Abstract
Astroglial glutamate transporter EAAT2/GLT1 prevents glutamate-induced excitotoxicity in the central nervous system. Expression of EAAT2/GLT1 is dynamically regulated by neurons. The pathogenesis of amyotrophic lateral sclerosis (ALS) involves astroglial dysfunction, including dramatic loss of EAAT2/GLT1. DNA methylation of gene promoters represents one of the most important epigenetic mechanisms in regulating gene expression. The involvement of DNA methylation in the regulation of astroglial EAAT2/GLT1 expression in different conditions, especially in ALS has not been explored. In this study, we established a procedure to selectively isolate a pure astrocyte population in vitro and in vivo from BAC GLT1 eGFP mice using an eGFP-based fluorescence-activated cell sorting approach. Astrocytes isolated from this procedure are GFAP+ and GLT1+ and respond to neuronal stimulation, enabling direct methylation analysis of GLT1 promoter in these astrocytes. To investigate the role of DNA methylation in physiological and pathological EAAT2/GLT1 expression, methylation status of the EAAT2/GLT1 promoter was analyzed in astrocytes from in vitro and in vivo paradigms or postmortem ALS motor cortex by bisulfite sequencing method. DNA demethylation on selective CpG sites of the GLT1 promoter was highly correlated to increased GLT1 mRNA levels in astrocytes in response to neuronal stimulation; however, low level of methylation was found on CpG sites of EAAT2 promoter from postmortem motor cortex of human amyotrophic lateral sclerosis patients. In summary, hypermethylation on selective CpG sites of the GLT1 promoter is involved in repression of GLT1 promoter activation, but this regulation does not play a role in astroglial dysfunction of EAAT2 expression in patients with ALS.
Keywords: epigenetic, astrocyte, GLT1
INTRODUCTION
Dysfunction of nonneuronal cells such as astrocytes and microglia has been repeatedly implicated in several neurodegenerative diseases (Boillee et al., 2006; Yamanaka et al., 2008). Selective expression of disease-causing mutant proteins and their removal from astrocytes has been shown to alter disease progression in transgenic models of amyotrophic lateral sclerosis (ALS), Huntington's disease, and spinocerebellar ataxia (Lobsiger and Cleveland, 2007). Fetal cultured astrocytes that overexpress disease-causing mutant proteins directly induce neuronal cell death by secreting unidentified toxic factors (Di Giorgio et al., 2007; Nagai et al., 2007). Although intensive efforts to identify the molecular pathways underlining astroglial dysfunction in neurodegeneration are underway, loss of astroglial glutamate transporters and subsequent excitotoxicity has been well characterized in various neurodegenerative diseases (Danbolt, 2001; Rothstein et al., 1996, 1992).
Glutamate transporters, especially astroglial glutamate transporter EAAT2 (rodent analog, GLT1) are fundamental in the maintenance of synaptic neurotransmitter glutamate homeostasis and are critical for preventing CNS excitotoxicity. Experimental removal of GLT1 has been shown to lead to severe neurological disorders (Rothstein et al., 1996; Tanaka, 1997). As one of the unique astroglial functional proteins, GLT1 protein is predominantly and abundantly expressed in astroglia across adult CNS (Furuta et al., 1997; Regan et al., 2007), although GLT1 mRNA but not GLT1 protein has also been detected in a small subset of embryonic and adult neurons (Chen et al., 2002, 2004). Physiological induction of GLT1 in astrocytes is strongly dependent upon neuronal signals. Early in vitro studies have shown that GLT1 expression (both mRNA and protein) levels are low in cultured astrocytes but are greatly induced with the presence of neurons (Swanson et al., 1997; Yang et al., 2009). GLT1 mRNA and protein expression is also greatly induced in astrocytes throughout the CNS in early postnatal development when synaptogenesis occurs (Furuta et al., 1997). By using a microfluidic culture platform, individual neuronal axons were found to be sufficient and essential for the transcriptional activation of GLT1 expression (Yang et al., 2009). Subsequent EAAT2/GLT1 promoter analysis identified kappa B-motif binding phosphoprotein (KBBP) as the neuron-dependent activator for astroglial GLT1 expression (Yang et al., 2009). In addition, NF-kB and C-Myc have also been involved in the transcriptional regulation of EAAT2 promoter in a glioma H4 cell line (Sitcheran et al., 2005).
Methylation of cytosine residues on CpG islands of functional gene promoters is an important epigenetic mechanism in regulating gene transcriptional activation/depression in response to physiological and pathological stimuli. Gene-specific hypermethylation of CpG islands are usually associated with transcriptional repression of genes, e.g., cancer cells where many tumor suppressor genes are silenced by the hypermethylation through unclear mechanisms. Epigenetic regulation is also implicated in the pathogenesis of neurodegenerative diseases. Sporadic loss-of-function mutations in the methyl-cytosine binding protein 2 (MeCP2) cause Rett Syndrome (Amir et al., 1999). In fact, loss of MeCP2 function in astrocytes was recently shown to contribute to abnormal morphology of neuronal dendrites (Ballas et al., 2009). The contribution of epigenetic regulation to the pathogenesis of other neurodegenerative diseases is not known.
Severe downregulation of astroglial EAAT2/GLT1 protein has been found in patients and rodent models of various neurodegenerative diseases, including ALS and Huntington's disease (Lievens et al., 2001; Rothstein et al., 1992). Our recent findings suggest that transcriptional dysregulation of GLT1 promoter leads to greatly reduced GLT1 mRNA levels and subsequent loss of GLT1 protein in SOD1 G93A transgenic mice (Yang et al., 2009). The potential association of DNA methylation in the transcriptional dysregulation of EAAT2/GLT1 promoter in neurodegeneration has not been explored. Although DNA methylation plays a role in transcriptional regulation, methylation status of the EAAT2/GLT1 promoter in astrocytes in transcriptionally activated conditions, e.g. postnatal development period or in response to neuronal stimulation has not been explored. Interestingly, the EAAT2 promoter appears to be hypermethylated in several human glioma cell lines, which correlates with their low expression of EAAT2 protein (Zschocke et al., 2007).
