Abstract
The paramyxovirus family contains established human pathogens such as measles virus and human respiratory syncytial virus, and emerging pathogens including the Hendra and Nipah viruses and the recently identified human metapneumovirus. Two major envelope glycoproteins, the attachment protein and the fusion protein, promote the processes of viral attachment and virus-cell membrane fusion required for entry. While common mechanisms of fusion protein proteolytic activation and the mechanism of membrane fusion promotion have been shown in recent years, considerable diversity exists in the family related to receptor binding and the potential mechanisms of fusion triggering.
Introduction and overview
The paramyxovirus family is composed of enveloped, negative-stranded RNA viruses, many of which are major human pathogens [1]. Members of this family include human respiratory syncytial virus (hRSV), the leading cause of viral lower respiratory tract infections in infants and children worldwide, and measles virus, which remains a significant source of morbidity and mortality in developing countries. In recent years, a number of new paramyxoviruses have been recognized, including the Hendra and Nipah viruses, which are highly pathogenic in humans and are the only identified zoonotic members of the paramyxovirus family [2].
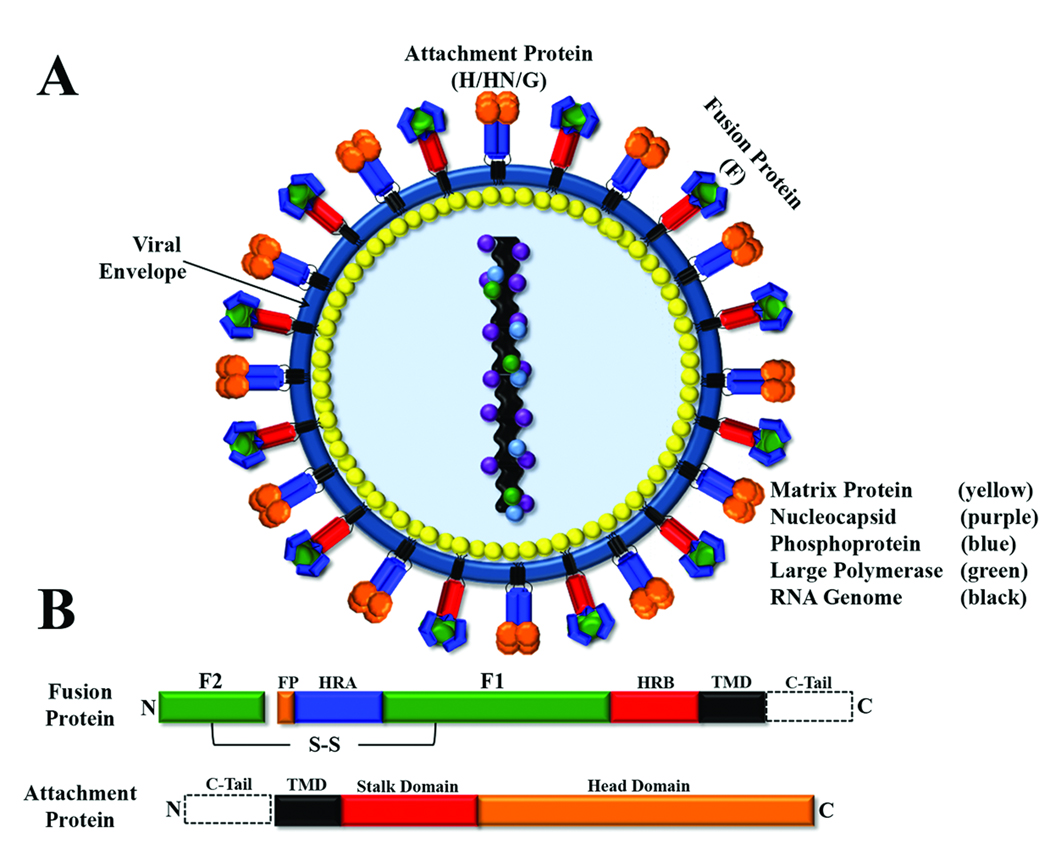
Paramyxoviruses contain between six and ten genes, encoding proteins involved in critical processes including transcription/replication (L, large polymerase; N, nucleocapsid and P, phosphoprotein), assembly (M, matrix protein) and viral entry. Paramyxovirus entry into target cells is mediated by two glycoproteins present on the viral membrane: the attachment protein (termed HN for hemagglutinin-neuraminidase, H for hemagglutinin, or G for glycoprotein, depending on the virus) and the fusion (F) protein (Figure 1A). Recent examination by cryo-EM indicated that these glycoproteins are packed in a dense layer on the viral surface [3]. Primary adsorption of the virus to the target cell is generally promoted by the attachment protein, with sialic acid residues or cell surface proteins serving as receptors. The F protein is then responsible for fusion of the viral membrane with a host cell membrane. Paramyxovirus F proteins are trimeric type I integral membrane proteins initially synthesized as non-fusogenic F0 precursors which require subsequent cleavage into the fusogenic disulfide-linked F1+F2 heterodimer (Figure 1B). This cleavage event places the conserved fusion peptide at the N-terminus of the newly created F1 subunit, priming the protein for fusion activity. Most paramyxoviruses require their homotypic attachment protein for membrane fusion activity, suggesting a role for F-attachment protein interactions in control of fusion [4–9]. The Hendra and Nipah F proteins interchangeably utilize the Hendra and Nipah G proteins in the fusion process, and this fully functional bidirectional heterotypic fusion activity is unique among paramyxoviruses [10]. Interestingly, some paramyxovirus fusion proteins can promote membrane fusion in the absence of their homotypic attachment protein [8,11,12], making the role of paramyxovirus attachment proteins in membrane fusion unclear and potentially virus specific. Despite varying sequence homology among paramyxoviruses and the diverse requirement for the attachment protein, the positional conservation of a number of structural elements suggests a similar mechanism of fusion. Membrane fusion is thought to be driven by very large conformational changes [13] following triggering of the F protein, leading to exposure and insertion of the fusion peptide into the target membrane and subsequent fusion of the viral and cellular membranes.
