Abstract
Divalent cations play critical structural and functional roles in many RNAs. While the hepatitis delta virus (HDV) ribozyme can undergo self-cleavage in the presence of molar concentrations of monovalent cations, divalent cations such as Mg2+ are required for efficient catalysis under physiological conditions. Moreover, the cleavage reaction can be inhibited with Co(NH3)63+, an analog of Mg(H2O)62+. Here, the binding of Mg2+ and Co(NH3)63+ to the HDV ribozyme are studied by Raman microscopic analysis of crystals. Raman difference spectra acquired at different metal ion conditions reveal changes in the ribozyme. When Mg2+ alone is introduced to the ribozyme, inner sphere coordination of Mg(H2O)x2+ (x≤5) to non-bridging PO2− oxygen, and changes in base stretches and phosphodiester group conformation are observed. In addition, binding of Mg2+ induces deprotonation of a cytosine assigned to the general acid C75, consistent with solution studies. When Co(NH3)63+ alone is introduced, deprotonation of C75 is again observed, as are distinctive changes in base vibrational ring modes and phosphodiester backbone conformation. In contrast to Mg2+ binding, Co(NH3)63+ binding does not perturb PO2− group vibrations, consistent with its ability to make only outer sphere contacts. Surprisingly, competitive binding studies reveal that Co(NH3)63+ ions displace some inner sphere-coordinated magnesium species, including ions coordinated to PO2− groups or the N7 of a guanine, likely G1 at the active site. These observations contrast with the tenet that Co(NH3)63+ ions displace only outer sphere magnesium ions. Overall, our data support two classes of inner sphere Mg2+-PO2− binding sites: sites that Co(NH3)63+ can displace, and others it cannot.
Metal ions play essential roles in the structure and function of ribozymes. Effects of metal ions on the small nucleolytic HDV ribozyme (Figure 1) have been studied for a number of years (1–9). Self-cleavage of the HDV ribozyme has only a weak dependence on divalent ion identity. For instance, self-cleavage is efficient in the presence millimolar amounts of Mg2+, Ca2+, Mn2+, Sr2+, Ba2+, or Co2+ (1, 5). In addition, self-cleavage can occur in the complete absence of divalent cations if molar concentrations of monovalent cations such as Na+, Li+ and NH4+ are provided (3, 4, 6, 10, 11). Nonetheless, the reaction catalyzed in the presence of 1 M monovalent cations alone is 25- to 100-fold less efficient (4–6), and when extrapolated to physiological monovalent concentrations (~150 mM ionic strength) is ~500-fold less efficient (4). These observations suggest critical structural and functional roles for divalent cations in the ribozyme (3, 6).
Figure 1.
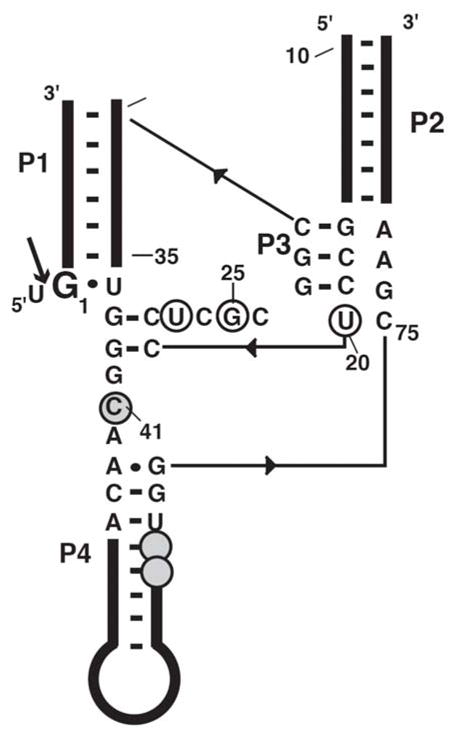
Secondary structure of the genomic HDV ribozyme. The RNAs used in this study are a 62 nt ribozyme annealed to a 9 nt inhibitor strand containing a methoxyuridine at position −1 to prevent cleavage. The ribozyme cleavage site is indicated with an arrow. Nucleotides involved in binding Co(NH3)63+ ion 1 (Figure 6A) are circled, and nucleotides involved in binding Co(NH3)63+ ion 2 (Figure 6C) are circled and shaded.
The catalytic activity of the HDV ribozyme is inhibited by competition of Co(NH3)63+ ions for Mg2+ ions (3). Co(NH3)63+ mimics hexhydrated Mg2+ in its size and coordination geometry and is exchange-inert (kexchange, NH3 ~10−10 s−1); as such, it can make only outer sphere interactions with RNA (2, 12–14). On the basis of these characteristics, it was initially suggested that the catalytic Mg2+ ion bound within the HDV ribozyme active site is in the hexahydrated form. Recent Raman crystallographic and biochemical studies, however, indicate that this ion is directly coordinated to the N7 of the cleavage site guanosine as an inner sphere ion and that it is likely only penta or tetra-coordinated to water molecules (9).
For several decades, Raman spectroscopy has provided invaluable insights into interactions between metal ions and nucleic acids (15–21). The ability of Raman to probe metal-nucleic acid interactions is further enabled by Raman difference spectroscopy (21, 22). In this approach, the spectrum of the RNA without metal ions is subtracted from the spectrum of RNA with metal ions. Features in the difference spectrum provide detailed information on metal ion binding sites, changes in base stacking interactions, and backbone conformational changes owing to metal ion binding. This approach is particularly powerful when single crystals are used since Raman of single RNA or protein crystals provides data of unprecedented quality owing to high sample concentration in crystals, low background interference, and high spectral resolution (8, 23–25).
