Abstract
Evidence has been presented (Kukar et al. 2008 Nature 453, 925–929) that certain γ–secretase modulators (GSMs) target the 99 residue C-terminal domain (C99) of the amyloid precursor protein, a substrate of γ-secretase, but not the protease complex itself. Here, NMR results demonstrate a lack of specific binding of these GSMs to monodisperse C99 in LMPG micelles. In addition, results indicate that C99 was likely to have been aggregated in some of the key experiments of the previous work, and that binding of GSMs to these C99 aggregates is also of a non-specific nature.
Among the therapeutic targets for Alzheimer’s disease (AD), the amyloid pathway has long been paramount (1). Familial early-onset AD (FAD) is associated with autosomal dominant mutations in the amyloid precursor protein (APP) and in the catalytic subunits (presenilin 1 and presenilin 2) of the intramembrane protease that processes it, γ-secretase (2). According to the amyloid hypothesis, oligomeric forms of Aβ are the principal agents underlying disease pathogenesis (1). The Aβ peptide is generated by proteolysis of APP. Cleavage of APP by β-secretase yields C992, which is then heterogeneously processed by γ-secretase to generate Aβ species with a variety of lengths, principally Aβ40 (3). Familial AD is associated with an increase in the Aβ42/Aβ40 ratio, with Aβ42 being the primary species deposited in the brain parenchyma of most individuals with AD (4). Because Aβ is thought to be central to the pathogenesis of AD, inhibiting its production is a potential therapeutic strategy (1). Although significant progress has been made in the identification and development of potent γ-secretase inhibitors, their clinical application has been limited by significant toxicities resulting from interference with processing of other γ-secretase substrates, particularly Notch (5). Indeed, γ-secretase is a highly promiscuous protease with more than sixty identified targets (6).
The discovery that a subset of non-steroidal anti-inflammatory drugs could selectively reduce Aβ42 production without abrogating Notch cleavage suggested an alternative therapeutic strategy for AD (7). The Aβ42-lowering activity of these γ-secretase modulators (GSMs) was recapitulated in cell-free assays of γ-secretase activity. Several groups have produced data suggesting that GSMs interact allosterically with presenilin, thereby modifying the enzyme’s conformation (8–10). Moreover, GSMs were observed to influence the cleavage of an unrelated substrate by signal peptide peptidase – an enzyme with homology to the presenilin subunit of γ-secretase, suggesting that the modulators interact with the enzyme rather than substrate (11). Corroborating the premise that GSMs are enzyme-targeting was the finding that certain NSAID GSMs can also influence the precise γ-secretase cleavage site of Notch (12). Okochi et al have shown that Notch cleavage is modulated but not inhibited by NSAID GSMs, providing a plausible explanation for the lack of adverse Notch-related toxicities of such compounds (12).
A recent report from the Golde laboratory, however, postulates that GSMs specifically target APP and its C-terminal derivatives, providing an alternative explanation to the apparent specificity that GSMs exert on cleavage of C99 (13). The authors demonstrated that application of biotinylated photoactivatable affinity probe derivatives of certain GSMs – namely fenofibrate (an Aβ42-raising GSM) and tarenflurbil (an Aβ42-lowering GSM) – to CHAPSO detergent extracts from human neuroglioma H4 cells failed to label any core γ-secretase subunits, but instead produced covalent conjugates with C83, the product of α-secretase cleavage of APP. While labeling of C99 did not appear to take place in those same extracts (see Fig. 1e and Supporting Fig. 2b in (13)), purified recombinant C99 in CHAPSO-containing solutions could be modified by derivatized fenofibrate and tarenflurbil, an interaction subject to complete abrogation by a number of other GSMs. Photoaffinity labeling of purified C99 was localized to residues 29–36 (GAIIGLMV, where Gly29 corresponds to Gly700 in full length APP770).