Figure 1. Schematic of paramyxovirus virion and surface glycoproteins.
A) Schematic of a paramyxovirus; viral membrane shown in blue. B) Conserved domains of paramyxovirus fusion and attachment proteins. Domain abbreviations: fusion peptide (FP, orange); heptad repeat A (HRA, blue); heptad repeat B (HRB, red); transmembrane domain (TMD, black); cytoplasmic tail (C-Tail, dotted box); disulfide bond (S-S).
Attachment proteins and receptors
For the majority of paramyxoviruses, interaction of the attachment protein with a cellular receptor is necessary for virus binding to target cells, and for the triggering of F protein-promoted fusion. All paramyxovirus attachment proteins characterized to date are type II integral membrane proteins that form homotetramers [1,14] (Figure 1B). Attachment protein nomenclature is defined by two characteristics: the ability or inability to bind sialic acid, and the presence or absence of neuramidase activity (or the ability to cleave sialic acid). The Respirovirus, Rubulavirus and Avulavirus attachment proteins are denoted HN, because they bind sialic acid-containing glycoproteins or glycolipids on the cell surface (hemagglutinin activity (H)) and also remove sialic acid from carbohydrates on viral glycoproteins and other cell surface molecules (neuraminidase activity (N)), thus preventing viral self-agglutination during budding [15]. The HN proteins differ in their binding affinity for varying sialic acid-containing molecules [15], likely contributing to their differing pathogenesis. The Morbillivirus attachment proteins (H) lack neuraminidase activity and utilize protein cellular receptors instead of sialic acid. Measles virus H binds to CD46 or SLAM (signal lymphocyte-activating molecule) receptors [16,17], potentially accounting for the restriction of measles infection to higher primates. Downregulation of CD46 and SLAM in infected cells presumably prevents viral aggregation during budding [18]. The Pneumovirus and Henipavirus attachment proteins lack both hemagglutinin and neuraminidase activity, and are therefore termed G (for Glycoprotein). The Hendra and Nipah G proteins have been shown to bind EphrinB2 and EphrinB3 cellular receptors [19,20]. The hRSV G protein has been shown to bind heparin [21] and cell surface proteoglycans [22].
The crystal structures of a number of paramyxovirus attachment proteins have been determined, including the HN proteins from Newcastle Disease virus (NDV), parainfluenza virus 5 (PIV5) and human parainfluenza virus 3 (hPIV3), the H protein from measles virus and the G protein from Nipah virus [23–29]. In all cases, a C-terminal globular head which contains the receptor binding and the enzymatic activity site is observed to sit on top of a membrane-proximal stalk domain. The globular head is composed of four identical monomers arranged with 4-fold symmetry, each of the monomers consisting of a six-blade β-propeller fold [23–28]. For the majority of HN proteins, a single binding site on top of the globular head domain has both hemagglutinin and neuraminidase activity [24]. However, NDV HN has been demonstrated to contain two sialic acid binding sites, one in the globular head, and one at an interface between two dimers [28]. Interestingly, for measles virus H protein, the CD46/SLAM binding sites are located toward the sides of the H protein β-barrel [26,29]. This altered placement of the receptor binding domain led to the suggestion that differences in sialic acid versus protein receptor binding may lead to different mechanisms of fusion initiation [30]. However, the binding site for ephrinB2/B3 on Nipah G was recently shown to reside at the top of the globular head domain, in a similar position to HN protein sialic acid binding sites, and a co-complex with ephrin-B3 revealed extensive protein-protein interactions, including insertion of a portion of ephrin-B3 into the central cavity of Nipah G [27]. Thus, conserved positioning of the binding site is seen for at least some protein-binding and sialic-acid binding attachment proteins.