Although we have not previously published data on cobalt-hexamine binding to HDV, in order to understand the data and discussion in the present paper we must summarize the findings of three recent publications from our laboratories. In Gong et al. (2008) we provided the first evidence for a direct relationship between magnesium hydrate binding inner sphere to the oxygens of -PO2− backbone groups and the up-shift in the Raman symmetrical stretching frequency of these groups. This involved titrating Mg2+ hydrate into single crystals of the HDV ribozyme and identifying the Raman mode of bound magnesium penta- or tetra hydrate using isotopic substitutions. There is a simple linear relationship between the intensity of the up-shifted (by 17 cm−1) -PO2− mode and the intensity of the magnesium penta- or tetra- hydrate mode that appears near 320 cm−1. This uniquely establishes that direct contact between the Mg2+ ion and the -PO2− group brings about the observed up-shift in the latter’s Raman frequency. Other papers have dealt with the active site properties of HDV. In Gong et al. (2007) we measured directly the pKa of the active site cytosine 75 (C75) in crystals of HDV. This was made possible by the fact that we could identify the Raman feature from a single neutral cytosine in HDV, occurring near 1528 cm−1. Its identity as C75 was confirmed by the finding that the variant of HDV possessing C75 replaced by uridine showed no evidence for a titration in the Raman spectra of the corresponding crystals. The results on the crystals received additional support from the observation that the crystal pKa values obtained in the presence of 20 and 2 mM Mg2+, 6.15 and 6.40 respectively, agree well with the values derived from HDV activity studies in solution. The active site studies were extended recently in Chen et al. (2009) where solution kinetic and parallel Raman crystallographic studies examined the effects of pH on the rate and Mg2+ binding properties of wild-type and 7-deazaguanosine mutants of the HDV ribozyme. These data suggested that a previously unobserved hydrated magnesium ion interacts with N7 of the cleavage site G•U wobble base pair.
Herein, we use Raman crystallography to study interactions of Mg2+ and Co(NH3)63+ ions with the HDV ribozyme. These ions were chosen because of their unique physicochemical characteristics, spectrally disperse Raman features, and well-documented effects on HDV ribozyme function in solution. We find that introduction of either Mg2+ or Co(NH3)63+ results in deprotonation of a cytosine, previously identified as C75, as well as perturbation of the N7 of a guanine, previously identified as G1. C75 directly participates in catalysis, acting as the general acid in the cleavage reaction. Together, these data suggest that the binding sites of these two ions must be in proximity to C75 within the ribozyme active site, and likely at least partially overlap. Moreover, these results indicate that inactivation of a ribozyme’s activity by Co(NH3)63+ does not necessarily imply that the displaced Mg2+ ions were hexahydrated. Raman spectra also reveal that the molecular conformation of the HDV ribozyme in the presence of Co(NH3)63+ is slightly different from that in the presence of Mg2+, which indicates that inhibition by Co(NH3)63+ could, in part, involve rearrangement of the active tertiary conformation.
MATERIALS AND METHODS
Chemicals
Chemicals used in the present study were purchased from Sigma. All stabilizing buffer solutions were prepared using 50 mM Na-cacodylate adjusted to the appropriate pH using HCl. Both Mg2+ and Co(NH3)63+ binding experiments were therefore performed in the presence of 50 mM Na+ ions. Experiments were conducted at pH 6.0, except competitive binding experiments between Mg2+ and Co(NH3)63+, which were at pH 7.0 to allow comparison with previous experiments in solution (3).
Preparation and crystallization of the HDV ribozyme
HDV ribozyme crystals used for Raman crystallography studies were prepared as described (9, 26). For the present study, the cleavage reaction was inhibited by modifying the catalytic 2′-OH on the U-1 group to a 2′-OCH3.
Raman spectroscopy of HDV ribozyme crystals
Raman spectra were nonresonant and obtained at room temperature (22 °C) as described (25, 26). All bound Mg2+ ions in the original HDV ribozyme crystals were removed by soaking in 50 mM EDTA five times, as described in Gong et al. (8). Crystals were then transferred to 5 μL drops containing stabilizing buffer (50% MPD, 2 mM spermine in 50 mM Na-cacodylate buffer, pH 6.0) and washed with this buffer 3–5 times to remove the EDTA from the crystals. Spectra in the absence of Mg2+ were then obtained. To obtain Raman spectra of the HDV crystal with varying Mg2+ and Co(NH3)63+ concentrations, crystals were then soaked in 5 μL stabilizing buffer containing the desired cation salt for 3 times over the course of 2 h to ensure that metal ion-binding equilibrium had been reached. Equilibrium was determined by recording Raman spectra at several different times following buffer exchange.
Raman difference spectra and data analysis were performed using GRAMS/32 software (Galactic Industries, Inc.). In general, data were treated by first subtracting out the buffer from each spectrum according to the equation (25),
Binding analysis of Mg2+ in HDV ribozyme crystals
Mg2+ binding experiments were performed at pH 6.0 at concentrations of Mg2+ ranging from 0 to 30 mM. Standardized Raman intensity of neutral cytosine band at 1528 cm−1, assigned to C75, was plotted as a function of Mg2+ concentration. The Hill coefficient was obtained by fitting to a Hill-type equation (eq 1, see Supporting Information for derivation),
| (1) |
where Iobs is the observed intensity at 1528 cm−1; IIS is the intensity of an internal standard band at 725 cm−1, which represents an adenine ring mode that is insensitive to metal binding in HDV; I0 is the intensity of the 1528 cm−1 band at 0 mM Mg2+; Imax is the intensity of the 1528 cm−1 band at saturating Mg2+; n is Hill coefficient; and KD,Mg2+ is the concentration of Mg2+ corresponding to half occupation of the Mg2+ binding sites in the ribozyme.
Analysis of Co(NH3)63+ binding in HDV ribozyme crystals
Competitive binding of Co(NH3)63+ (0 – 1 mM) in the presence of 0 – 5 mM Mg2+ at pH 7.0 was studied using Raman crystallography. The standardized intensity of the band at 441 cm−1, representing the Co-N stretching of Co(NH3)63+, was plotted as function of Co(NH3)63+ concentration and fit to eq 2 (see Supporting Information for derivation),
| (2) |
where Iobs is the observed intensity at 441cm−1; IIS is the intensity of the internal standard band at 725 cm−1; Imax is the intensity of the 441 cm−1 band at saturating Co(NH3)63+; n is the Hill coefficient for Co(NH3)63+ binding; and Ki,app is the concentration of Co(NH3)63+ corresponding to half occupation of the Co(NH3)63+ binding sites. This equation is essentially identical to eq 1, but with Io = 0, which is the case at 441 cm−1 (see Figure 7A).
Figure 7.
Quantitative assessment of competition between Co(NH3)63+ and Mg2+. (A) Co(NH3)63+ binding profiles in HDV ribozyme at Mg2+ concentrations of 0 mM (○), 1 mM (●), 2 mM (□), 3 mM (■) 4 mM (△) and 5 mM (▲). The y-axis is the ratio of Raman intensity of Co-N stretching of Co(NH3)63+ at 441 cm−1 to the intensity of adenine ring mode at 725 cm−1 (internal standard). Data were fit to eq 2, from which the apparent binding constants of Co(NH3)63+ (Ki, app) were obtained in the presence of different [Mg2+]. All experiments were performed at pH 7.0. (B) Secondary plot of K1.5i, app versus [Mg2+]1.5 fit to eq 3. This gave values of 47 μM and 1.7 mM for Ki, Co(NH3)63+ and KD,Mg2+, respectively.
