Abstract
Mdm2 inhibitors represent a promising class of p53 activating compounds that may be useful in cancer treatment and prevention. However, the consequences of pharmacological p53 activation are not entirely clear. We observed that Nutlin-3 triggered a DNA damage response in azoxymethane-induced mouse AJ02-NM0 colon cancer cells, characterized by the phosphorylation of H2AX (at Ser-139) and p53 (at Ser-15). The DNA damage response was highest in cells showing robust p53 stabilization, it could be triggered by the active but not the inactive Nutlin-3 enantiomer, and it was also activated by another pharmacological Mdm2 inhibitor (Caylin). Quantification of γH2AX-positive cells following Nutlin-3 exposure showed that approximately 17% of cells in late S and G2/M were mounting a DNA damage response (compared to a ~50% response to 5-fluorouracil). Nutlin-3 treatment caused the formation of double strand DNA strand breaks, promoted the formation of micronuclei, accentuated strand breakage induced by doxorubicin and sensitized the mouse colon cancer cells to DNA break-inducing topoisomerase II inhibitors. Although the HCT116 colon cancer cells did not mount a significant DNA damage response following Nutlin-3 treatment, Nutlin-3 enhanced the DNA damage response to the nucleotide synthesis inhibitor hydroxyurea in a p53-dependent manner. Finally, p21 deletion also sensitized HCT116 cells to the Nutlin-3-induced DNA damage response, suggesting that cell cycle checkpoint abnormalities may promote this response. We propose that p53 activation by Mdm2 inhibitors can result in the slowing of double stranded DNA repair. Although this effect may suppress illegitimate homologous recombination repair, it may also increase the risk of clastogenic events.
Keywords: p53, Mdm2 inhibitors, Nutlin-3, double strand DNA breaks, γH2AX, doxorubicin
1. Introduction
p53 is activated following DNA damage through the phosphorylation of specific N-terminal serine residues, which prevents p53 from interacting with its negative regulator, Mdm2 [1–3]. Gene expression changes induced by p53 lead either to cell cycle arrest, which enables cells to repair DNA damage, or to apoptosis[4]. In addition to facilitating the repair or removal of damaged cells, p53 can also suppress cancer development after oncogene activation. Cells expressing activated oncogenes can initiate a checkpoint pathway that culminates in the expression of p19ARF (p14 in humans), which activates p53 by binding and neutralizing Mdm2 [5–9]. Interestingly, oncogene activation has also been reported to induce DNA replication stress, which results in prematurely terminated DNA replication forks, double-strand breaks and p53 activation through the ATM pathway [10–14]. The oncogene-induced activation of p53 through these two mechanisms appears to suppress carcinogenesis; in a number of mouse genetic models p53 activation in cancer cells can trigger tumor regression [15–17]. Understanding how p53 is regulated in normal and transformed cells could provide important insight into how its activity could be manipulated for cancer treatment and prevention.
Mdm2 inhibitors have been developed that may be able to reinforce the anti-cancer activities of p53 in cancers and pre-cancerous lesions [18, 19]. One potential advantage of the Mdm2 inhibitors is that they activate p53 directly, unlike most other chemotherapeutic compounds that work through the formation of DNA lesions and strand breaks. This property of Mdm2 inhibitors may reduce their general toxicity and the risk of therapy-induced neoplasms. The availability of relatively non-toxic p53 activators also raises the possibility that these compounds could be employed as cancer preventive agents to treat high-risk individuals with pre-cancerous lesions, prior to the mutational loss of a functional p53. However, it is not entirely clear how p53 activation though this direct pharmacological mechanism compares to that mediated by a DNA damage response, either on a cellular or tissue-level basis.
To determine the cellular consequences of p53 activation in colon cancers, we have been studying the mouse AOM model of colon cancer. Lesions formed in this model are generally non-invasive, genetically stable, possess a sequence-normal p53 gene, and thus appear to be roughly equivalent to late adenomas in humans [20–24]. In addition to possessing a sequence normal p53 gene, these lesions express the p19ARF protein, which indicates that at least this portion of the oncogene checkpoint pathway has been mobilized [20]. Although this checkpoint pathway may be important for slowing the progression of AOM-induced lesions, it appears to be insufficient for preventing tumor formation. The elevated expression of the Mdm2 in AOM-induced tumors appears to be partly responsible for suppressing p53 activity [20]. Treatment of these lesions ex vivo with the Mdm2 inhibitor Nutlin-3 generates a robust and selective activation of p53 target genes, relative to normal adjacent tissue [25]. These findings suggest that Mdm2 inhibitors may provide an effective method for cancer prevention or treatment in this model, and potentially in early human colonic lesions that possess an intact p53 gene.
In this present report, we find that p53 activation by Mdm2 inhibitors can themselves induce a DNA damage response under certain situations. We discuss the potential mechanism for the induction of this DNA damage response, and the implications of these findings for the potential clinical application of Mdm2 inhibitors.
2. Material and Methods
2.1. Cell culture and treatments
AJ02-NM0 cells were cultured in RPMI 1640 with Glutamax (Invitrogen, Carlsbad, CA) supplemented with 5% (v/v) fetal bovine serum (Lonza, Rockland, ME ), 5% (v/v) heat-inactivated horse serum (Invitrogen, Carlsbad, CA), 1% (v/v) insulin-transferrin-selenium (Gibco, Grand Island, NY), 100 mM non-essential amino acids (Invitrogen, Carlsbad, CA) and antibiotic–antimycotic (Invitrogen, Carlsbad, CA). Doxorubicin and 5-fluorouracil (5-FU) were purchased from Sigma (St. Louis, MO) and used at a final concentration of 500 nM and 100 µM, respectively. The Mdm2 inhibitors, Nutlin-3 and Caylin were purchased from Cayman Chemical (Ann Arbor, MI), and stored frozen as a 10 mM stock solution in DMSO.
2.2. Cell fractionation
AJ02-NM0 cells were washed twice with cold phosphate buffer saline (PBS) and incubated in lysis buffer A [10 mM Hepes, pH 7.6, 15 mM KCl and 2 mM MgCl2 plus 0.1% (v/v) Nonidet P-40] supplemented with proteinase/phosphatase inhibitor cocktails (Sigma, St. Louis, MO) and 1 mM DTT for 8 min on ice. The cells were then scraped into tubes and centrifuged for 10 min at 4° C (14,000 rpm). The supernatant was the cytoplasmic extract and the resulting nuclear pellets were rinsed with the above buffer A without NP-40. Nuclear extracts were prepared by resuspending nuclear pellets with a high-salt buffer C [20 mM Hepes, pH 7.6, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 5% glycerol and proteinase/phosphatase inhibitor cocktails (Sigma, St. Louis, MO)], incubating on ice for 40 min and then centrifuging for 10 min at 4° C. Total protein in the extracts was quantified using the Bio-Rad protein assay (BioRad, Hercules, CA).
2.3. Immunoblotting and immunofluorescence
For immunoblotting studies, 10 µg of protein was denatured under reducing conditions, separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose by voltage gradient transfer. The resulting blots were blocked with 5% (w/v) non-fat dry milk in PBS + 0.1% (v/v) Tween. Specific proteins were detected with appropriate antibodies using enhanced chemiluminescence detection (Santa Cruz Biotechnology, Santa Cruz, CA) as recommended by the manufacturer. Immunoblotting antibodies were p53 (Ab-1) and phospho-p53 Ser-15 (Ab-3) from Calbiochem (San Diego, CA). The anti-actin antibody (I-19) used was from Santa Cruz Biotechnogy (Santa Cruz, CA).
For immunofluorescence, AJ02-NM0 cells were washed with cold PBS and fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature. The PFA was removed, and permeabilizing reagent (0.5% Triton X-100) was added to the cells for 10 min at room temperature. Cells were then incubated in 5% serum for 30 min, to block non-specific antibody binding. After blocking, the cells were incubated with primary antibody at a 1:100 dilution followed by incubation with FITC conjugated or Cy-3 conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells were counterstained with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) (10µg/ml) (Invitrogen, Eugene, Oregon). Fluorescent imaging was performed on an inverted microscope (Eclipse TE 300; Nikon, Melville, NY, USA) using a 20X objective. Images were acquired using a CCD camera (Quantix 57, Roper Scientific, Tuscon, AZ). The H2AX rabbit polyclonal antibody (sc-101696) from Santa Cruz Biotechnology was used for these studies.