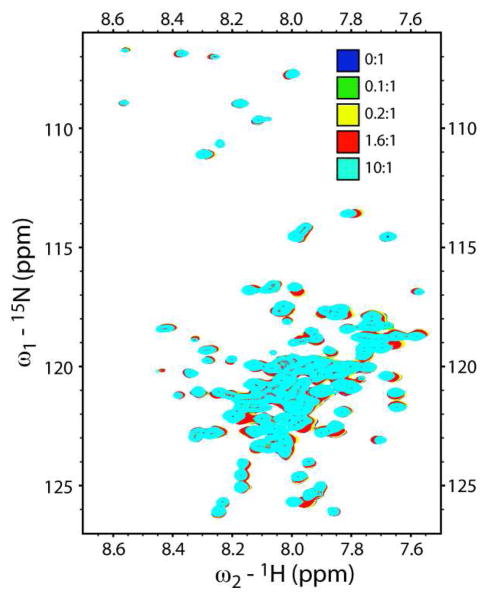
To further investigate the putative binding of GSMs to APP and its derivatives, we monitored the spectroscopic response of purified and monodisperse U-15N-C99 to addition of GSMs in a membrane-like environment. Human C99 was expressed and labeled in E. coli and purified into micelles composed of LMPG, a phospholipid-derived detergent that generally maintains membrane proteins in native-like structural and functional states (14–17). Previous studies of U-15N-C99 incorporated in LMPG micelles demonstrated specific and saturable changes in 1H,15N-TROSY-HSQC spectra when titrated with a cholesterol derivative (17). However, titrations of U-15NC99 in LMPG micelles with the GSMs tarenflurbil, indomethacin, fenofibrate, and sulindac sulfide revealed no evidence for specific binding, even at compound concentrations in the mM range (Fig. 1 and Figs. S1-S5). For example, while titration of tarenflurbil induces modest chemical shift perturbations in the 1H,15N-TROSY-HSQC of C99 (Fig. 1), the concentration-dependence of the changes observed is not consistent with avid and specific binding of the protein by the GSMs (Fig. S1). Moreover, the C99 NMR resonances that undergo GSM-induced shifts tend to differ from GSM to GSM, as might be expected for non-specific interactions. In no case were the peaks from residues in the putative GSM binding site (residues 29–36) seen to be among those that shifted the most in response to addition of GSMs. In addition, experiments of a reciprocal nature were performed in which titrations of U-15N-C99 in LMPG micelles with tarenflurbil were monitored using the 19F NMR signal from this GSM. Figure S6 reveals that the 19F signal from tarenflurbil neither shifts nor is significantly broadened relative to free ligand over a range of GSM:C99 molar ratios from 0.1 to 1 (at ca. 250μM C99), indicating a lack of significant binding. These results indicate that non-aggregated C99 in LMPG micelles exhibits no specific avidity for GSMs and related compounds.
Figure 1.
Titration of U-15N-C99 with tarenflurbil as monitored using 600 MHz 1H,15N-TROSY NMR. Samples contained 1 mM U-15N-C99 in 200 mM LMPG micelles at pH 6.5 and 45 degrees C and the indicated molar ratios of tarenflurbil to C99. The modest changes observed indicate only non-specific interactions.
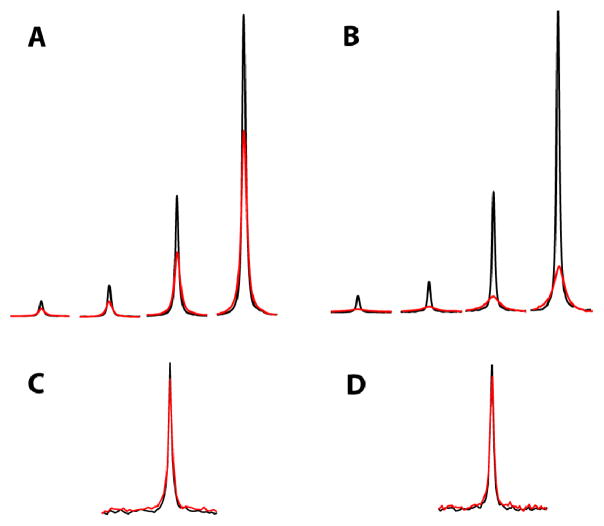
We also used 19F NMR spectroscopy to test for binding of GSMs to C99 in a solution containing CHAPSO as the detergent, conditions that more faithfully replicate the photoaffinity crosslinking experiments of Kukar et al involving purified Flag-tagged C99 (c.f., Fig. 1c and Supplementary Fig. 4 in (13)). Solutions containing 5μM C99 were titrated individually with tarenflurbil, celecoxib, sulindac sulphone (negative control), and a known γ-secretase inhibitor (negative control). In the presence of C99, significant broadening of the 19F NMR signals from both tarenflurbil and celecoxib is observed (Figs. 2A–B). This broadening is not seen in CHAPSO-containing samples lacking C99 or in those experiments involving titration with negative control compounds (Figs 2C–D). It is notable that peak linewidths remain nearly constant throughout the titrations (Figs. 2A–B) despite the presence of a significant molar excess of GSM over C99 at all points. A similar result was obtained when 200 μM C99 was titrated with tarenflurbil over a range of 25 μM to 2 mM. In this case the 19F signal of tarenflurbil is dramatically broadened in the presence of protein, but the linewidth then decreases by only 33% over the full range of the titration (Figure S7). These results are not consistent with a saturable binding process, but instead suggests that C99 somehow facilitates and/or nucleates deposition of GSM into aggregates of a potentially colloidal nature (hence the broader but still observable resonances), which have properties that are largely independent of either the C99 or the GSM concentration. A similar phenomenon was observed for tarenflurbil when it was titrated into samples containing KCNE1, an unrelated ion channel modulatory membrane protein (18). Linewidths broadened significantly and failed to exhibit behavior indicative of saturation (Fig. S8), suggesting that KCNE1 in 0.5% CHAPSO also triggers the formation of tarenflurbil aggregates.