The Co(NH3)63+ inhibition constant Ki,Co(NH3)63+, and the Mg2+ dissociation constant KD,Mg2+ were determined by a secondary plot of Kni,app versus [Mg2+]n (n fixed to 1.5 or 2, as appropriate) and fitting to eq 3 (see Supporting Information for derivation),
| (3) |
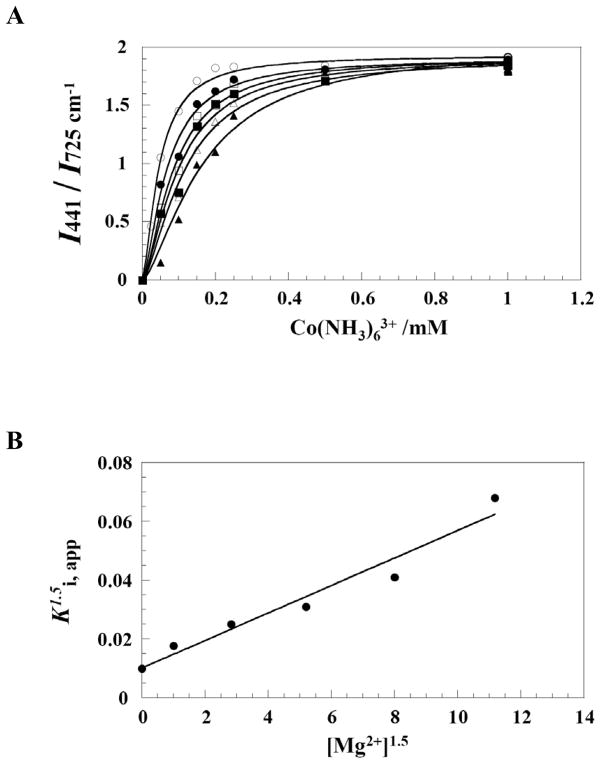
Apparent binding constants of Co(NH3)63+ (Ki,app) in 0, 1, 2, 3, 4 and 5 mM Mg2+ were obtained by fitting Co(NH3)63+ titration data to eq 2. Initially, we allowed the value of n to float and fit all the data individually. Fitting gave values for n between 1.2 and 1.6 (Table 2). Next, we fit all data forcing n = 1, 1.5, 2, or 3. Only n = 1.5 or 2 gave good fits to the data, supporting binding of at least two Co(NH3)63+ ions to the ribozyme. Data for n = 1.5 are provided in the Results, while data for n = 2 are in the Supporting Information.
Table 2.
Comparison of binding parameters from Raman crystallographic and biochemical studiesa
| Raman crystallographic | biochemical | |||||
|---|---|---|---|---|---|---|
| Condition | KD,Mg2+(mM) | Ki, Co(NH3)63+(μM) | nHill | KD,Mg2+(mM) | Ki, Co(NH3)63+(μM) | nHill |
| Mg2+ aloneb | 7.4 ± 0.2 | — | 8.0 ± 1.6 | 9.8c | — | 1.3 |
| Competitive binding of Co(NH3)63+ and Mg2+ d | 1.7 ± 0.4 | 47 ± 9 | 1.5e | 2.4f | 45 ± 10 | 1.4g |
| 2.0 ± 0.3 | 54 ± 8 | 2h | ||||
HDV single crystal used in this study has 2′-OCH3 substitution on the U-1 group.
Mg2+-HDV interaction was studied at pH 6.0.
Value measured at pH 6.0 (3).
Competitive binding of Co(NH3)63+ and Mg2+ was performed at pH 7.0.
The nHill for both Co(NH3)63+ and Mg2+ binding was held constantly at 1.5 for all analysis (see text) since the floating values of nHill in the presence of 0, 1, 2, 3, 4 and 5 mM Mg2+ are 1.4 ± 0.2, 1.4 ± 0.3, 1.2 ± 0.3, 1.4 ± 0.4, 1.6 ± 0.2, 1.4 ± 0.2, respectively (average R2 was 0.9814).
Value measured at pH 7.0 (3).
This value is for Mg2+ binding at pH 7.0 (3). The Hill value for Co(NH3)63+ was not measured explicitly in that study.
nHill = 2 also gave good curve fit using eq 2 (Figure S1A, see Supporting Information) (average R2 was 0.9844). However, nHill = 1 and nHill = 3 did not (data not shown) (average R2 values were 0.9642 and 0.9544, respectively).
RESULTS
Overview
The Results are divided into eight sections. The first four sections describe how Mg2+ alone: 1) interacts with the phosphate groups and nucleotide bases of the ribozyme, 2) induces conformational changes within the RNA, 3) causes cytosine deprotonation, and 4) binds in a reversible manner. The next two sections describe how Co(NH3)63+ alone: 5) interacts with the ribozyme and causes cytosine deprotonation, and 6) induces unique conformational changes of the ribozyme. The final two sections describe competition between Mg2+ and Co(NH3)63+ in 7) structural and 8) quantitative fashions.
Mg2+ interactions with the HDV Ribozyme
The effect of Mg2+ on the HDV ribozyme was studied by recording Raman spectra of a ribozyme crystal in the presence and absence of Mg2+. As described (8), the condition of [HDV no Mg2+] was obtained by replacing the surrounding buffer containing 20 mM Mg2+ through 5 independent 50 mM EDTA soaks. This process “soaks out” the Mg2+ inside the solvent channels of the crystal. The resulting Raman difference spectrum of [HDV + Mg2+] minus [HDV no Mg2+] is shown in Figure 2A, where features above the baseline arise from [HDV + Mg2+] and features below the baseline arise from [HDV no Mg2+]. Importantly, the spectrum of the stabilization buffer was subtracted from each spectrum first, as described in the Materials and Methods. Spectral features were assigned on the basis of literature studies (15, 16, 27, 28) and our recent work (8, 25, 26) and are provided in Table 1.
Figure 2.

Effects of Mg2+ on HDV ribozyme conformation. (A) Raman difference spectrum of [HDV + 20 mM Mg2+] minus [HDV no Mg2+] (pH 6.0) was collected. [HDV no Mg2+] condition was obtained by removing Mg2+ using 50 mM EDTA. The spectrum of the stabilization buffer was subtracted from each spectrum prior to subtraction (see Materials and Methods). Experimental details are described in Gong et al. (8). (B) Raman difference spectrum of [original crystal with 20 mM Mg2+] minus [crystal re-soaked in 20 mM Mg2+] (pH 6.0). To obtain spectrum of [crystal re-soaked in 20 mM Mg2+], a crystal of [HDV no Mg2+] was resoaked in 20 mM Mg2+-containing stabilization buffer after Mg2+ had been soaked out by EDTA.