2.4. Cell Cycle Analysis
Cells were fixed with 4% (w/v) PFA at room temperature for 10 minutes and permeabilized with PBS plus 0.5% (v/v) Triton-X100 for 10 minutes. The cells were washed with PBS and blocked with 5% (v/v) serum in PBS. Cells were then stained for γH2AX using a p-Histone H2AX (Ser-139) primary antibody (Santa Cruz Biotechnology) at 1:100 dilution in 5% (v/v) serum and a FITC conjugated secondary antibody (Jackson Immuno Labs) at 1:200 dilution in 5% serum. Following staining, cells were harvested by trypsinization and stained with 30 µg/ml propidium iodide with 0.3 mg/ml RNase A. At least 6,000 cells were evaluated for fluorescence using a Becton Dickinson FACSCalibur flow cytometer (San Jose, CA).
2.5. Cell viability assay
Cells were plated in triplicate into 96-well plates and treated with doxorubicin (0 to 1.0 µM), etoposide (0 to 300 µM) or 5-fluorouracil (5FU; 0 to200 µM) plus 0.2% (w/v) DMSO (vehicle control) or 20 µM Nutlin-3. After a 40-h exposure, cytotoxicity was assessed using the CellTiter 96 Aqueous One kit (Promega, Madison, WI) according to manufacturer’s protocol. The resulting absorbance values were averaged and expressed as a fraction of control vehicle.
2.6. DNA analysis
Field inversion gel electrophoresis (FIGE) was used to resolve high molecular weight fragments resulting from DNA cleavage in whole cells. Agarose plugs containing cells were digested for 24 h at 56° C in buffer A (0.5M EDTA, 1% sarkosyl, 1mg/ml Proteinase K). The plugs were washed six times in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and stored at 4° C. Plugs (approximately 0.5–1 ×107 cells) were loaded onto a 14 cm long gel casted with 1% (w/v) SeaKemR Gold Agarose (Lonza, Rockland, ME) in 0.5 × TBE (45 mM Tris, 45 mM borate, 1 mM EDTA) and electrophoresis was carried out at 22° C using the FIGE Mapper electrophoresis system with a buffer circulation pump (BioRad, CA, USA). The total run time was 14 hours with forward and reverse voltages of 140V and 80V, respectively. The forward switch time was increased linearly to 24 sec, and the reverse switch time was increased linearly to 8 sec over the 14 hr run. Two sets of standards were used; the Yeast Chromosome PFG Marker (225 – 1,900 kb) and the MidRange PFG Marker I (15 – 300 kb) [New England Biolabs, MA]. The gel was stained with ethidium bromide for visualization and photography.
2.7. Micronulei Assay
AJ02-NM0 cells were grown in 35mm culture dishes for 24 hours to near confluency. Cells were then treated with DMSO (0.2%) or Nutlin-3 (20µM) for 24 hours. The media was removed and replaced with media containing the inhibitor of microfilament formation, cytochalasin-B (1µg/µl) (Enzo Life Sciences, Farmingdale, NY). After 24 hours cells were fixed with 4% PFA and stained with DAPI (10µg/ml). Approximately 700 binucleated cells per treatment were evaluated for the presence of micronuclei.
2.8. Caspase 3 assay
Cells cultured and treated on 24 well plates were washed with cold PBS and lysed in 100 µl of buffer containing 10 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1 mM EDTA and 0.01% Triton X-100 using a single freeze-thaw cycle. 50 µl of this extract was combined with 50 µl 2X Reaction buffer containing 20 mM PIPES, pH 7.4, 4 mM EDTA, 0.2% CHAPS and 0.2 mM of caspase 3 substrate Z-DEVD-AMC (Enzo Life Sciences International, Plymouth Meeting, PA). The increase in fluorescence was determined over the course of one hour. The change is fluorescence was then normalized to the total protein content of the extract.
3. Results
3.1. Nutlin-3 causes p53 phosphorylation in AJ02-NM0 cells
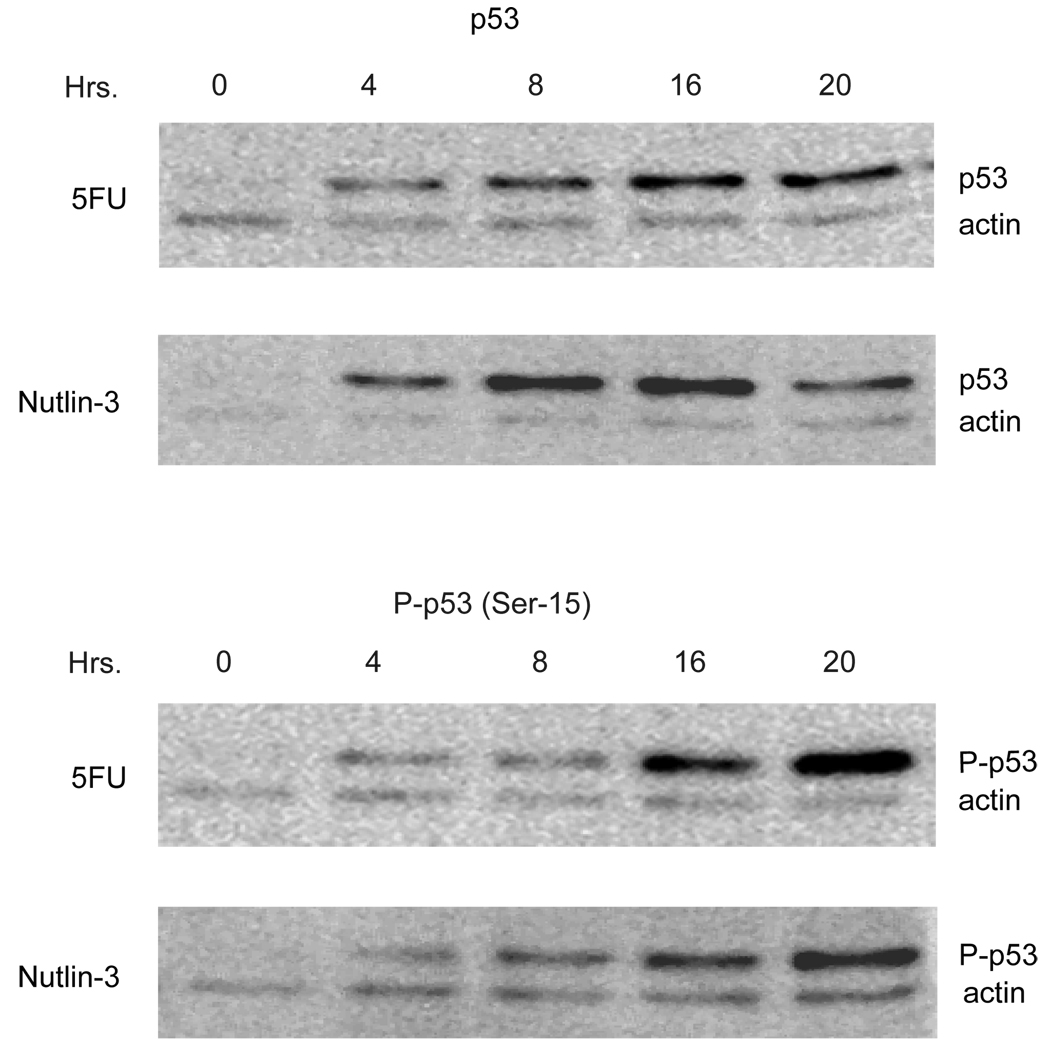
The activation of p53 in cancer cells with Mdm2 inhibitors, such as Nutlin-3, has been described to occur with a limited number of post-translational p53 modifications [26]. In particular, it has been found that the ATM target site on human p53, Ser-15, is not phosphorylated after treatment of a number of cancer cell lines following Nutlin-3 stimulation [26]. To further develop the mouse AOM model to test the efficacy of Mdm2 inhibitors, we determined whether p53 activation likewise occurred in the absence of Ser-15 phosphorylation in cells derived from an AOM-induced colon tumor [27]. As shown in Figure 1, treatment of AJ02-NM0 cells with Nutlin-3 generated a rapid stabilization of p53. Unexpectedly, an antibody specific for p53 phosphorylated at Ser-15 showed that this modification was in fact occurring. The level of p53 phosphorylation induced by Nutlin-3 was comparable to that obtained following treatment with 5-fluorouracil (5FU), a thymidine synthesis inhibitor that is known to trigger a DNA damage response as a result of uracil incorporation into the DNA. The extent of p53 phosphorylation appeared to be most pronounced at the later time points, so subsequent analysis employed a 24 hour Nutlin-3 exposure.
