Abstract
High-level expression of the human growth hormone (hGH) gene is limited to somatotrope and lactosomatotrope cells of the anterior pituitary. We previously identified a locus control region (LCR) for the hGH gene composed of four tissue-specific DNase I-hypersensitive sites (HS) located between −14.6 kb and −32 kb 5′ to the hGH transcription start site that is responsible for establishing a physiologically regulated chromatin domain for hGH transgene expression in mouse pituitary. In the present study we demonstrated that the LCR mediates somatotrope and lactosomatotrope restriction on an otherwise weakly and diffusely expressed hGH transgene. The subregion of the LCR containing the two pituitary-specific HS, HSI and HSII (−14.6 to −16.2 kb relative to the hGH promoter and denoted HSI,II), was found to be sufficient for mediating somatotrope and lactosomatotrope restriction, for appropriately timed induction of hGH transgene expression between embryonic days 15.5 and 16.5, and for selective extinction of hGH expression in mature lactotropes. When studied by cell transfection, the HSI,II fragment selectively enhanced transcription in a presomatotrope-derived cell line, although at levels (2- to 3-fold) well below that seen in vivo. The LCR activity of the HSI,II element was therefore localized by scoring transgene expression in fetal founder pituitaries at embryonic day 18.5. The data from these studies indicated that a 404-bp segment of the HSI,II region encodes a critical subset of LCR functions, including the establishment of a productive chromatin environment, cell-specific restriction and enhancement of expression, and appropriately timed induction of the hGH transgene during embryonic development.
The steps of chromatin activation preceding gene transcription have been studied intensively. Alterations in chromatin structure are thought to enhance access of trans-acting factors to their respective transcriptional regulatory sequences (1). Experimental approaches to identify chromatin-activating elements have relied on their ability to overcome site-of-integration or position effects after random insertion into a host genome and to establish an autonomous and transcriptionally competent environment for transgene expression (2). The elements that underlie this dominantly acting activity have been referred to as locus control regions (LCRs) (5). LCR components, often located at substantial distances from their corresponding promoter(s), can be identified by their abilities to establish DNase I-hypersensitive sites (HS) in the chromatin of appropriate cell types. In previous studies we identified four HS located 5′ to the human growth hormone (hGH) gene cluster that form selectively in pituitary chromatin and together function as an LCR after germ-line transformation into the mouse genome (3).
The hGH-N gene is the most 5′ member of a cluster of five structurally related genes sharing greater than 95% sequence identity (4). The hGH-N gene is expressed in the somatotrope and lactosomatotrope cells of the anterior pituitary, whereas the remaining four genes—hCS-L, hCS-A, hGH-V, and hCS-B—are expressed in the syncytiotrophoblastic layer of the placenta (5, 6). hGH-N, hCS-A, and hCS-B are expressed at very high levels, constituting 2–3% of total mRNA in their respective tissues. The basis for the high-level expression, cell specificity, and mutually exclusive tissue expression profiles of these genes is poorly understood. These distinct tissue specificities cannot be explained by differences in the activities of the conserved promoters (7) and instead may reflect the respective HS patterns in chromatin 5′ of the hGH gene cluster; four HS (HSI, HSII, HSIII, and HSV as numbered from 3′ to 5′) are formed in the chromatin of somatotrope cells, and a partially overlapping set of three HS (HSIII, HSIV, and HSV) form in the chromatin of placental syncytiotrophoblasts. The previously demonstrated selective formation of HSI and HSII (denoted HSI,II) in the pituitary and their in vivo enhancer function suggest that they might play a major role in the somatotrope-restricted induction of hGH gene expression. In the present paper we establish the role of the full LCR and the sufficiency of the HSI,II LCR subcomponents as they relate to pituitary cell-type restriction and developmental regulation and establish an in vivo assay that will allow detailed mapping of the corresponding elements.
MATERIALS AND METHODS
Immunohistochemical Staining.
Antibodies specific for hGH were identified by scanning an epitope-defined library of anti-hGH monoclonal antibodies (mAbs) (8). mAb7 and mAb9 (kindly supplied by Genentech) recognize epitopes encompassing residues 61–66 and 120–143 in mature hGH, respectively, which are divergent from mouse growth hormone (mGH). Only mAb9 was specific for human somatotropes [defined by staining with cross-reacting anti-rat growth hormone (rGH)]. Anti-rGH and anti-rat prolactin (rPrl) were obtained from the National Hormone and Pituitary Program and anti-Pit-1 was from Berkeley Antibody (Richmond, CA). Mouse pituitaries were fixed in 10% buffered formalin and embedded in paraffin, and 4- to 5-μm sections were stained with hematoxylin and eosin. Anti-rGH was used at a 1:2,500 dilution and mAb9 at 1:1,500; incubations were for 24 hr at room temperature. Specificity of immunostaining was documented by nonimmune sera and by competition with purified hGH (Dako). All reactions were visualized with streptavidin-biotin-peroxidase (Autoprobe III detection system; Biomeda, Foster City, CA) and were revealed with the chromogen 3,3′-diaminobenzidine (DAB) (9). Double staining experiments were performed as described previously (10). Pit-1 was localized after microwave antigen retrieval (11) with a 1:2,000 dilution of anti-Pit-1 and detected with the avidin–biotin–peroxidase Vectastain Elite detection system (Vector Laboratories), using cobalt DAB.