Table 1.
Positions (cm−1) and assignment of Raman difference bands observed in Raman difference spectra of [HDV + Mg2+] minus [HDV no Mg2+]
| Wavenumbuer/cm−1 | Assignment |
|---|---|
| 1719 | C=O |
| 1679 | C=O |
| 1570/1557 | G (A?) |
| 1528 | C |
| 1491/1480 | G |
| 1453 | C-H deformation |
| 1320 | G |
| 1265 (broad) | C+ |
| 1197 | G |
| 1117/1101 | PO2− |
| 930/915 | Ribose backbone |
| 877 | Ribose backbone |
| 825/812 | -O-P-O- |
| 779 | C (U?) |
| 788 | C+ (U?) |
| 671 | G |
C, neutral cytosine; C+, protonated cytosine; C=O, carbonyl group. The features assigned to the bases are due to ring modes.
As shown in Figure 2A, the most intense differential band is centered near 1110 cm−1.1 The negative feature at 1101 cm−1, which has been unambiguously assigned to symmetric stretching of PO2− (27, 28), shifts to 1117 cm−1 owing to Mg2+ - PO2− interactions. We recently assigned this differential spectral feature to inner sphere interactions between Mg2+ and PO2− groups on the basis of Mg-hydrates bound to the HDV ribozyme, model compound studies, isotope effects, and quantum-mechanical calculations (8).
Differential bands are also observed near 1719, 1570/1557, 1491/1480, 1320, and 671 cm−1 (Figure 2A) and are likely due to effects of Mg2+ on the nucleobases. Importantly, these effects are not due simply to changes in ionic strength, as a Raman difference spectrum of [300 mM Na+] minus [50 mM Na+] does not show these features (data not shown). The negative band at 1719 cm−1 is assigned to the stretching vibrations of the carbonyl groups of G, C and U involved in inter-base hydrogen bonds (27–29). The intensity loss of this band likely reflects subtle change in inter-base hydrogen binding interactions in the presence of Mg2+. The differential band at 1570/1557 cm−1 is attributed to G ring modes with possibility of minor contribution from A and represents perturbations of purines upon Mg2+ binding, the details of which are not yet understood. These changes are also consistent with the observation of negative bands at 1320 and 671 cm−1, as these two bands have been unambiguously assigned to ring breathing of G bases. The intensity decrease of these two bands in the presence of Mg2+ may also indicate alteration of G stacking (30, 31).
The differential band at 1491/1480 cm−1 is assigned mainly to a G mode with a strong component from N7-C8 vibrational motion (16, 32). Previous Raman studies showed that the band perturbation at this region indicates binding of electrophilic agents (Mg2+ in this case) at N7 of G (16, 33). In our recent work, we observed a similar spectral feature in the pH difference spectrum of [HDV pH 7.5] minus [HDV pH 5.0] (9). This feature is lost upon site-specific mutation at N7 of G1 using 7-deaza substitution, supporting the conclusion that the band perturbation near 1485 cm−1 results, at least in part, from an inner sphere contact between the active site Mg2+ and the N7 of G1 (9).
Mg2+ binding induces ribozyme conformational changes
The differential feature observed at 825/812 cm−1 in Figure 2A can be assigned to symmetric stretches of phosphodiester backbone modes (-O-P-O-), which are sensitive to conformational changes (27, 28). The negative band of 812 cm−1 resulting from canonical A-form RNA shifts to higher frequency (825 cm−1) in the presence of Mg2+. In other nucleic acids, this kind of shift has been considered an indicator of conformational changes upon Mg2+ binding, where canonical A-type C3′-endo-anti ribose puckers change to C2′-endo-anti (34).
The nonhelical regions of HDV (bulge, loop, etc.) are likely candidates for generating these conformational perturbations (34, 35). A semi-quantitative analysis on the basis of band intensity decrease of -O-P-O- indicates that the deviation is ~3 % the intensity of the peak in the parent spectrum, suggesting that the conformational change is small. These changes may be localized to C41 and C75, or they may represent small effects spread over the entire ribozyme (see Discussion).
Mg2+ binding induces cytosine deprotonation
Raman Mg2+ difference spectra reveal features that arise from neutral and protonated cytosine, including a positive band at 1528, a negative band at 1265, and a differential band at 788/779 cm−1 (Figure 2A). These spectral features are readily assigned on the basis of work of ourselves (9) and others (27). The negative 1265 cm−1 band has been assigned to protonated cytosine, indicating that cytosine deprotonates at pH 6.0 when Mg2+ is added. Complementarily, the positive 1528 cm−1 band denotes the presence of additional neutral cytosine in the presence of 20 mM Mg2+.
We previously demonstrated that there is just a single cytosine base that is protonatable in this pH range and assigned it as the catalytic cytosine (C75) on the basis of mutational analysis (26). The Raman difference spectrum of [HDV + Mg2+] minus [HDV no Mg2+] (Figure 2A) thus provides strong evidence that addition of 20 mM Mg2+ leads to the deprotonation of C75.
The extent of cytosine deprotonation can be quantitated via the absolute intensity of the 1528 cm−1 band in the Raman difference spectrum. Using band integrations as previously described (26), we determined that exposing the pH 6.0 crystal to 20 mM Mg2+ creates a population increase of neutral cytosine of about 10 mM. Since pH 6.0 is slightly below the pKa value of 6.15, previously identified by Raman crystallography under these Mg2+ conditions, and because the concentration of HDV ribozyme in the crystal is about 26 mM (26), it appears that adding Mg2+ at constant pH drives deprotonation of a single cytosine, the active site cytosine residue (C75).2 This interpretation is consistent with solution kinetics and Raman crystallographic data, which demonstrated that the pKa of C75 is inversely correlated with Mg2+ concentration (3, 26).
To semi-quantitatively analyze the relationship of Mg2+ binding and C75 protonation, we measured the standardized intensity of the neutral C75 band at 1528 cm−1 as a function of added Mg2+ concentration. The ratio of the 1528 to 725 cm−1 intensities in the Mg2+ concentration range from 0 to 30 mM are plotted in Figure 3 and fit to eq 1. (The 725 cm−1 band, an adenosine ring mode, was chosen since it is insensitive to Mg2+ binding, as confirmed by absence of the 725 cm−1 band in Figure 2A.3) Analysis gave a Hill coefficient of 8.0 ± 1.6 and a dissociation constant KD,Mg2+ of 7.4 mM for bound magnesium (Table 2). The latter value is in good agreement with biochemical studies at pH 6.0 (KD,Mg2+ ~9.8 mM) (3).