Figure 1.
Treatment of AJ02-NM0 cells with Nutlin-3 stabilizes p53 and leads to the phosphorylation of p53 at Ser-15. AJ02-NM0 cells were treated with Nutlin-3 (20µM) or 5-FU (100µM) for the indicated lengths of time. Nuclear extracts were then prepared and immunoblotted for p53 or p53 phosphorylated at Ser-15. Immunoblots were also probed with actin, which served as a loading control.
3.2. Nutlin-3 causes H2AX phosphorylation in AJ02-NM0 cells
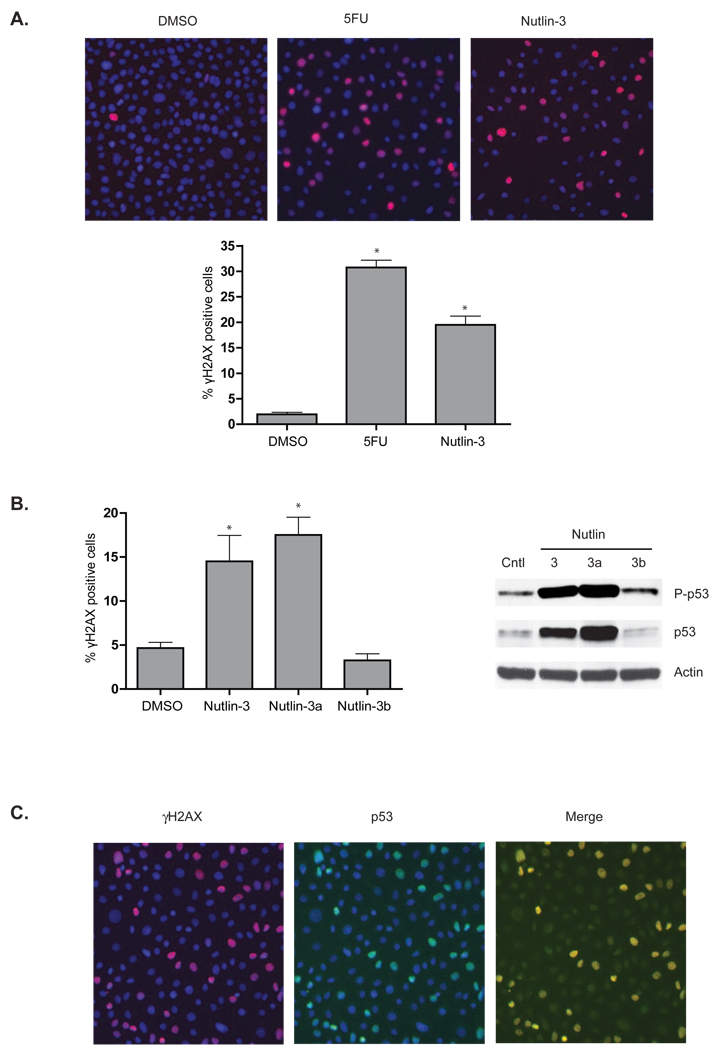
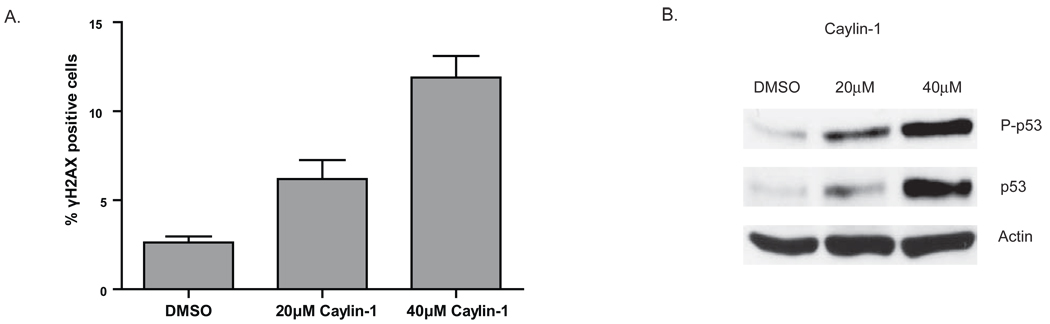
Phosphorylation of histone H2AX at Ser-139 to form γH2AX following DNA damage was used to further examine the impact of Nutlin-3 on the DNA damage response [28]. As shown in Figure 2A, Nutlin-3 increased the number of AJ02-NM0 cells expressing γH2AX several fold, consistent with Nutlin-3 inducing a DNA damage response. This degree of activation was similar to that obtained from 5FU treatment (Figure 2A). We also compared the ability of the active and inactive Nutlin-3 enantiomers to increase γH2AX staining of AJ02-NM0 cells [29]. The active Mdm2 inhibitor Nutlin-3a was able to induce γH2AX staining and p53 phosphorylation, whereas the inactive enantiomer Nutlin-3b was not (Figure 2B). To further examine the relationship between γH2AX and p53 expression, cells were co-stained for these two proteins (Figure 2C). Interestingly, p53 activation was highest in cells that expressed γH2AX, suggesting a connection between p53 stabilization and the DNA damage response. Finally, another Mdm2 inhibitor, Caylin, was also found to activate H2AX and p53 phosphorylation (Figure 3), supporting the role of Mdm2 inhibition in this process.
Figure 2.
A) Nutlin-3 elicits a DNA damage response comparable to 5-FU. AJ02-NM0 cells were treated with DMSO (vehicle control), 5FU, or Nutlin-3 as indicated for 24 hours and analyzed for γH2AX by immunofluorescence (red). Nuclei were counter-stained with DAPI (blue). The top panel shows representative fluorescence images, and the bottom panel shows the fraction of cells staining positively for γH2AX. The asterisks indicate p < 0.01 as determined by an ANOVA with a Bonferroni’s post test. B) The active Nutlin-3 enantiomer (Nutlin-3a) is required for induction of a DNA damage response. AJ02-NM0 cells were treated with DMSO, racemic Nutlin-3, active Nutlin-3a or inactive Nutlin-3b, as indicated. Cells were then processed for γH2AX expression as described in panel 2A and immunoblot for P-p53 (Ser-15). The graph to the left shows the fraction of γH2AX positive cells while the immunoblot on the right shows p53 stabilzation and phosphorylation. Both γH2AX staining and p53 phosphorylation requires the presence of the active enantiomer Nutlin-3a. The asterisks indicate p < 0.01 as determined by an ANOVA with a Bonferroni’s post test. C) Activation of γH2AX is localized to cells in which p53 has been stabilized. AJ02-NM0 cells were treated with Nutlin-3 for 24 hours and analyzed for γH2AX (red) and p53 (green) by immunofluorescence. Images were merged using Image J software.
Figure 3.
A) An alternative inhibitor of Mdm2, Caylin-1, also induces the phosphorylation of H2AX to form γH2AX. AJ02-NM0 cells were treated with DMSO, 20µM Caylin-1, or 40µM Caylin-1 for 24 hours and analyzed for γH2AX by immunofluorescence. The graph shows the fraction of cells staining positively for γH2AX. The asterisks indicate p < 0.01 as determined by an ANOVA with a Bonferroni’s post test. B) Treatment of AJ02-NM0 cells with Caylin-1 leads to the stabilization and phosphorylation of p53. AJ02-NM0 cells were treated with Caylin-1 for 24 hours at the indicated concentrations. Nuclear extracts were then prepared and immunoblotted for p53 or p53 phosphorylated at Ser-15. Immunoblots were also probed with actin, which served as a loading control.