Construction of Recombinant Plasmids.
Restriction and modification enzymes for the constructions were obtained from New England Biolabs. phGHN-CAT: To generate phGHN-BSKSII, the hGH-N gene [2.6-kb EcoRI fragment R2 of Chen et al. (12) extending 494 bp 5′ to the transcription start site and 530 bp 3′ to the polyadenylation site] was cloned into Bluescript II KS+ (Stratagene). The hGH-N promoter was released as a 494-bp EcoRI/BamHI fragment, blunt-ended with the Klenow fragment of DNA polymerase, ligated to SpeI linkers, and cloned in the native orientation at the XbaI site of the pCAT-basic vector (Promega) 5′ to the chloramphenicol acetyltransferase (CAT) coding region, generating phGHN-CAT. pHSI,II-hGHN-CAT: A 1.6-kb BglII fragment (coordinates −14.56 to −16.16 kb relative to the hGH-N transcription initiation site) encompassing HSI,II was isolated from the K2B cosmid (3), ligated to SalI linkers, and subcloned in the native orientation at the SalI site of phGHN-CAT 5′ to the hGH-N promoter, generating pHSI,II-hGHN-CAT. The ptk-neor, phGHN-neor, pHSI,II-tk-neor, pHSI,II-hGHN-neor, and pHSI,II-hGHN-BSKSII vectors were assembled from the hGH- and HSI,II-containing plasmids. Deletion derivatives of pHSI,II-hGHN-BSKSII: These derivatives were generated by PCR using the 1,602-bp HSI,II BglII fragment as a template: 5′-deletion termini of F10 (1,217 bp), F12 (816 bp), F13 (614 bp), F14 (405 bp), and 3′-deletion termini of F17 (1,059 bp) and F18 (853 bp). All amplicons were cloned into phGHN-BSKSII digested with HindIII and ClaI (further details and sequences of oligomers are available on request).
Tissue Culture.
GH3 (13), GHFT1 (14), JEG3 (15), and NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (GH3, GHFT1, JEG3) or calf serum (NIH 3T3), 2 mM glutamine, 100 international units/ml penicillin, and 100 μg/ml streptomycin.
Transfections and Expression Assays.
One picomole (for stable transfections) or 2 pmol (for transient transfections) of supercoiled test DNA and 1 pmol of pCMVβ (cytomegalovirus promoter/β-galactosidase gene; CLONTECH) were transfected by Ca3(PO4)2 coprecipitation. β-Galactosidase and CAT assays were performed 36–48 hr after transfection (16). The CAT activities were quantitated by PhosphorImager (Molecular Dynamics), normalized to β-galactosidase values. For the colony formation assay (17), the Ca3(PO4)2 transfection was followed by incubation of the cells with 300 μg/ml (GH3), 400 μg/ml (JEG3 and NIH 3T3), or 500 μg/ml (GHFT1) G418 (Life Technologies) for 2–4 weeks. Colonies were stained with 0.1% methylene blue and 0.2% crystal violet in 70% ethanol and counted. All data are the result of at least three independent experiments performed in duplicate.
Transient Transgenic Assay.
DNA inserts were released from vector sequences with BssHII digestion, resolved on agarose gels, recovered with glass beads (QiaexII; Qiagen, Santa Clarita, CA), purified by Elutip (Schleicher and Schuell) and brought to a concentration of 2 ng/ml in microfiltered 10 mM Tris⋅HCl, pH 7.6/0.1 mM Na2EDTA. One picogram of DNA was microinjected into fertilized C57BL/6J × SJL oocytes. Twenty-two eggs were reimplanted into each foster female, and the foster mothers were sacrificed 18.5 days post coitus (embryonic day 18.5, E18.5). Placental DNA was analyzed for transgene DNA by dot blotting. The rostral half of each embryo was bisected by an off-center sagittal incision, fixed, embedded in paraffin, and the pituitary was located by serial sectioning.
Sequence Analysis.