Figure 3.

Dependence of C75 ionization on Mg2+ concentration. HDV crystals at pH 6.0 show a Mg2+ concentration-dependent change of Raman intensity of neutral C75 peak (1528 cm−1). The y-axis is the ratio of Raman intensity of neutral C75 at 1528 cm−1 to the intensity of adenine ring mode at 725 cm−1 (internal standard). The intensity of the latter is independent of Mg2+ concentration.
Binding of Mg2+ is reversible
As mentioned, the difference spectrum in Figure 2A was generated by treating a 20 mM Mg2+-containing crystal with 50 mM EDTA five times and subtracting its Raman spectrum from that of the original crystal in the presence of 20 mM Mg2+. Subsequently, we re-exposed the EDTA-soaked crystal to a solution of 20 mM Mg2+ in order to “re-soak in” Mg2+. In this case, the crystal was soaked three times in the 20 mM Mg2+-containing stabilization buffer. The difference spectrum of [original crystal with 20 mM Mg2+] minus [crystal re-soaked in 20 mM Mg2+] is essentially flat (Figure 2B). Absence of hysteresis suggests that soaking Mg2+ in and out does not damage the crystals and that binding of Mg2+ is reversible.
Co(NH3)63+ interactions with the HDV ribozyme and induction of cytosine deprotonation
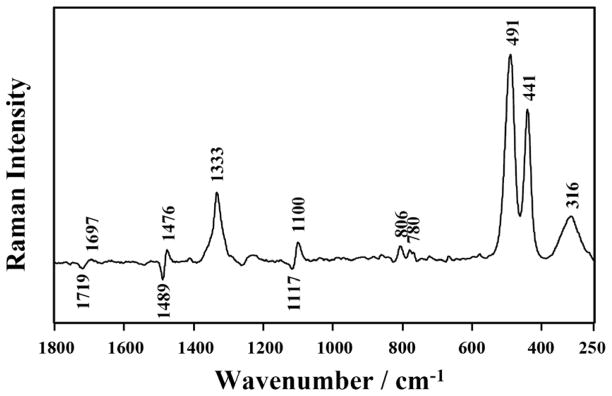
Next, we turn our attention to the interactions of Co(NH3)63+ with the HDV ribozyme crystals. Effects of Co(NH3)63+ binding were revealed by the Raman difference spectrum of [HDV + 0.1 mM Co(NH3)63+] minus [HDV no Co(NH3)63+] (Figure 4). An apparent concentration of 0.1 mM Co(NH3)63+ was established in the crystals by soaking them three times in 0.1 mM Co(NH3)63+- containing stabilization buffer. Intense positive bands observed in the spectral region 250—1800 cm−1 are due mainly to bound Co(NH3)63+, including N-H3 bending near 1333 cm−1, Co-N stretching at 491 and 441 cm−1, and N-Co-N deformation at 315 cm−1. These features are essentially unchanged compared to the corresponding modes seen for the unbound cobalt hexamine. Importantly, these signals arise from bound rather than free Co(NH3)63+ because the Raman spectrum of the stabilization buffer is subtracted from the parent [HDV + 0.1 mM Co(NH3)63+] spectrum (see Materials and Methods, “Raman spectroscopy of HDV ribozyme crystals”). Thus, negative peaks from unbound Co(NH3)63+ do not appear in Figures 4 and 5.
Figure 4.
Effects of Co(NH3)63+ on HDV ribozyme conformation. Raman difference spectrum of [HDV + 0.1 mM Co(NH3)63+] minus [HDV no Co(NH3)63+] (pH 6.0) was collected. Both [HDV + 0.1 mM Co(NH3)63+] and [HDV no Co(NH3)63+] spectra were collected in the absence of added Mg2+. The spectrum of the stabilization buffer was subtracted from each spectrum first. Main band assignments are provided in Table 1 except bands resulting from Co(NH3)63+: 1333 cm−1 (N-H3 bending of Co(NH3)63+), 491 and 441 cm−1 (Co-N stretching of Co(NH3)63+) and 315 cm−1 (N-Co-N deformation of Co(NH3)63+).
Figure 5.
Competitive binding of Mg2+ and Co(NH3)63+ in the HDV ribozyme. Raman difference spectrum of [HDV + 20mM Mg2+ + 5mM Co(NH3)63+] minus [HDV + 20mM Mg2+] at pH 7.0 was collected. The spectrum of the stabilization buffer was subtracted from each spectrum first. The differential peak around 1100 cm−1 directly indicates the loss of inner sphere bound Mg2+ upon Co(NH3)63+ binding. Band assignments are described in Table 1 and caption of Figure 4.
Binding of Co(NH3)63+ also perturbs several vibrational modes of the HDV ribozyme. The differential band of 1484/1475 cm−1 in Figure 4 represents perturbation of one or more guanine ring modes upon Co(NH3)63+ binding. The guanine base(s) is in an environment in the absence of Co(NH3)63+ that gives rise to the band of frequency of 1484 cm−1, which is similar to the frequency of guanine ring in aqueous GTP (27). In the presence of Co(NH3)63+ this band is reduced in intensity and/or shifts providing a less intensity feature at 1475 cm−1. A similar effect gives rise to a negative band at 1575 cm−1, which is due to guanine or possibly adenine (Table 1). The negative band at 1720 cm−1 indicates modest perturbation to interbase hydrogen-bonding in the presence of the metal ions, while the positive band at 777 cm−1 is assigned to C and/or U, and indicates a small change in the environment of these bases.
While binding of Co(NH3)63+ perturbs a number of vibrational modes of the ribozyme, it does not perturb PO2− bands of the ribozyme. In contrast to Mg2+ binding, which gives a strong PO2− differential band near 1100 cm−1 (Figure 2A), the Raman stabilization buffer-corrected difference spectrum of [HDV + Co(NH3)63+] minus [HDV no Co(NH3)63+] does not show peak perturbations in this region. This observation is consistent with our recent work that supported the ~ 20 cm−1 upshift of the 1100 cm−1 PO2− peak as being due to inner sphere interactions with Mg2+ (8). Thus, as expected, Co(NH3)63+ does not form inner sphere interactions with the ribozyme.