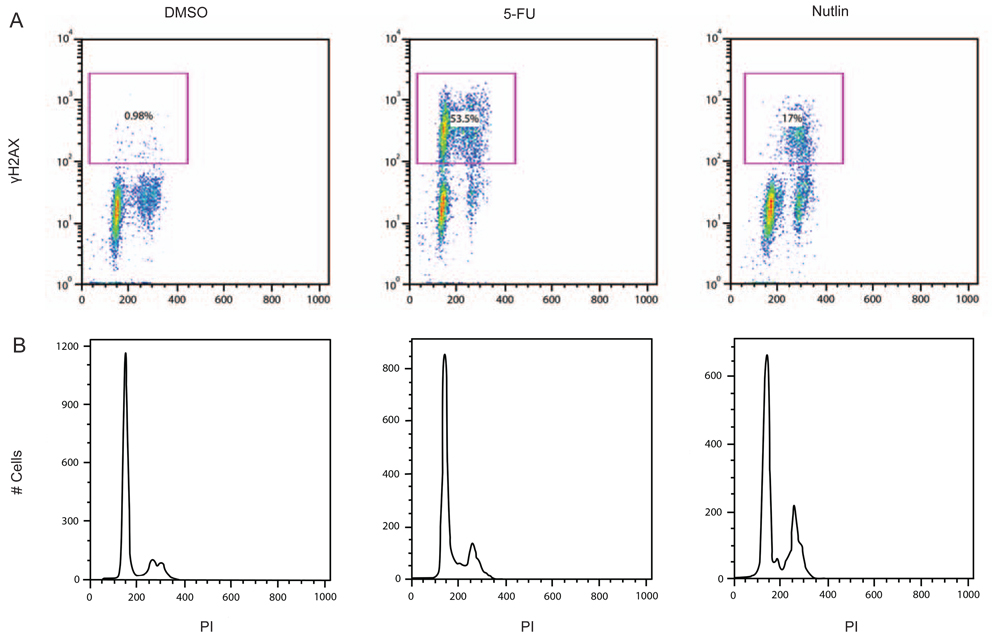
To determine whether the Nutlin-3 activation of γH2AX was cell cycle dependent, cells were immuno-stained for γH2AX expression and DNA content (by propidium iodide) and analyzed by flow cytometry. Figure 4A shows the 2-dimensional plots relating γH2AX expression and DNA content and Figure 4B shows the cell cycle distribution. As shown in Figure 4A, 5FU treatment increased the γH2AX staining in approximately half of the cells. Positively staining cells were apparent in both the diploid and tetraploid populations of the culture. When Nutlin-3-treated cells were analyzed, increased γH2AX expression was observed in approximately 17% of the cells. Interestingly, most of the cells expressing γH2AX following Nutlin-3 treatment appeared to be in late S and G2/M. The cell cycle distribution trace of Nutlin-3-treated cells showed an accumulation of cells at late S and G2/M, suggesting a degree of arrest at this cell cycle phase (Figure 4B).
Figure 4.
A) Nutlin-3 induces a DNA damage response in late S and G2/M. AJ02-NM0 cells were treated with DMSO (vehicle control), 5-FU, or Nutlin-3, as indicated, for 20 hours. Immunofluorescent staining was performed to detect γH2AX levels, which was quantified by flow cytometry. Cells were also analyzed for DNA content by propidium iodide staining. Cells within the square analysis region shown were defined as γH2AX positive. B) Nutlin-3 increases AJ02-NM0 cell accumulation in the late S and G2/M. AJ02-NM0 cells were treated as described in 3A, with the results for propidium iodide DNA staining shown.
3.3. Nutlin-3 treatment generates double strand DNA breaks in AJ02-NM0 cells
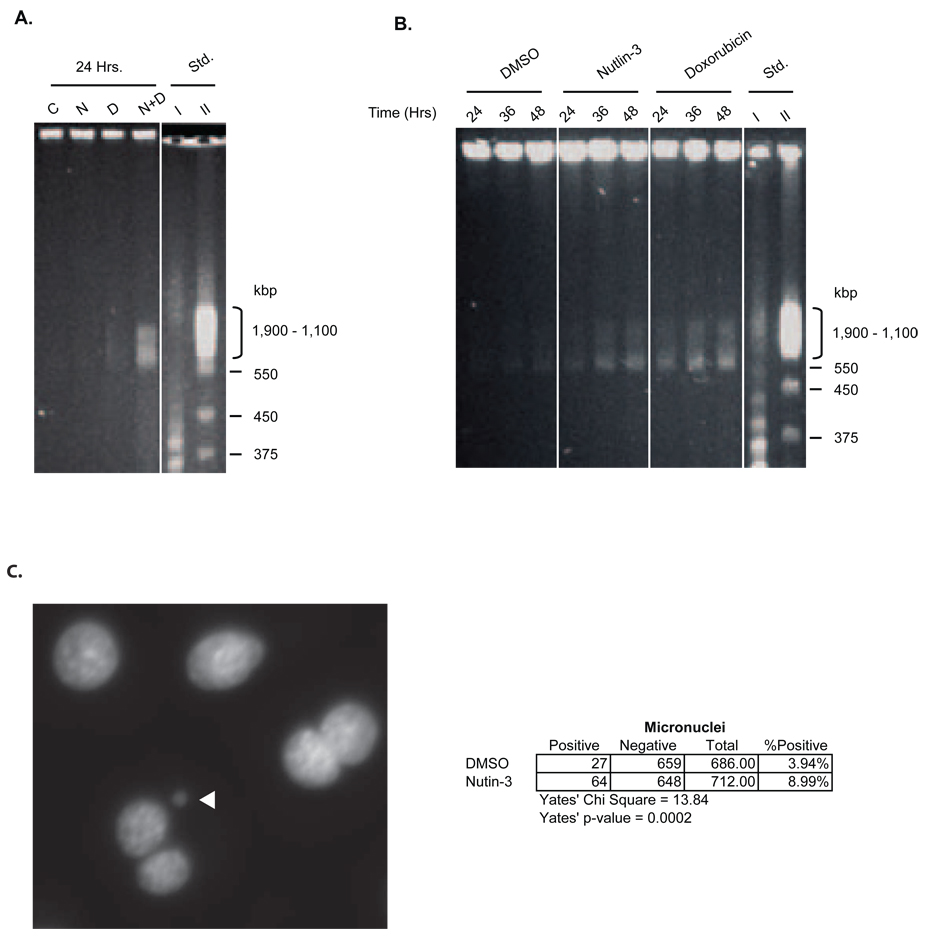
To determine the effect of Nutlin-3 on double strand DNA break formation, genomic DNA from AJ02-NM0 cells was analyzed using field inversion gel electrophoresis (FIGE). Figure 5A shows the results of such an analysis when AJ02-NM0 cells were treated with Nutlin-3, the toposiomerase II inhibitor doxorubicin, or a combination of both agents for 24 hours [30]. Double strand breaks between 500–2000 kbp were apparent in cells treated with the combination of Nutlin-3 and doxorubicin, indicating that Nutlin-3 may promote or stabilize DNA breaks generated by the topoisomerase inhibitor. The increase in DNA strand breakage by the combination treatment was approximately 5-times that observed in control cells. To determine whether Nutlin-3 could promote strand breakage on its own, cells were treated for additional time points, and DNA from a greater number of cells was analyzed by FIGE to increase sensitivity. As shown in Figure 5B, Nutlin-3 and doxorubicin accentuated DNA breakage relative to control cells (approximately 50 and 100%, respectively), consistent with an ability of both agents to generate or stabilize DNA breaks. The enhanced formation of micronuclei following Nutlin-3 exposure provides additional evidence for chromatin breakage (Figure 5C).
Figure 5.
