Abstract
Claudin proteins are frequently overexpressed in various tumors such as breast, prostate and ovarian cancer. While their functions in cancer have not been completely elucidated, roles in survival, adhesion, and invasion have been suggested. In order to clarify the roles of claudins in ovarian cancer, we have performed gene expression profiling of ovarian surface epithelial cells overexpressing claudin-4 and compared the expression patterns to the parental, non-expressing cells. Claudin-4 expression leads to the differential expression of several genes, including many that have previously been implicated in angiogenesis. In particular, angiogenic cytokines, such as IL-8, were found elevated while genes of the angiostatic interferon pathway were found down-regulated. In vitro assays show that claudin-4-expressing cells produce factors that can stimulate angiogenesis as measured by tube formation and migration in HUVEC cells. In addition, an in vivo mouse dorsal skinfold assay confirms that cells expressing claudin-4 secrete factors that can mediate angiogenesis in the dorsal skin of mice. Our data suggest a novel function for claudin-4 in cancer and provide an additional rationale for its common overexpression in human tumors.
Keywords: Ovarian Cancer, expression profiling, claudin, interleukin, interferon, angiogenesis
Introduction
Ovarian cancer is the fifth cause of cancer-related deaths in women in the US.1 Advanced ovarian cancer has a particularly poor prognosis and, unfortunately, the lack of obvious symptoms and screening strategies results in late diagnosis for most ovarian cancer patients. While the exact molecular pathways involved in ovarian tumorigenesis have not been completely elucidated, recent work in the field has begun to shed some light on the fundamental molecular mechanisms involved in this disease.2,3 Molecular genetics evidence suggest a two-pathway model for ovarian tumorigenesis with one pathway involved in the development of low grade tumors (Type I Pathway), and the other in the generation of serous high grade tumors (Type II pathway).2 Epithelial ovarian tumors are believed to arise from ovarian surface epithelial (OSE) cells covering the ovary 4-6 and the relatively undifferentiated OSE cells can undergo metaplastic transformation, possibly through aberrant activation of certain HOX genes, to give rise to the various histological subtypes of ovarian carcinoma.7 In addition to the changes important in histological differentiation, pathways important for increased growth, evasion from apoptosis and senescence, as well as events related to angiogenesis and metastasis have been shown aberrantly regulated in ovarian cancer.3
Among the molecular features observed in ovarian cancer is the frequent overexpression of several members of the claudin family of tight junction proteins.8 In particular, we and others have shown that claudin-3 and claudin-4 are frequently elevated in ovarian cancer.9-18 Certain claudins have been suggested as prognostic markers in various cancers and, in ovarian cancer, claudin-3 and claudin-7 levels are inversely correlated with survival.17,18 Claudin-3 and claudin-4 expression has been shown to be associated with poor clinical outcome in endometrial cancer 19 and renal cell carcinoma.20
While claudins play crucial roles in the formation and function of tight junction in normal epithelial and endothelial cells,21 the functional consequences of overexpressed claudin-3 and -4 in ovarian cancer have not been completely elucidated. Several lines of evidence suggest that these proteins may be important for invasion, motility, and survival.22-27 Consistent with these findings, other claudins have also been associated with invasiveness and motility in cancer.28,29
In this report, we have used microarray analysis to identify molecular changes that occur in claudin-4-overexpressing cells. We find that several genes that have previously been implicated in the regulation of angiogenesis are altered following claudin-4 overexpression. Importantly, we show that these genes may be functionally relevant, since cells expressing claudin-4 exhibit elevated angiogenesis properties as measured by both in vitro and in vivo assays. Our data suggest a novel function for overexpressed claudin-4 in cancer and may explain the frequent presence of this protein in a number of human cancers.
Results
Gene expression alterations in response to claudin-4 expression
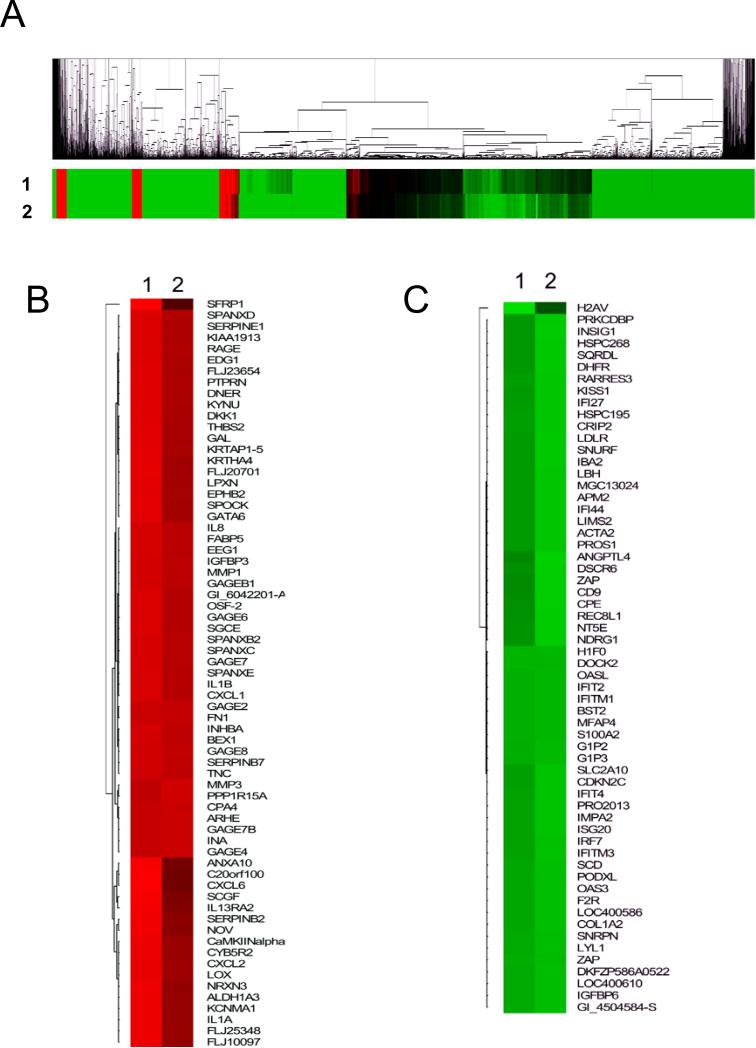
The HOSE-B cells were generated from immortalization of normal ovarian surface epithelial cells 30 and lack claudin-4 expression. Because we have previously observed changes in the behavior of HOSE-B cells following forced claudin-4 expression,23 we hypothesized that claudin-4 expression may lead to significant changes in gene expression. In order to test this hypothesis, the Human Illumina Expression BeadChip system was used to perform gene expression profiling on claudin-4-overexpressing cell lines HOSE-CLDN4-1 and HOSE-CLDN4-2, as well as two non-expressing control lines. These cell lines have been described previously.23 Microarray analysis revealed that a large number of genes were altered following expression of claudin-4 (Figure 1). Overall, the expression patterns in the two independent claudin-4-expressing clones were very similar and the vast majority of genes altered were decreased (Figure 1A). Genes up-regulated more than 2-fold (Figure 1B) or downregulated more than 3-fold (Figure 1C) were identified and clustered. Table 1 and Table 2 list the top 25 most upregulated and downregulated genes, respectively, in cells overexpressing claudin-4.
