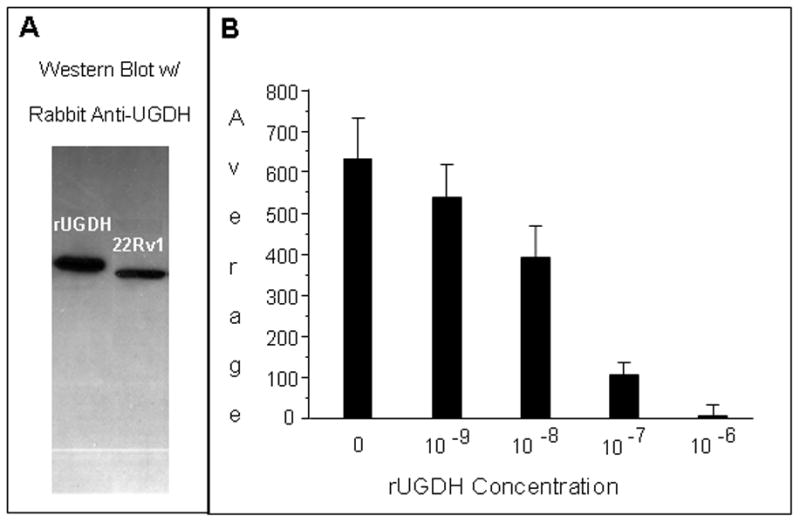
(A) A Western Blot of whole cell lysate of 22Rv1 prostate cancer cells and purified recombinant UGDH (rUGDH) was probed with rabbit anti-UGDH serum, yielding a single band. (B) Slide specimens of a benign hyperplastic prostate gland were probed with anti-UGDH serum alone or in combination with the indicated concentrations of rUGDH. After secondary labeling with Alexa Fluor-conjugated goat anti-rabbit IgG antibodies, fluorescence images were captured, and prostate acini were partitioned. Background-corrected MPI was determined for each acinus and average MPI was calculated for 50 to 70 acini per slide specimen.
As a requirement for reproducible analysis, performance of the imaging system was evaluated at the start of each image acquisition session with standard fluorescent microspheres. The AMPI of 200 to 400 microspheres per slide was determined with three slide preparations of microspheres. Fluorescence emission of the standard microspheres varied less than 5% among the image capture sessions required for this study, as exemplified by the results of four of these system evaluation routines (Figure 2).
Figure 2. Stability of the imaging system and reproducibility of UGDH quantification across analytical runs.
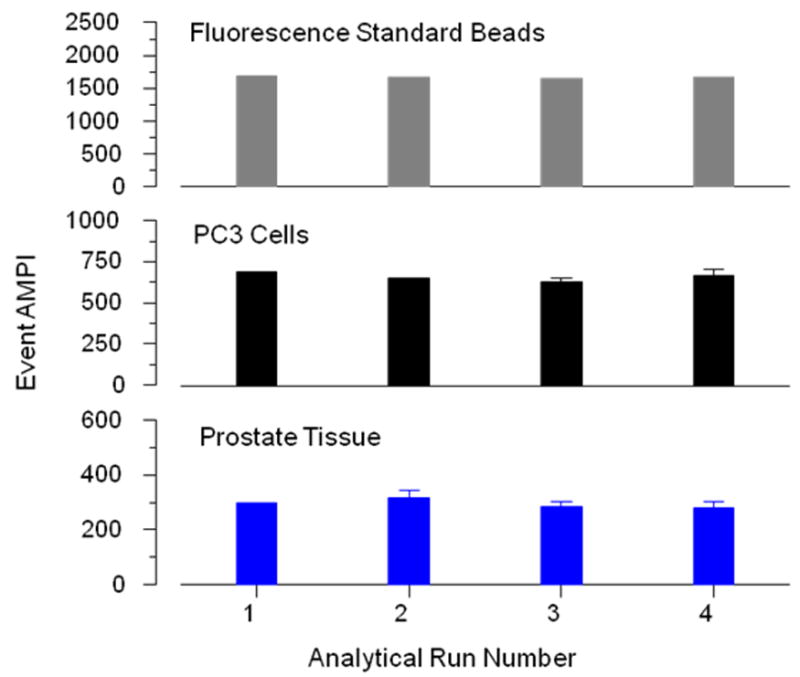
System stability was determined by quantifying the fluorescence emission of standard microspheres in a set of three slides; prior to each imaging/image capture session. Fluorescence was corrected for background and expressed as AMPI of 200–300 microspheres. AMPI are given in grayscale units (gsu) on a 12-bit scale (0–4095 units). The data of four representative analytical runs are presented. Six slide specimens of standard PC3 cells and two slide specimens of a benign prostate gland were incorporated into each analytical run. Both standards were probed with rabbit anti-UGDH anti-serum and labeled with Alexa Fluor 568-conjugated goat anti-rabbit IgG. Background corrected fluorescence emission was expressed as AMPI of 400–700 single cells and 100–200 prostatic acini, per slide. Each bar represents the mean of the AMPI of three slides (microspheres), six slides (PC3 cells) or two slides (prostate tissue). Data of four representative analytical runs are presented. The standard errors for each set of three microsphere slides are 1, 10, 2, and 12 gsu; each set of six PC3 slides are 13, 19, 29 and 32 gsu; and each set of two prostate tissue slides are 15, 25, 18 and 24 gsu, for analytical run 1, 2, 3, and 4, respectively.
Reproducibility of the UGDH measurements within and across analytical runs was determined with PC3 cells (six slides per run) and prostate tissue standards (two slides per run) incorporated into each analytical run. Negative controls were included for background correction. Representative data from four analytical runs are presented in Figure 2. Within each run, the AMPIs of individual slides differed from the means of replicate slides by an average of 7% for PC3 cells and 8% for prostatic acini. Across runs, the means of replicate slides differed from the mean of the AMPIs of all slides by an average of 5% for PC3 cells and 8% for acini.
Validation of Quantitative Fluorescence Imaging Analysis (QFIA) of UGDH in Prostate Specimens by Reverse-Phase Protein Array Analysis (RPPA)
Correlation of UGDH content in slide specimens of methacarn-fixed, paraffin-embedded prostate tissues, determined by both QFIA and RPPA
The study design required parallel analyses of adjacent serial sections, minimizing differences in protein expression between the slide specimens to be analyzed. RPPA was selected for validation of QFIA because it is an established method for quantification of proteins in laser-dissected slide specimens and can be applied to individual slide specimens.34,35 Since our goal was to analyze UGDH in paraffin-embedded, archived specimens, we implemented this stage of validation with methacarn-fixed, paraffin-embedded prostate specimens. Proteins may be extracted, by standard extraction procedures, with high efficiency from tissues fixed with methacarn, unlike tissues fixed with the crosslinking fixative formaldehyde.36,37 Most of the UGDH signal was seen in the prostate epithelium; consequently, we determined total UGDH signal in paired slide specimens of each tissue sample.