While many of the above features are expected when most RNAs are exposed to Co(NH3)63+, one feature is unique to the HDV ribozyme. Deprotonation of C75 is induced when Co(NH3)63+, like Mg2+, is soaked into HDV ribozyme crystals. Evidence of cytosine deprotonation includes spectral features at 1526, 1258 and 777 cm−1 (Figure 4), as described above for Mg2+. This observation suggests that Co(NH3)63+, like the Mg(H2O)52+ coordinated to the N7 of G1, binds in the active site close enough to C75 that it exhibits electrostatic repulsion with the bound proton.
Co(NH3)63+ binding induces ribozyme conformational changes
For Mg2+ binding, a positive differential band is observed at 825 cm−1 (Figure 2A), while for Co(NH3)63+ binding, a positive differential band is observed at 805 cm−1 (Figure 4). These differences support the appearance of slightly more canonical A-type RNA structures in the presence of Co(NH3)63+. The estimated change of intensity of -O-P-O- band at 805 cm−1 is ~2% of the parent band, which suggests that the main increase of canonical A-type RNA conformation (which has a 3′-endo sugar pucker) is very small.
Overall, comparison of the differential bands located at O-P-O- region in Figures 2A and 4 provides spectroscopic evidence that small conformational changes are induced by both Co(NH3)63+ and Mg2+ binding, but that the nature of these conformational changes is different: Mg2+ induces an increase in the mean amount of C2′-endo in the sugar backbone, while Co(NH3)63+ induces a decrease. These conformations are discussed in detail under “Mg2+ binding induces ribozyme conformational changes” above.
Competitive binding of Co(NH3)63+ and Mg2+: Structural analysis
Now that the separate effects of Mg2+ and Co(NH3)63+ have been presented, we consider interactions between these ions. Competitive binding between Co(NH3)63+ and Mg2+ in HDV crystals can be probed using a Raman difference experiment: [HDV + Mg2+ + Co(NH3)63+] minus [HDV + Mg2+], each with appropriate stabilization buffers subtracted (Figure 5). Most of the spectral features observed in this difference spectrum are similar to those for Co(NH3)63+ binding alone (Figure 4). Since the Raman spectrum of [HDV + Mg2+ + Co(NH3)63+] was recorded in the presence of 5 mM Co(NH3)63+ and 20 mM Mg2+, this observation supports the binding affinity of Co(NH3)63+ being significantly higher than that of Mg2+. This observation is consistent with solution experiments, which indicated ~50-fold tighter binding of Co(NH3)63+ (3). This observation is confirmed quantitatively in the crystal by analyzing the inhibition constant of Co(NH3)63+ (Ki,Co(NH3)63+) and the dissociation constant of Mg2+ (KD,Mg2+), as described below.
A significant feature of Figure 5 is the differential band of PO2− in 1100 cm−1 region, which is characterized by the positive band at 1100 with a trough at 1117 cm−1. This differential is the inverse of that observed in Figure 2A for inner sphere Mg2+ binding. The trough at 1117 cm−1 indicates less Mg2+ coordinated to the PO2− oxygen in the presence of Co(NH3)63+. Similarly, the positive feature at 1100 cm−1 reflects appearance of a new, unperturbed PO2− population. These observations comprise strong evidence that Co(NH3)63+ has displaced inner sphere Mg2+-PO2− complexes in the HDV ribozyme.
Using the intensity of the 1100 cm−1 differential in Figure 5 relative to the parent peak at 1100 cm−1 it is possible to assess that 1 to 1.5 Mg2+-PO2− inner sphere contacts are displaced by Co(NH3)63+ (8). This value is consistent with crystallographic studies of the HDV ribozyme, in which one high-occupancy Co(NH3)63+ binding site that overlaps with an inner sphere bound Mg2+ was observed (Figure 6A, B); we note that a second Co(NH3)63+ binding site was also observed, but it overlaps with an outer sphere bound Mg2+ ion (Figure 6C, D). We also note that the number of Mg2+ ions displaced is less than 5 (8) –the number of Mg2+ ions observed in the absence of Co(NH3)63+–indicating that Co(NH3)63+ cannot effectively displace all of the Mg2+ ions (see Discussion).
Figure 6.
Cobalt hexamine binding sites in the HDV ribozyme overlap with Mg2+ binding sites. The crystal structure of the inactive C75U ribozyme bound to a substrate was determined in the presence of Co(NH3)63+ (pdb ID 1SJF, shown in green) and in the presence of Mg2+ (pdb ID 1SJ3, shown in blue) (37). Co(NH3)63+ ion near the active site (A) (“site 1”), displaces a Mg2+ ion with in inner sphere ligand from U75 (B, highlighted in magenta). A second Co(NH3)63+ ion binds in the major groove of P4, near C41 (C) (“site 2”). This Co(NH3)63+ ion displaces a Mg(H2O)62+ ion bound to the RNA purely through second-shell ligands (D).
A further observation in the competition difference spectrum is the differential near 1489/1476 cm−1 (Figure 5). As observed from inspection of Figures 2A and 4, Mg2+ and Co(NH3)63+ ions give rise to differentials with opposite signs in this region, with the differential in Mg2+ being characteristic of coordination to the N7 of G1. Thus, the data in Figure 5 support the ability of a Co(NH3)63+ ion to displace the catalytic inner sphere Mg2+ ion at the N7 of G1.
Competitive binding of Co(NH3)63+ and Mg2+: Quantitative analysis
Lastly, a quantitative analysis of competition between Co(NH3)63+ and Mg2+ was performed. This was achieved by measuring the intensity ratio of the Co-N stretching band at 441 cm−1 referenced to 725 cm−1 (the same internal standard used in Figure 3) as a function of Co(NH3)63+ concentration. This titration was carried out in the background of 0, 1, 2, 3, 4 or 5 mM Mg2+ at pH 7.0 (Figure 7A).
Fitting the Co(NH3)63+ titration data to eq 2 using a Hill coefficient of 1.5 (see Materials and Methods) gave Ki, apps for Co(NH3)63+ of 0.046, 0.069, 0.085, 0.099, 0.119 and 0.167 mM, respectively. A secondary plot of K1.5i, app versus [Mg2+]1.5 (Figure 7B) using eq 3 afforded the apparent inhibition constant for Co(NH3)63+ (Ki, Co(NH3)63+) of 47 μM and the apparent dissociation constant of Mg2+ (KD,Mg2+) of 1.7 mM, respectively (Table 2). (This Ki is defined in eq 3 and is likely intermediate between the actual Kis for the two Co(NH3)63+ suggested from nHill.) These values are in agreement with previous biochemical results at pH 7.0 of 45 μM and 2.4 mM for Ki, Co(NH3)63 and KD,Mg2+, respectively (3), as well as the Hill value of 1.4 for Mg2+, suggesting that the conformation of the RNA in the crystal is catalytically relevant. In addition, the Hill constant of 1.5 supports binding of at least two Co(NH3)63+ ions in place of Mg2+ ions, consistent with spectral data presented above. A Hill coefficient of 2 also gave reasonable fits to the data, while Hill coefficients of 1 or 3 were less good, consistent with this interpretation (see Supporting Information).