Figure 1.
Clustering analysis of gene expression following claudin-4 overexpression. Gene expression patterns for claudin-4 overexpressing clones HOSE-CLDN4-1 (1) and HOSE-CLDN4-2 (2) were obtained using the Illumina Sentrix Beadchip microarray. Gene expression levels are shown relative to HOSE-EV1 and HOSE-EV2, two cell lines lacking claudin-4 expression. A. All genes expressed above background levels (a total of 8,793 genes) were used for clustering analysis and the results shown as a heat map. B. Clustering analysis is shown for a subgroup of genes elevated more than 3-fold in claudin-4 expressing cells. C. A similar analysis is shown for genes downregulated more than 3.5 fold in claudin-4 expressing cells. In the heat maps, the red-coded areas correspond to elevated genes while the green color represents decreased genes.
Table 1.
Genes elevated in claudin-4-expressing cells
| Gene Symbol | Accession | Gene Names | Fold in CLDN4-1 | Fold in CLDN4-2 | Av. fold change |
|---|---|---|---|---|---|
| MMP1 | NM_002421.2 | Matrix metalloproteinase 1 (interstitial collagenase) | 93.0 | 95.1 | 94.0 |
| MMP3 | NM_002422.2 | Matrix metalloproteinase 3 (stromelysin 1, progelatinase) | 32.1 | 36.9 | 34.5 |
| IL1B | NM_000576.2 | Interleukin 1, beta | 24.8 | 24.8 | 24.8 |
| GAGEB1 | NM_003785.2 | G antigen, family B, 1 | 18.9 | 19.4 | 19.1 |
| CXCL8 (IL8) | NM_000584.2 | C-X-C chemokine-interleukin 8 (IL-8) | 18.0 | 18.3 | 18.2 |
| SPANXE | NM_145665.1 | SPANX family, member E | 11.6 | 11.7 | 11.7 |
| FABP5 | NM_001444.1 | Fatty acid binding protein 5 | 10.9 | 11.1 | 11.0 |
| CXCL1 | NM_001511.1 | C-X-C chemokine-growth-related oncogene alpha (GROα) | 9.6 | 9.6 | 9.6 |
| DNER | NM_139072.2 | Delta-notch-like EGF repeat-containing transmembrane | 9.3 | 9.0 | 9.1 |
| SPANXB2 | NM_145664.1 | SPANX family, member B2 | 8.6 | 8.7 | 8.7 |
| GAGE6 | NM_001476.1 | G antigen 6 | 8.4 | 8.4 | 8.4 |
| SERPINB2 | NM_002575.1 | Serine (or cysteine) proteinase inhibitor, clade B 2 | 8.5 | 7.3 | 7.9 |
| POSTN | NM_006475.1 | Periostin | 7.0 | 7.0 | 7.0 |
| KYNU | NM_003937.1 | Kynureninase | 6.6 | 6.5 | 6.5 |
| INHBA | NM_002192.1 | Inhibin, beta A (activin A) | 5.7 | 6.0 | 5.8 |
| CAMK2A | NM_018584.4 | Calcium/calmodulin-dependent protein kinase II alpha | 6.0 | 5.4 | 5.7 |
| CXCL2 | NM_002089.1 | C-X-C chemokine-growth-related oncogene beta (GROβ) | 5.8 | 5.4 | 5.6 |
| PTPRN | NM_002846.2 | Protein tyrosine phosphatase, receptor type, N | 5.2 | 5.1 | 5.2 |
| SPOCK1 | NM_004598.2 | Sparc/osteonectin, cwcv and kazal-like proteoglycan | 5.0 | 4.7 | 4.8 |
| TMEM200A | NM_052913.2 | Transmembrane protein 200A | 4.8 | 4.8 | 4.8 |
| SPANXD | NM_032417.2 | SPANX family, member D | 4.6 | 4.6 | 4.6 |
| GAGE7B | NM_001477.1 | G antigen 7B | 4.3 | 4.8 | 4.6 |
| SPANXC | NM_022661.2 | SPANX family, member C | 4.4 | 4.6 | 4.5 |
| KRTHA4 | NM_021013.3 | Keratin, hair, acidic, 4 | 4.6 | 4.3 | 4.5 |
| ALDH1A3 | NM_000693.1 | Aldehyde dehydrogenase 1 family, member A3 | 4.5 | 4.1 | 4.3 |
Table 2.