The 1.6-kb HSI,II region was sequenced by the DNA Core of the University of Pennsylvania School of Veterinary Medicine (GenBank no. AF039413) and was scanned for potential transcription factor binding sites [Signal Scan v4.0 utilizing tfd (18) and Transfac (19)] and for matrix-attachment sites [mar-finder search engine (20)]. Significant levels of matrix associated potential (0.75 unit) are as previously defined (20).
RESULTS
The Full LCR Directed hGH Transgene Expression to Somatotropes.
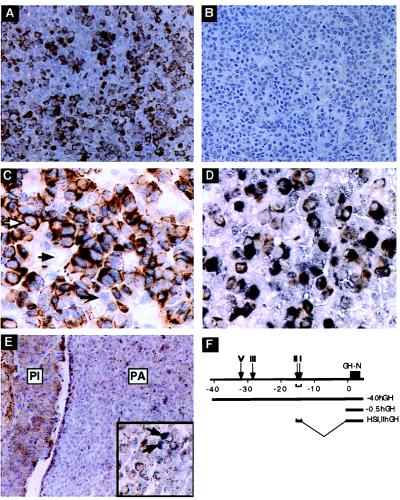
The full hGH LCR directs high-level expression of the hGH gene to the mouse pituitary in a site-of-integration-independent and copy-number-dependent fashion (3). To determine whether this expression is restricted to somatotropes, the pituitary from a representative mouse carrying the −40hGH transgene (line 149C; ref. 3) was studied by immunostaining using mAb9, an hGH-specific antibody. The −40hGH transgene encompasses all defined HS of the LCR (Fig. 1F). mAb9 staining was specific to the transgenic pituitary (compare Fig. 1 A and B) and identified a subset of cells in the −40hGH pituitary that appeared to be somatotropes by number and distribution. To characterize this subset of cells, double-immunostaining was carried out by using mAb9 in combination with an antibody to the nuclear transcription factor Pit-1. Pit-1 expression is restricted to somatotrope, lactotrope, and thyrotrope cells of the anterior pituitary (11). All mAb9-positive cells were also positive for Pit-1 nuclear staining, consistent with a somatotrope identity (Fig. 1C); Pit-1-positive cells with nonstaining cytoplasm represented the lactotropes and thyrotropes, whereas cells that failed to stain with either antibody represented corticotropes and gonadotropes. The identity of the mAb9-positive cells as somatotropes was confirmed by double staining using mAb9 in combination with an antiserum that recognized mGH to define the somatotrope population (Fig. 1D). These data demonstrated that expression of the −40hGH transgene, containing the full set of LCR HS, was restricted to the somatotrope lineage.
Figure 1.
−40hGH transgene expression in mouse pituitary was restricted to somatotrope cells. (A) Representative section of a pituitary from a −40hGH (F) adult transgenic mouse. A subset of cells stained positively with mAb9, which is specific for hGH (brown cytoplasmic stain). (B) A nontransgenic mouse pituitary stained with mAb9 was negative, indicating the specificity of mAb9 for hGH. (C) −40hGH pituitary cells were doubly immunostained with mAb9 and anti-Pit-1 (blue-gray nuclear stain). mAb9 positivity (brown) was detected only in the cytoplasm of cells that also exhibited nuclear staining for Pit-1 (white arrow). Pit-1-positive cells unreactive with mAb9 represent lactotrope or thyrotrope populations (black, barbed arrow). Other cells were negative for both stains (solid black arrow). (D) Double stain of the −40hGH pituitary with mAb9 and anti-rGH colocalized mGH (brown) and hGH using mAb9 (charcoal black) in the same cells, resulting in a dirty brown appearance. (E) Representative section of a pituitary from a −0.5hGH adult transgenic mouse. The weakly staining mAb-positive cells (brown cytoplasmic stain) were widely and nonspecifically distributed throughout the anterior (PA, pars anterior) and intermediate (PI, pars intermedia) lobes of the pituitary. The higher-power (×170) Inset shows a double stain for mGH and hGH; the reactivity showed distinct mGH-positive somatotropes (charcoal black; white arrows) in contrast to the diffuse and generalized positivity for hGH (mAb9; brown) visible in most of the cells. (F) The hGH gene cluster, pituitary HS, and the hGH-N transgenes used in these studies. (A, B, D, and E were counterstained with hematoxylin; C had no counterstain. A, B, and E, ×70; C and D ×170.)