Concordance of the UGDH measurements determined by RPPA and QFIA of slide-mounted sections was evaluated with specimens from two BH glands and three cancerous glands, previously screened by QFIA and selected to provide a wide range of UGDH expression. Consistency of the protein arraying procedure is apparent in the infrared scans of the UGDH standard and extracts of the slide specimens (Figure 3A), which show similar fluorescence intensity and spot size among triplicate spots. For all specimens, integrated fluorescence was a linear function of extract dilution (Figure 3B), permitting quantitative comparisons of UGDH measurements obtained by the two analytical methods. Integrated fluorescence intensity of each tissue section, determined by QFIA, was plotted as a function of integrated intensity of the paired tissue section, determined by RPPA (Figure 4A). The data exhibited a linear relationship with a correlation coefficient of 0.97, indicating that the QFIA measurements were directly proportional to the UGDH content of the tissue specimens.
Figure 3. Reverse-Phase Protein Array analysis of UGDH in slide specimens of prostate tissue.
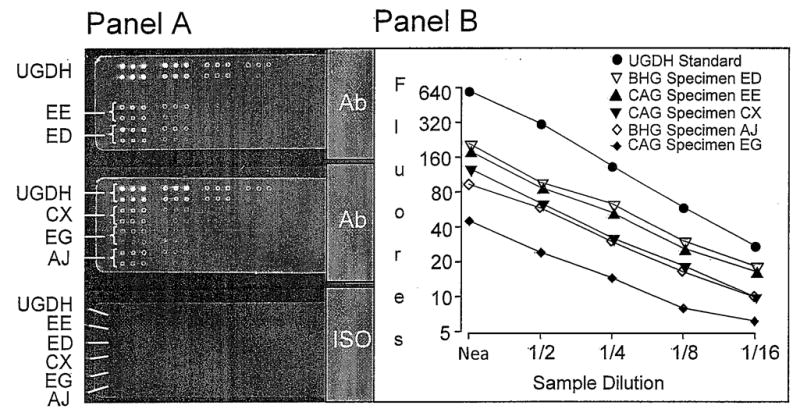
Slices of two benign hyperplastic (BHG; specimens ED and AJ) and three cancerous (CAG; specimens EE, CX, and EG) glands were fixed in methacarn, embedded in paraffin and sectioned at four microns. One slide specimen of each gland was shaved from its slide and deposited in extraction buffer. Panel A. Serial two-fold dilutions of the extracts and human rUGDH standard (10 μg/100 μL) were spotted (40–50 nL/spot) in triplicate to nitrocellulose slides that were probed with anti-UGDH antiserum (Ab) and labeled with IRDye 800 CW conjugated goat anti-rabbit IgG. Slides were scanned with an infrared imaging system. Each specimen (EE, ED, CX, EG, AJ) and recombinant UGDH is represented by two rows of spots. Top row, dilutions of 1/1, 1/4, 1/16, and 1/64; bottom row, dilutions of 1/2, 1/8, 1/32, and 1/128. A control slide (ISO), probed with pre-immune rabbit serum and labeled with IRDye 800 CW conjugated secondary Ab, served as a background control. Panel B. The background-corrected fluorescence of each spot was expressed as integrated fluorescence intensity and plotted against sample dilution.
Figure 4. Validation of QFIA for expression profiling of UGDH in archived prostate specimens.
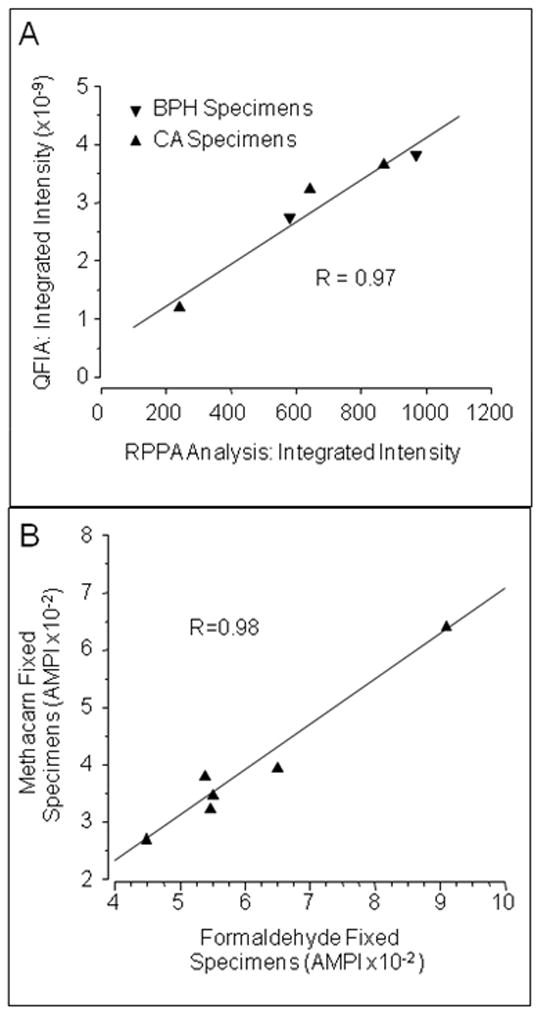
Panel A. Tissue slices from two cancerous (CA) and three benign hyperplastic (BPH) prostate glands were fixed in methacarn, embedded in paraffin and sectioned at four microns. UGDH expression in adjacent serial sections was determined, in parallel, by QFIA and RPPA and expressed as integrated fluorescence intensity. Measurements by these two methods exhibited a linear correlation with an R-value of 0.97. Panel B. Tissue slices from cancerous and non-cancerous glands were fixed in 4% formaldehyde or in methacarn, embedded in paraffin and incorporated into a tissue microarray that was sectioned at four microns. UGDH in normal prostatic acini of two BH glands, normal-appearing acini of one cancer bearing gland and cancerous acini of three cancerous glands was quantified by QFIA and expressed as AMPI. Measurements of AMPI in the acini of methacarn-fixed and formaldehyde-fixed tissue slices exhibited a linear correlation with an R-value of 0.98.