Genes decreased in claudin-4-expressing cells
| Gene Symbol | Accession | Gene Names | Fold in CLDN4-1 | Fold in CLDN4-2 | Av fold change |
|---|---|---|---|---|---|
| H1F0 | NM_005318.2 | H1 histone family, member 0 | 12.5 | 12.3 | 12.4 |
| G1P2 | NM_005101.1 | Interferon, alpha-inducible protein | 7.0 | 7.3 | 7.1 |
| OASL | NM_198213.1 | 2'-5'-oligoadenylate synthetase-like | 6.5 | 6.6 | 6.5 |
| DOCK2 | NM_004946.1 | Dedicator of cytokinesis 2 | 5.9 | 5.5 | 5.7 |
| G1P3 | NM_002038.2 | Interferon, alpha-inducible protein | 5.6 | 5.7 | 5.6 |
| MFAP4 | NM_002404.1 | Microfibrillar-associated protein 4 | 5.2 | 5.1 | 5.1 |
| IFIT2 | NM_001547.3 | Interferon-induced prot. with tetratricopeptide repeats 2 | 4.8 | 4.7 | 4.8 |
| S100A2 | NM_005978.3 | S100 calcium binding protein A2 | 4.5 | 4.4 | 4.4 |
| F2R | NM_001992.2 | Coagulation factor II (thrombin) receptor | 4.0 | 4.4 | 4.2 |
| SCD | NM_005063.3 | Stearoyl-CoA desaturase (delta-9-desaturase) | 4.0 | 4.4 | 4.2 |
| LOC400586 | XM_375424.1 | Similar to stearoyl-CoA desaturase | 3.9 | 4.2 | 4.0 |
| COL1A2 | NM_000089.2 | Collagen, type I, alpha 2 | 3.8 | 4.0 | 3.9 |
| IFITM1 | NM_003641.2 | Interferon induced transmembrane protein 1 (9-27) | 3.8 | 3.7 | 3.7 |
| SQRDL | NM_021199.1 | Sulfide quinone reductase-like | 3.4 | 4.2 | 3.7 |
| LOC400610 | XM_375478.1 | Similar to RIKEN cDNA 1100001G20 | 3.7 | 3.7 | 3.7 |
| IGFBP6 | NM_002178.1 | Insulin-like growth factor binding protein 6 | 3.7 | 3.7 | 3.7 |
| NT5E | NM_002526.1 | Ecto-5-prime-nucleotidase | 3.2 | 4.0 | 3.6 |
| PODXL | NM_005397.2 | Podocalyxin-like | 3.5 | 3.6 | 3.5 |
| ZC3HAV1 | NM_024625.3 | Zinc finger CCCH type, antiviral Protein 1 | 3.5 | 3.4 | 3.5 |
| IFIT1 | NM_001548.1 | Interferon-induced prot. with tetratricopeptide repeats 1 | 3.4 | 3.5 | 3.5 |
| BST2 | NM_004335.2 | Bone marrow stromal cell antigen 2 | 3.5 | 3.3 | 3.4 |
| APM2 | NM_006829.1 | Adipose specific 2 | 3.2 | 3.6 | 3.4 |
| CXXC5 | NM_016463.5 | CXXC finger 5 | 3.2 | 3.5 | 3.3 |
| ISG20 | NM_002201.4 | Interferon stimulated gene 20kDa | 3.1 | 3.3 | 3.2 |
| KISS1 | NM_002256.2 | KISS-1 metastasis-suppressor | 3.0 | 3.4 | 3.2 |
Several of the genes identified have previously been implicated in proliferation/survival (SPOCK1, CAMK2, DNER), migration (MMP1, MMP3), or angiogenesis (CXCL8 (IL-8), IL1B, CXCL1, CXCL2, POSTN). The upregulation of several genes known to encode cytokines that promote angiogenesis was particularly intriguing. In addition, several of the downregulated genes are involved in interferon signaling (Figure 1C, Table 2). For example, interferon inducible genes G1P2, G1P3, IFIT2, IFITM1, IFIT1, OASL, and ISG20 were all among the top 25 most highly downregulated genes in claudin-4 expressing HOSE-B cells. Interferons and some of their targets have previously been shown to exhibit angiostatic activity.31-33
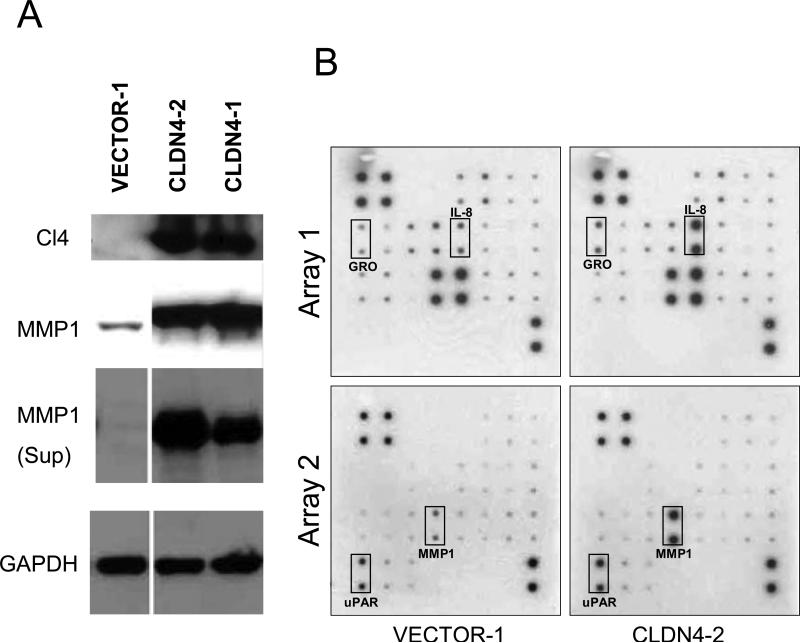
Interestingly the most highly elevated gene in claudin-4 expressing HOSE-B cells was MMP1 (Table 1). This is consistent with previous work showing that claudins may be involved in invasion/motility and that MMPs may play a role in this process. 23,26,29,34 Moreover, MMPs may have important roles in angiogenesis.35 In order to validate our finding that MMP1 mRNA is elevated in claudin-4 expressing cells, we investigated the levels of MMP1 protein in HOSE-EV1 and HOSE-CLDN4 cells by immunoblotting (Figure 2A). We found that MMP1 protein was greatly increased in both the cell lysates and the media of claudin-4 expressing cells.
Figure 2.
Validation of Illumina array results by Western analysis and antibody arrays. A. Two cell lines overexpressing claudin-4 (CLDN4-1 and CLDN4-2) were assayed by Western for MMP1 expression. MMP1 was found elevated in both the cell lysates and the supernatants of claudin-4 expressing cells. B. Conditioned media from claudin-4-expressing HOSE-B cells (CLDN4-2) were used for angiogenesis antibody arrays. IL-8, GRO (CXCL1), uPAR, and MMP1 expression were increased significantly in claudin-4 expressing cells.
Because many of the genes identified as altered in claudin-4 expressing cells are cytokines that have previously been implicated in inflammation-induced angiogenesis, we used the RayBio human angiogenesis antibody arrays to investigate the expression of several proteins involved in angiogenesis (Figure 2B). This experiment confirmed that several angiogenesis proteins were elevated in claudin-4 expressing HOSE-B cells compared to non-expressors. Indeed, the GRO, IL-8, uPAR, and MMP-1 were all shown to be elevated in HOSE-CLDN4 media using the antibody arrays.