To demonstrate the necessity of the LCR for establishing somatotrope specificity, the expression pattern of the −0.5hGH transgene was studied next. This hGH transgene contains 0.5 kb of 5′-flanking region that includes Pit-1 and Zn-15 (21) binding sites, but lacks all LCR elements (Fig. 1F). In contrast to the −40hGH transgene, the −0.5hGH transgene demonstrated marked line-to-line variability in hGH expression; most lines did not express hGH at all (3). The adult pituitary from a −0.5hGH expressing hGH (line 493D; ref. 3) was immunostained; mAb9 stain in the pituitary was faint, consistent with the level of mRNA expression (3). Of note, the staining was diffusely distributed throughout the anterior lobe and was also present in the intermediate lobes, which lacks somatotropes. In addition, mAb9 staining did not colocalize with mGH-positive somatotropes (Fig. 1E and Inset). Thus the hGH promoter alone was insufficient to establish somatotrope restriction; such restriction depended on the presence of the LCR in the construct.
A Genomic Fragment Containing HSI and HSII Activated and Restricted hGH Transgene Expression to Somatotropes.
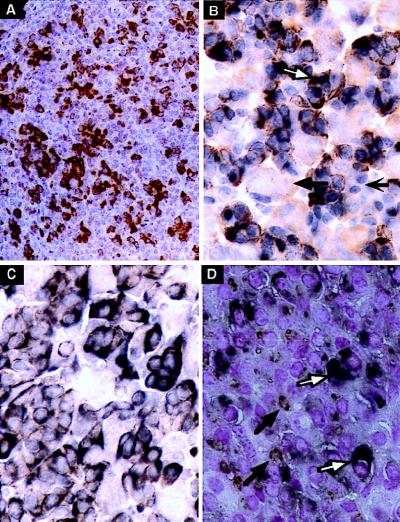
HSI,II formation is specific to pituitary chromatin; in contrast, HSIII and HSV are activated in both pituitary and placental cells and HSIV is specific to the placenta (3). Immunostaining of a pituitary from a mouse carrying the HSI,IIhGH transgene was carried out to determine whether HSI and HSII were sufficient to reproduce the somatotrope-restriction (Fig. 2 A, C, and D). HSI,II line 499D was used for these experiments because it contains a single copy of the transgene and is expressed at relatively moderate levels; most of the other HSI,II lines are expressed at supraphysiologic levels which result in sterility or morbidity (3). mAb9 staining revealed the same pattern as with the full LCR (Fig. 2A vs. Fig. 1A), but staining was of higher intensity. Double immunostaining confirmed that these mAb9-positive cells constituted a subset of the Pit-1-positive cell population (Fig. 2B), and combined immunostaining for mGH and hGH resulted in double staining of the vast majority of positive cells (Fig. 2C). Therefore, HSI,II was sufficient to restrict high-level hGH transgene expression to somatotropes.
Figure 2.
The HSI,IIhGH transgene was expressed in somatotropes but silenced in lactotropes. (A) Representative section of a pituitary from an HSI,IIhGH (Fig. 1E) adult transgenic mouse stained with mAb9. The number and distribution of mAb9-positive cells (brown cytoplasm) corresponded to that seen with the −40hGH transgene (Fig. 1A) but the staining is more intense because of the higher level of transgene expression. (B) Following double immunostaining, mAb9 positivity (brown) was detected only in the cytoplasm of cells also positive with anti-Pit-1 (blue nuclei). A representative pituitary cell positive for both Pit-1 and hGH (white arrow), another pituitary cell-type positive for Pit-1 but not hGH (barbed black arrow), and a cell negative for both (black arrow) are indicated. (C) Double staining colocalized mGH (brown) and hGH (mAb9; charcoal black) in the cytoplasm of the same cells, yielding a dirty brown reaction product (compare with the pure brown reaction product in A). (D) Double staining was also carried out with mAb9 (charcoal black diffuse cytoplasmic stain; white arrows) and anti-Prl (granular rust-colored stain with juxtanuclear Golgi pattern of reactivity; black arrows). Mature lactotropes that expressed mPrl but not hGH, indicating appropriate silencing of the HSI,IIhGH transgene, were clearly identifiable (black arrow) and were distinct from cells expressing hGH (white arrow). (A and C were counterstained with hematoxylin, B was not counterstained, and in D the nuclei were counterstained with nuclear fast red. A, ×70; B–D, ×170.)
Somatotropes and lactotropes pass through a Prl+/GH+ precursor lactosomatotrope stage; subsequent differentiation results in a Prl+/GH− subset of cells representing mature lactotropes (22). Double staining for mPrl and hGH was carried out to determine whether the HSI,IIhGH transgene could be appropriately silenced at this step in differentiation. While many cells stained with both mAb9 and anti-PRL, a subset stained only with anti-Prl (Fig. 2D). These data demonstrated that HSI,II mediated appropriate cell-specific induction as well as silencing of hGH transgene expression in mature lactotropes, thus restricting expression to the somatotrope and somatolactotrope lineages.
Sequence Analysis of the HSI,II Region.