Correlation of QFIA measurements of UGDH in acini/epithelium of prostate specimens fixed in methacarn or formaldehyde
Concordance of QFIA and RPPA data was evaluated with methacarn-fixed slide specimens, because proteins may be extracted by standard extraction methods, with high yield.36 More complex procedures are required for lower-yield extractions of proteins from tissues fixed with the cross-linking fixative formaldehyde without additional specific procedures.38 Since we were interested in applying QFIA to archived tissues routinely fixed with formaldehyde, it was necessary to evaluate the concordance of QFIA measurements of UGDH in methacarn-fixed and formaldehyde-fixed tissues. We used a tissue microarray (TMA) that incorporated both methacarn- and formaldehyde-fixed specimens from the same surgically removed cancerous and non-cancerous prostate glands. The TMA permitted a direct comparison of expression in the NA of three BH glands, the NAA of two cancer-bearing glands, and the CA of one cancer-bearing gland. Prostate acinar epithelium was analyzed for this study. The AMPIs of acinar epithelium in tissues fixed with formaldehyde were plotted as a function of the AMPIs of acinar epithelium in tissues fixed with methacarn, and the correlation coefficient was calculated (Figure 4B). AMPIs of formaldehyde-fixed and methacarn-fixed tissues exhibited a linear relationship with a correlation coefficient of 0.98. We concluded that QFIA of UGDH in slide specimens produced measurements that were directly proportional to UGDH expression, as determined by RPPA validation, and that QFIA may be applied to quantification of the protein in archived prostate specimens fixed in a formaldehyde-based fixative, as determined by parallel QFIA of methacarn- and formaldehyde-fixed tissues.
Application of QFIA to an Evaluation of UGDH Expression in Archived Prostate Biopsies
The preceding work provided the foundation for the application of QFIA to expression profiling of UGDH in acinar epithelium of archived prostate biopsies. UGDH was analyzed in a cross-sectional case-control study that included 32 cancer cases and 32 controls matched on the basis of age (±5 years) and year of biopsy (±3 years). Clinical and demographic data are summarized in Table 1. AMPIs corrected for background fluorescence are presented in Table 2. Expression in CA compared to NA was significantly higher (P<0.01) in 22 of the 32 matched pairs of cases and controls.
Table 2.
UGDH expression in the acini/epithelial cells of biopsy specimens
| # Average of corrected MPI ± STDEV | Δ p values | |||||
|---|---|---|---|---|---|---|
| Prostate Cancer Cases |
||||||
| Patient prostate core biopsies in matched pairs | Non Prostate Cancer Control Cases Normal Acini (NA) | Cancerous Acini (CA) | Normal Appearing Acini (NAA) | ¥ NA vs. CA | Φ CA vs. NAA | † NA vs. NAA |
| 1 | 458.2 ± 33.5 | 445.5 ± 137.6 | 606.0 ± 358.6 | >0.05 | <0.01 | <0.01 |
| 2 | 479.5 ± 67.2 | 352.4 ± 13.3 | 446.1 ± 24.1 | <0.01 | <0.01 | >0.05 |
| 3 | 470.7 ± 86.2 | 744.2 ± 26.6 | 487.2 ± 52.2 | <0.01 | <0.01 | >0.05 |
| 4 | 453.0 ± 142.3 | 880.8 ± 43.8 | 424.15 ± 75.0 | <0.01 | <0.01 | <0.01 |
| 5 | 605.3 ± 0.7 | 684.4 ± 61.2 | 449.0 ± 37.2 | <0.01 | <0.01 | <0.01 |
| 6 | 451.0 ± 5.3 | 845.1 ± 179.7 | 588.6 ± 35.7 | <0.01 | <0.01 | <0.01 |
| 7 | 737.2 ± 49.4 | 414.0 ± 25.2 | 618.4 ± 79.3 | <0.01 | <0.01 | <0.01 |
| 8 | 779.3 ± 10.8 | 705.6 ± 163.3 | 348.3 ± 75.1 | >0.05 | <0.01 | <0.01 |
| 9 | 574.1 ± 4.0 | 876.1 ± 43.4 | 660.2 ± 155.0 | <0.01 | <0.01 | 0.02 |
| 10 | 633.1 ± 50.2 | 653.2 ± 7.5 | 435.8 ± 17.0 | >0.05 | <0.01 | <0.01 |
| 11 | 430.7 ± 114.7 | 843 ± 43.4 | 488.9 ± 79.3 | <0.01 | <0.01 | <0.01 |
| 12 | 451.2 ± 10.1 | 552.6 ± 6.1 | 293.5 ± 14.9 | <0.01 | <0.01 | <0.01 |
| 13 | 548.6 ± 61.4 | 611.1 ± 55.0 | 490.3 ± 46.9 | <0.01 | <0.01 | >0.05 |
| 14 | 568.5 ± 4.3 | 1113.0 ± 30.9 | 563.7 ± 89.8 | <0.01 | <0.01 | >0.05 |
| 15 | 498.3 ± 71.6 | 735.5 ± 150.7 | 436.3 ± 121.9 | <0.01 | <0.01 | <0.01 |
| 16 | 512.1 ± 9.0 | 452.1 ± 2.0 | 292.5 ± 0.4 | <0.01 | <0.01 | <0.01 |
| 17 | 605.4 ± 35.3 | 909.3 ± 15.2 | 571.0 ± 11.4 | <0.01 | <0.01 | >0.05 |
| 18 | 591.5 ± 104.4 | 718.1 ± 99.6 | 392.3 ± 5.3 | <0.01 | <0.01 | <0.01 |
| 19 | 636.5 ± 105.6 | 536.15 ± 60.5 | 409.4 ± 74.0 | <0.01 | <0.01 | <0.01 |
| 20 | 392.2 ± 23.3 | 503.7 ± 88.9 | 485.0 ± 113.4 | <0.01 | >0.05 | <0.01 |
| 21 | 469.6 ± 8.1 | 742.2 ± 42.1 | 565.8 ± 22.9 | <0.01 | <0.01 | <0.01 |
| 22 | 762.5 ± 92.7 | 961.0 ± 22.1 | 447.0 ± 93.6 | <0.01 | <0.01 | <0.01 |
| 23 | 424.9 ± 12.8 | 813.5 ± 29.3 | 474.5 ± 76.5 | <0.01 | <0.01 | <0.01 |
| 24 | 676.1 ± 19.8 | 407.7 ± 47.2 | 470.2 ± 40.1 | <0.01 | <0.01 | <0.01 |