A) FIGE showing dsDNA breaks in AJ02-NM0 cells treated with DMSO vehicle control (C), Nutlin-3 (N), doxorubicin (D), or a Nutlin-3/doxorubicin combination (N+D). Cells were treated for 24 hours prior embedding and FIGE analysis. B) FIGE analysis shows dsDNA breaks in Nutlin-3 and doxorubicin treated AJ02-NM0 cells. Cells were treated with DMSO, Nutlin-3, or doxorubicin for 24, 36, or 48 hours. Cells were embedded in agarose plugs and DNA was resolved by FIGE. C) Increased formation of micronuclei following treatment with Nutlin-3 indicates the induction of dsDNA damage. AJ02-NM0 cells were treated with DMSO (vehicle control) or Nutlin-3 for 24 hours. The cells were then treatment with cytochalasin-B for 24 hours to prevent cytokinesis. The panel to the left is a representative image of a micronucleus from Nutlin-3 treated AJ02-NM0 cells. The chart on the right shows the increased formation of micronuclei following treatment with Nutlin-3 compared to treatment with DMSO.
3.4. Nutlin-3 sensitizes AJ02-NM0 cells to topoisomerase II inhibitors
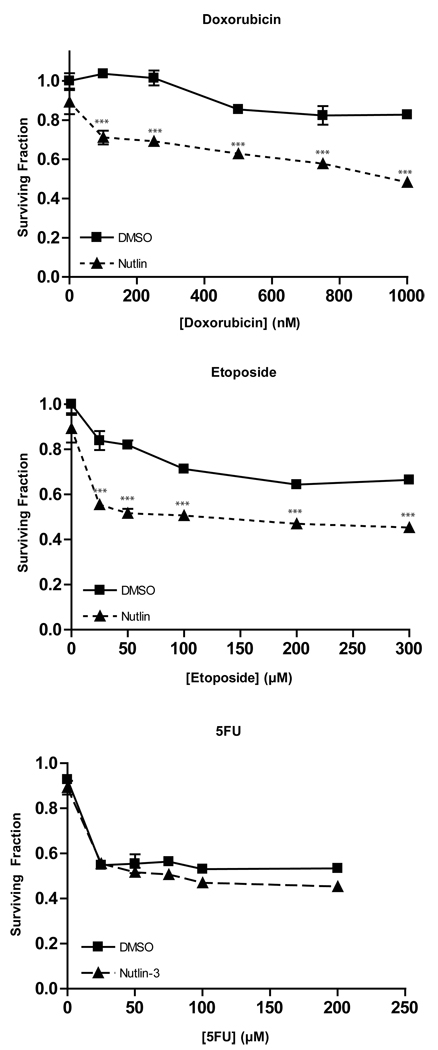
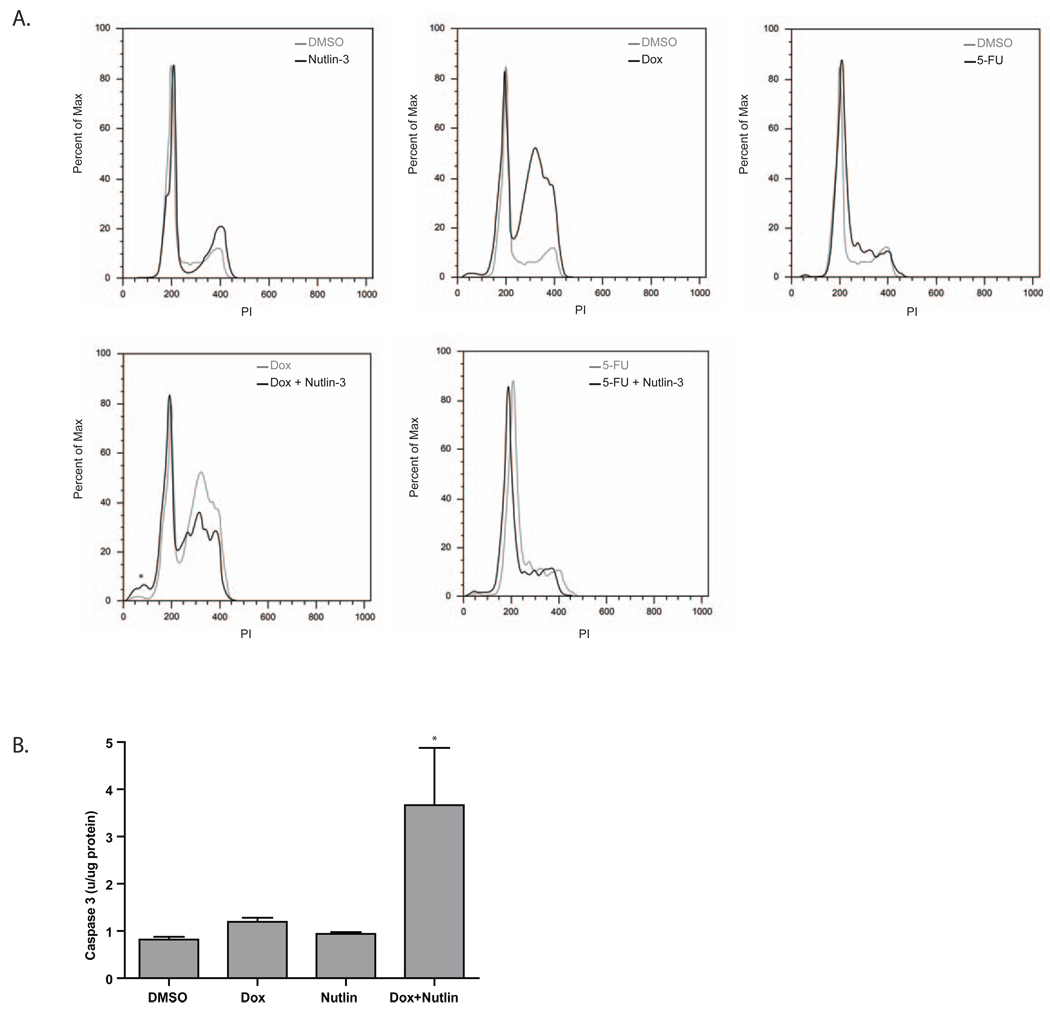
The cooperation between doxorubicin and Nutlin-3 in promoting DNA breakage prompted us to determine the effect of these agents on the growth of AJ02-NM0 cells. As shown in Figure 6, growth of the AJ02-NM0 cells was only marginally affected by doxorubicin, even when this drug was present at micromolar concentrations. However, the presence of Nutlin-3 served to sensitize AJ02-NM0 cells to doxorubicin. Similarly, Nutlin-3 sensitized AJ02-NM0 cells to another topoisomerase inhibitor, etoposide (Figure 6)[31–33]. Growth inhibition by 5FU was not significantly enhanced by Nutlin-3, possibly because this agent primarily causes uracil incoporation, rather than double strand DNA breaks [34, 35]. Flow cytometeric analysis was performed to compare the effect of Nutlin-3 on cell cycle distribution changes induced by doxorubicin and 5FU. As shown in Figure 7A, the individual treatments with 5FU, doxorubicin and Nutlin-3 all generated characteristic changes in the cell cycle distribution, with Nutlin-3 and doxorubicin causing cells to collect later in the cell cycle than 5FU. The doxorubicin/Nutlin-3 combination caused a small increase in the number of sub-diploid cells in the population, whereas the Nutlin-3 and 5FU combination looked much like treatment with 5FU alone. The sub-diploid cells formed following the doxorubicin/Nutlin-3 combination were likely to be apoptotic, as supported by the fact that caspase 3 activation was also observed following treatment with this drug combination (Figure 7B). The interaction between Nutlin-3 and doxorubicin in this cell cycle analysis reflects the growth inhibitory effect observed in Figure 6.
Figure 6.
Nutlin-3 sensitizes AJ02-NM0 cells to treatment with the topoisomerase II inhibitors doxorubicin and etoposide, but not the thymidylate synthase inhibitor, 5FU. AJ02-NM0 cells were treated for 24 hours with the indicated drugs, with Nutlin-3 (20µM) or DMSO vehicle control (as indicated). Cell viability was measured using the CellTiter 96 Aqueous One kit (Promega). The mean viability of triplicate cultures are shown ± SD. *** indicates p < 0.001 as determined by a two-way ANOVA with a Bonferroni’s post test.