In this study, GLT1 expressing astrocytes were selectively isolated and the methylation status of GLT1 promoter from in vitro astrocyte cultures and from adult brain was analyzed. We found that DNA methylation-based epigenetic regulation is involved in neuron-dependent GLT1 promoter activation, but this regulation does not appear to be operational in diseased astroglia, with altered expression of EAAT2, from patients with ALS.
MATERIALS AND METHODS
Cultures and Transfection
Cortical astrocyte cultures were prepared from P2−4 mouse pups of BAC GLT1 eGFP transgenic mice. Cortical neuronal cultures were prepared from E14−16 mouse embryos of wild type mice. Co-culture of astrocytes and neurons was performed by plating neurons directly on the top of the confluent astrocyte cultures.
Immunostaining and Microscopy
Cultures were directly fixed in culture dishes with 4% paraformaldehyde (30 min). After three rinses in PBS, cells were treated with blocking buffer (0.4% BSA, 5% goat-serum, and 0.2% Triton-X 100 in PBS) for 20 min at room temperature. Primary antibodies GFAP (mouse, 1:1,000) were incubated overnight at 4°C in blocking buffer. Anti-mouse Alexa 555 conjugated antibody (1:1,000) was added for 90 min at room temperature. Nuclear counterstain was performed with hoechst33342; eGFP+ cells collected from FACS were also examined by direct brightfield visualization at the time of fluorescent microscopy.
Preparation of Cell Suspension from In Vitro Cultures or from Brain Tissue
For FACS of in vivo brain tissue, adult brains from BAC GLT1 eGFP transgenic reporter mice (8- to 12-weeks old) were used. Mice were anesthetized with pentobarbital (50 mg/kg, i.p.), perfused with cold Hanks buffer (Invitrogen, Carlsbad, CA), and decapitated. The brain was immediately dissected in cold Hanks buffer containing glutamate receptor antagonists, 3 mM DNQX and 100 mM APV (Sigma, St. Louis, MO), and cut into small pieces. Cell suspension was prepared as described in the neural tissue dissociation kit (Miltenyi biotech, Auburn, CA). Briefly, small pieces of tissue were treated with papain enzymatic mix (37°C, 15 min) and then digested with DNase I (37°C, 10 min), followed by careful trituration. Cell mixtures were then filtered through a cell strainer (70 mm) and resuspended in cold HBSS solution (5−10 × 106 cells/mL) for FACS. The whole cell suspension procedure was completed in 1−2 h. For cell suspension preparations of in vitro astrocyte cultures or neuron astrocyte co-cultures, cultured cells were gently scraped and spun down (1,000g, 5 min). The cell pellet was then resuspended in cold HBSS solution.
FAC Sorting of Cell Suspension
Cells were sorted using MoFlo MLS high-speed cell sorter (Beckman coulter) with Summit version 4.3 software. GFP and Propidium iodide (PI) were all excited by a 488 nm laser, and emissions were collected by 530/30 nm and 575/26 nm discrimination filters, respectively. The signals were manually compensated, and cells were sorted into cold HBSS. The whole FAC sorting process was completed within 2−3 h.
RNA Isolation and QRT-PCR
Total RNA from FAC sorted cells (0.5−1 × 106) was prepared by using Absolutely RNA Miniprep Kit from Stratagene (Stratagene, La Jolla, CA). Total RNA were then converted to cDNA using high archive cDNA synthesis kit (Applied Biosystems, Foster city, CA). The relative abundance of GLT1, GFAP, and βIII-tubulin mRNA was determined by using TaqMan premade GLT1, GFAP, and βIII-tubulin probes (Applied Biosystems, Foster city, CA). Ribosomal 18s rRNA was used as endogenous control for the normalization of RNA quantity.
Preparation of Site-Specific Methylated GLT1 Promoter and Luciferase Reporter Assay
Mouse GLT1 promoter (1kb) was amplified from genomic DNA by using primers: F: 5′-atatatatctcgag tgctcgccctcggggaa-3′; R: 5′-atatatataagctttgcggggatcctg cacc-3′. GLT1 promoter was then cloned into pGL4.14 luciferase reporter at Xho I/Hind III sites and named as pGL4.14-PGLT1. Primers with methyl group addition on selective CpG sites on GLT1 promoter were then used to amplify linear GLT1 promoter and luciferase reporter fragment from pGL4.14-PGLT1. Primer sequences are summarized in Fig. 4C. Purified linear GLT1 promoter luciferase reporter together with beta galactosidase was transfected into HEK cells. Luciferase and β-gal assay was performed 48h after transfection.
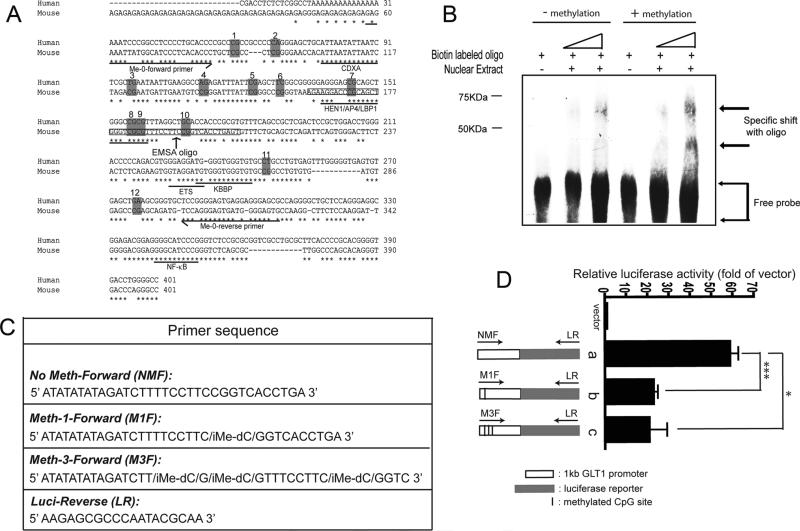
Fig. 4.