| 25 | 598.4 ± 75.6 | 561.0 ± 86.3 | 407.4 ± 34.6 | >0.05 | <0.01 | <0.01 |
| 26 | 571.6 ± 89.4 | 475.8 ± 28.1 | 272.5 ± 35.0 | <0.01 | <0.01 | <0.01 |
| 27 | 422.4 ± 42.8 | 661.3 ± 4.9 | 392.6 ± 4.6 | <0.01 | <0.01 | >0.05 |
| 28 | 557.8 ± 60.7 | 647.4 ± 14.2 | 447.6 ± 61.7 | <0.01 | <0.01 | <0.01 |
| 29 | 447.7 ± 43.6 | 633.2 ± 15.4 | 501.7 ± 1.8 | <0.01 | <0.01 | <0.01 |
| 30 | 578.3 ± 15.3 | 448.5 ± 42.4 | 475.4 ± 37.1 | <0.01 | <0.01 | <0.01 |
| 31 | 552.9 ± 82.9 | 792.5 ± 45.4 | 463.3 ± 58.2 | <0.01 | <0.01 | <0.01 |
| 32 | 478.6 ± 41.8 | 570.9 ± 281.3 | 439.3 ± 1.0 | <0.01 | <0.01 | <0.01 |
Significance of differences was determined by a two-tailed Student’s t-test between two samples with unequal variance
Average of the background corrected Mean pixel intensity (MPI) of UGDH labeling in individual acini
Cancerous acini (CA) compared to non-cancerous acini (NA) of the matched controls
Normal appearing acini (NAA) in cancerous glands compared to cancerous acini (CA) of the same gland
NAA in cancerous glands compared to non-cancerous acini (NA) of the matched controls
The mean ± standard error of the AMPIs of CA for the 32 cases was 665 ± 32 compared with 544 ± 18 (P<0.003), respectively, for NA of the matched controls. Surprisingly, UGDH expression in NAA relative to NA was significantly lower. The mean ± standard error of the AMPIs of NAA for all cases was 464 ± 16 (P<0.003 versus NA of matched controls).” Grayscale images acquired from a representative pair of patients depict these differences in AMPIs of NA, NAA and CA (CA>NA>NAA) and identify acinar epithelial cells as the main site of UGDH expression (Figure 5A). Discrimination of cancer cases and non-cancer controls, on the basis of UGDH expression in prostatic acini, was visualized with cumulative frequency plots of AMPIs of the NA of each control and the CA and NAA of each case. The percent of individuals at or below designated AMPI values was plotted as a function of AMPI (Figure 5B). The frequency plots were well separated and illustrated quantitative differences in UGDH expression. To quantify the observed discrimination of cases and controls, we analyzed ROC plots of the UGDH expression data. First, we compared cases and controls on the basis of UGDH expression in CA and NA of the matched pairs (Figure 6, plot A). At a cut point of 700 gsu, UGDH expression classified 42% (95% CI, 23–61%) of cancer cases and only 9% (95% CI, 0–19%) of controls as cancer positive. Then, we compared cases and controls on the basis of UGDH expression in the NAA and NA of the matched pairs (Figure 6, plot B). At a cut point of 410 gsu, UGDH classified 35% (95% CI, 17–52%) of cancer cases and only 9% (95% CI, 0–20%) of controls as cancer positive. These data support the application of QFIA to archived biopsies for protein expression profiling and identification of candidate biomarkers of disease.
Figure 5. UGDH expression determined by QFIA of archived prostate biopsies is significantly different among three classes of prostatic acini; normal acini (NA) of non-cancerous glands and normal appearing acini (NAA) and cancerous acini/epithelium (CA) of cancerous glands.
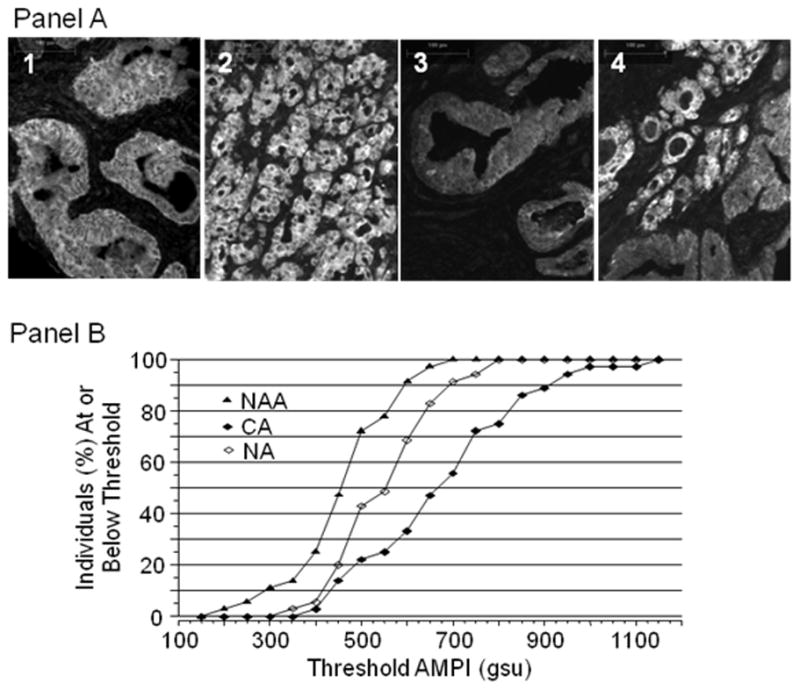
Panel A. Grayscale images of prostatic acini labeled with anti-UGDH anti-serum and Alexa Fluor 568 conjugated goat anti-rabbit IgG were captured with a 10x objective. Normal acini (image 1) exhibited fluorescence intensity intermediate between cancerous acini (image 2) and normal appearing acini (image 3). Both cancerous and normal appearing acini were captured in image 4. Panel B. UGDH was quantified in the CA and NAA of biopsies from 32 prostate cancer patients and in the NA of biopsies from 32 age-matched controls without clinical cancer. The average of the background-corrected mean pixel intensity (AMPI) for acini/epithelium within each class was determined for each case and control. The percentage of cases or controls at or below the selected threshold AMPIs was plotted for each class of acinus/epithelium.
Figure 6. Receiver operating characteristic plots of UGDH expression determined as AMPI of the CA and NAA of cancer patients, in relation to UGDH expression in the NA of the matched controls.
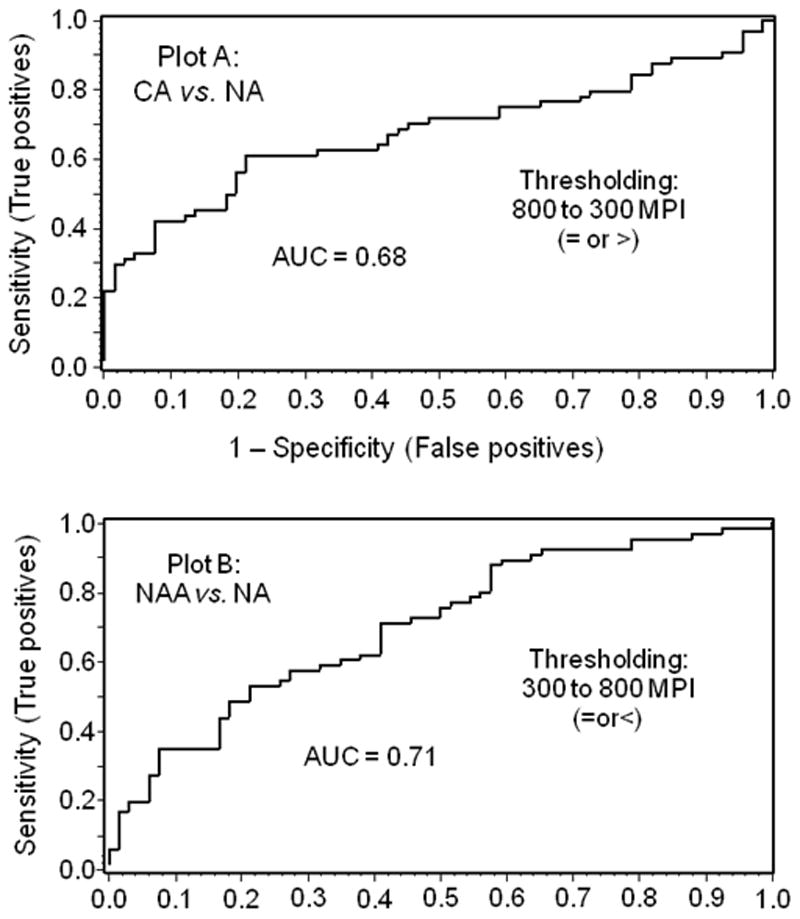
The fraction of controls with NA values at or below selected AMPIs and that of cases with NAA values at or below the same AMPIs were plotted as 1-specificity and sensitivity, respectively (Plot A). The fraction of controls with NA values at or above selected AMPIs and that of cases with CA at or above the same AMPIs were plotted as 1-specificity and sensitivity, respectively (Plot B).