Figure 7.
A) The effect of Nutlin-3 on the cell cycle when administered alone, or in combination with doxorubicin or 5FU. AJ02-NM0 cells were treated with Nutlin-3, 5FU or doxorubicin for 20 hours and the effect on the cell cycle distribution was analyzed by flow cytometry. The top panels show the impact of these agents relative to vehicle (DMSO)-treated control cells. The bottom panels show the cell cycle distribution of AJ02-NM0 cells treated with Nutlin-3 in combination with doxorubicin or 5FU. The asterisk indicates the sub-diploid cell population appearing in the cells treated with the doxorubicin plus Nutlin-3 combination. B) Increased caspase-3 activity indicates increased apoptosis following combination treatment of AJ02-NM0 cells with doxorubicin and Nutlin-3. AJ02-NM0 cells were treated with DMSO, doxorubicin, Nutlin-3, or a combination of doxorubicin and Nutlin-3 for 24 hours. Protein extracts were prepared and analyzed for caspase-3 activity using the Z-DEVD-AMC substrate.
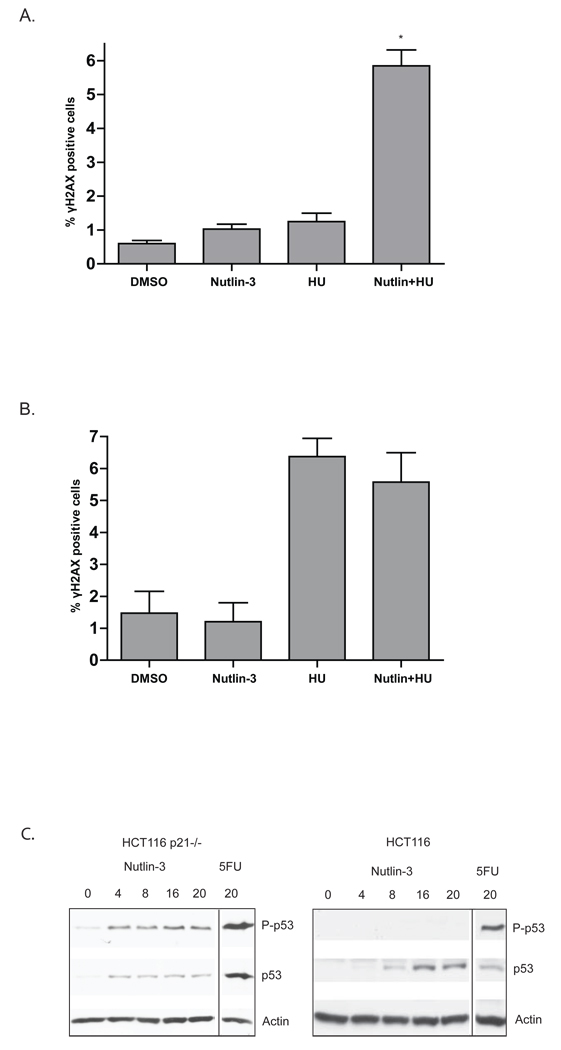
3.5. Nutlin-3 promotes H2AX phosphorylation in HCT116 cells in the presence of hydroxyurea
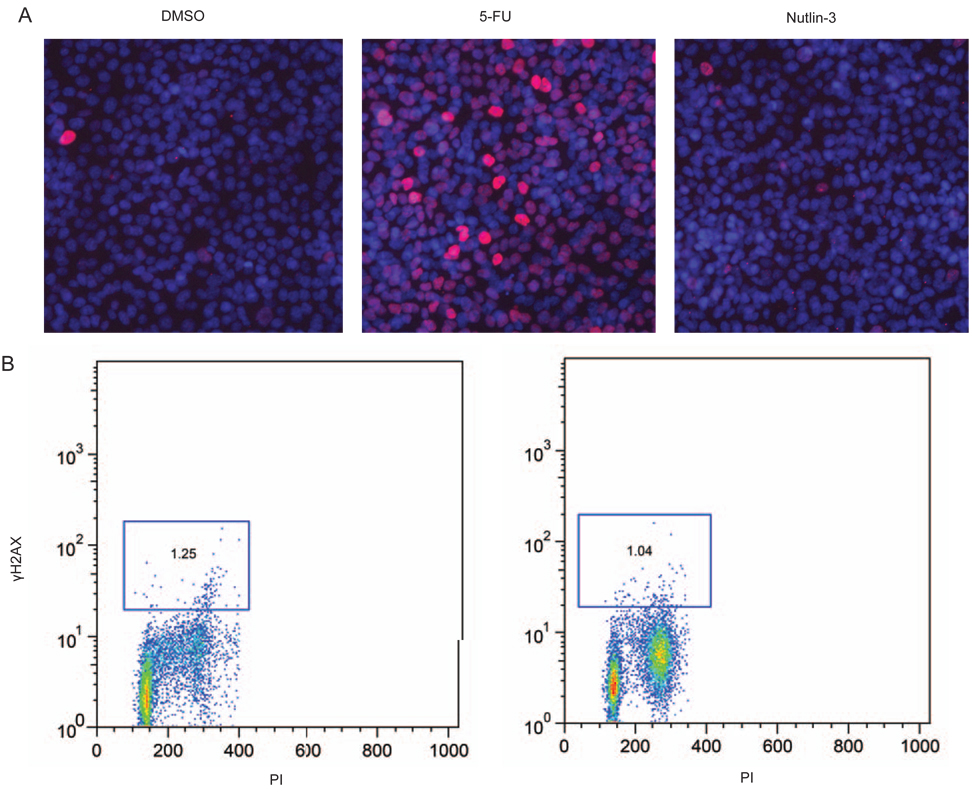
The analysis of other cancer cell lines suggests that the DNA damage response pathway is not always activated by Nutlin-3 [26]. We therefore determined the impact of Nutlin-3 on γH2AX expression in the HCT116 colon cancer cell line. As shown in Figure 8 and 9, Nutlin-3 serves to stabilize p53 in HCT116 cells, but does not increase the number of γH2AX positive cells (Figures 8A and 8B) nor lead to p53 phosphorylation at serine 15 (Figure 9C). Given that γH2AX expression was related to the cell cycle in AJ02-NM0 cells, occurring primarily at late S and G2/M, we determined whether cell cycle inhibitors may promote the DNA damage response in HCT116 cells. As shown in Figure 9A, combining Nutlin-3 with hydroxyurea (HU) increased γH2AX staining of HCT116 cells. To assess the role of p53 in this response, H2AX phosphorylation was assessed in a line HCT116 line with a deleted p53 gene (Figure 9B). Although the p53 null cells were more sensitive to HU than the parental line, Nutlin-3 did not further enhance the response. Finally, we found that HCT116 cells with a deletion in the Cdk inhibitor p21 were likewise sensitized to Nutlin-3, as indicated by an enhanced serine-15 phosphorylation following Nutlin-3 treatment (Figure 9C). The potential role of Nutlin-3 activated p53 in slowing the repair of double strand DNA breaks that form during S phase or unregulated cell cycle progression is discussed.
Figure 8.
A) Nutlin-3 does not induce a DNA damage response in HCT116 cells. HCT116 cells were treated for 24 hours with 5FU or Nutlin-3 as indicated and stained for γH2AX (red). Nuclei were counterstained with DAPI (blue). B) HCT116 cells treated with 5FU, or Nutlin-3 were quantified for γH2AX staining by flow cytometry. Cells within the square analysis region are defined as γH2AX positive.
Figure 9.
A) Treatment of HCT116 cells with Nutlin-3 in the presence of hydroxyurea enhances γH2AX expression. HCT116 cells were treated for 24 hours with DMSO vehicle (Control), Nutlin-3, hydroxyurea (HU), or a combination of Nutlin-3 and hydroxyurea as indicated. Cells were analyzed for γH2AX by immunofluorescence. The graph shows the quantification of γH2AX-positive cells. B) HCT116 cells with a targeted deletion of the p53 gene were treated as described in Figure 9A. The fraction of γH2AX positive cells appearing in the culture was determined and is shown in the graph. For both Figure 9A and 9B, the asterisks indicate p < 0.01 as determined by an ANOVA with a Bonferroni’s post test. C) Increased phosphorylation of p53 is exhibited in HCT116 p21 −/− cells compared to HCT116 cells following treatment with Nutlin-3 and 5FU. HCT116 with or without a targeted deletion of the p21 gene were treated with 5FU or Nutlin-3 for 24 hours. Nuclear extracts were then prepared and immunoblotted for p53 or p53 phosphorylated at Ser-15.