Methylation induced functional change of the GLT1 promoter. (A) Sequence analysis of putative transcriptional factors in Me-0 region of GLT1 promoter and human EAAT promoter. CDXA, chicken homeodomain protein; HEN1, Hua enhancer 1; AP4, activating enhancer binding protein 4; LBP1, upstream binding protein 1; KBBP, kappa B-motif binding phosphoprotein; NF-kB, nuclear factor kappa B. Sequences were analyzed in sequence alignment program Mulan (http://mulan.dcode.org). (B) Methylation induced change in the nuclear factor binding EMSA assay of the methylated and unmethylated oligos was performed with nuclear extract from adult mouse cortex. (C) Primer sequence used for making a linear GLT1 promoter and luciferase reporter fragment from pGL4.14-PGLT1. iMe-dC: addition of methyl group on the specific cysteine site. (D) GLT1 promoter activity with site-specific methylation on selective CpG sites. a, wild type GLT1 promoter reporter; b, GLT1 promoter reporter with methylation on #10 CpG site; c, GLT1 promoter reporter with methylation on #8, #9, and #10 CpG sites. (n = 6, ***P < 0.001, *P < 0.05, One-way ANOVA and Bonferroni test).
Luciferase and β-gal Assay
Astrocytes/tissues were collected and lysated in 1× cell culture lysis buffer (Promega, Madison, WI). Lysates (50 μL for luciferase and 50 μL for β-gal) were transfered to a 96-well assay plate and substrate (100 μL for luciferase and 50 μL for β-gal) was added to the assay plate and the activity measured in a FluoSTAR OPTIMA luminometer (BMG labtech, Durham, NC).
Isolation of Genomic DNA and Bisulfite Sequencing Analysis
For genomic DNA isolation, cell pellet (0.5−1 × 106) or homogenized human tissue (0.5 mg) was resuspended in 200−600 μL of nuclear lysis buffer (10 mM Tris/HCl pH 8.0, 400 mM NaCl, and 10 mM EDTA) and was disrupted by supplementing SDS to a final 1%. Proteinase K (4 mg/mL) was then added, and incubated for 3 h at 37°C, followed by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. Bisulfite conversion of genomic DNA was performed as described in EZ-DNA methylation-gold kit (Zymo research, Orange, CA). Briefly, 200−400 ng of genomic DNA was mixed with freshly prepared bisulfite reagent and was incubated for 3 h in programmed temperature cycles. After the bisulfite treatment, converted genomic DNA was column purified. Region of interest was amplified by specific bisulfite sequencing primers and was cloned into TOPO TA vector (Invitrogen, Carlsbad, CA) for sequencing analysis. Individual bacteria clones were then selected for the preparation of the plasmid and subsequent sequencing. The frequency of the various methylation patterns recovered in the bacterial clones will provide a close approximation of the overall frequency of the methylation patterns in the native samples.
Promoter CpG Analysis and Design of the Bisulfate-Sequencing Primers
The GC content, distribution of CpG sites, and predicted bisulfite converted sequence on the full genomic promoter of EAAT2 or GLT1 was analyzed by the Web-based program MethPrimer (http://www.urogene.org/methprimer). On the basis of the distribution of the CpG sites and predicted bisulfite converted sequence, bisulfite-sequencing primers were designed in Vector NTI (Invitrogen, Carlsbad, CA) to amplify each region of interest.
Electrophoretic Mobility Shift Assay (EMSA)
Oligos (45mer) from −833 to −788 of GLT1 promoter were directly synthesized with biotin modification added to both 5′ ends. Cytosine of selective CpG site was also directly methylated during synthesis. Nuclear extracts were prepared by using NE-PER Nuclear and Cytoplasmic Extraction Reagent (Pierce Biotechnology, Rockford, IL) from freshly dissected cortex of adult wild type mice. Gel shift was performed with procedures described in Lightshift Chemiluminescent EMSA kit (Pierce Biotechnology, Rockford, IL). In each reaction, 2 mL (50 fmol/mL) biotin labelled oligos and 5 mg nuclear extract were added.
Western Blot Analysis
FAC sorted cells were lysed in a buffer containing 62.5 mM Tris, 2% SDS, 10% sucrose, and protease inhibitor cocktail (Roche). Homogenates were briefly sonicated and were incubated at 37°C for 30 min. Postmortem human tissue (0.5 mg) was homogenized in 1 mL of homogenization buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, protease inhibitor cocktail) using a electric homogenizer, and the homogenate was sonicated and subsequently centrifuged at 14,000g for 10 min. The supernatants were separated by 4−12% gradient PAGE gel and transferred to PVDF membrane. Blots were placed in blocking solution (5% nonfat milk in TBST) for 1 h at room temperature and then incubated with EAAT2/GLT1 antibody (C-terminal polyclonal, 1:5,000) or anti-actin (1:2,000, Sigma) diluted in TBST overnight at 4°C. After extensive washing, the blots were incubated with horseradish peroxidase conjugated antibodies (1:5,000, Sigma) for 1 h and washed again. Immunoreactive bands were detected using the ECL procedure.