The DNA sequence of the 1.6-kb region was determined as a basis for subsequent deletional mapping. This sequence was searched for binding sites corresponding to factors important in the establishment of the somatotrope lineage; by this computer-based search (see Materials and Methods) only a single variant putative Pit-1 binding site (GATGCAT; ref. 23) was detected near the 5′ terminus. In addition, a highly significant peak of matrix attachment potential (20) was located, centered 900 bp from the 5′ end.
Transient and Stable Transfections of HSI,II-Containing Reporter Constructs Failed to Accurately Model in Vivo Enhancer Activity.
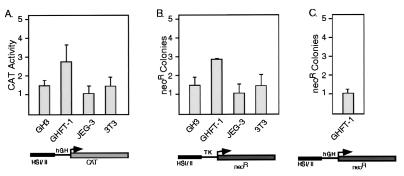
Cell transfection studies were carried out to functionally map the HSI,II region. Four cell lines were studied: lactosomatotrope-derived GH3, presomatotrope-derived GHFT1, JEG3 choriocarcinoma cells, and NIH 3T3 fibroblasts. HSI,II enhancement of the hGH gene promoter was seen only in GHFT1 cells (Fig. 3A). The 2- to 3-fold enhancement was markedly less than the 100-fold enhancement previously seen in HSI,IIhGH transgenic pituitaries (3). A set of stable transfections was next carried out using a tk promoter-driven neor reporter gene. The number of neor cell colonies generated by this assay has been reported in other systems to reflect the ability of the transfected construct to set up transcriptionally competent chromatin domains (17). The HSI,II fragment demonstrated a low level of activity (2.8-fold) in GHFT1 cells (Fig. 3B). Substituting the hGH-N promoter resulted in even less activity (Fig. 3C). It should be noted that the HSI,IIhGH transgene and the HSI,IIhGHpCAT and HSI,IIhGHpneor genes used in these transfections differed only with respect to their reporter segments. These data demonstrated that expression of these transfected genes in developmentally static cell lines failed to model the markedly positive in vivo effect of HSI,II on the hGH gene in the transgenic pituitary. Therefore the HSI,II element differed functionally from a classical enhancer.
Figure 3.
HSI,II demonstrated minimal function following transient and stable transfections into pituitary and placental cell lines. (A) Transient transfection studies. The hGH-N promoter was fused to CAT with or without the linked HSI,II fragment (diagrammed below the histogram). The effect of HSI,II on CAT expression was determined after transfections into GH3, GHFT1, JEG3, and NIH 3T3 cells. The ordinate represents the ratio of the pHSI,II-hGH-CAT expression to phGHN-CAT. (B) Stable transfections and neor (neomycin-resistant) colony assay. The tk (thymidine kinase) promoter was linked to the neor gene in the presence or absence of HSI,II. The ordinate represents the ratio of neor colonies generated by pHSI,II-tk-neor to those generated with ptk-neor. (C) Stable transfections and neor colony assay in GHFT1 cells using the hGH gene promoter linked to the neor gene in the presence or absence of HSI,II. The ordinate represents the ratio of neor colonies generated by pHSI,II-hGHN-neor to those generated with phGHN-neor. Error bars span one standard deviation.
HSI,II Induced Expression of the hGH Transgene in Somatotropes at the Appropriate Developmental Time.
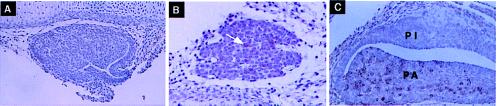
A modified transgenic approach using pituitaries from fetal founder mice was established to provide a biologically valid assay for LCR activity. To validate this approach it was necessary to demonstrate that HSI,II activated hGH gene expression at the appropriate time during embryogenesis. Endogenous mouse Pit-1 is induced at E13.5 and mGH mRNA expression is first detected between E15.5 and E17.5 (24). Immunostains of E15.5, E16.5, and E18.5 transgenic embryo pituitaries from the HSI,IIhGH line (Fig. 4) revealed that the HSI,IIhGH transgene was activated between E15.5 and E16.5, with maximal activation at E18.5, in parallel with the endogenous mGH gene. By double immunostaining, the transgene was activated only in somatotropes (see below). Therefore, HSI,II contributed to proper developmental induction of the hGH transgene in somatotropes in vivo.
Figure 4.
HSI,IIhGH transgene expression was induced between E15.5 and E16.5. Representative pituitary sections from HSI,IIhGH transgenic fetal mice stained with mAb9 are shown. Sections obtained at E15.5, E16.5, and E18.5 were stained with mAb9. No staining was detectable at E15.5 (A), a few cells were positive by E16.5 (B; white arrow), and many cells were positive by E18.5 (C), whereas the pars intermedia (PI) remained negative. (×70.)