4. Discussion
Mdm2 inhibitors are a new class of p53 activating agents that may prove to be of considerable value for cancer treatment. In addition, their ability to activate p53 without first damaging DNA suggests a low genotoxic potential. Therefore, they may be useful for the treatment of relatively early lesions, prior to p53 mutation. However, little is known about the consequences activating p53 through the pharmacological inhibition of Mdm2. For instance, p53 activated in this manner lacks many p53 modifications resulting from a DNA damage response [26], and these post-translational modifications may be important for regulating some aspects of p53’s functions. We have been employing the mouse AOM colon cancer model as a pre-clinical model of Nutlin-3 efficacy [25]. This model is particularly well-suited for studying Nutlin-3 since colon tumors in this model are typically p53-normal. Our previous analysis indicated that Mdm2 is the primary p53 regulator in these tumors and that Mdm2 expression is frequently elevated relative to normal mucosa. Interestingly, ex vivo analysis of tumor and normal tissue showed that tumors had an elevated level of sensitivity to Nutlin-3, although the molecular basis of this heightened sensitivity is not clear. In this study we find that Nutlin-3 induces double strand DNA breaks in AOM-induced colon cancer cells. This finding has a number of implications regarding the safety and efficacy of pharmacological Mdm2 inhibitors.
It would seem counterintuitive that p53 activation by Nutlin-3 would show an enhancement of DNA breakage since p53 typically stabilizes genome integrity. However, it has been well documented that p53 slows DNA repair through the homologous recombination repair pathway [36, 37]. Specifically, p53 regulates the Rad51 strand exchange step of this repair pathway [38, 39]. It is not entirely clear what the advantage of slowing this step of the repair process might be, but it has been proposed that this may enhance the fidelity of repair by ensuring that heteroduplexes are not formed [40]. Excessive or low-fidelity repair through this pathway can lead to gene conversion events, genome rearrangements and potentially loss of heterozygosity [36]. Interestingly, double strand DNA breaks occur spontaneously during DNA replication, but these breaks are usually resolved rapidly through homologous recombination and do not persist long enough to induce a DNA damage responses [41]. It should be noted that the ability of p53 to slow the homologous recombination repair appears to be restricted to this particular DNA repair pathway: other forms of repair, such as base and nucleotide excision repair do not appear to be affected.
We propose that the pharmacological inhibition of Mdm2 and activation of p53 during S phase specifically slows homologous recombination repair of the breaks that arise during S phase in sensitive cell lines, which in turn activates the DNA damage response pathway. This model is consistent with the finding that γH2AX staining is pronounced in late S phase, when the replication associated breaks are predicted to occur. In addition, the sensitization of AJ02-NM0 cells to doxorubicin and etoposide by Nutlin-3 could result from slowing the repair of double strand breaks induced by these agents. Finally, γH2AX staining can be accentuated with the deoxyribose nucleotide reductase inhibitor HU, or by deletion of the p21 Cdk inhibitor. In the case of HU, interfering with DNA replication through reducing nucleotide pools could accentuate DNA breakage at replication forks [42]. Cells with a compromised ability to regulate cell cycle progression, such as through reduced p21 expression, may also be sensitized to the strand breakage effects of Nutlin-3. This latter possibility is supported by a recent report showing that sensitivity to Nutlin-3 is enhanced by high expression levels of E2F-1. Elevated E2F-1 expression can also perturb cell cycle checkpoint regulation [43]. If Mdm2 inhibitors can in fact slow homologous repair, it is not clear whether this would have a beneficial or harmful affect on patients treated with this type of agent. However, it will be important to assess the extent of this effect if Nutlin-3 or other Mdm2 inhibitors advance to clinical trials.
Activation of a DNA damage response following Nutlin-3 treatment may generate a positive feedback loop that further accentuates the actions of p53. The phosphorylation of p53 at Ser-15 following Nutlin-3 treatment could further suppress Mdm2 binding and prevent p53 from associating with other inhibitory proteins, such as MdmX [44–46]. In addition, a DNA damage response causes a number of other post-translational modifications on p53, which can enhance its ability to activate target genes and promote apoptosis [47–49]. The sensitivity of cells to the mobilization of a DNA damage response following Nutlin-3 treatment may therefore alter their sensitivity to this agent. Since a DNA damage response induced by Nutlin-3 occurs in late S and G2/M, proliferating cells may be more sensitive to this activity. We previously reported that the ex vivo treatment of mouse colonic tissues with Nutlin-3 generated a significantly more robust p53 response in AOM-induced tumors than in normal adjacent mucosa [25]. In these experiments, tumors and normal tissue were removed from AOM-treated mice, bisected, and cultured in control or Nutlin-3 containing medium. It was found that tumors maintained in Nutlin-3 medium significantly activated the expression of p53 target genes including Mdm2, p21, GADD45 and Bax, whereas no activation was observed in the normal tissue. We propose that the ability of Nutlin-3 to induce a DNA damage response in the AOM-induced tumors may be responsible for the selective gene activation in tumors; AOM tumor cells may be more prone to the Nutlin-3 activation of a DNA damage response due to their higher proliferative index and/or because of more frequent DNA replication errors [11, 50].
Although p53 is readily activated in response to DNA damage through a well-orchestrated series of events that feature a number of post-translational p53 modifications, Mdm2 inhibitors in some regard function in a manner similar to the p19 and p14 ARF proteins. These proteins are expressed following oncogene activation, bind to Mdm2 and inhibit its interaction with p53, and can also localize Mdm2 away from p53 by recruiting it into the nucleolus [5, 7, 51–53]. Interestingly, some reports have indicated that the oncogene activation of p53 entails an induction of the DNA damage response [11, 14, 54]. We propose that the induction of the DNA damage response by oncogenes may occur following p53 stabilization through an interference with homologous recombination repair during S phase. If this is in fact the case, Mdm2 inhibitors may be useful for accentuating the oncogene checkpoint. The use of Mdm2 inhibitors in cancer prevention paradigms, for example to clear microscopic preneoplastic lesions such as microadenomas and difficult to detect flat adenomas, would seem a promising application.
Although our present model focuses on the contribution of p53 on the Nutlin-3 induced DNA damage response, other Nutlin-3-induced changes may be playing an important role. For example, Mdm2 itself has been reported to increase in spontaneous chromosome breaks in fibroblasts independent of their p53 status [55, 56], and it is possible that Nutlin-3 binding of Mdm2 accentuates this activity. Nutlin-3 may also affect the activity of other Mdm2-target proteins involved in DNA repair, such as Tip60 [57–59]. Understanding how cellular functions are modulated by pharmacological Mdm2 inhibitors will be important for optimizing the use of this family of therapeutic agents.