DISCUSSION
Metal ions serve a variety of catalytic and structural roles in RNA. Divalent ions are particularly important as they can neutralize more charge with less entropy loss than monovalent ions. In the case of the HDV ribozyme, crystal structures have revealed the presence of many divalent metal ion binding sites (36, 37). These include fully hydrated magnesium ions found in the major groove and a partially hydrated magnesium ion that bridges the close approach of the negatively charged RNA backbone. A metal ion also appears to be recruited to the active site in the presence of the inactivating C75U mutation (37).
Co(NH3)63+ has been used as a mechanistic probe in both protein and RNA enzymes. As Jou and Cowan pointed out, Co(NH3)63+ is similar in size to Mg(H2O)62+, Co3+ is slow to exchange its ligands, and Co(NH3)63+ can hydrogen bond similarly to Mg(H2O)62+ (38). In some cases, protein and RNA enzymatic activity has been shown to be retained when Co(NH3)63+ is added into the reaction, while in other cases activity it is lost. Retention of activity has been used to infer a functional role for fully hydrated metal ions that do not coordinate directly to macromolecular ligands in the reaction. Examples include the proteins RNase H and exonuclease III (38, 39), as well as the hairpin ribozyme and an acyl-transferase ribozyme (12–14). In these instances, the inference was that the hydrated magnesium ion serves an electrostatic or structural role, but does not participate in proton transfer. Loss of activity through inhibition of the competitive type has also been used to suggest a functional role for an outer sphere metal ion in an enzymatic reaction, but in a more active role wherein the ability of the metal ion to deprotonate is lost (3). This follows because Co(NH3)63+ is similar in size and charge to Mg(H2O)62+ but lacks exchangeable ligands and the ability to participate in proton transfer under physiologically relevant conditions. In the case of the HDV ribozyme, the inference was that a fully hydrated magnesium ion serves as a general base in the reaction. Our results in the present study serve as a paradigm shifter because they show that Co(NH3)63+ can compete for Mg(H2O)4,52+ sites too. As such, retention of activity upon Co(NH3)63+ addition does not necessarily imply that the displaced magnesium was outer sphere bound (although they do show activity is at least compatible with an outer sphere coordination); likewise, loss of activity upon Co(NH3)63+ addition does not necessarily imply that the displaced magnesium was outer sphere bound. Apparently, the stronger Coulombic attractions and plasticity of the macromolecule allow the outer sphere trivalent ion to bind into inner sphere sites.
In the present study, we used Raman spectroscopic data to analyze the nature of Mg2+ ion interaction with the HDV ribozyme by studying Mg2+ binding, Co(NH3)63+ binding, and their competition. A key finding of our studies is that Co(NH3)63+, which binds in an outer sphere fashion, can displace several Mg2+ ions with inner sphere ligands, including the Mg2+ ion at G1. Importantly, the qualitative and quantitative results observed herein are fully consistent with solution data, supporting catalytic relevance of the interactions.
Nature of the interactions between metal ions and RNA
The backbone of RNA is negatively charged, and many magnesium binding sites within folded RNAs involve direct coordination between divalent metal ions and non-bridging phosphate oxygens. Our earlier observations showed that inner sphere Mg2+—PO2− contacts in HDV give rise to a strong spectral perturbation of PO2− symmetric stretch peak near 1100 cm−1, which take the form of a significant intensity loss of the PO2− peak combined with an ~20 cm−1 higher frequency shift (8) Similar effects have been noted in model compounds (Christian, Gong, Carey, unpublished observations). The inner sphere nature of this ligand was confirmed by a positive correlation between the perturbation to the PO2− stretch and the appearance of a Mg(H2O)x2+ ion (x≤5) mode near 320 cm−1. The present work builds on these findings, and the data in Figure 2 show that the changes in HDV upon Mg2+ binding extend beyond the PO2− groups to include changes in base ring modes and in the conformation of the ribose groups within the backbone.
The changes brought about by Co(NH3)63+ binding in HDV crystals are markedly different than those observed upon Mg2+ ion binding. Since the cobalt complex cannot make inner sphere contact to the phosphate oxygens, no band perturbation near 1100 cm−1 is observed (Figure 4). Moreover, when 5 mM Co(NH3)63+ is allowed to compete with 20 mM Mg2+ a 1117/1100 cm−1 differential is revealed in the Raman difference data (Figure 5). This feature was quantitated to show that Co(NH3)63+ displaces Mg(H2O)x2+ ion (x≤5) at approximately 1 – 1.5 inner sphere sites PO2− sites in the HDV ribozyme.
RNAs attract a cloud of diffusely bound positive ions whose binding is weak (millimolar) and likely highly dynamic (40, 41), and thus, it can be envisaged that Co(NH3)63+ can out-compete Mg2+ at these sites on an electrostatic basis. This contrasts with Mg(H2O)x2+(x≤5) ions that are bound within discrete electronegative binding pockets formed by the RNA and are often visible in X-ray crystal structures. Thus, competition between Mg(H2O)x2+ and Co(NH3)63+ may occur at 1–2 individual sites or may result from the mean of lower occupancies at more sites. Previous studies have suggested that there are 5–6 Mg2+ ions within the in the HDV ribozyme that coordinate to -PO2− and could contribute to this differential (8). Our finding that Co(NH3)63+ can displace Mg(H2O)x2+ with inner sphere contacts at the oxygens of PO2− is at variance with the generally stated view that Co(NH3)63+ can only displace Mg2+ at outer sphere sites, although it is also clear that Co(NH3)63+ cannot displace all inner sphere Mg2+ ions.