RESULTS
Selective Isolation of eGFP+ Astrocytes from Neuron-Astrocyte Co-Cultures and Mouse Adult Brain
The typical in vitro neuron astrocyte co-culture system limits a mechanistic study of neuron-dependent GLT1 regulation as biochemical analysis of neuron-stimulated astrocytes is interfered by the presence of co-cultured neurons. Although neuron conditioned medium (NCM) is sufficient to induce GLT1 expression, it may only partially represent the regulatory signals as neuron-astrocyte membrane contact may also play a role in neuron-dependent activation of GLT1 (Yang et al., 2009). We recently developed a BAC GLT1 eGFP transgenic reporter mouse line that expresses an eGFP reporter driven by the GLT1 genomic promoter (Regan et al., 2007), which allows selective isolation of astrocytes based on the eGFP expression by FAC sorting. By using this transgenic mouse line, we tested the possibility to selectively isolate astrocytes from BAC GLT1 eGFP mice using the eGFP based FACS approach. Primary astrocytes derived from BAC GLT1 eGFP were sorted following co-culture with neurons for 7 days, as well as from adult mouse brain (Fig. 1A). Both eGFP+ and eGFP- cells were observed in FAC sorting, which is consistent with previous observations that other cell types are also present in primary astrocyte cultures. Postsorting examination of cells under light and fluorescence microscopy revealed that almost all collected cells from either in vitro co-culture or in vivo adult brain are eGFP+ (Fig. 1B). These eGFP+ cells also exhibited typical polygonal morphology of astrocytes and were GFAP immunoreactive after cultured in astrocyte medium (Fig. 1C). However, eGFP- cells were often found as clumped or irregular shapes, and were GFAP- or showed atypical astrocyte morphology after cultured in astrocyte medium (data not shown). Measurement of GFAP mRNA levels by QRT-PCR in both eGFP+ and eGFP- cells also revealed that very low levels of GFAP mRNA was expressed in eGFP- cells (Fig. 1D), regardless the presence of neurons, further confirming that eGFP+ cells are the astrocytes that express typical astroglial markers GFAP and GLT1. In addition, the relative quantity of βIII-tubulin mRNA levels, a neuronal gene, was also measured in eGFP+ cells and neuronal cultures by QRT-PCR and minimal levels of βIII-tubulin expression in eGFP+ cells were found (Fig. 1E). This suggests that neuronal contamination in eGFP+ cells was very minimal. The percentage of eGFP+ cells in the collected cells from in vitro co-culture or in vivo adult brain was 98.2 and 96.8% respectively (Fig. 1F,G), suggesting that this approach is highly effective in isolating astrocytes.
Fig. 1.
Selective isolation of eGFP+ astrocytes from neuron astrocyte co-cultures and mouse adult brain. (A) Diagram for the isolation of GLT1+ astrocytes under different conditions by FACS. (B) Postsorting examination of eGFP+ cells under bright and fluorescence microscopy. Scale bar, 50 μm (C) EGFP+ cells collected from FACS are polygonal GFAP+ astrocytes. Scale bar, 100 μm (D) Relative GFAP mRNA quantity in eGFP+ and eGFP- cells determined by QRT-PCR (n = 3−6, ***P < 0.001, One-way ANOVA and Bonferroni). (E) Relative βIII-tubulin mRNA abundance in eGFP+ cells and neuronal cultures determined by QRT-PCR (n = 6−9, ***P < 0.001, unpaired t-test was used). (F) Purity percentage of eGFP+ cells from in vitro astrocyte co-culture by FACS. (G) Purity percentage of eGFP+ cells from mouse adult brain by FACS.
Expression of GLT1 mRNA and Protein in Isolated Astrocytes
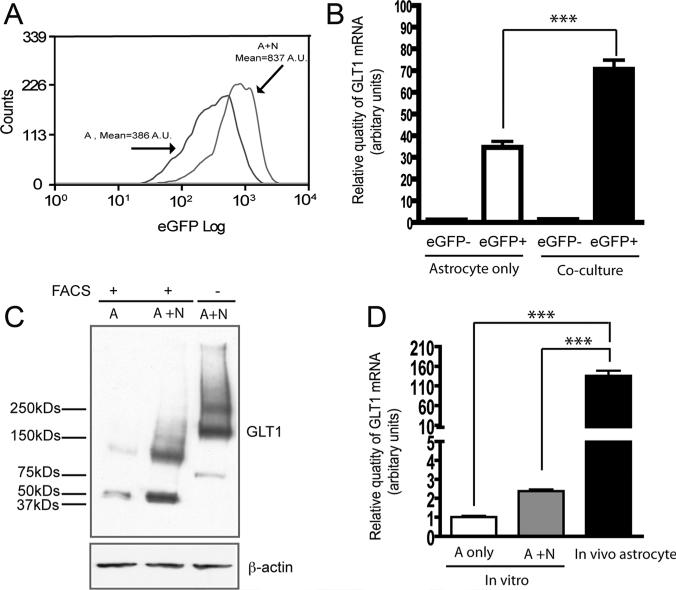
Direct overlay of the eGFP histogram from astrocyte alone and astrocyte neuron co-culture samples showed that eGFP intensity increased from 386 (A.U.) to 837 (A.U.) in eGFP+ cells in response to neuronal stimulation (Fig. 2A). In addition, QRT-PCR analysis also found that GLT1 mRNA levels in eGFP+ cells isolated from co-cultures were 2 fold higher than that in eGFP+ cells isolated from astrocyte cultures alone (Fig. 2B), while GLT1 mRNA levels in eGFP- cells isolated from both conditions were undetectable (Fig. 2B), further suggesting that eGFP- cells in primary astrocyte cultures are likely to be other cell types. The GLT1 protein levels in eGFP+ cells isolated from co-cultures are comparable to the GLT1 levels in astrocytes from normal co-cultures without FAC sorting, and also showed clear increase compared with that in eGFP+ cells from astrocyte culture alone. However, the molecular weight of GLT1 protein, including monomers and multimers, shifted downward to 25 kD (monomer) or 50 kD (dimer) less than the molecular weight of GLT1 protein from normal co-cultures without sorting (Fig. 2C). The difference in the molecular weight of GLT1 protein may be caused by the proteolytic cleavage of extracellular domain of GLT1 protein during FAC sorting process, though exact mechanisms are unclear. We further found that GLT1 mRNA levels in eGFP+ cells isolated from mouse adult brain are 50- to 100-fold higher than that in eGFP+ cells isolated from in vitro astrocyte cultures (Fig. 2D), which is consistent with the GLT1 protein level differences observed from in vitro cultures and in vivo adult brain tissue. The analysis of GLT1 mRNA and protein expression levels in eGFP+ cells isolated from different conditions clearly indicates that these eGFP+ cells respond very similarly as wild type astrocytes to neuronal stimulation. Thus, it is feasible to examine the correlation between DNA methylation changes on the GLT1 promoter and the GLT1 mRNA increases in response to neuronal stimulation using these cells.