The range of PSA values of the cancer patients was higher (1.3 to 70.5 ng/mL) than that (0.8 to 13.7 ng/mL) of control patients. To further evaluate the ability of UGDH expression to differentiate CA or NAA from NA, we compared the group means of UGDH fluorescence intensity (AMPI) for PC (N=21) and control (N=32) patients with similar range of PSA values (< 14.0 ng/mL). Mean values for CA and NAA were significantly different (P<0.003) from the mean for NA (Figure 7). Interestingly, the means ± standard errors for NAA and CA (445.1 ± 17.9 and 651.8 ± 42.8 gsu, respectively) in PC patients from these groups were similar to values obtained for all PC patients (463.8 ± 16.3 and 665.3 ± 32.5 gsu for NAA and CA, respectively, N=32), possessing similar differences (P<0.003) from the mean for NA. We conclude that the ability of UGDH expression to differentiate CA or NAA from NA was independent of the PSA values. Differences between CA and NA in UGDH expression also remained similar when cancer cases were subdivided to low grade (Gleason score 3–6) and high grade (Gleason score 7–9) cases demonstrating independence from the grade of alteration in UGDH expression in CA vs. NAA (data not shown). Taken collectively, the data demonstrate (1) increased UGDH expression in the CA and decreased expression in the NAA of the same prostate relative to the NA of non-cancerous prostates, and (2) the ability of QFIA to quantify UGDH within acini of the non-malignant field in a biopsy specimen with a potential to improve diagnosis of PC.
Figure 7. Comparative analysis of UGDH expression in normal acini (NA), cancerous acini (CA) and normal appearing acini (NAA) of specimens grouped by patient PSA level.
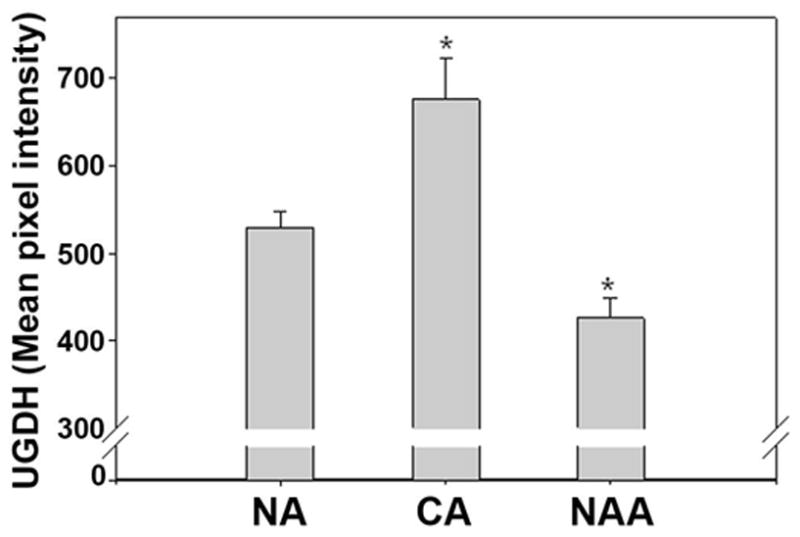
Averages (AMPI) and standard errors were compared to evaluate correlation between UGDH expression in NA, CA and NAA obtained from 28 control patients (NA) and a cohort of patients (n=21) with PC selected for a similar range of PSA (<14 ng/ml). *P<0.003 vs. NA.
DISCUSSION
Quantification of heterogeneous proteins in prostate tissues in the context of a tumor has significant application to studies of human pathogenesis and molecular epidemiology. In a previous study, β-catenin was quantified by QFIA within compartments of prostate tissue biopsy specimens.10 In the premalignant field, β-catenin was a strong biomarker for prostate cancer. The objective of the current study was to quantify UGDH as a second potential biomarker for detecting individuals at risk. The advantage of QFIA is the ability to precisely quantify protein profiles in small human biopsy specimens using fluorescently-labeled antibodies without laser microdissection.10 Although antibodies have been widely used in flow cytometry and conventional immunohistochemistry for decades, recent engineering advances clearly demonstrated the power of imaging technology in combination with quantitative immunohistochemistry.10 Rao, et.al,39–41 and Rimm,42,43 et.al. have confirmed the development of a quantitative fluorescence method (AQUA) for detecting biomarkers in tissue microarrays. Additional instrumentation has been developed to promote the rapid development of similar methodological approaches.44,45 This report exemplifies some of the technical aspects of QFIA validation and focuses on UGDH as a field disease/effect marker in prostate cancer biopsies.
Our study results indicate a UGDH expression differential between the malignant and the premalignant fields that may be a component of both the early and late stages of prostate carcinogenesis. In malignant acini, overexpression of UGDH may drive overproduction of HA and other proteoglycans, which are synthesized from the UDP-glucuronic acid product of UGDH. Excess HA in conjunction with elevation of its processing enzyme promotes prostate tumor growth12,46 through increased tumor cell cycling and intrinsic tumor cell motility. HA ligation of CD44 receptors activates various rho-family GTPases, PI3-kinase/Akt,47,48 and tyrosine phosphorylation of c-Met, the receptor for hepatocyte growth factor/scattering factor that controls prostate tumor cell proliferation, motility and invasion.49 In addition to the direct stimulation of tumor cells through cell surface receptors, HA may also promote cancer progression via activation of cancer stem-like cells. Recent studies in breast and ovarian tumor cell lines demonstrated a link between HA-CD44 interaction and activation of signaling via both Nanog (embryonic transcription factor expressed in stem cells) and ankyrin (cytoskeletal protein).47 Since prostate cancer stem-like cells express CD4427,49 and Nanog,26 HA may thereby stimulate stem cell migration from the tumor site. The androgen-induced stimulation of UGDH expression in the LNCaP prostate tumor cell line did not result in overproduction of HA.33 This does not preclude a link between UGDH and HA within malignant compartments in clinical prostate cancer.