Analysis of Transgenic Fetal Pituitaries Sublocalized HSI,II Activity to a 404-bp Region.
On the basis of the above data, we developed a “transient-transgenic” assay using E18.5 fetal pituitaries to map region(s) of HSI,II important in hGH gene activation. To validate the reliability of this assay, litters were generated by crossing wild-type females to transgenic males from the −40hGH (line 149C) or the HSI,IIhGH (line 449D) lines; E18.5 embryos were harvested and assayed for hGH expression. Of 29 successive embryos, 16 were transgenic; 15 of these 16 transgenic embryos scored positively for pituitary expression of hGH by mAb9 staining, and all nontransgenic littermates scored negatively (not shown). Thus expression of the hGH-N transgene, both in the context of the complete LCR (−40hGH) and in the context of the isolated HSI,II fragment (HSI,IIhGH), could be reliably scored by mAb9 staining at E18.5.
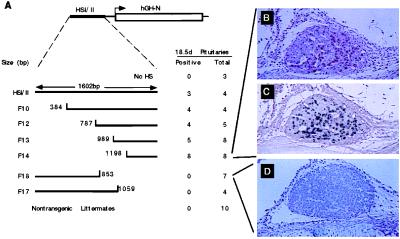
The above controls validated functional mapping of the HSI,II fragment by analysis of founder embryos at E18.5. A series of 5′ and 3′ deletions of the 1,602-bp HSI,II fragment were generated by site-specific PCR. Each deletion fragment of HSI,II was linked in its native orientation to the hGH gene (−0.5hGH), and the resultant constructs were microinjected into fertilized oocytes (Fig. 5A). Founder transgenic embryos were identified and their pituitaries were immunostained with mAb9. Embryos containing only the −0.5hGH transgene were uniformly negative for mAb9 staining. In contrast, linking the HSI,II fragment to this minimal gene resulted in hGH expression in 3 of 4 embryos. Four deletion constructs that consecutively deleted 384 through 1,198 bp from the 5′ terminus of HSI,II retained full activity. In contrast, all pituitaries isolated from embryos containing the transgene with the 749-bp 3′ deletion (F18) or a less extensive 543-bp 3′ deletion (F17) were negative for mAb9 staining. Immunostains of the pituitaries from embryos containing hGH linked to the minimal 3′-terminal fragment of HSI,II (F14) showed the usual distribution of staining (Fig. 5B), and dual immunostaining for hGH and endogenous mGH demonstrated colocalization of the two antigens (Fig. 5C), indicating somatotrope-specific expression. In contrast, mAb9 immunostaining of an F18 pituitary demonstrated a total lack of hGH expression (Fig. 5D). Thus the 404-bp F14 fragment was sufficient to consistently establish a productive chromatin domain and to restrict expression in the embryonic somatotrope.
Figure 5.
HSI,II activity sublocalized to a 404-bp fragment. (A) The E18.5 fetal pituitary transgene expression assay. The structure of the HSI,IIhGH transgene is shown on the first line. The lines below this diagram represent each of the 7 HSI,II segments studied, as well as the nontransgenic littermate controls. The first transgene contained only the hGH-N gene and promoter (No HS; equivalent to −0.5hGH), and the remainder contained HSI,II or subsegments of this element linked to the −0.5hGH. The full-length 1,602-bp HSI,II fragment (HSI,II), and 6 transgenes with 5′-terminal or 3′-terminal deletions (F10, F12, F13, F14, F18, and F17) are depicted. The base positions of the deletion termini are indicated. Pituitaries from transgenic E18.5 embryos were stained with mAb9. To the right of each transgene are the number of pituitaries positive by mAb9 staining as well the total number of transgenic embryos with that construct. Nontransgenic fetuses were assayed in parallel with their transgenic littermates (Nontransgenic Littermates). (B) The pituitary of an F14 fetus stained with mAb9 showed scattered cells immunoreactive for hGH. (C) A subsequent section of the pituitary shown in B was doubly stained for hGH (brown) and mGH (charcoal black). There were no cells showing only brown staining (compare with brown color of frame B), indicating that all high-expressing cells also produced mGH. (D) An F18 pituitary stained with mAb9 was negative for hGH expression. (B–D, ×60.)