Claudin-4 expression increases angiogenesis in HUVEC assays
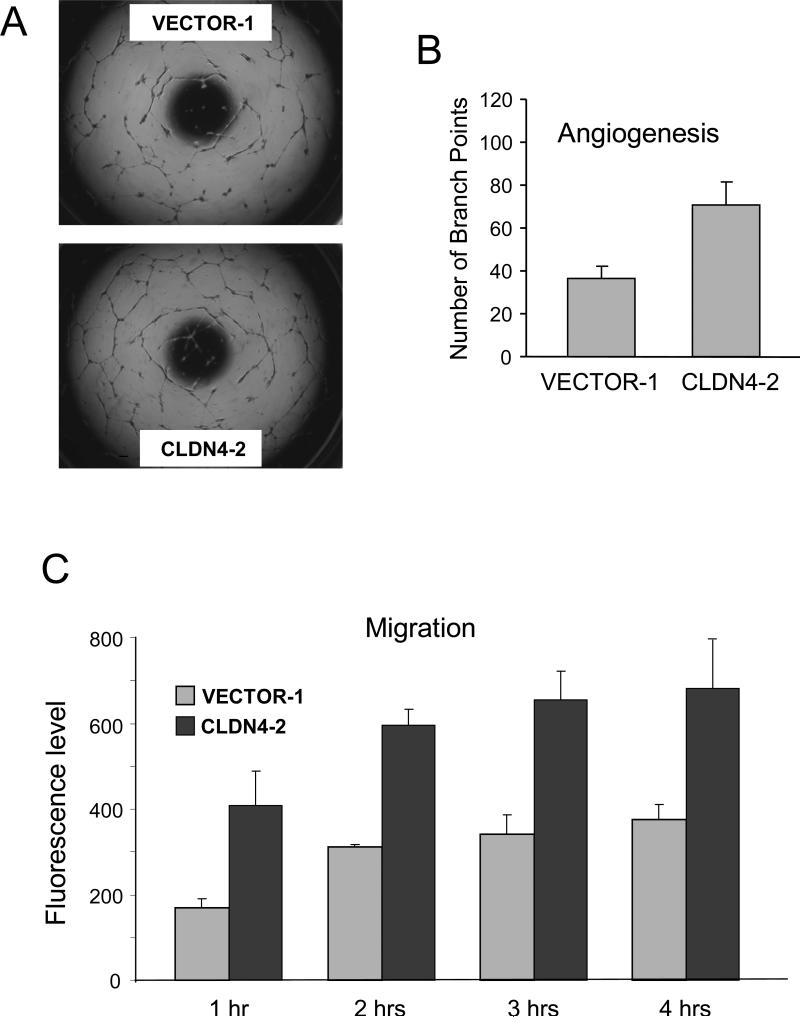
Because the claudin-4-mediated expression pattern changes suggested a possible angiogenic role for this protein, we next sought to functionally test this hypothesis. We first investigated whether claudin-4 overexpression could have angiogenic consequences using an in vitro HUVEC model system. HUVECs grown in the presence of conditioned media obtained from cells expressing claudin-4 exhibited an increase in tube formation compared to HUVECs grown in the presence of control cells (Figure 3A). In fact, the numbers of tubes formed was almost doubled when media from claudin-4 expressing cells added to the HUVEC (Figure 3B). Similarly, the claudin-4 media significantly increased HUVEC cell migration (Figure 3C), another indication that claudin-4 expression could promote angiogenesis.
Figure 3.
Claudin-4 promotes HUVEC angiogenesis. A. Pictures show HUVEC cultures following incubation with conditioned media from HOSE-B cells expressing claudin-4 (CLDN4-2) or non expressing control-tranfected cells (VECTOR-1). HUVEC incubated with claudin-4 expressing cells show an increase in tube formation. B. Tube formation was quantitated by counting the number of branch points in a given frame. The graph shows the average of 3 experiments. C. Effects of claudin-4 expression on HUVEC migration. The graph shows the migration (fluorescence levels) following claudin-4 expression at four different time points. The data shown is the average of 3 independent experiments. Media from claudin-4 expressing cells lead to a significant increase in HUVEC migration.
In vivo angiogenesis is increased by claudin-4 expression
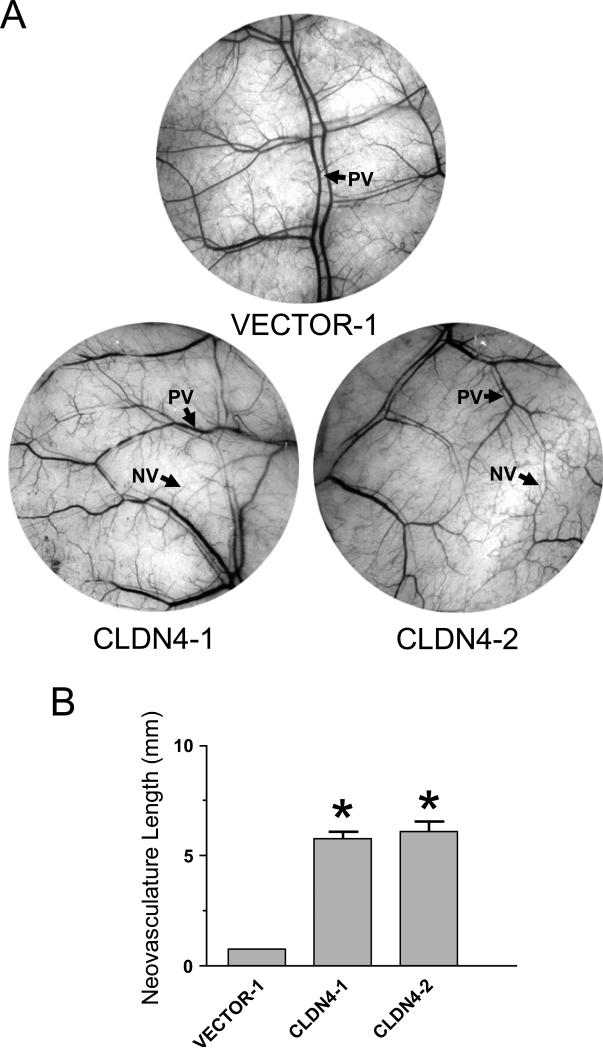
To extend the results of the in vitro angiogenesis studies to in vivo conditions, we performed angiogenesis experiments using the mouse dorsal skinfold assay. This assay allows the measurements of neovasculature formed in the skin of mice in response to factors secreted by the cells present in an implanted chamber.36 In the presence of HOSE-B cells expressing claudin-4, we observed a significant increase in skin blood vessels compared to what was observed in the presence of cells that do not express this protein (Figure 4A). When the length of the vasculature was quantitated, both claudin-4-expressing HOSE-B clones exhibited more than five-fold increase in neovasculature compared to skin that had been exposed to non-expressing cells (Figure 4B).