Fig. 2.
Expression of GLT1 mRNA and protein in isolated eGFP+ astrocytes. (A) Histogram overlay of eGFP+ cells from astrocytes cultured alone and astrocyte neuron co-cultures. A, astrocyte alone; A+N, astrocyte and neuron co-culture. (B) Relative GLT1 mRNA levels in eGFP+ and eGFP- cells isolated from astrocyte cultured alone and astrocyte neuron co-cultures (n = 9, ***P < 0.001, one-way ANOVA and Bonferroni test was used). (C) GLT1 protein expression levels in eGFP+ cells from astrocytes cultured alone and astrocyte neuron co-cultures (n = 3) A, astrocyte alone; A+N, astrocyte and neuron co-culture. (D) Relative GLT1 mRNA levels in eGFP+ cells isolated from in vitro astrocyte cultures and mouse adult brain. (n = 9, ***P < 0.001, One-way ANOVA and Bonferroni).
Methylation Profile of CpG Sites on GLT1 Promoter from Neuron Astrocyte Co-cultures and Mouse Adult Brain
Full-length GLT1 genomic promoter was first analyzed using the MethPrimer program that shows the distribution of individual CpG site on the sequence (Li and Dahiya, 2002). CpG sites analysis showed that the GLT1 promoter has a typical distribution of CpG in a functional promoter, i.e., dense CpG sites in the proximal region of the promoter (within 1 kb upstream of the transcription start site) and 5′UTR (Fig. 3A), while very few CpG sites were seen in further upstream promoter regions (data not shown). CpG distribution pattern on the GLT1 promoter is consistent with that on the human EAAT2 promoter described before (Zschocke et al., 2007). To examine the methylation pattern on the GLT1 promoter, special bisulfite sequencing primers were designed to selectively amplify the individual region of interest on the promoter: Me-0, 1, 2, 3 (Fig. 3A), which covers most of the CpG sites on the proximal region of GLT1 promoter and 5′UTR. The number of CpG sites in each region is shown in Fig. 3A. Methylation profile of these 83 CpG sites on the GLT1 promoter was examined in eGFP+ astrocytes collected by FAC sorting from either astrocyte cultures alone or astrocyte neuron co-cultures, or mouse adult brain (8- to 12-weeks old) of BAC GLT1 eGFP reporter mice.
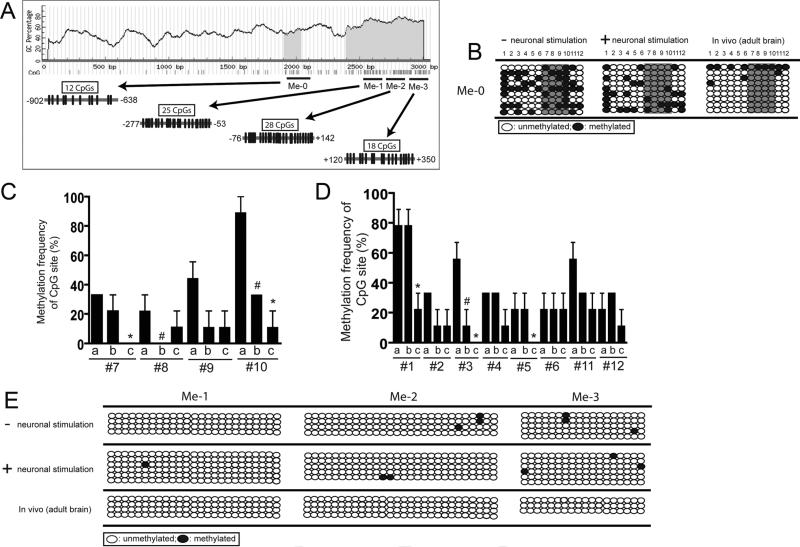
Fig. 3.
Methylation profile of CpG sites on GLT1 promoter in astrocytes under physiological conditions. (A) Distribution of CpG sites and location of bisulfite primers on the proximal GLT1 promoter and 5′UTR. Predicated CpG island area was highlighted as grey. Black circle, methylated; clear circle, unmethylated. (B) Methylation status change of CpG sites in Me-0 region on GLT1 promoter of astrocytes under different conditions. Sequences from multiple clones (n = 9 from 3 replicated experiments for each condition) were examined. (C) Quantitative analysis of the methylation status of selective CpG sites (from #7 to #10) on GLT1 promoter under different conditions. (D) Quantitative analysis of the methylation status of selective CpG sites (from #1 to #6 and #11 to #12) on GLT1 promoter under different conditions. 0%, unmethylated; 100%, methylated; a, untreated astrocytes; b, astrocytes co-cultured with neurons; c, astrocytes isolated from adult mouse brain (n = 9, *P < 0.05, between a and c; #P < 0.05, between a and b) (E) Methylation status of CpG sites in Me-1, 2, 3 regions of GLT1 promoter in astrocytes under different conditions. Sequences from multiple clones (n ≥ 4 from two replicate experiments for each condition) were examined.