Interestingly, recent data suggests that the Pneumovirus attachment protein may not be obligatory for attachment and entry in all cases. An attenuated hRSV missing the G protein or hRSV and bovine respiratory syncytial virus (bRSV) recombinants lacking the G protein were found to replicate in cell culture [31–33], indicating that the RSV F protein can provide sufficient binding to allow viral entry. Similarly, the G protein from the recently identified human metapneumovirus (HMPV) has been shown to be dispensible for growth in both cell culture and animal models [34]. The hRSV F protein has been shown to bind to heparin [35], though a recombinant hRSV virus lacking the G protein has been found to be less dependent on glycosaminoglycans (GAGs) for attachment than the wild type virus [36], suggesting interactions with a receptor in addition to GAGs. No specific receptor for the RSV F protein has been identified, but a recent study indicates a role for α5β1 integrin-HMPV F protein interactions in HMPV entry [37]. Finally, studies have shown that the human asialoglycoprotein receptor (ASGP-R, a mammalian lectin) may be an attachment factor for the Sendai F protein [38]. Thus, it seems possible that the process of paramyxovirus attachment may be more complex than had previously been thought, potentially involving interactions beyond those of the well-characterized attachment protein-receptor. Interaction between the F protein and the target cell might allow for a final selection step prior to triggering fusion.
Proteolytic processing of paramyxovirus F proteins
Proteolytic processing of the non-fusogenic precursor forms (F0) of paramyxovirus fusion proteins into the disulfide-linked heterodimer F1+F2 is essential for formation of fusogenically active proteins, as it primes the protein for fusion by positioning the fusion peptide at the newly formed N-terminus of F1 [39]. While the requirement for proteolytic processing is conserved among paramyxoviruses, the protease responsible for cleavage of the F0 precursor varies. Many paramyxovirus F proteins are cleaved during transport through the trans Golgi network by the ubiquitous subtilisin-like cellular protease, furin [40]. Furin-mediated proteolytic cleavage occurs following R-X-K/R-R sequences and has been demonstrated to occur in the F proteins of several paramyxoviruses including hRSV [41], PIV5 [40] and mumps virus [42]. Interestingly, hRSV F has recently been shown to undergo two N-terminal furin-mediated cleavage events, both of which are required for fusion promotion [43,44]. The Hendra and Nipah F proteins, however, lack the R-X-K/R-R consensus sequence for furin mediated cleavage. Instead, both the Hendra and Nipah F proteins are cleaved by the endosomal/lysosomal protease cathepsin L following a single basic residue in the N-terminal sequences VGDVK109 and VGDVR109, respectively [45–47]. Finally, some viral F proteins, including F proteins from HMPV [48,49] and Sendai virus [50], are cleaved by tissue-specific extracellular proteases such as tryptase Clara and mini-plasmin. Despite containing a minimal furin cleavage sequence (R-X-X-R), HMPV is not cleaved intracellularly but requires exogenous protease addition for activation [51,52], though intracellular cleavage has been observed in laboratory-expanded strains [52].
Regardless of the protease responsible for F cleavage, this step is essential for both virulence and pathogenicity. The presence of single or multiple basic residues has been demonstrated to modulate proteolytic processing and thus acts in determining pathogen virulence. NDV F proteins containing multiple basic residues in proximity to the cleavage site are more virulent and exhibit higher levels of dissemination throughout the host as compared to their F counterparts containing only one basic residue [53,54]. Proteolytic cleavage of F proteins can also result in structural rearrangement as peptide antibodies directed to the PIV5 heptad repeats recognized primarily the uncleaved form [55]. Interestingly, insertion of both multi-basic cleavage sites present in RSV F into Sendai F leads to a decreased dependency on the Sendai attachment protein and increased cell-cell fusion [56]. Thus cleavage of viral F proteins constitutes a pivotal point in the viral life cycle affecting both pathogenesis and virulence, most likely by reducing the energy required to promote the structural rearrangements of the protein needed for membrane fusion activity.
Triggering of membrane fusion
Many viral fusion proteins contain both receptor-binding and fusion activities, suggesting a straightforward model for how fusion is triggered by receptor binding. However, the separation of these two functions in paramyxoviruses makes control of fusion triggering more complex. Fusion-associated conformational changes in the F protein are thought to be irreversible, leading to a non-fusogenically active post-fusion form of the protein. Thus, it is extremely important that triggering is properly regulated both spatially and temporally [57]. The majority of paramyxovirus F proteins promote membrane fusion at neutral pH, with the exception of F proteins from certain HMPV strains shown to be triggered by exposure to low pH [11,58]. Thus, alterations in pH are not the universal trigger for paramyxovirus F protein fusion. Substantial evidence suggests that for most members of the family, fusion triggering involves specific interactions of the cleaved, metastable F protein with its homotypic attachment protein [59–64]. Upon receptor binding, the attachment protein “transmits” a signal to the F protein, potentially through conformational changes in the attachment protein and/or changes in the F protein-attachment protein interaction. Structural analysis of the NDV HN protein suggested significant conformational changes upon ligand binding [23,28], but similar changes were not observed in the PIV5 or hPIV3 HN following sialic acid binding [24,25], or in Nipah G following ephrin B3 binding [27]. Thus, a model where receptor engagement results in subtle rearrangements and reposition of the fusion and attachment proteins has been proposed [27].