Acknowledgements
This work was supported by NIH grant R21 CA125592 to CG. We are grateful to Dr. Vogelstein and Dr. Kinzler for providing the HCT116 cell lines used in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997 Oct 31;91(3):325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 2.Bean LJ, Stark GR. Regulation of the accumulation and function of p53 by phosphorylation of two residues within the domain that binds to Mdm2. J Biol Chem. 2002 Jan 18;277(3):1864–1871. doi: 10.1074/jbc.M108881200. [DOI] [PubMed] [Google Scholar]
- 3.Araki R, Fukumura R, Fujimori A, Taya Y, Shiloh Y, Kurimasa A, et al. Enhanced phosphorylation of p53 serine 18 following DNA damage in DNA-dependent protein kinase catalytic subunit-deficient cells. Cancer Res. 1999 Aug 1;59(15):3543–3546. [PubMed] [Google Scholar]
- 4.Lowe SW, Jacks T, Housman DE, Ruley HE. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2026–2030. doi: 10.1073/pnas.91.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999 Jan 4;18(1):22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamijo T, van de Kamp E, Chong MJ, Zindy F, Diehl JA, Sherr CJ, et al. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res. 1999 May 15;59(10):2464–2469. [PubMed] [Google Scholar]
- 7.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Contribution of two independent MDM2-binding domains in p14(ARF) to p53 stabilization. Curr Biol. 2000 May 4;10(9):539–542. doi: 10.1016/s0960-9822(00)00472-3. [DOI] [PubMed] [Google Scholar]
- 8.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998 Sep 1;17(17):5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999 Jun 8;96(12):6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008 May 22;27(23):3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 11.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007 Dec 10;26(56):7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of 'conditional haploinsufficiency'. Cell Cycle. 2007 Oct 1;6(19):2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- 13.Mallette FA, Ferbeyre G. The DNA damage signaling pathway connects oncogenic stress to cellular senescence. Cell Cycle. 2007 Aug 1;6(15):1831–1836. doi: 10.4161/cc.6.15.4516. [DOI] [PubMed] [Google Scholar]
- 14.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006 Nov 30;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 15.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007 Feb 8;445(7128):656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006 Dec 29;127(7):1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007 Feb 8;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 18.Patel S, Player MR. Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin Investig Drugs. 2008 Dec;17(12):1865–1882. doi: 10.1517/13543780802493366. [DOI] [PubMed] [Google Scholar]
- 19.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007 Jan;13(1):23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Nambiar PR, Giardina C, Guda K, Aizu W, Raja R, Rosenberg DW. Role of the alternating reading frame (P19)-p53 pathway in an in vivo murine colon tumor model. Cancer Res. 2002 Jul 1;62(13):3667–3674. [PubMed] [Google Scholar]
- 21.Walchle C, Diwan BA, Shiao YH, Calvert RJ. Microsatellite instability is infrequent in azoxymethane-induced rat intestinal tumors: An assessment by capillary electrophoresis. Toxicol Appl Pharmacol. 1999 May 15;157(1):9–15. doi: 10.1006/taap.1999.8662. [DOI] [PubMed] [Google Scholar]
- 22.Erdman SH, Wu HD, Hixson LJ, Ahnen DJ, Gerner EW. Assessment of mutations in Ki-ras and p53 in colon cancers from azoxymethane- and dimethylhydrazine-treated rats. Mol Carcinog. 1997 Jun;19(2):137–144. doi: 10.1002/(sici)1098-2744(199707)19:2<137::aid-mc8>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Shivapurkar N, Belinsky SA, Wolf DC, Tang Z, Alabaster O. Absence of p53 gene mutations in rat colon carcinomas induced by azoxymethane. Cancer Lett. 1995 Sep 4;96(1):63–70. doi: 10.1016/0304-3835(95)03947-u. [DOI] [PubMed] [Google Scholar]
- 24.Guda K, Upender MB, Belinsky G, Flynn C, Nakanishi M, Marino JN, et al. Carcinogen-induced colon tumors in mice are chromosomally stable and are characterized by low-level microsatellite instability. Oncogene. 2004 May 6;23(21):3813–3821. doi: 10.1038/sj.onc.1207489. [DOI] [PubMed] [Google Scholar]
- 25.Aizu W, Belinsky GS, Flynn C, Noonan EJ, Boes CC, Godman CA, et al. Circumvention and reactivation of the p53 oncogene checkpoint in mouse colon tumors. Biochem Pharmacol. 2006 Oct 16;72(8):981–991. doi: 10.1016/j.bcp.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, et al. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004 Dec 17;279(51):53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 27.Belinsky GS, Claffey KP, Nambiar PR, Guda K, Rosenberg DW. Vascular endothelial growth factor and enhanced angiogenesis do not promote metastatic conversion of a newly established azoxymethane-induced colon cancer cell line. Mol Carcinog. 2005 Jun;43(2):65–74. doi: 10.1002/mc.20079. [DOI] [PubMed] [Google Scholar]
- 28.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001 Nov 9;276(45):42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 29.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004 Feb 6;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 30.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984 Oct 26;226(4673):466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 31.Rowe T, Kupfer G, Ross W. Inhibition of epipodophyllotoxin cytotoxicity by interference with topoisomerase-mediated DNA cleavage. Biochem Pharmacol. 1985 Jul 15;34(14):2483–2487. doi: 10.1016/0006-2952(85)90530-1. [DOI] [PubMed] [Google Scholar]
- 32.Long BH, Musial ST, Brattain MG. Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 1985 Jul;45(7):3106–3112. [PubMed] [Google Scholar]
- 33.Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984 Dec;44(12 Pt 1):5857–5860. [PubMed] [Google Scholar]
- 34.Sartorelli AC, Creasey WA. The antineoplastic and biochemical effects of some 5-fluoropyrimidines. Cancer Res. 1967 Nov;27(11):2201–2206. [PubMed] [Google Scholar]
- 35.Goldberg AR, Machledt JH, Jr, Pardee AB. On the action of fluorouracil on leukemia cells. Cancer Res. 1966 Aug;26(8):1611–1615. [PubMed] [Google Scholar]
- 36.Bertrand P, Saintigny Y, Lopez BS. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 2004 Jun;20(6):235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006 Jun;13(6):1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 38.Akyuz N, Boehden GS, Susse S, Rimek A, Preuss U, Scheidtmann KH, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002 Sep;22(17):6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saintigny Y, Rouillard D, Chaput B, Soussi T, Lopez BS. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene. 1999 Jun 17;18(24):3553–3563. doi: 10.1038/sj.onc.1202941. [DOI] [PubMed] [Google Scholar]
- 40.Dudenhoffer C, Rohaly G, Will K, Deppert W, Wiesmuller L. Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol Cell Biol. 1998 Sep;18(9):5332–5342. doi: 10.1128/mcb.18.9.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrini JH. DNA replication reaches the breaking point. Cell. 2009 Apr 17;137(2):211–212. doi: 10.1016/j.cell.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001 Aug 2;412(6846):557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa M, Aonuma M, Lee SH, Fukutake S, McCormick F. E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene. 2008 Sep 11;27(40):5303–5314. doi: 10.1038/onc.2008.164. [DOI] [PubMed] [Google Scholar]
- 44.Jackson MW, Berberich SJ. Constitutive mdmx expression during cell growth, differentiation, and DNA damage. DNA Cell Biol. 1999 Sep;18(9):693–700. doi: 10.1089/104454999314971. [DOI] [PubMed] [Google Scholar]
- 45.Bottger V, Bottger A, Garcia-Echeverria C, Ramos YF, van der Eb AJ, Jochemsen AG, et al. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene. 1999 Jan 7;18(1):189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 46.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006 Nov 3;281(44):33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi H, Woods NT, Piluso LG, Lee HH, Chen J, Bhalla KN, et al. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J Biol Chem. 2009 Apr 24;284(17):11171–11183. doi: 10.1074/jbc.M809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle. 2007 Nov 1;6(21):2594–2598. doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006 Jun;13(6):941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 50.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007 Jan 1;21(1):43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bothner B, Lewis WS, DiGiammarino EL, Weber JD, Bothner SJ, Kriwacki RW. Defining the molecular basis of Arf and Hdm2 interactions. J Mol Biol. 2001 Nov 23;314(2):263–277. doi: 10.1006/jmbi.2001.5110. [DOI] [PubMed] [Google Scholar]
- 52.Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002 May;22(10):3497–3508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998 Jul 7;95(14):8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005 Apr 14;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 55.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008 Aug;28(15):4862–4874. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci. 2009 Jun;34(6):279–286. doi: 10.1016/j.tibs.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 57.Legube G, Linares LK, Tyteca S, Caron C, Scheffner M, Chevillard-Briet M. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 2004 Oct 22;279(43):44825–44833. doi: 10.1074/jbc.M407478200. [DOI] [PubMed] [Google Scholar]
- 58.Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D, et al. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 2002 Apr 2;21(7):1704–1712. doi: 10.1093/emboj/21.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, et al. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006 Jun;26(11):4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]