As shown in Fig. 3B, dynamic changes of the methylation status in individual CpG sites in the Me-0 region was found under different conditions. Extensive overall methylation on CpG sites in this region was found in astrocytes isolated from astrocytes cultured alone. The overall methylation levels were reduced in astrocytes isolated from astrocyte-neuron co-cultures, and a very low level of overall methylation of CpG site was found in this region in astrocytes isolated from mouse adult brain. The quantitative analysis of the methylation status of CpG sites on Me-0 region of GLT1 promoter is summarized in Fig. 3C (CpG sites from #7 to #10) and Fig. 3D (CpG sites from #1 to #6 and #11, #12). In particular, CpG sites from #7 to #10 in this region are hypermethylated in astrocytes without neuronal stimulation, but not in astrocytes with neuronal stimulation. Moreover, #1 CpG site is only hypermethylated in in vitro astrocytes, regardless of the presence of neurons, but not in in vivo astrocytes. In contrast, very low levels of methylation were found on other regions of the GLT1 promoter (Me-1, 2, 3) in astrocytes in vitro and in vivo (Fig. 3E), suggesting that the methylation status of CpG sites is selectively altered under different conditions. The gradually decreased methylation levels of CpG sites in Me-0 region of GLT1 promoter in astrocytes correlate with gradually increased GLT1 mRNA levels under these conditions described above (Fig. 2B,D), implicating that CpG methylation, especially the methylation on CpG sites from #7 to #10 in Me-0 region may play an inhibitory role in neuron-dependent transcriptional regulation of GLT1 promoter.
Methylation Induced Functional Change of the GLT1 Promoter
Sequence alignment of rodent GLT1 promoter and human EAAT2 promoter in Me-0 region found several putative transcription factor binding sites (Fig. 4A). In particular, the CpG sites from #7 to #10 are located in the putative binding site of HEN1 (Hua enhancer 1)/AP4 (activating enhancer binding protein 4)/LBP1 (upstream binding protein 1) factors. Functional EMSA binding assay with oligos that have either methylated or unmethylated CpG sites from #7 to #10 showed specific binding with nuclear extracts prepared from mouse adult cortex (Fig. 4B). Interestingly, a unique shift band was only observed in the reaction with the methylated oligo, suggesting that methylation on these CpG sites may recruit additional nuclear factors to bind to the oligo for subsequent transcriptional repression. To demonstrate whether methylation on selective CpG sites changes the GLT1 promoter activity, linear mouse GLT1 promoter luciferase reporter with site-specific methylation on selective CpG sites were prepared and transfected into HEK cells. Primers with site-specific methylation on selective CpG sites were directly chemically synthesized from Integrated DNA Technologies (IDT, Coralville, IA) (Fig. 4C). As shown in Fig. 4D, selective methylation of #10 CpG site decreased the GLT1 promoter activity. Methylation of 3 CpG sites, #8, #9, and #10, did not further reduce GLT1 promoter activity. The functional promoter assay results further indicated that selective methylation on GLT1 promoter, especially the #10 site in Me-0 region of GLT1 promoter may result in transcriptional repression of GLT1 promoter.
Methylation Profile of Selective CpG Sites on EAAT2 Promoter in Motor Cortex of Postmortem ALS Patient
Severe loss of EAAT2 protein has been found in the motor cortex and spinal cord of postmortem ALS patients. Although early studies found no alteration of EAAT2 mRNA levels in these ALS tissues, these studies were comprised by bulk tissue homogenization and low sensitivity of detection approaches (Bristol and Roth-stein, 1996). Our recent studies clearly demonstrated reduced GLT1 mRNA in the lumbar spinal cord of rodent models of ALS using in situ hybridization analysis and more sensitive QRT-PCR approaches, suggesting that transcriptional dysfunction of GLT1 promoter contributes to the GLT1 protein loss observed in disease (Yang et al., 2009). The association between potential EAAT2 promoter dysregulation and the methylation status of EAAT2 promoter remains unexplored in ALS and other neurodegenerative disorders.