The requirement for a homotypic attachment protein for fusion triggering suggests a specific interaction between the fusion and attachment proteins, and considerable research has focused on characterizing the physical interaction between these key proteins. Both co-immunoprecipitation studies and antibody-induced co-capping analyses have demonstrated interactions for the fusion and attachment proteins from a number of paramyxoviruses [59,60,62,64,65]. Numerous studies indicate that the membrane proximal stalk domain of the attachment protein is important for interaction with the fusion protein [6,9,65–68], but residues present in the globular head domain [60,69,70] or the transmembrane domain [14,71] have also been implicated. Studies have also indicated a role for the F protein TM-proximal heptad repeat B region [72] or a region within the F protein globular head [73] in these critical glycoprotein interactions.
Triggering of F protein-promoted membrane fusion is clearly also modulated by factors beyond the attachment protein. A number of F protein mutations have been shown to affect fusion triggering and/or the requirement for a homotypic attachment protein. The NDV F protein requires its homotypic HN protein, but a single amino acid change (L289A) [12] can remove this requirement in some cell types [74]. Substitution of the extended hRSV cleavage-site into the Sendai F protein can modulate attachment protein dependence [56]. Mutations in the cytoplasmic tail of the SER virus have also been found to confer HN independence to this F protein [75]. Several specific regions in paramyxovirus F proteins have also been implicated in triggering, including the linker region immediately preceding heptad repeat B [76,77], portions of heptad repeat A [78] and a conserved region of F2 that interacts with heptad repeat A in the prefusion form [79]. The F protein from the PIV5 strain WR, which normally requires the presence of an HN protein for function, can promote HN-independent membrane fusion when present at elevated temperature [80], suggesting that the requirement for HN triggering of F can also be replaced by conditions which destabilize the F protein. For the HMPV F protein, low pH can efficiently trigger fusion for some strains, and no requirement for an attachment protein is observed [11,58]. Additionally, hRSV, PIV5 strain W3A, and Sendai virus F proteins can also mediate membrane fusion even in the absence of their attachment protein [36,38,81], suggesting that their F proteins have a lower energy requirement to transition from their metastable state [39], and do not require the presence of an attachment protein to stabilize the prefusion form.
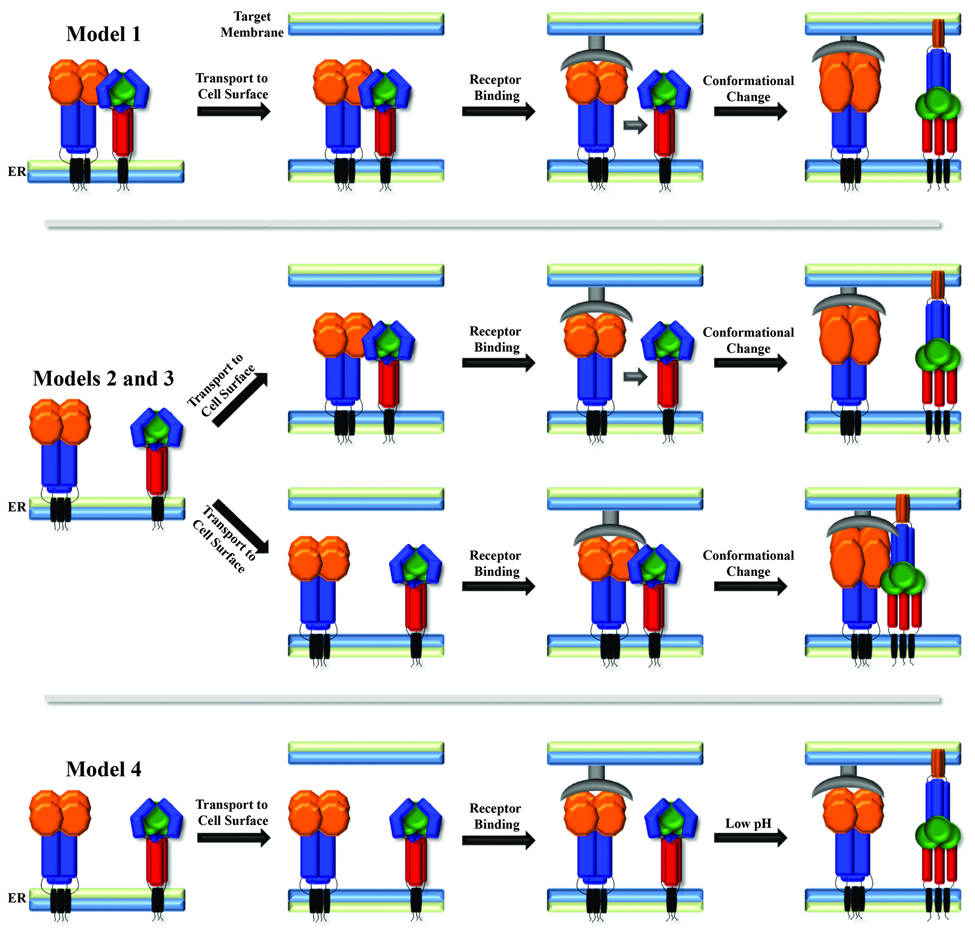
The time and place where the fusion and attachment proteins interact is critical to understanding the mechanism of fusion control, but the details of these interactions are still under investigation, and may vary between viruses. One proposed model (Figure 2, Model 1) suggests that the initial interaction between the two glycoproteins occurs within the endoplasmic reticulum (ER) at the time of synthesis, potentially allowing the attachment protein to hold the F protein in its prefusion conformation until after receptor binding. Studies of measles virus [82,83] and NDV [62] support this model, but recent studies of the Henipavirus glycoproteins suggest differential trafficking through the secretory pathway [84,85]. In addition, fusion proteins which do not require their attachment protein for function do not fit this model, as they clearly maintain their prefusion state independently. The fusion and attachment proteins may instead traffic separately through the secretory pathway, arriving at the cell surface independently. Interaction could then occur, with subsequent disruption of the F protein-attachment protein interaction by receptor binding leading to fusion triggering (Figure 2, Model 2). Recent studies of Hendra and Nipah fusion support this model, as it was shown that G mutations that inhibit F-G interaction also inhibit the fusion process [66], and that fusion promotion also correlates inversely with F-G avidity [59,60]. Alternatively, interaction between the two proteins may