Figure 4.
Claudin-4 promote angiogenesis in vivo. Cells expressing claudin-4 (CLDN4-1 and CLDN4-2) or lacking claudin expression (VECTOR-1) were implanted in mouse skin and the resulting changes in vasculature observed. A. Representative pictures of mouse skin showing blood vessels 10 days after the implantation of the chamber containing the indicated cells. The presence of claudin-4-expressing cells (CLDN4-1 and CLDN4-2) leads to an increase in neovasculature (NV). Preexisting vasculature is indicated (PV). B. Changes in neovasculature resulting from exposure to the different cell lines were quantitated by evaluating the length of new blood vessels in each microscope field. Results shown are the average of 3 mice.
Discussion
The aberrant expression of claudin proteins in cancer has been well documented but the exact functions of these proteins in tumorigenesis remain under investigation. Based on experiments involving the forced expression or knockdown of specific claudin proteins, we and others have suggested that these proteins may have roles in survival, invasion, and motility.22-27 These roles would be consistent with claudin contributing to the emergence of metastasis in advanced tumors. Because, the changes in claudin levels leads to significant alteration in the behavior of the cells, we hypothesized that claudin overexpression may lead to measurable changes in the global patterns of gene expression. We further hypothesized that knowledge of these changes may suggest pathways by which claudins may influence cell behavior in cancer. We therefore used our cell line model consisting of overexpression of claudin-4 in immortalized normal ovarian HOSE-B cells in order to identify changes in gene expression. Overexpression of claudins in these cells may mimic the appearance of these proteins during ovarian cancer progression as the majority of epithelial ovarian cancers express claudin-4.
Genes expression profiling was performed on two HOSE-B clones overexpressing claudin-4 and both the clones exhibited a remarkable similarity in gene expression (Figure 1A). Many of the genes identified as altered in these clones have been implicated in cell growth, survival, and invasion, consistent with previous functional studies suggesting roles for claudins in these processes. For example, SPOCK, CAMK2, and DNER have been implicated in cell proliferation and survival. Similarly, the overexpression of MMPs may be related to the ability of claudins to affect invasion and motility.23,26,29 However, our attention was drawn to a number of genes and pathways that have previously been implicated in angiogenesis. Indeed, several cytokines, (IL-8, IL-1B, CXCL1, CXCL2) which have previously been implicated in angiogenesis 37 were found elevated in claudin-4 expressing cells. MMPs have also been implicated in promoting angiogenesis.35 Conversely, several genes of the interferon pathway, which has been shown to inhibit angiogenesis,31,33 were found downregulated by claudin-4 expression. The gene expression changes following claudin-4 expression therefore suggest specific alterations in pathways that would favor a more angiogenic environment.
Among the several pro-angiogenic cytokines found elevated in claudin-4-expressing cells, IL-8 is particularly interesting, as it has been implicated in angiogenesis and other tumorigenic processes in several cancers, including ovarian cancer.38-43 IL-8 binding to its receptors (CXCR1/2) has been shown to result in downstream signaling that promotes cell survival, proliferation, invasion, and angiogenesis.44 In particular, it is believed that IL-8 production by the cancer cells can result in significant changes in the microenvironment, leading to increases in proliferation and survival of the cancer cells through autocrine mechanisms, as well as increases in angiogenesis through IL-8 action on neighboring endothelial cells. Interestingly, it has been reported that the presence of IL-8 can lead to EGFR activation in endothelial cells.45 The EGFR downstream signaling cascade, which includes the MAPK pathway, is known to be crucial in the angiogenic response of endothelial cells.46 In addition, IL-8 was recently shown to induce endothelial cells permeability through VEGFR2 activation in a VEGF-independent fashion.47 This suggests that IL-8 may, in certain cases, be able to substitute for some VEGF functions, which may explain why we do not see significant changes in VEGF in our system (data not shown). Overall our finding that IL-8, together with other pro-angiogenic cytokines (such as IL-1B, CXCL1, and CXCL2) are highly elevated following claudin-4 expression supports our hypothesis that claudin-4 may promote angiogenesis in ovarian tumors.
IL-8 is overexpressed in a number of malignancies and is often associated with poor clinical outcome.40 In particular, in melanoma, IL-8 expression was shown to be associated with increased microvessel density and an IL-8 antibody could reduce tumor growth and metastasis in a mouse model of melanoma.41 In ovarian cancer, IL-8 expression is associated with poor clinical outcome.43 IL-8 knockdown using siRNA showed a significant therapeutic benefit in terms of tumor size in three different ovarian xenograft mouse models.43 Importantly, tumors treated with IL-8 siRNA had significantly lower microvessel density compared to control treatment. In addition, MMP2 and MMP9 were reduced following IL-8 silencing. This is interesting considering the fact that we previously showed that claudin-3 and claudin-4 overexpression led to an increase in these MMPs.23 In addition to their roles in invasion and metastasis, these MMPS have been shown to promote angiogenesis. Overall, our findings suggest that IL-8 overexpression may be linked to claudin proteins. The mechanism by which claudin-4 overexpression leads to the expression of IL-8 and other pro-angiogenic cytokines is currently under investigation.
Remarkably, several IFN-regulated genes are downregulated in the claudin-4 expressing cells, implying a decrease in IFN production or signaling by these cells. IFNs have long been known to inhibit angiogenesis, and in fact, were the first angiogenesis inhibitors identified.31,32,48 The antiangiogenic effects of IFNs have been demonstrated both in animal models and in the clinic.49 The strong effect of IFNs on angiogenesis likely stems from their ability to modulate the tumor cell angiogenic signal 31 as well as their direct effects on endothelial cells.48,50 The effects of IFNs on endothelial cells are believed to be mediated through inhibition of bFGF and other FGF family members.49,51 In combination with the increase in pro-angiogenic cytokines described above, the apparent decrease in anti-angiogenic IFNs following claudin-4 expression suggests a role for this protein in the regulation of the balance between angiogenic and angiostatic cytokines.
In order to functionally investigate the significance of these changes in gene expression, we performed in vitro and in vivo angiogenesis assays. Consistent with our hypothesis that claudin-4 may alter cytokine secretion from the tumor cells to favor a more angiogenic environment, we found that media from claudin-4 expressing cells was much more efficient at mediating tube formation in HUVEC than media from control cells (Figure 3). Interestingly, IL-8 is known to be sufficient to mediate angiogenesis phenotypes in HUVECs and HMECs cells.52-54 However, the changes in expression of multiple cytokines in our system, as well as the complex interactions between the angiogenic and angiostatic factors may explain why we were unsuccessful at inhibiting the angiogenesis effects of the media using an anti-IL-8 antibody (data not shown). We are currently attempting to identify the exact combination of factors that may be sufficient to mediate tumor angiogenesis following claudin-4 overexpression. Similarly, in vivo angiogenesis assays using a mouse skinfold approach clearly demonstrated that claudin-4 expressing cells represented a more pro-angiogenic environment compared to non-expressing cells (Figure 4).