Figure 2.
19F NMR titrations of 5uM APP(C99)-Flag with GSMs at concentrations of 25, 50, 200, and 500μM – Right to Left) tarenflurbil (A) or celecoxib (B). Black: control series lacking C99. Red: samples contained 5μM C99. Changes in chemical shifts occurring during the titrations of (A) and (B) were negligible. (C) and (D) represent single point titrations of 5μM C99 with 25uM sulindac sulphone or 25μM of a known γ-secretase inhibitor, respectively, as negative controls. Buffers contained 50mM HEPES (deuterated), pH 7.4, and the C99 sample buffer contained 0.5% CHAPSO (8 mM). Spectra were recorded at 25 degrees C.
The above results failed to provide evidence for avid complexation of GSMs with purified C99 in CHAPSO-containing solutions. These experiments were designed to approximate the 0.25% CHAPSO conditions used in studies of purified C99 by Kukar et al. (c.f., Fig. 1c and Supplementary Fig. 4 in (13)). Given that 0.25% CHAPSO is below the critical micelle concentration of this detergent (which is 0.5%, or equivalently 8 mM), we investigated the physical state of C99 present in our samples. Shown in Figure S9 is a comparison of the 1H NMR spectra of 5 μM solutions of ubiquitin and C99 in 0.5% CHAPSO. No peaks can be detected from C99, indicating that even dilute C99 is present only in the form of very high molecular weight aggregates. In the experience of the Sanders lab, these results are not surprising because such low concentrations of CHAPSO and related bile salt-derivative detergents usually fail to solubilize membrane proteins in the absence of significant amounts of lipids or other detergents, particularly at concentrations so near to the critical micelle concentration.
In conclusion, we have demonstrated that monodisperse C99 lacks specific affinity for an array of GSMs under conditions in which the protein is solubilized in non-denaturing detergent micelles and also under conditions in which the protein is aggregated. Surprisingly, under the latter conditions, it appears that C99 facilitates the formation of GSM-containing aggregates through an unknown mechanism. An unrelated aggregated membrane protein, KCNE1, was also able to trigger a similar phenomenon. One possible explanation is that the GSMs associated non-specifically and non-stoichiometrically with aggregated C99 and KCNE1, perhaps in a manner analogous to either the association of dyes such as Congo red with amyloid deposits (19, 20) or to the association of certain fluorescent dyes with molten globular proteins (21). Alternatively, given that the NMR signals from these aggregates are broad but still visible, it is possible that the presence of protein induces the formation of GSM aggregates that are colloidal in nature. It has previously been shown that the formation of such aggregates involving small drug-like compounds is not unusual and that such aggregates often appear to inhibit enzymes (22). In summary, our data are not consistent with the formation of an avid and stoichiometric complex between GSM and C99, but instead suggest non-specific interactions. This conclusion applies both to conditions that mimic those used for purified C99 in the previous work (CHAPSO-containing solutions) and also under more ideal membrane mimetic conditions (LMPG micelles).