not occur until after the attachment protein binds its receptor (Figure 2, Model 3). Interactions between the NDV F and HN protein have been demonstrated only in the presence of receptor, and mutations which alter receptor binding decrease both fusion and F-HN interactions [86,87], supporting this model. Finally, the attachment protein is not required to interact with F for fusion promotion in some cases, although receptor binding likely facilitates the process by bringing the two membranes into close proximity (Figure 2, Model 4). The HMPV F protein has replaced the requirement for an attachment protein with a low pH-induced triggering [11], with electrostatic repulsion in the HRB linker domain shown to be critical for the triggering process [77]. It is unclear which factors drive triggering of other attachment protein-independent paramyxovirus fusion proteins.
Figure 2. Potential mechanisms of paramyxovirus fusion protein triggering.
Attachment protein shown with orange head domain and blue stalk, fusion protein shown in blue/green head domain and red stalk region, receptor shown in grey.
Paramyxovirus F protein-mediated membrane fusion
Fusion between the viral envelope and cell membrane presents a daunting challenge for enveloped viruses. To drive membrane merger, the virus must provide sufficient energy to deform opposing bilayers, ultimately resulting in the formation of a fusion pore and the release of the viral genome inside the cell (Figure 3A). Promotion of this energetically demanding process is driven by viral fusion proteins, including HIV env (envelope protein), influenza HA and the paramyxovirus F proteins, which act as molecular machines driving fusion through a series of dramatic conformational changes (reviewed in [88]). Despite little sequence homology between these disparate class I fusion proteins, all share common features including glycosylation, trimerization, the need for proteolytic cleavage and conserved sequence motifs [39]. Thus, it is likely that they mediate membrane fusion through very similar mechanisms.
Figure 3. Models of lipid and protein fusion intermediates.
A) Lipid intermediates culminating in the formation of a full fusion pore. B) Proposed fusion protein intermediates with subsequent formation of the post-fusion six-helix bundle. Fusion peptide,orange; heptad repeat A, blue; heptad repeat B, red; transmembrane domain, black.
Paramyxovirus F proteins, like other class I fusion proteins, are present in their metastable, prefusion conformation prior to fusion activation [88]. Following proteolytic processing and triggering, a series of conformational changes lead to the formation of a more stable, post-fusion form of the protein, with the energy released utilized to drive the fusion process. Understanding of paramyxovirus F protein-mediated membrane fusion has increased greatly with the recent crystal structures of the prefusion form of the PIV5 F protein [89] and of the postulated postfusion forms of the NDV and hPIV3 F proteins [90–92]. Despite these advances, many important questions related to key intermediates remain. Research to date on a number of paramyxovirus F proteins suggests a model for membrane fusion which demonstrates the importance of key conserved regions within the F protein (Figure 3B). In the prefusion form, the heptad repeat A domains (HRA, blue) are separated, the hydrophobic fusion peptide (FP) is buried, and the heptad repeat B regions HRB regions (HRB, red) interact in a coiled-coil conformation. Following triggering, conformational changes result in the release of the fusion peptide, formation of a long HRA coiled-coil, and subsequent insertion of the fusion peptide into the target membrane [93]. The HRB regions separate, and subsequent refolding leads to formation of a hairpin structure which positions HRB in an anti-parallel fashion within the grooves of the HRA trimeric coiled-coil. It is hypothesized that the formation of this six-helix bundle complex provides at least a portion of the energy needed for the merging of the lipid bilayers [13]. Subsequently, the fusion pore expands, and this expansion step is hypothesized to be the most energetically costly stage of the membrane fusion process [94].