DISCUSSION
GH gene expression has been previously characterized in detail, and pituitary-specific cis/trans interactions central to its tissue-specific expression have been identified. Pit-1, a POU-homeodomain protein, was identified initially as a nuclear factor in a somatotrope cell line. It binds to a highly conserved element in the GH promoter and is essential for the activation of GH gene transcription (25). Genetic studies have confirmed the importance of this factor to the expression of GH in somatotrope cells. Prl in lactotrope and TSHβ in certain thyrotrope cells (26, 27). Additional factors such as the widely distributed Zn-15 and Sp1 were subsequently defined as important in hGH gene activity (21, 13). Despite these detailed studies, it is clear that these promoter-proximal elements alone are insufficient for activation of hGH gene expression or for its somatrope restriction in vivo. In previous studies (3) we demonstrated that the hGH gene with as much as 7.5 kb of 5′-flanking sequences is insufficient to establish consistent expression in transgenic mouse pituitaries; of 10 such lines studied, 5 failed to express and three expressed at only trace levels. In contrast, the hGH-N transgene with 40 kb of contiguous 5′-flanking sequences which include all four of the HS formed in pituitary chromatin (−40hGH; Fig. 1F) is expressed in a reproducible and copy-number-dependent manner; 5 of 5 lines expressed at levels (per transgene) between 10% and 100% of the endogenous mGH gene. These mice have normal growth curves, suggesting that the −40hGH transgene is also under normal physiologic regulation. Thus, although the mouse has a single endogenous GH gene as compared with the multilocus GH cluster seen in humans, there appears to be sufficient conservation of trans-acting factors in the somatotrope lineage for the activation of the hGH LCR. These previously published studies (3) suggested that the mouse might be an appropriate model for the study of hGH gene expression and hGH LCR function.
In the present study we extended our characterization of hGH LCR activity to the cellular level. The normal adult anterior pituitary contains six classical hormone-producing cell types: lactosomatotrope, somatotrope, lactotrope, thyrotrope, corticotrope, and gonadotrope. Expression from the −40hGH transgene was shown to be appropriately restricted to somatotropes (Fig. 1). Parallel studies demonstrated that HSI,II was fully sufficient for this cell-type restriction as well as developmentally accurate induction (Figs. 2 and 4). In addition, HSI,II mediated selective silencing of the hGH-N transgene during the time interval when the lactosomatotrope precursor lineage differentiated into mature lactotropes (Fig. 2D). Thus, while it is clear that HSI,II on its own does not constitute a full LCR because it does not mediate copy-number dependence and is expressed at supraphysiologic levels (3), it appeared sufficient to overcome site-of-integration effects and to mediate pituitary cell type and developmental specificity in the present study.
Deletion mapping of HSI,II activity was carried out with pituitaries from E18.5 transgenic founders. Assays of this activity in cell culture were avoided because of the weak enhancement observed (Fig. 3). Pituitaries from three groups of these founders that did not contain the 3′ terminus of HSI,II were negative for transgene expression (total of 0/14), and pituitaries from five groups containing this region expressed in the majority of pituitaries (total of 24/29; Fig. 5A). The five nonexpressing transgenic pituitaries in this second group most likely resulted from transgene chimerism, known to occur in about 30% of transgenic founders (28). The results of the transient-transgenic analyses of these eight groups of embryos were internally consistent in sublocalizing the functionally active sequences to the F14 fragment, a 404-bp segment of the HSI,II region located 15 kb 5′ to the hGH-N gene promoter. In addition to having the ability to establish consistent pituitary expression, F14 was sufficient to induce expression by E18.5 and to restrict hGH gene expression to somatotropes (Fig. 5 B and C). This finding suggested that F14 contained the important cis determinants for in vivo activity.
In the present report we focused on HSI and HSII of the hGH LCR. These two sites were dealt with as a single element because they are very closely spaced and are both specifically present in pituitary but not placenta. The major effect of the HSI,II region on hGH-N gene expression in vivo was not reflected in either transiently or stably transfected cultured cells (Fig. 3). It is remarkable therefore that the HSI,II subregion of the hGH LCR appeared to retain the activity necessary for a wide spectrum of LCR functions. The establishment of a physiologically regulated transgene, which requires additional elements present in the −40hGH transgene, may modulate HSI,II activity. The inability of established pituitary-derived cell lines to recapitulate the in vivo activity of HSI,II and the lack of recognizable binding sites to somatotrope-restricted transcription factors in the active F14 subregion suggest that hGH LCR action, or at least the action of the HSI,II subcomponent, may reflect novel cis–trans interactions distinct from those previously defined for the more proximal GH promoter regions.
Acknowledgments
The authors thank Drs. James Wells and Germaine Fuh of Genentech for mAb7 and mAb9; Drs. Pei Fu He and Jean Richa for help in generating the transgenic embryos; Dr. Pamela Mellon for the gift of GHFT1 cells; Mrs. Catherine Grabowski, Ms. Sue Campbell, and Mr. Kelvin So for assistance in immunocytochemistry studies; and Ms. Jessie Harper for secretarial assistance. This work was supported by National Institutes of Health Grant HD25147 (N.E.C. and S.A.L.). S.A.L. is an Investigator in the Howard Hughes Medical Institute.