Overall, our data show that claudin-4 expression leads to significant changes in gene expression, including changes in a number of soluble factors that are known to mediate angiogenesis. We were able to show that these changes in the environment of claudin-4 expressing cells were accompanied by functional changes in angiogenesis using both in vitro and in vivo assays. Claudin-4 is overexpressed in several tumors and has been suggested to have roles in cell proliferation, motility, invasion, and survival. The current data suggest yet another role for claudin-4 in cancer through its ability to reorganize the cellular environment to favor angiogenesis
Materials and Methods
Cell Culture
HOSE-B cell clones stably expressing claudin-4 (HOSE-CLDN4-1, HOSE-CLDN4-2) as well as corresponding non-expressing vector-transfected cell lines (HOSE-EV1 and HOSE-EV2) were previously generated in our laboratory.23 These cell lines were cultured in RPMI medium (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum, 5 ng/ml epidermal growth factor, antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin), 300 μg/ml G418, and 350 μg/ml Zeocin.
For the preparation of lysates, the cells were cultured to 80% confluency, washed with HBSS (Invitrogen), and whole cell lysates were made using lysis buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS). Protein concentration was determined using the BCA assay kit (Pierce, Rockford, IL). For the preparation of conditioned culture media, cells were grown to 80% confluency and then incubated with medium without fetal bovine serum for 24 hours. The culture media were collected and centrifuged at 1,000 rpm for 10 min to remove cells/debris, and were further concentrated by Centriplus YM-10 (Millipore, Billerica, MA).
Immunoblotting
Fifteen μg of lysate proteins or 10 μl of concentrated culture media were separated by 10-20% SDS-PAGE (Tris-Glycine gels, Invitrogen Life Technologies, Carlsbad, CA), and transferred to PVDF membranes (Millipore, Bedford, MA). The membranes were blocked with 5% nonfat dry milk, washed in TBST buffer and probed with the primary antibody. The primary antibodies used for this work are: claudin-4 (Invitrogen), MMP1 (R&D Systems), and GAPDH (Abcam Inc.). After washing in TBST, membranes were incubated with HRP-conjugated secondary antibody (anti-mouse IgG: 1:10,000; Amersham Biosciences Corp, Piscataway, NJ). Detection was carried out using the ECL plus kit (Amersham Biosciences Corp).
RNA isolation for microarrays
The claudin-4-expressing cell lines HOSE-CLDN4-1 and HOSE-CLDN4-2, as well as the two non-expressing control lines (HOSE-EV1 and HOSE-EV2) were cultured to 50% confluence in duplicate. Total RNA was then isolated using the RNAeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's recommendation. A total of 8 RNAs (4 cell lines in duplicate) were prepared for microarray analysis. The RNA quality and quantity was assessed using an Agilent 2100 bio-analyzer and the RNA 6000 Nano Chip kit.
Illumina oligonucleotide microarray
Total RNA was used to generate biotin labeled cRNA using the Illumina TotalPrep RNA Amplification Kit (Ambion; Austin, TX, cat # IL1791). Briefly, 0.5 μg of total RNA was first converted into single-stranded cDNA with reverse transcriptase using an oligo-dT primer containing the T7 RNA polymerase promoter site and then copied to produce double-stranded cDNA molecules. The double stranded cDNA was purified and concentrated with the supplied columns and used in an overnight in vitro transcription reaction where single-stranded RNA (cRNA) was generated and labeled by incorporation of biotin-16-UTP. A total of 0.75 μg of biotin-labeled cRNA was hybridized at 58 degrees C for 16 hours to Illumina's Sentrix HumanRef-8 Expression BeadChips (Illumina, San Diego, CA). Each BeadChip has 24,000 well-annotated RefSeq transcripts with an average of 30-fold redundancy. The arrays were washed, blocked and the labeled cRNA was detected by staining with streptavidin-Cy3. The arrays were scanned using an Illumina BeadStation 500X Genetic Analysis Systems scanner and the image data extracted using Illumina BeadStudio software, version 3.0. Gene expression data was transferred to an excel spreadsheet and analyzed using the Z-score method.55 Changes in gene expression were compared for each individual gene by comparing array results for the HOSE-EV and HOSE-CLDN4 clones. Clustering analysis and gene expression visualization were done with the Dchip software.56
Angiogenesis Antibody Membrane Arrays
RayBio human angiogenesis antibody membrane array I and II (RayBiotech, Inc., Norcross, GA) were used to screen for angiogenic factors induced by claudin-4 expression in HOSE-B cells. The screen was conducted according to the manufacturer's recommendations. In brief, the array membranes were sequentially incubated with blocking buffer, 1 ml conditioned media, biotin-conjugated primary antibody, and HRP-conjugated streptavidin. The positive protein spots were detected by chemiluminescence (Amersham ECL) according to the manufacturer's protocol.
In Vitro Angiogenesis Assay
The assay was performed using the Chemicon International Inc (Billerica, MA) In Vitro Angiogenesis Assay Kit. Briefly, ECMatrix gel (50 μl) was added to a 96-well plate and incubated at 37°C for 1.5 hours to allow the gel solution to solidify. Human umbilical vein endothelial cells (HUVECs) were starved for 4 hours and 1×104 cells were seeded onto the ECMatrix in 100 μl of McCoy 5A medium containing 0.2% BSA. Conditioned media from HOSE cells with or without claudin-4 expression were concentrated 10-fold and 50 μl added to HUVECs. After 16 hours incubation, the tube-like structures were quantified by counting branch points in whole microscope fields.
Migration Assay
HUVECs were grown to about 80% confluency, serum-starved for 3 hours, and labeled with 5 μM calcein AM for 1 hour in serum-free medium. Cells were then trypsinized and 5×104 cells were added to the upper chamber of a fluoroblok insert (8 μm pore size and 24-well format; BD Biosciences) in 0.2% BSA-McCoy 5A medium. Conditioned media from HOSE-B cells expressing claudin-4 or without expression (cells transfected with empty vector) were added to the lower chamber. The assay plate was incubated at 37°C for 1, 2, 3, or 4 hours and the migrated cells were measured by a Cytofluor-4000. Experiments were repeated three separate times in triplicate.