Route of paramyxovirus entry
Enveloped viruses can enter cells either via receptor-mediated endocytosis or by direct fusion between the viral envelope and the plasma membrane. Viruses that require low pH for fusion, such as influenza virus and vesicular stomatitis virus (VSV), utilize the cellular endocytic machinery to enter cells, as vesicles from the major endocytic pathways converge into acidified endosomes [95]. Other viruses such as Ebola require endocytosis to expose their fusion proteins to pH-dependent proteases before membrane fusion can occur [96,97]. In these cases, viral-cell fusion occurs somewhere within the endocytic pathway. Viruses with pH-independent fusion proteins, such as paramyxoviruses and retroviruses, are generally thought to enter cells at the plasma membrane, as the majority of viruses from these families can efficiently infect cells in the presence of agents such as ammonium chloride that raise the endosomal pH. However, recent studies suggest that some viruses with pH independent fusion proteins may still utilize endosomal entry routes [98]. Most paramyxovirus F proteins can induce cell-cell fusion when expressed on the cell surface at neutral pH, leading to the formation of giant multinucleated cells termed syncytia. These experiments clearly indicate that the triggering for most paramyxovirus F proteins is pH-independent, with the exception of the HMPV F protein [11]. However, these experiments do not directly address the site of virus-cell fusion.
Though paramyxoviruses have generally been thought to enter at the plasma membrane, recent evidence points towards a more complex mechanism of cell entry for at least some members of the family. Internalization of viral particles prior to fusion has been noted for Sendai virus [99] and Nipah virus [100]. Chemical agents that sequester cholesterol have recently been shown to disrupt NDV infection, indicating that this paramyxovirus could be utilizing caveolin-mediated endocytosis as an entry pathway [101]. Endocytosis has also been implicated in hRSV entry, as hRSV infection was decreased in cells expressing siRNAs against key components of the clatrhin-mediated endocytosis pathway, namely the clathrin light chain, the clathrin-adapter complex, dynamin 3, and the small GTPase Rab5A. Further experiments utilizing chemical inhibitors as well as dominant negative proteins further supported the hypothesis that hRSV may at least partially utilize clathrin-dependent endocytosis to establish an active infection [102]. Recent work indicates that HMPV may utilize the cellular endocytic machinery for entry, as treatment with chlorpromazine, an inhibitor of clathrin-mediated endocytosis, conferred protection against this virus. Furthermore, dynasore, a small molecule inhibitor of dynamin, a protein required in the final step of vesicle formation in both clathrin-and caveolin-mediated endocytosis, was highly effective at blocking HMPV infection, reducing infection levels by up to 90% [77]. For some strains, HMPV F protein triggering is strongly stimulated by low pH [11], suggesting a role for the lower endosomal pH in entry, and inhibitors of endosomal acidification like bafilomycin A1, concanamycin, ammonium chloride, and monensin have all shown some efficacy at preventing HMPV infection [77]. Thus, reports to date indicate that at least some members of the paramyxovirus family utilize endocytic entry routes. Endosomal entry could potentially protect viruses from the host immune system and provide unique environments, in addition to lowered pH, that assist in productive infection. Further work is needed to more fully characterize the entry pathways utilized by paramyxoviruses.
Acknowledgements
We thank the members of the Dutch laboratory for careful reading of the manuscript. This work was supported by NIAID/NIH grants R01AI051517 and R21AI074783 to RED.
References
- 1.Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott: Williams and Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 2.Eaton BT, Broder CC, Middleton D, Wang LF. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig K, Schade B, Bottcher C, Korte T, Ohlwein N, Baljinnyam B, Veit M, Herrmann A. Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions. J Virol. 2008;82:3775–3781. doi: 10.1128/JVI.02154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong S, Compans RW. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 1999;80:107–115. doi: 10.1099/0022-1317-80-1-107. [DOI] [PubMed] [Google Scholar]
- 5.Cattaneo R, Rose JK. Cell fusion by the envelope glycoproteins of persistent measles viruses which cause lethal human brain disease. J. Virol. 1993;67:1493–1502. doi: 10.1128/jvi.67.3.1493-1502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 7.Ebata SN, Cote MJ, Kang CY, Dimock K. The fusion and hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus 3 are both required for fusion. Virology. 1991;183:437–441. doi: 10.1016/0042-6822(91)90162-5. [DOI] [PubMed] [Google Scholar]
- 8.Horvath CM, Lamb RA. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossart KN, Wang LF, Flora MN, Chua KB, Lam SK, Eaton BT, Broder CC. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 2002;76:11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schowalter RM, Smith SE, Dutch RE. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 2006;80:10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sergel TA, McGinnes LW, Morrison TG. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol. 2000;74:5101–5107. doi: 10.1128/jvi.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 14.McGinnes L, Sergel T, Morrison T. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology. 1993;196:101–110. doi: 10.1006/viro.1993.1458. [DOI] [PubMed] [Google Scholar]
- 15.Villar E, Barroso IM. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj J. 2006;23:5–17. doi: 10.1007/s10719-006-5433-0. [DOI] [PubMed] [Google Scholar]