In the premalignant field concept, genetic and epigenetic changes sequentially drive prostate epithelial cells toward the metastatic phenotype. Pre-cancerous lesions express molecular markers proximal to the tumor prior to discernable morphological patterns of malignancy. Field cancerization has been confirmed for several cancers since Slaughter suggested this concept in 1953.8 Recent studies reported alterations in the mitochondrial genome, physical DNA conformation changes, methylation of GSTP1 and RARβ2, expression of β-catenin in normal appearing cells adjacent to prostate tumor when compared to cancer cells or non-cancerous cells, supporting Slaughter’s hypothesis.5–7, 9, 10, 50 This concept has resulted in the identification of field disease/effect markers which have significantly improved diagnosis and risk assessment among individuals in a prospective longitudinal study who had been occupationally exposed to carcinogens.50 Such a profile for prostate cancer would assist in determining who is at risk for prostate cancer and who requires rebiopsy, but only longitudinal studies can distinguish field effect from field disease. This is feasible in bladder cancer screening because the entire organ can be viewed cystoscopically and sampled by bladder wash or biopsy at the initiation of the study to assure there is no occult tumor causing a field effect change.51
Besides regulating HA production, the activity of UGDH controls the level of intracellular androgen by supplying UGT2B15 and UGT2B17 isozymes with the requisite UDP-glucuronate precursor for glucuronidation and inactivation of androgen.29–32 Ligation of the androgen receptor (AR) within tumor cells by intracellular androgen results in enhanced cell growth, proliferation, stress responses, and apoptosis.52 Decreased expression of UGDH in NAA in cancer patients would restrict glucuronate availability, thus increasing active androgen levels and potential for increased AR signaling. Genetic polymorphisms that reduce or eliminate UGT2B15 or UGT2B17 activity have been associated with higher risk of PC.53 Similarly, accumulation of active androgens and increased proliferation was observed in LNCaP cells after decreasing the level of glucuronidation by these enzymes with specific siRNA.32 Altogether, these data support a role for decreased hormone glucuronidation in early development of PC.
Low expression of UGDH in the premalignant field may influence additional pathways of carcinogenesis by a similar mechanism. For example, lipoxygenase-1 (15-LOX-1) is overexpressed in prostate carcinomas54,55 and enhances malignant potential by upregulating IGF-1 receptor and activating the Akt pathway.56,57 The active product of lipoxygenation by 15-LOX-1, 13-(S)hydroxyoctadecadienoic acid (13-HODE)58 is also a substrate for UGT2B-mediated glucuronidation in the prostate. Therefore, decreased UGDH expression in the pre-malignant field may promote accumulation of 13-HODE, contributing to carcinogenesis. Since UGDH expression has the potential to drive UGT2B equilibria in the direction of hormone and xenobiotic inactivation,33 its loss may have broad consequences for increasing the response to androgens and xenobiotics that contribute to tumorigenesis.
The causes of UGDH down regulation are unknown and may include multiple mechanisms: (1) hypermethylation or other epigenetic modification of the UGDH gene; (2) activation of UGDH promoter-specific transcriptional repressor proteins, which bind a putative peroxisome proliferators-activated receptor-α, PPARα, response element and/or PPARα agonists;59–60 (3) loss of Sp1 activated transcription of UGDH during metabolic stress.61–63 Although precise mechanisms are not known, this does not detract from the utility of UGDH as a candidate component of a biomarker panel to detect individuals at risk for PC.64 UGDH is a weaker marker than a β-catenin, and the ROC plot for UGDH is similar to those for PSA. However, the strength of the multiplexing approach, which measures biomarker proteins as continuous variables, resides in the ability to optimize the ROC plots for multiple biomarkers quantified in the same tissue section.
The data presented here indicate that UGDH may potentially add at least 14% to a biomarker profile, setting the ROC at 98% specificity. Combination of tissue biomarkers such as UGDH with serum biomarkers such as PSA in a nomogram approach may improve individual risk assessment. Whether the quantitative profiling of UGDH expression on a biopsy, alone or in combination with other potential biomarkers of a nonmalignant field, may supplement use of PSA blood testing to diagnose prostate malignancy remains to be determined. Although UGDH alone is not a highly sensitive biomarker, the results indicate that it may be useful as a component of a biomarker panel to detect the premalignant field.64,65 Our studies warrant an independent validation of UGDH alone, and in concert with other biomarkers such as β-catenin10 using prostate biopsy specimens to assess the value of premalignant field characterization in prostate cancer risk assessment.
Acknowledgments
Grant sponsor: DOD Idea Development Award #DAMD17-02-1-0121 (2002-2005), and a Nebraska Department of Health Institutional LB595 Grant for Cancer and Smoking Disease Research (to GPH), NIH R01 CA106584 (to MAS), NCI Cancer Center Support Grant P30 CA36727.
Authors appreciate Ms. Lynette Smith, M.S., College of Public Health, UNMC for the help with statistical evaluation of the results and Ms. Cheryl Abboud for technical assistance with the manuscript.
Abbreviations
- UGDH
UDP-glucose dehydrogenase
- PC
prostate cancer
- NA
normal acini
- NAA
normal-appearing acini of cancerous gland
- BPH
benign prostatic hyperplasia
- BH
benign hyperplastic
- CA
cancerous acini
- QFIA
quantitative fluorescence imaging analysis
- ROC
receiver operating characteristic
- AMPI
average mean pixel intensity
- TMA
tissue microarray
- RPPA
reverse-phase protein array analysis
- AR
androgen receptor
Footnotes
Novelty and Impact: This paper describes for the first time that quantitative alteration in expression of UGDH within malignant and premalignant fields may be used as a candidate biomarker of prostate cancer. Thus, early diagnosis of this disease may be advanced by quantitative profiling of normal appearing acini in a premalignant field using validated quantitative fluorescence imaging analysis.