In vivo Dorsal Skinfold Angiogenesis assay
Dorsal skinfolds of nude mice were exposed to the factors secreted by claudin-expressing cells by placing the claudin-expressing (or control) cells in diffusion chambers and implanting them to skinfolds as described previously.36,57,58 Briefly, the diffusion chambers were prepared by cementing 0.45-μm membranes on both sides of the rim of an “O” ring (Millipore, Bedford, MA). The chambers were then sterilized by UV radiation for 20 minutes and wetted with sterile PBS. HOSE-EV1, HOSE-CLDN4-1, or HOSE-CLDN4-2 cells (2 × 106 cells per 125 μL of sterile PBS) were injected individually into each chamber. A dorsal air sac in an anesthetized mouse was prepared by injecting 10 ml of air in the skinfold. The air sac was then opened to implant the chamber underneath the skin, and the incision was sutured. After 10 days, the animals were euthanized, and carefully skinned around the implanted chambers. The skinfolds covering the chambers were carefully removed, stretched on a glass slide and photographed under visible light microscope (20X). The length of newly formed vessels was measured and plotted against a PC-3M variant cell lines. Sterile small-animal surgical techniques were followed during the entire procedure. The protocol was approved by the institutional Animal Care and Use Committee.
Statistics
The Z-score method was used for the analysis of microrarray data. 55 One-way ANOVA and Newman-Keuls tests were used to determine significance of the changes in blood vessel lengths in the mouse skinfolds. Student's t-tests were used to estimate statistical significance of the in vitro angiogenesis (tube formation) changes as well as the changes in HUVEC migration.
Acknowledgements
We thank members of our laboratory for helpful comments on the manuscript. This research was supported entirely by the Intramural Research Program of the National institutes of Health, National Institute on Aging.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–8. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landen CN, Jr., Birrer MJ, Sood AK. Early Events in the Pathogenesis of Epithelial Ovarian Cancer. J Clin Oncol. 2008 doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 4.Auersperg N, Maines-Bandiera SL, Dyck HG, Kruk PA. Characterization of cultured human ovarian surface epithelial cells: phenotypic plasticity and premalignant changes. Lab Invest. 1994;71:510–8. [PubMed] [Google Scholar]
- 5.Testa JR, Getts LA, Salazar H, Liu Z, Handel LM, Godwin AK, et al. Spontaneous transformation of rat ovarian surface epithelial cells results in well to poorly differentiated tumors with a parallel range of cytogenetic complexity. Cancer Res. 1994;54:2778–84. [PubMed] [Google Scholar]
- 6.Feeley KM, Wells M. Precursor lesions of ovarian epithelial malignancy. Histopathology. 2001;38:87–95. doi: 10.1046/j.1365-2559.2001.01042.x. [DOI] [PubMed] [Google Scholar]
- 7.Naora H. Developmental patterning in the wrong context: the paradox of epithelial ovarian cancers. Cell Cycle. 2005;4:1033–5. doi: 10.4161/cc.4.8.1906. [DOI] [PubMed] [Google Scholar]
- 8.Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603–6. doi: 10.1158/0008-5472.CAN-05-2782. [DOI] [PubMed] [Google Scholar]
- 9.Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, et al. Large-Scale Serial Analysis of Gene Expression Reveals Genes Differentially Expressed in Ovarian Cancer. Cancer Research. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 10.Rangel LBA, Agarwal R, D'Souza T, Pizer ES, Alò PL, Lancaster WD, et al. Tight Junction Proteins Claudin-3 and Claudin-4 Are Frequently Overexpressed in Ovarian Cancer but Not in Ovarian Cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 11.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, et al. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: Identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 12.Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR, Jr., et al. Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol. 2004;165:397–414. doi: 10.1016/S0002-9440(10)63306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–36. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- 14.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 15.Bignotti E, Tassi RA, Calza S, Ravaggi A, Romani C, Rossi E, et al. Differential gene expression profiles between tumor biopsies and short-term primary cultures of ovarian serous carcinomas: identification of novel molecular biomarkers for early diagnosis and therapy. Gynecol Oncol. 2006;103:405–16. doi: 10.1016/j.ygyno.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, Brannstrom M, Janson PO, Sundfeldt K. Differences in expression patterns of the tight junction proteins,claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int J Cancer. 2006;118:1884–91. doi: 10.1002/ijc.21506. [DOI] [PubMed] [Google Scholar]
- 17.Choi YL, Kim J, Kwon MJ, Choi JS, Kim TJ, Bae DS, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol. 2007;22:1185–95. doi: 10.14670/HH-22.1185. [DOI] [PubMed] [Google Scholar]
- 18.Kleinberg L, Holth A, Trope CG, Reich R, Davidson B. Claudin upregulation in ovarian carcinoma effusions is associated with poor survival. Hum Pathol. 2008;39:747–57. doi: 10.1016/j.humpath.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Konecny GE, Agarwal R, Keeney GA, Winterhoff B, Jones MB, Mariani A, et al. Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol Oncol. 2008;109:263–9. doi: 10.1016/j.ygyno.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechpammer M, Resnick MB, Sabo E, Yakirevich E, W OG, K TS, et al. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod Pathol. 2008 doi: 10.1038/modpathol.2008.116. [DOI] [PubMed] [Google Scholar]
- 21.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 22.Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–71. [PubMed] [Google Scholar]
- 23.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–85. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 24.Sobel G, Paska C, Szabo I, Kiss A, Kadar A, Schaff Z. Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum Pathol. 2005;36:162–9. doi: 10.1016/j.humpath.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Usami Y, Chiba H, Nakayama F, Ueda J, Matsuda Y, Sawada N, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37:569–77. doi: 10.1016/j.humpath.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 26.Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–56. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 27.Oshima T, Kunisaki C, Yoshihara K, Yamada R, Yamamoto N, Sato T, et al. Reduced expression of the claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol Rep. 2008;19:953–9. [PubMed] [Google Scholar]