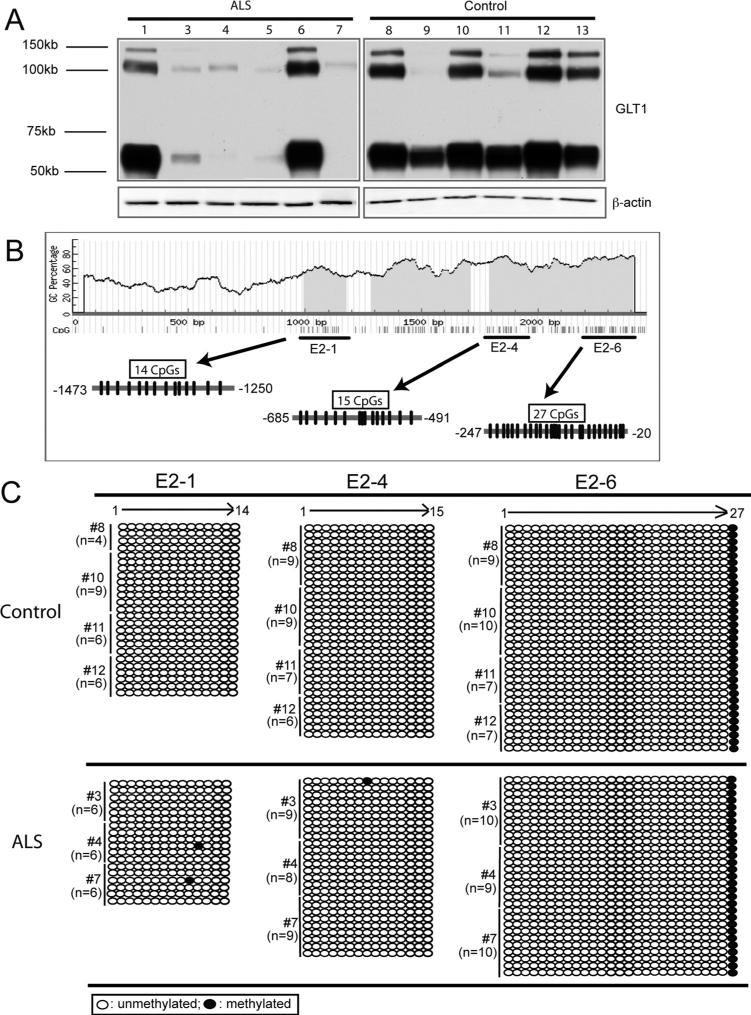
Postmortem motor cortex tissues from patients with ALS were collected for protein and methylation analysis. GLT1 mRNA levels from these tissues were not analyzed as total RNA prepared from these tissues failed to pass quality control, possibly due to long-term storage and repeated partial thaw-freeze cycle of tissue. The expression levels of GLT1 protein from age-matched control and patient with ALS tissues were shown in Fig. 4A. Severe loss of GLT1 protein was found in the motor cortex of most of the patient with ALS samples, while abundant GLT1 protein was found in the motor cortex of age-matched control samples. To examine the methylation change of CpG sites on the EAAT2 promoter, special bisulfite sequencing primers were designed to selectively amplify the region of interest on EAAT2 promoter, namely as E2−1, 4, 6 individually (Fig. 4B). The selection of the region of interest was based on the hypermethylation pattern of selective CpG sites on EAAT2 promoter in glioma cell lines described previously (Zschocke et al., 2007) and the methylation analysis of GLT1 promoter mentioned earlier. The number of CpG sites in each region was shown in Fig. 4B. High-quality genomic DNA from multiple patient with ALS samples (#3, #4, and #7) and age-matched control samples (#8, #10, #11, and #12) were treated with bisulfite and subject to the methylation analysis. Multiple bacteria clones of each EAAT2 promoter region (E2−1, 4, and 6) were sequenced, and the methylation status of each CpG site was recorded from the sequence. As shown in Fig. 4C, low levels of overall methylation were found on the regions examined on EAAT2 promoter in both patients with ALS and control samples. In E2−1 region, 1 of 18 bacteria clones examined found methylated on the #10 and #11 CpG sites in ALS samples, respectively, but no methylation was found in control samples. In E2−4 region, 1 of 26 clones examined found methylated on the #8 CpG site in ALS sample with no methylation found in control samples. In E2−6 regions, only #27 CpG site was found methylated in all bacterial clones examined from all ALS and control samples.
DISCUSSION
This study provides the first DNA methylation-based epigenetic analysis of the astroglial GLT1 promoter under physiological conditions and the first attempt to probe cell specific epigenetic changes in ALS. In this study, DNA methylation change of 83 CpG sites on the astroglial GLT1 promoter in response to neuronal stimulation in vitro and in vivo was investigated by selectively isolating GLT1+ astrocytes through FAC sorting. We found that neuron-induced demethylation of certain CpG sites on GLT1 promoter is closely associated with the neuron-dependent GLT1 mRNA increase in astrocytes. This suggests that neurons can act to modify epigenetic regulation of the astroglial transporter genes. However, methylation pattern of selected 56 CpG sites on the EAAT2 promoter from motor cortex of both ALS and age-matched control patients were not significantly different. In addition, almost all CpG sites were hypomethylated in all the samples examined. Thus, despite the ability of neurons to regulate methylation of the astroglial transporter—in a disease involving transporter dysregulation—methylation of this promoter does not appear to be pathophysiologically relevant.
The understanding of astroglial functions in the CNS has been partially limited by the difficulty to isolate specific astrocyte populations from adult CNS for biochemical or molecular analysis. By FAC sorting the eGFP+ cells from BAC GLT1 eGFP mice, astrocytes were successfully isolated from in vitro cultures and in vivo adult brain. This approach provides the ability to cross correlate in vitro astroglial analysis to endogenous astroglia in adult brain, and to examine the molecular changes of astrocytes in response to different physiological stimuli. In addition, this also allows investigation of the molecular changes or identification of disease related genes in diseased astrocytes by profiling the gene expression from disease-transgenic mouse models mated to BAC GLT1 eGFP reporter mice. Similar antibody-based fluorescence-activated cell sorting (FACS) procedures have achieved success in isolating astrocytes from young adult mice brain tissue in recent astroglial gene profiling studies (Lovatt et al., 2007).