References
- 1.Yatani R, Chigusa I, Akazaki K, et al. Geographic pathology of latent prostatic carcinoma. Int J Cancer. 1982;29(6):611. doi: 10.1002/ijc.2910290602. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Loeb S, Catalona WJ. What to do with an abnormal PSA test. Oncologist. 2008;13:299. doi: 10.1634/theoncologist.2007-0139. [DOI] [PubMed] [Google Scholar]
- 4.Welch HG, Fisher ES, Gottlieb DJ, et al. Detection of prostate cancer via biopsy in the Medicare-SEER population during the PSA era. J Natl Cancer Inst. 2007;99:1395. doi: 10.1093/jnci/djm119. [DOI] [PubMed] [Google Scholar]
- 5.Dakubo GD, Jakupciak JP, Birch-Machin MA, et al. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grover AC, Tangrea MA, Woodson KG, et al. Tumor-associated endothelial cells display GSTP1 and RARbeta2 promoter methylation in human prostate cancer. J Transl Med. 2006;4:13. doi: 10.1186/1479-5876-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald SA, Greaves LC, Gutierrez-Gonzalez L, et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology. 2008;134:500. doi: 10.1053/j.gastro.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 8.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 9.Ushijima T. Epigenetic field for cancerization. J Biochem Mol Biol. 2007;40:142. doi: 10.5483/bmbrep.2007.40.2.142. [DOI] [PubMed] [Google Scholar]
- 10.Huang D, Casale GP, Tian J, et al. Quantitative fluorescence imaging analysis for cancer biomarker discovery: application to beta-catenin in archived prostate specimens. Cancer Epidemiol Biomarkers Prev. 2007;16:1371. doi: 10.1158/1055-9965.EPI-06-0718. [DOI] [PubMed] [Google Scholar]
- 11.Lokeshwar VB, Rubinowicz D, Schroeder GL, et al. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J Biol Chem. 2001;276:11922. doi: 10.1074/jbc.M008432200. [DOI] [PubMed] [Google Scholar]
- 12.Tammi RH, Kultti A, Kosma VM, et al. Hyaluronan in human tumors: Pathobiological and prognostic messages from cell-associated and stromal hyaluronan. Semin Cancer Biol. 2008 doi: 10.1016/j.semcancer.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Ekici S, Cerwinka WH, Duncan R, et al. Comparison of the prognostic potential of hyaluronic acid, hyaluronidase (HYAL-1), CD44v6 and microvessel density for prostate cancer. Int J Cancer. 2004;112:121. doi: 10.1002/ijc.20368. [DOI] [PubMed] [Google Scholar]
- 14.Posey JT, Soloway MS, Ekici S, et al. Evaluation of the prognostic potential of hyaluronic acid and hyaluronidase (HYAL1) for prostate cancer. Cancer Res. 2003;63:2638. [PubMed] [Google Scholar]
- 15.Bharadwaj AG, Rector K, Simpson MA. Inducible hyaluronan production reveals differential effects on prostate tumor cell growth and tumor angiogenesis. J Biol Chem. 2007;282:20561. doi: 10.1074/jbc.M702964200. [DOI] [PubMed] [Google Scholar]
- 16.Ricciardelli C, Russell DL, Ween MP, et al. Formation of hyaluronan- and versican-rich pericellular matrix by prostate cancer cells promotes cell motility. J Biol Chem. 2007;282:10814. doi: 10.1074/jbc.M606991200. [DOI] [PubMed] [Google Scholar]
- 17.Simpson MA, Wilson CM, McCarthy JB. Inhibition of prostate tumor cell hyaluronan synthesis impairs subcutaneous growth and vascularization in immunocompromised mice. Am J Pathol. 2002;161:849. doi: 10.1016/S0002-9440(10)64245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draffin JE, McFarlane S, Hill A, et al. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 2004;64:5702. doi: 10.1158/0008-5472.CAN-04-0389. [DOI] [PubMed] [Google Scholar]
- 19.Lin SL, Chang D, Chiang A, et al. Androgen receptor regulates CD168 expression and signaling in prostate cancer. Carcinogenesis. 2008;29:282. doi: 10.1093/carcin/bgm259. [DOI] [PubMed] [Google Scholar]
- 20.Miyake H, Hara I, Okamoto I, et al. Interaction between CD44 and hyaluronic acid regulates human prostate cancer development. J Urol. 1998;160:1562. [PubMed] [Google Scholar]
- 21.Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 22.Maitland NJ, Collins A. A tumour stem cell hypothesis for the origins of prostate cancer. BJU Int. 2005;96:1219. doi: 10.1111/j.1464-410X.2005.05744.x. [DOI] [PubMed] [Google Scholar]
- 23.Kasper S. Characterizing the prostate stem cell. J Urol. 2007;178:375. doi: 10.1016/j.juro.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 24.Lin VK, Wang SY, Vazquez DV, et al. Prostatic stromal cells derived from benign prostatic hyperplasia specimens possess stem cell like property. Prostate. 2007;67:1265. doi: 10.1002/pros.20599. [DOI] [PubMed] [Google Scholar]
- 25.Wei C, Guomin W, Yujun L, et al. Cancer stem-like cells in human prostate carcinoma cells DU145: the seeds of the cell line? Cancer Biol Ther. 2007;6:763. doi: 10.4161/cbt.6.5.3996. [DOI] [PubMed] [Google Scholar]
- 26.Gu G, Yuan J, Wills M, et al. Prostate cancer cells with stem cell characteristics reconstitute the original human tumor in vivo. Cancer Res. 2007;67:4807. doi: 10.1158/0008-5472.CAN-06-4608. [DOI] [PubMed] [Google Scholar]
- 27.Hurt EM, Kawasaki BT, Klarmann GJ, et al. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin SL, Chang D, Ying SY. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28:310. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- 29.Chouinard S, Yueh MF, Tukey RH, et al. Inactivation by UDP-glucuronosyltransferase enzymes: The end of androgen signaling. J Steroid Biochem Mol Biol. 2008;109:247. doi: 10.1016/j.jsbmb.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Chouinard S, Pelletier G, Belanger A, et al. Cellular specific expression of the androgen-conjugating enzymes UGT2B15 and UGT2B17 in the human prostate epithelium. Endocr Res. 2004;30:717. doi: 10.1081/erc-200044014. [DOI] [PubMed] [Google Scholar]
- 31.Chouinard S, Pelletier G, Belanger A, et al. Isoform-specific regulation of uridine diphosphate-glucuronosyltransferase 2B enzymes in the human prostate: differential consequences for androgen and bioactive lipid inactivation. Endocrinology. 2006;147:5431. doi: 10.1210/en.2006-0229. [DOI] [PubMed] [Google Scholar]
- 32.Chouinard S, Barbier O, Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J Biol Chem. 2007;282:33466. doi: 10.1074/jbc.M703370200. [DOI] [PubMed] [Google Scholar]
- 33.Wei Q, Galbenus R, Raza A, et al. Androgen-stimulated UDP-glucose dehydrogenase expression limits prostate androgen availability without impacting hyaluronan levels. Cancer Res. 2009;69:2332. doi: 10.1158/0008-5472.CAN-08-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paweletz CP, Charboneau L, Bichsel VE, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 35.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 36.Lalani EN, Golding M, Hudson M, et al. Protein extraction and western blotting from methacarn-fixed tissue. J Pathol. 1995;177:323. doi: 10.1002/path.1711770316. [DOI] [PubMed] [Google Scholar]