ABBREVIATIONS
- LCR
locus control region
- HS
DNase I-hypersensitive sites
- hGH
mGH, and rGH, human, mouse, and rat growth hormone
- Prl
prolactin
- CAT
chloramphenicol acetyltransferase
- En
embryonic day n
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF039413).
References
- 1.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 2.Grosveld F, Blom van Assendelft G, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 3.Jones B K, Monks B R, Liebhaber S A, Cooke N E. Mol Cell Biol. 1995;15:7010–7021. doi: 10.1128/mcb.15.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeburg P H. DNA. 1982;1:239–249. doi: 10.1089/dna.1.1982.1.239. [DOI] [PubMed] [Google Scholar]
- 5.McWilliams D, Boime I. Endocrinology. 1980;107:761–765. doi: 10.1210/endo-107-3-761. [DOI] [PubMed] [Google Scholar]
- 6.Liebhaber S A, Urbanek M, Ray J, Tuan R S, Cooke N E. J Clin Invest. 1989;83:1985–1991. doi: 10.1172/JCI114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke N E, Liebhaber S L. Vitam Horm. 1995;50:385–459. doi: 10.1016/s0083-6729(08)60659-7. [DOI] [PubMed] [Google Scholar]
- 8.Jin L, Fendly B M, Wells J A. J Mol Biol. 1992;226:851–865. doi: 10.1016/0022-2836(92)90636-x. [DOI] [PubMed] [Google Scholar]
- 9.Hsu S M, Soban E. J Histochem Cytochem. 1982;30:1079–1082. doi: 10.1177/30.10.6182185. [DOI] [PubMed] [Google Scholar]
- 10.Puy S L, Asa S L. Neuroendocrinology. 1996;63:349–355. doi: 10.1159/000126975. [DOI] [PubMed] [Google Scholar]
- 11.Asa S L, Puy L A, Lew A M, Sundmark V C, Elsholtz H P. J Clin Endocrinol Metab. 1993;77:1275–1280. doi: 10.1210/jcem.77.5.8077321. [DOI] [PubMed] [Google Scholar]
- 12.Chen E Y, Liao Y-C, Smith D H, Barrera-Saldaña H A, Gelinas R E, Seeburg P. Genomics. 1989;4:479–497. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
- 13.Tansey W P, Catanzaro D F. J Biol Chem. 1991;266:9805–9813. [PubMed] [Google Scholar]
- 14.Lew D, Brady H, Klausing K, Yaginuma K, Theill L E, Stauber C, Karin M, Mellon P L. Genes Dev. 1993;7:683–693. doi: 10.1101/gad.7.4.683. [DOI] [PubMed] [Google Scholar]
- 15.Kohler P O, Bridson W E. J Clin Endocrinol Metab. 1971;32:683–687. doi: 10.1210/jcem-32-5-683. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Moon A M, Ley T J. Proc Natl Acad Sci USA. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh D. Nucleic Acids Res. 1993;21:3117–3118. doi: 10.1093/nar/21.13.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingender E, Dietze P, H, K, Knuppel R. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh G B, Kramer J A, Krawetz S A. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipkin S M, Näär A M, Kalla K A, Sack R A, Rosenfeld M G. Genes Dev. 1993;7:1674–1687. doi: 10.1101/gad.7.9.1674. [DOI] [PubMed] [Google Scholar]
- 22.Borelli E, Heyman R A, Arias C, Sawchenko P E, Evans R M. Nature (London) 1989;339:538–541. doi: 10.1038/339538a0. [DOI] [PubMed] [Google Scholar]
- 23.Peers B, Voz M L, Monget P, Marthy-Hartert M, Berwaer M, Belayew A, Martial J A. Mol Cell Biol. 1990;10:4690–4700. doi: 10.1128/mcb.10.9.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Japon M A, Rubinstein M, Low M J. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 25.Nelson C, Albert V R, Elsholtz H P, Lu L I-W, Rosenfeld M G. Science. 1988;239:1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Crenshaw E B, Rawson E J, Simmons D M, Swanson L W, Rosenfeld M G. Nature (London) 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 27.Pfäffle R W, DiMattia G E, Parks J S, Brown M R, Wit J M, Jansen M, Van der Nat H, Van den Brande J L, Rosenfeld M G, Ingraham H A. Science. 1992;257:1118–1121. doi: 10.1126/science.257.5073.1118. [DOI] [PubMed] [Google Scholar]
- 28.Wilkie T M, Brinster R L, Palmiter R D. Dev Biol. 1986;118:9–18. doi: 10.1016/0012-1606(86)90068-0. [DOI] [PubMed] [Google Scholar]