- 16.Dorig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 17.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 18.Welstead GG, Hsu EC, Iorio C, Bolotin S, Richardson CD. Mechanism of CD150 (SLAM) down regulation from the host cell surface by measles virus hemagglutinin protein. J Virol. 2004;78:9666–9674. doi: 10.1128/JVI.78.18.9666-9674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonaparte MI, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 21.Krusat T, Streckert HJ. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 22.Escribano-Romero E, Rawling J, Garcia-Barreno B, Melero JA. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J Virol. 2004;78:3524–3532. doi: 10.1128/JVI.78.7.3524-3532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 24.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 26.Hashiguchi T, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaitsev V, von Itzstein M, Groves D, Kiefel M, Takimoto T, Portner A, Taylor G. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 30.Iorio RM, Mahon PJ. Paramyxoviruses: different receptors - different mechanisms of fusion. Trends Microbiol. 2008;16:135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karron RA, et al. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karger A, Schmidt U, Buchholz UJ. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J Gen Virol. 2001;82:631–640. doi: 10.1099/0022-1317-82-3-631. [DOI] [PubMed] [Google Scholar]
- 33.Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75:6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J Virol. 2005;79:12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Techaarpornkul S, Collins PL, Peeples ME. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002;294:296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 37.Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, Williams JV. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc Natl Acad Sci U S A. 2009;106:1566–1571. doi: 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leyrer S, Bitzer M, Lauer U, Kramer J, Neubert WJ, Sedlmeier R. Sendai virus-like particles devoid of haemagglutinin-neuraminidase protein infect cells via the human asialoglycoprotein receptor. J. Gen. Virol. 1998;79:683–687. doi: 10.1099/0022-1317-79-4-683. [DOI] [PubMed] [Google Scholar]
- 39.Dutch RE, Jardetsky TS, Lamb RA. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Bioscience Reports. 2000;20:597–612. doi: 10.1023/a:1010467106305. [DOI] [PubMed] [Google Scholar]
- 40.Garten W, Hallenberger S, Ortmann D, Schafer W, Vey M, Angliker H, Shaw E, Klenk HD. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 41.Ortmann D, Ohuchi M, Angliker H, Shaw E, Garten W, Klenk H-D. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and KEX2. J. Virol. 1994;68:2772–2776. doi: 10.1128/jvi.68.4.2772-2776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe M, Hirano A, Stenglein S, Nelson J, Thomas G, Wong TC. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 1995;69:3206–3210. doi: 10.1128/jvi.69.5.3206-3210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Begona Ruiz-Arguello M, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298:317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Reyes L, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diederich S, Moll M, Klenk HD, Maisner A. The nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem. 2005;280:29899–29903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 46.Pager CT, Craft WW, Jr, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol. 2004;78:12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami M, Towatari T, Ohuchi M, Shiota M, Akao M, Okumura Y, Parry MA, Kido H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem. 2001;268:2847–2855. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- 51.Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. Modification of the trypsin-dependent cleavage activation site of the human metapneumovirus fusion protein to be trypsin independent does not increase replication or spread in rodents or nonhuman primates. J Virol. 2006;80:5798–5806. doi: 10.1128/JVI.00294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schickli JH, Kaur J, Ulbrandt N, Spaete RR, Tang RS. An S101P substitution in the putative cleavage motif of the human metapneumovirus fusion protein is a major determinant for trypsin-independent growth in vero cells and does not alter tissue tropism in hamsters. J Virol. 2005;79:10678–10689. doi: 10.1128/JVI.79.16.10678-10689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagai Y, Klenk H-D. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology. 1977;77:125–134. doi: 10.1016/0042-6822(77)90412-3. [DOI] [PubMed] [Google Scholar]
- 54.Nagai Y, Klenk H-D, Rott R. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. J. Virol. 1976;20:501–508. doi: 10.1016/0042-6822(76)90178-1. [DOI] [PubMed] [Google Scholar]
- 55.Dutch RE, Hagglund RN, Nagel MA, Paterson RG, Lamb RA. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology. 2001;281:138–150. doi: 10.1006/viro.2000.0817. [DOI] [PubMed] [Google Scholar]
- 56.Rawling J, Garcia-Barreno B, Melero JA. Insertion of the two cleavage sites of the respiratory syncytial virus fusion protein in Sendai virus fusion protein leads to enhanced cell-cell fusion and a decreased dependency on the HN attachment protein for activity. J Virol. 2008;82:5986–5998. doi: 10.1128/JVI.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 58.Herfst S, Mas V, Ver LS, Wierda RJ, Osterhaus AD, Fouchier RA, Melero JA. Low pH induced membrane fusion mediated by human metapneumoviruses F protein is a rare, strain dependent phenomenon. J Virol. 2008;82:8891–8895. doi: 10.1128/JVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aguilar HC, Matreyek KA, Choi DY, Filone CM, Young S, Lee B. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol. 2007;81:4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bishop KA, et al. Identification of hendra virus g glycoprotein residues that are critical for receptor binding. J Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol. 2002;76:5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone-Hulslander J, Morrison TG. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 1997;71:6287–6295. doi: 10.1128/jvi.71.9.6287-6295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J Virol. 2002;76:13028–13033. doi: 10.1128/JVI.76.24.13028-13033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Q, Hu X, Compans RW. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 1997;71:650–656. doi: 10.1128/jvi.71.1.650-656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bishop KA, et al. Residues in the stalk domain of the hendra virus g glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82:11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Porotto M, Murrell M, Greengard O, Moscona A. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J Virol. 2003;77:3647–3654. doi: 10.1128/JVI.77.6.3647-3654.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sergel T, McGinnes LW, Peeples ME, Morrison TG. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology. 1993;193:717–726. doi: 10.1006/viro.1993.1180. [DOI] [PubMed] [Google Scholar]
- 69.Aguilar HC, Ataman ZA, Aspericueta V, Fang AQ, Stroud M, Negrete OA, Kammerer RA, Lee B. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F) J Biol Chem. 2009;284:1628–1635. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mirza AM, Deng R, Iorio RM. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidae glycoprotein: effects on antigenic structure and function. J. Virol. 1994;68:5093–5099. doi: 10.1128/jvi.68.8.5093-5099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bousse T, Takimoto T, Gorman WL, Takahashi T, Portner A. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology. 1994;204:506–514. doi: 10.1006/viro.1994.1564. [DOI] [PubMed] [Google Scholar]
- 72.Gravel KA, Morrison TG. Interacting domains of the HN and F proteins of newcastle disease virus. J Virol. 2003;77:11040–11049. doi: 10.1128/JVI.77.20.11040-11049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J, Melanson VR, Mirza AM, Iorio RM. Decreased dependence on receptor recognition for the fusion promotion activity of L289A-mutated newcastle disease virus fusion protein correlates with a monoclonal antibody-detected conformational change. J Virol. 2005;79:1180–1190. doi: 10.1128/JVI.79.2.1180-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seth S, Vincent A, Compans RW. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J Virol. 2003;77:167–178. doi: 10.1128/JVI.77.1.167-178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell CJ, Kantor KL, Jardetzky TS, Lamb RA. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J Cell Biol. 2003;163:363–374. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol. 2009;83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luque LE, Russell CJ. Spring-loaded heptad repeat residues regulate the expression and activation of paramyxovirus fusion protein. J Virol. 2007;81:3130–3141. doi: 10.1128/JVI.02464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gardner AE, Dutch RE. A conserved region in the F(2) subunit of paramyxovirus fusion proteins is involved in fusion regulation. J Virol. 2007;81:8303–8314. doi: 10.1128/JVI.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 81.Feldmann H, Kretzschmar E, Klingeborn B, Rott R, Klenk H-D, Garten W. The structure of serotype H10 hemagglutinin of influenza A virus: Comparison of an apathogenic avian and a mammalian strain pathogenic for mink. Virology. 1988;165:577–583. doi: 10.1016/0042-6822(88)90586-7. [DOI] [PubMed] [Google Scholar]
- 82.Corey EA, Iorio RM. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology. 2009;383:1–5. doi: 10.1016/j.virol.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 84.Whitman SD, Dutch RE. Surface density of the Hendra G protein modulates Hendra F protein-promoted membrane fusion: Role for Hendra G protein trafficking and degradation. Virology. 2007;363:419–429. doi: 10.1016/j.virol.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitman SD, Smith EC, Dutch RE. Differential rates of protein folding and cellular trafficking for the Hendra virus F and G proteins: implications for F-G complex formation. J Virol. 2009;83:8998–9001. doi: 10.1128/JVI.00414-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, Smith BJ, Colman PM, Lawrence MC. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Camb) 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 91.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 92.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asano K, Asano A. Why is a specific amino acid sequence of F glycoprotein required for the membrane fusion reaction between envelope of HVJ (Sendai virus) and target cell membranes? Biochem. International. 1985;10:115–122. [PubMed] [Google Scholar]
- 94.Chernomordik LV, Zimmerberg J, Kozlov MM. Membranes of the world unite! J Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pelkmans L, Bürli T, Zerial M, Helenius A. Caveolin-Stabilized Membrane Domains as Multifunctional Transport and Sorting Devices in Endocytic Membrane Traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 96.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal Proteolysis of the Ebola Virus Glycoprotein Is Necessary for Infection. Science. 2005 doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rasmusson BJ, Flanagan TD, Turco SJ, Epand RM, Petersen NO. Fusion of Sendai virus and individual host cells and inhibition of fusion by lipophosphoglycan measured with image correlation spectroscopy. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1998;1404:338–352. doi: 10.1016/s0167-4889(98)00082-2. [DOI] [PubMed] [Google Scholar]
- 100.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375:391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cantin C, Holguera J, Ferreira L, Villar E, Munoz-Barroso I. Newcastle disease virus may enter cells by caveolae-mediated endocytosis. J Gen Virol. 2007;88:559–569. doi: 10.1099/vir.0.82150-0. [DOI] [PubMed] [Google Scholar]
- 102.Kolokoltsov AA, Deniger D, Fleming EH, Roberts NJ, Jr, Karpilow JM, Davey RA. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J Virol. 2007;81:7786–7800. doi: 10.1128/JVI.02780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]