- 37.Shibutani M, Uneyama C, Miyazaki K, et al. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest. 2000;80:199. doi: 10.1038/labinvest.3780023. [DOI] [PubMed] [Google Scholar]
- 38.Kroll J, Becker KF, Kuphal S, et al. Isolation of high quality protein samples from punches of formalin fixed and paraffin embedded tissue blocks. Histol Histopathol. 2008;23:391. doi: 10.14670/HH-23.391. [DOI] [PubMed] [Google Scholar]
- 39.Hemstreet GP, Bonner RB, Rao JY, et al. G-actin levels as a marker for malignancy within the prostate. J Urol. 1993;194:334A. [Google Scholar]
- 40.Rao JY, Hemstreet GP, III, Hurst RE, et al. Alterations in phenotypic biochemical markers in bladder epithelium during tumorigenesis. Proc Natl Acad Sci U S A. 1993;90:8287. doi: 10.1073/pnas.90.17.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao JY, Hemstreet GP, Bonner RB, Hurst RE, Qiu WR, Reznikoff CA. Nuclear actin as a biomarker for bladder cancer risk assessment. Journal of Urology. 1994;151(5):349A. [Google Scholar]
- 42.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 43.McCabe A, Dolled-Filhart M, Camp RL, et al. Automated quantitative analysis (AQUA) of in situ protein expression, antibody concentration, and prognosis. J Natl Cancer Inst. 2005;97:1808. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 44.Kramer G, Steiner GE, Neumayer C, et al. Over-expression of anti-CD75 reactive proteins on distal and collecting renal tubular epithelial cells in calcium-oxalate stone-forming kidneys in Egypt. BJU Int. 2004;93:822. doi: 10.1111/j.1464-410x.2003.04751.x. [DOI] [PubMed] [Google Scholar]
- 45.Streit M, Ecker RC, Osterreicher K, et al. 3D parallel coordinate systems--a new data visualization method in the context of microscopy-based multicolor tissue cytometry. Cytometry A. 2006;69:601. doi: 10.1002/cyto.a.20288. [DOI] [PubMed] [Google Scholar]
- 46.Simpson MA, Lokeshwar VB. Hyaluronan and hyaluronidase in genitourinary tumors. Front Biosci. 2008;13:5664. doi: 10.2741/3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourguignon LY, Peyrollier K, Xia W, et al. Hyaluronan-CD44 interaction activates stem cell marker, nanog, stat-3-mediated MDR1 gene expression and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008 doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itano N, Kimata K. Altered hyaluronan biosynthesis in cancer progression. Semin Cancer Biol. 2008 doi: 10.1016/j.semcancer.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki M, Kobayashi H, Kanayama N, et al. CD44 stimulation by fragmented hyaluronic acid induces upregulation and tyrosine phosphorylation of c-Met receptor protein in human chondrosarcoma cells. Biochim Biophys Acta. 1591;37:2002. doi: 10.1016/s0167-4889(02)00246-x. [DOI] [PubMed] [Google Scholar]
- 50.Hanson JA, Gillespie JW, Grover A, et al. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 51.Hemstreet GP, III, Yin S, Ma Z, et al. Biomarker risk assessment and bladder cancer detection in a cohort exposed to benzidine. J Natl Cancer Inst. 2001;93:427. doi: 10.1093/jnci/93.6.427. [DOI] [PubMed] [Google Scholar]
- 52.Bolton EC, So AY, Chaivorapol C, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park J, Chen L, Shade K, et al. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171:2484. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
- 54.Kelavkar UP, Glasgow W, Olson SJ, et al. Overexpression of 12/15-lipoxygenase, an ortholog of human 15-lipoxygenase-1, in the prostate tumors of TRAMP mice. Neoplasia. 2004;6:821. doi: 10.1593/neo.04286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelavkar UP, Harya NS, Hutzley J, et al. DNA methylation paradigm shift: 15-lipoxygenase-1 upregulation in prostatic intraepithelial neoplasia and prostate cancer by atypical promoter hypermethylation. Prostaglandins Other Lipid Mediat. 2007;82:185. doi: 10.1016/j.prostaglandins.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Hsi LC, Wilson LC, Eling TE. Opposing effects of 15-lipoxygenase-1 and -2 metabolites on MAPK signaling in prostate. Alteration in peroxisome proliferator-activated receptor gamma. J Biol Chem. 2002;277:40549. doi: 10.1074/jbc.M203522200. [DOI] [PubMed] [Google Scholar]
- 57.Kelavkar UP, Cohen C. 15-lipoxygenase-1 expression upregulates and activates insulin-like growth factor-1 receptor in prostate cancer cells. Neoplasia. 2004;6:41. doi: 10.1016/s1476-5586(04)80052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spindler SA, Sarkar FH, Sakr WA, et al. Production of 13-hydroxyoctadecadienoic acid (13-HODE) by prostate tumors and cell lines. Biochem Biophys Res Commun. 1997;239:775. doi: 10.1006/bbrc.1997.7471. [DOI] [PubMed] [Google Scholar]
- 59.Bontemps Y, Vuillermoz B, Antonicelli F, et al. Specific protein-1 is a universal regulator of UDP-glucose dehydrogenase expression: its positive involvement in transforming growth factor-beta signaling and inhibition in hypoxia. J Biol Chem. 2003;278:21566. doi: 10.1074/jbc.M209366200. [DOI] [PubMed] [Google Scholar]
- 60.Vatsyayan J, Lin CT, Peng HL, et al. Identification of a cis-acting element responsible for negative regulation of the human UDP-glucose dehydrogenase gene expression. Biosci Biotechnol Biochem. 2006;70:401. doi: 10.1271/bbb.70.401. [DOI] [PubMed] [Google Scholar]
- 61.Vatsyayan J, Lee SJ, Chang HY. Effects of xenobiotics and peroxisome proliferator-activated receptor-alpha on the human UDPglucose dehydrogenase gene expression. J Biochem Mol Toxicol. 2005;19:279. doi: 10.1002/jbt.20099. [DOI] [PubMed] [Google Scholar]
- 62.Vatsyayan J, Peng HL, Chang HY. Analysis of human UDP-glucose dehydrogenase gene promoter: identification of an Sp1 binding site crucial for the expression of the large transcript. J Biochem. 2005;137:703. doi: 10.1093/jb/mvi082. [DOI] [PubMed] [Google Scholar]
- 63.Wei S, Chuang HC, Tsai WC, et al. Thiazolidinediones Mimic Glucose Starvation in Facilitating Sp1 Degradation Through the Upregulation of {beta}-TRCP. Mol Pharmacol. 2009 doi: 10.1124/mol.109.055376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemstreet GP, Wang W. Genotypic and phenotypic biomarker profiles for individual risk assessment and cancer detection (lessons from bladder cancer risk assessment in symptomatic patients and workers exposed to benzidine) Front Biosci. 2004;9:2671. doi: 10.2741/1426. [DOI] [PubMed] [Google Scholar]
- 65.Hemstreet GP, Hurst RE, Bonner RB. Selection and development of biomarkers for bladder cancer. In: Hanausek M, Walaszek Z, editors. Tumor Marker Protocols. Vol. 14. Totowa, NJ: Humana Press; 1998. p. 37. chapt. 3. [Google Scholar]