CpG site analysis revealed that the GLT1 promoter, especially the proximal sequence, contains many CpG sites. Our results showed that one area (Me-0) of CpG sites on the promoter was demethylated under neuronal influences in vitro, and the methylation level was further reduced in astrocytes collected from adult brain tissue. As a greater interaction between astrocytes and neurons are present in adult brain, the greater demethylation levels of GLT1 promoter likely reflected the greater degree of neuron-astrocyte interaction in vivo. More interestingly, the demethylation of the GLT1 promoter, especially the demethylation on CpG sites from #7 to #10 in Me-0 region, were highly correlated with the increase of GLT1 mRNA levels under these conditions. In particular, an in vitro versus in vivo comparison of the demethylation on GLT1 promoter in astroglia correlated with a 50- to 100-fold increase of GLT1 mRNA, implicating a strong role of DNA methylation based epigenetic regulation in the transcriptional activation of the GLT1 promoter. Artificial in vitro enzymatic methylation of all CpG sites on the EAAT2 promoter also resulted in complete loss of EAAT2 promoter activity in primary astrocytes (Zschocke et al., 2007). Moreover, gel shift pattern changes induced by the oligo with methylation of selective CpG sites also suggested that changes of these CpG sites are likely due to recruitment of nuclear factors to the GLT1 promoter. Sequence alignment of the rodent GLT1 promoter and human EAAT2 promoter revealed that putative binding sites of transcription factors HEN1, AP4, and LBP1 are located in the region of GLT1 promoter with dramatic methylation changes. Future studies will determine if the methylation status of these CpG sites alters binding of the nuclear factors. Notably, previous studies demonstrated that DNA methylation-induced binding changes of transcriptional factor STAT is involved in the transcriptional regulation of the GFAP promoter (Song and Ghosh, 2004). The involvement of other epigenetic features, including posttranslational histone modifications, including acetylation, methylation, and phosphorylation, in the regulation of GLT1 promoter activation in astrocytes remain to be investigated in future studies.
DNA-based epigenetic changes have been found in postmortem tissue of some neurological disorders. To date, these early studies have focused on tissue rather than cell specific targets. Our study investigated the possible involvement of epigenetic change in ALS by examining the DNA methylation of the astroglial EAAT2 promoter in both ALS and control motor cortex tissue. Although EAAT2 protein is markedly downregulated in ALS motor cortex, most of the CpG sites on EAAT2 promoter examined were hypomethylated, and no difference of the DNA methylation was found between ALS and control samples. Thus, the loss of EAAT2 protein in ALS does not appear to be the result of epigenetic regulation of the promoter. In contrast, the limitations of using human postmortem tissues could compromise the methylation analysis of EAAT2 promoter. First, the loss of EAAT2/GLT1 in patient with ALS or transgenic rodent models of ALS is focal, which makes it difficult to detect the methylation changes in tissue typically analyzed by bulk tissue homogenization methods. Second, the EAAT2 mRNA could not be determined as the RNA failed to pass quality control due to the repeated freeze-thaw cycle. Third, the complex heterogeneous nature, i.e., mixture of many cell types, of motor cortex samples could also prohibit the detection of cell specific or micro-region specific methylation changes in ALS. Fourth, although we specially chose samples with known normal and abnormal EAAT2 protein changes, our total sample size for control and patients with ALS was small. A large-scale whole genome DNA methylation analysis of the pathological tissues of larger number of patients with ALS and controls is needed in the future to reveal the possible epigenetic changes involved in ALS astroglia.
Fig. 5.
Methylation profile of CpG sites on EAAT2 promoter in motor cortex of postmortem ALS patient. (A) GLT1 protein expression in motor cortex of postmortem ALS patient and age-matched control (n = 6 for control and n = 6 for ALS). (B) Distribution of CpG sites and location of bisulfite primers on EAAT2 promoter. (C) Methylation status of selective CpG sites on EAAT2 promoter from ALS and control motor cortex samples. Black circle: methylated; clear circle: unmethylated. Sequences from multiple clones (n = 18−25 for E2−1 region, n = 26−31 for E2−4 region, n = 29−33 for E2−6 region) from patients with ALS (n = 3) and control (n = 4) tissue were examined.
Acknowledgments
Grant sponsor: NIH; Grant numbers: NS33958, NS036465; Grant sponsor: Muscular Dystrophy Association.
REFERENCES
- Amir RE, Van den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Yamanaka K, Lobsiger CS, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Bristol LA, Rothstein JD. Glutamate transporter gene expression in amyotrophic lateral sclerosis motor cortex. Ann Neurol. 1996;39:676–679. doi: 10.1002/ana.410390519. [DOI] [PubMed] [Google Scholar]
- Chen W, Aoki C, Mahadomrongkul V, et al. Expression of a variant form of the glutamate transporter GLT1 in neuronal cultures and in neurons and astrocytes in the rat brain. J Neurosci. 2002;22:2142–2152. doi: 10.1523/JNEUROSCI.22-06-02142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Mahadomrongkul V, Berger UV, et al. The glutamate transporter GLT1a is expressed in excitatory axon terminals of mature hippocampal neurons. J Neurosci. 2004;24:1136–1148. doi: 10.1523/JNEUROSCI.1586-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:101–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, et al. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Lievens JC, Woodman B, Mahal A, et al. Impaired glutamate uptake in the R6 Huntington's disease transgenic mice. Neurobiol Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- Lobsiger CS, Cleveland DW. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci. 2007;10:1355–1360. doi: 10.1038/nn1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovatt D, Sonnewald U, Waagepetersen HS, et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27:12255–12266. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, et al. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Martin LJ, Kuncl RW. Decreased glutamate transport by the brain and spinal cord in amyotrophic lateral sclerosis. N Engl J Med. 1992;326:1464–1468. doi: 10.1056/NEJM199205283262204. [DOI] [PubMed] [Google Scholar]
- Sitcheran R, Gupta P, Fisher PB, Baldwin AS. Positive and negative regulation of EAAT2 by NF-kappaB: A role for N-myc in TNFalpha-controlled repression. EMBO J. 2005;24:510–520. doi: 10.1038/sj.emboj.7600555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Swanson RA, Liu J, Miller JW, et al. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. Epilepsy and exacerbation of brain injury in mice lacking glutamate transporter GLT-1. Science. 1997;278:21. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Watkins A, et al. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61:880–894. doi: 10.1016/j.neuron.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschocke J, Allritz C, Engele J, Rein T. DNA methylation dependent silencing of the human glutamate transporter EAAT2 gene in glial cells. Glia. 2007;55:663–674. doi: 10.1002/glia.20497. [DOI] [PubMed] [Google Scholar]