Supplementary Material
Acknowledgments
We thank Dr. Bing Jap of the LBL for providing the recombinant C99 expression system used herein by the Sanders group. We would also like to acknowledge Jeff Lewis, Roger Fachini, and Trace Tsuruda for the expression and purification of the protein used at Amgen.
Abbreviations
- Aβ
amyloid beta polypeptide
- APP
amyloid precursor protein
- C99
99 residue C-terminal domain of the amyloid precursor protein
- CHAPSO
3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1- propanesulfonate
- GSM
γ-secretase modulator
- LMPG
lyso-myristoylphosphatidylglycerol
- NSAID
non-steroidal anti-inflammatory drug
- NMR
nuclear magnetic resonance
Note that the terminology C99 and C100 can be used interchangeably to describe the 99-residue C-terminal fragment of human APP. In this and previous work, the protein was expressed in E. coli, resulting in the addition of an N-terminal methionine hence the nomenclature “C99” and “C100” are synonymous.
Footnotes
This work was supported by grants from the US NIH (PO1 GM080513, to CRS) and from the Alzheimer’s Association (IIRG-07-59379, to CRS).
SUPPORTING INFORMATION AVAILABLE
Methodological details and Figures S1-S10. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Tanzi RE, Bertram L. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Esler WP, Wolfe MS. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- 4.Borchelt DR, Thinakaran G, Eckman CB, Lee MK, Davenport F, Ratovitsky T, Prada CM, Kim G, Seekins S, Yager D, Slunt HH, Wang R, Seeger M, Levey AI, Gandy SE, Copeland NG, Jenkins NA, Price DL, Younkin SG, Sisodia SS. Neuron. 1996;17:1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 5.Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, Engstrom L, Pinzon-Ortiz M, Fine JS, Lee HJ, Zhang L, Higgins GA, Parker EM. J Biol Chem. 2004;279:12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- 6.Beel AJ, Sanders CR. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weggen S, Eriksen JL, Sagi SA, Pietrzik CU, Ozols V, Fauq A, Golde TE, Koo EH. J Biol Chem. 2003;278:31831–31837. doi: 10.1074/jbc.M303592200. [DOI] [PubMed] [Google Scholar]
- 8.Lleo A, Berezovska O, Herl L, Raju S, Deng A, Bacskai BJ, Frosch MP, Irizarry M, Hyman BT. Nat Med. 2004;10:1065–1066. doi: 10.1038/nm1112. [DOI] [PubMed] [Google Scholar]
- 9.Czirr E, Leuchtenberger S, Dorner-Ciossek C, Schneider A, Jucker M, Koo EH, Pietrzik CU, Baumann K, Weggen S. J Biol Chem. 2007;282:24504–24513. doi: 10.1074/jbc.M700618200. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Hayashi I, Tominari Y, Rikimaru K, Morohashi Y, Kan T, Natsugari H, Fukuyama T, Tomita T, Iwatsubo T. J Biol Chem. 2003;278:18664–18670. doi: 10.1074/jbc.M301619200. [DOI] [PubMed] [Google Scholar]
- 11.Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, Wolfe MS. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 12.Okochi M, Fukumori A, Jiang J, Itoh N, Kimura R, Steiner H, Haass C, Tagami S, Takeda M. J Biol Chem. 2006;281:7890–7898. doi: 10.1074/jbc.M513250200. [DOI] [PubMed] [Google Scholar]
- 13.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, Liu Q, Scarborough GA. Anal Biochem. 1998;259:89–97. doi: 10.1006/abio.1998.2633. [DOI] [PubMed] [Google Scholar]
- 15.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 16.Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr, Sanders CR. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, Vanoye CG, Kim HJ, Sanders CR. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prento P. Biotech Histochem. 2009:1–20. [Google Scholar]
- 20.Inouye H, Kirschner DA. Subcell Biochem. 2005;38:203–224. doi: 10.1007/0-387-23226-5_10. [DOI] [PubMed] [Google Scholar]
- 21.Matulis D, Lovrien R. Biophys J. 1998;74:422–429. doi: 10.1016/S0006-3495(98)77799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.