- 28.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oku N, Sasabe E, Ueta E, Yamamoto T, Osaki T. Tight junction protein claudin-1 enhances the invasive activity of oral squamous cell carcinoma cells by promoting cleavage of laminin-5 gamma2 chain via matrix metalloproteinase (MMP)-2 and membrane-type MMP-1. Cancer Res. 2006;66:5251–7. doi: 10.1158/0008-5472.CAN-05-4478. [DOI] [PubMed] [Google Scholar]
- 30.Gregoire L, Munkarah A, Rabah R, Morris RT, Lancaster WD. Organotypic culture of human ovarian surface epithelial cells: a potential model for ovarian carcinogenesis. In Vitro Cell Dev Biol Anim. 1998;34:636–639. doi: 10.1007/s11626-996-0012-z. [DOI] [PubMed] [Google Scholar]
- 31.Sidky YA, Borden EC. Inhibition of angiogenesis by interferons: effects on tumor- and lymphocyte-induced vascular responses. Cancer Res. 1987;47:5155–61. [PubMed] [Google Scholar]
- 32.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov. 2007;6:273–86. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 33.Taylor KL, Leaman DW, Grane R, Mechti N, Borden EC, Lindner DJ. Identification of interferon-beta-stimulated genes that inhibit angiogenesis in vitro. J Interferon Cytokine Res. 2008;28:733–40. doi: 10.1089/jir.2008.0030. [DOI] [PubMed] [Google Scholar]
- 34.Miyamori H, Takino T, Kobayashi Y, Tokai H, Itoh Y, Seiki M, et al. Claudin promotes activation of pro-matrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. J Biol Chem. 2001;276:28204–11. doi: 10.1074/jbc.M103083200. [DOI] [PubMed] [Google Scholar]
- 35.Stetler-Stevenson WG. Matrix metalloproteinases in angiogenesis: a moving target for therapeutic intervention. J Clin Invest. 1999;103:1237–41. doi: 10.1172/JCI6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehr HA, Leunig M, Menger MD, Nolte D, Messmer K. Dorsal skinfold chamber technique for intravital microscopy in nude mice. Am J Pathol. 1993;143:1055–62. [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenkilde MM, Schwartz TW. The chemokine system -- a major regulator of angiogenesis in health and disease. Apmis. 2004;112:481–95. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 38.Arenberg DA, Kunkel SL, Polverini PJ, Glass M, Burdick MD, Strieter RM. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. J Clin Invest. 1996;97:2792–802. doi: 10.1172/JCI118734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bar-Eli M. Role of interleukin-8 in tumor growth and metastasis of human melanoma. Pathobiology. 1999;67:12–8. doi: 10.1159/000028045. [DOI] [PubMed] [Google Scholar]
- 40.Xie K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001;12:375–91. doi: 10.1016/s1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 41.Huang S, Mills L, Mian B, Tellez C, McCarty M, Yang XD, et al. Fully humanized neutralizing antibodies to interleukin-8 (ABX-IL8) inhibit angiogenesis, tumor growth, and metastasis of human melanoma. Am J Pathol. 2002;161:125–34. doi: 10.1016/S0002-9440(10)64164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rofstad EK, Halsor EF. Vascular endothelial growth factor, interleukin 8, platelet-derived endothelial cell growth factor, and basic fibroblast growth factor promote angiogenesis and metastasis in human melanoma xenografts. Cancer Res. 2000;60:4932–8. [PubMed] [Google Scholar]
- 43.Merritt WM, Lin YG, Spannuth WA, Fletcher MS, Kamat AA, Han LY, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100:359–72. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–41. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 45.Schraufstatter IU, Trieu K, Zhao M, Rose DM, Terkeltaub RA, Burger M. IL-8-mediated cell migration in endothelial cells depends on cathepsin B activity and transactivation of the epidermal growth factor receptor. J Immunol. 2003;171:6714–22. doi: 10.4049/jimmunol.171.12.6714. [DOI] [PubMed] [Google Scholar]
- 46.Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, et al. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J Immunol. 2000;165:5269–77. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 47.Petreaca ML, Yao M, Liu Y, Defea K, Martins-Green M. Transactivation of vascular endothelial growth factor receptor-2 by interleukin-8 (IL-8/CXCL8) is required for IL-8/CXCL8-induced endothelial permeability. Mol Biol Cell. 2007;18:5014–23. doi: 10.1091/mbc.E07-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brouty-Boye D, Zetter BR. Inhibition of cell motility by interferon. Science. 1980;208:516–8. doi: 10.1126/science.6154315. [DOI] [PubMed] [Google Scholar]
- 49.Lindner DJ. Interferons as antiangiogenic agents. Curr Oncol Rep. 2002;4:510–4. doi: 10.1007/s11912-002-0065-4. [DOI] [PubMed] [Google Scholar]
- 50.Dvorak HF, Gresser I. Microvascular injury in pathogenesis of interferon-induced necrosis of subcutaneous tumors in mice. J Natl Cancer Inst. 1989;81:497–502. doi: 10.1093/jnci/81.7.497. [DOI] [PubMed] [Google Scholar]
- 51.Dinney CP, Bielenberg DR, Perrotte P, Reich R, Eve BY, Bucana CD, et al. Inhibition of basic fibroblast growth factor expression, angiogenesis, and growth of human bladder carcinoma in mice by systemic interferon-alpha administration. Cancer Res. 1998;58:808–14. [PubMed] [Google Scholar]
- 52.Koch AE, Polverini PJ, Kunkel SL, Harlow LA, DiPietro LA, Elner VM, et al. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science. 1992;258:1798–801. doi: 10.1126/science.1281554. [DOI] [PubMed] [Google Scholar]
- 53.Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem. 1995;270:27348–57. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- 54.Salcedo R, Resau JH, Halverson D, Hudson EA, Dambach M, Powell D, et al. Differential expression and responsiveness of chemokine receptors (CXCR1-3) by human microvascular endothelial cells and umbilical vein endothelial cells. Faseb J. 2000;14:2055–64. doi: 10.1096/fj.99-0963com. [DOI] [PubMed] [Google Scholar]
- 55.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Wong WH. DNA-Chip Analyzer (dChip). In: Parmigiani G, Garrett E, Irizarry R, Zeger SL, editors. The analysis of gene expression data: methods and software. Springer; New York: 2003. pp. 120–141. [Google Scholar]
- 57.Leunig M, Yuan F, Menger MD, Boucher Y, Goetz AE, Messmer K, et al. Angiogenesis, microvascular architecture, microhemodynamics, and interstitial fluid pressure during early growth of human adenocarcinoma LS174T in SCID mice. Cancer Res. 1992;52:6553–60. [PubMed] [Google Scholar]
- 58.Chigurupati S, Kulkarni T, Thomas S, Shah G. Calcitonin stimulates multiple stages of angiogenesis by directly acting on endothelial cells. Cancer Res. 2005;65:8519–29. doi: 10.1158/0008-5472.CAN-05-0848. [DOI] [PubMed] [Google Scholar]