Raman difference spectra also reveal metal-dependent changes in the bases. In competition experiments between Mg2+ and Co(NH3)63+ a differential band corresponding to the guanosine base is observed around 1480 cm−1 (Figure 5). The negative limb at 1489 cm−1 is likely due to release of Mg2+ bound at the N7 of G (9) while the positive limb at 1476 is assigned to G free from any strong direct interaction with a metal. These data suggest that Co(NH3)63+ can displace a Mg(H2O)x2+ ion that makes an inner sphere contact to the N7 of a guanosine. In addition, at least two Mg2+ ions are displaced as suggested by the Hill coefficient of 1.5 for Co(NH3)63+ competition with Mg2+ ions bound (Figure 7). Thus, it is likely that at least one Mg2+ ion at an inner sphere complex with PO2− is displaced. As Mg2+ ions rarely make simultaneous contact to an N7 and a non-bridging phosphate oxygen, the Mg(H2O)x2+ ion displaced from the N7 of G is likely distinct from the ion(s) displaced at non-bridging phosphate oxygens.
Sugar pucker changes may be due to C41 and C75
The Raman spectra near 800 cm−1 are sensitive to changes in the phosphodiester backbone conformation. This region undergoes small and opposing changes when either Mg2+ or Co(NH3)63+ bind in HDV crystals. The data suggest that 2–3 % of the linkages undergo a change in the ribose moiety when Mg2+ binds. In addition, the changes are in opposite directions for the two ions, with Mg2+ binding slightly increasing the C2′-endo and cobalt hexamine increasing the C3′-endo conformations (27, 28, 34).
Analysis of the cleaved structure of the ribozyme by MC-annotate (42) reveals that only two HDV ribozyme nucleotides are in the C2′-endo-anti conformation: C41 and C75. Moreover, both of these protonatable residues have been implicated as being near Mg2+ ions (36, 43), consistent with appearance of these rare sugar puckers upon Mg2+ uptake. The crystal structure of the cleaved form of the ribozyme likely represents protonated C41 and deprotonated C75 (36, 44). This is because C41 protonation couples favorably with Mg2+ binding,4 while C75 couples unfavorably; moreover, the scissile phosphate is likely necessary for upward pKa shifting of C75 and is absent in this structure. Together, these and other observations suggest that Mg2+ binding drives deprotonation of C75 and protonation of C41, both of which give rise to C2′-endo-anti conformations. Since there are 71 nt in our crystallographic construct, changes at these two residues could account for all of the ~3% observed signal intensity change.
Quantitative analysis of metal ion and proton binding in the HDV ribozyme-comparison with other studies
Previous Raman crystallographic studies suggest that protonation of C75 and Mg2+ binding in the crystal form of the HDV ribozyme are similar to in solution (26). For example, pKa values of 6.15 and 6.4 in the presence of 20 and 2 mM Mg2+ measured for catalytic C75 in crystal (24) are consistent with solution pKas of 6.1 and 6.5 in 10 and 1.9 mM Mg2+ (3), respectively. Here, we found that the pH 7.0 inhibition constant for Co(NH3)63+ (Ki,Co(NH3)63+) and dissociation constant (KD,Mg2+) for Mg2+ are 47 μM and 1.7 mM in the crystal, in good agreement with values of 45 μM and 2.4 mM in solution (Table 2) (3). In addition, the Hill values for Mg2+ binding are in agreement in the crystal and solution with values of 1.5 and 1.4, respectively (Table 2). These data strongly support metal ion binding behavior in the crystal being essentially the same as in solution.
The Hill coefficient for Mg2+ binding obtained by measuring deprotonation of C75 as function of Mg2+ concentration was determined to be 8.0 ± 1.6 (Figure 3). This number differs from the Hill coefficient determined by examining the dependence of the rate constant for chemistry on the Mg2+ concentration (3). It is has been noted, however, that Hill coefficient >1 may not reflect the true number of metal binding sites (45). One possibility is that binding of the structural and catalytic ions are accompanied by the 5–6 additional ions observed in crystal structures of the ribozyme. Such ions could be silent to functional experiments in solution, which is consistent with their occupancy of non-active-site regions of the ribozyme such as within the major grooves of double helices.
Binding of either Mg2+ or Co(NH3)63+ induces deprotonation of C75
Solution kinetic studies indicate that the proton on C75 binds anticooperatively with a magnesium ion in the HDV ribozyme active site. In our previous characterization of the pKa of C75, this is revealed as a shift of the pKa of C75 toward neutrality as the magnesium ion concentration is reduced. In the present study, we observe related effects.
Introduction of the magnesium ion to the HDV ribozyme active site induces deprotonation of C75. At pH 6.0, binding of ≥ 10 mM Mg2+ (Figure 3) results in net gain of approximately one half of a neutral cytosine. Since this pH is close to the measured pKa of C75 and C75 is the only protonatable cytosine in this pH range, we interpret this finding in terms of protonation/deprotonation of the active site cytosine. As with the previous study, the values obtained by Raman crystallography agree with values obtained by kinetics studies in solution, suggesting that the conformation of the ribozyme in the crystal is relevant to the conformation of the ribozyme in solution.
In contrast to Mg2+ binding, introduction of Co(NH3)63+ to the HDV ribozyme is inhibitory; thus, it is difficult to measure the effects of this ion on pKa using solution kinetic methods. As Raman spectroscopy does not depend on ribozyme activity for readout, we used it to ask whether Co(NH3)63+ and Mg2+ have similar effects on the pKa of C75. As observed in Figure 4, binding of Co(NH3)63+ to the HDV ribozyme induces cytosine deprotonation. These data suggest that one of the binding sites for Co(NH3)63+ is within the HDV ribozyme active site such that the metal ion and the proton on C75 interact by electrostatic repulsion.
Structural basis for Co(NH3)63+ inhibition of the HDV ribozyme
A unique characteristic of the HDV ribozyme is its inhibition by Co(NH3)63+ ions. While many of the small ribozymes fold and function in the absence of Mg2+ and presence of high concentrations of monovalent cations, and the hairpin ribozyme functions in the presence of Co(NH3)63+ (12, 13), the HDV ribozyme is strongly inhibited by this Mg(H2O)62+ mimic. Binding of Co(NH3)63+ to the HDV ribozyme has been shown to be tight (Ki,Co(NH3)63+ ~45 μM) and competitive with Mg(H2O)62+ (KD,Mg2+ ~2.4 mM) (3). There are two models by which Co(NH3)63+ could have this effect on ribozyme activity: Model 1, in which there are overlapping binding sites for Co(NH3)63+ and catalytic Mg2+, or Model 2, in which Co(NH3)63+ induces inhibitory conformational changes.
In favor of Model 1, there is significant evidence that supports overlapping binding sites for Co(NH3)63+ and Mg(H2O)x2+ (x≤5) within the HDV ribozyme active site. Raman data provide direct evidence for displacement of inner sphere Mg2+ by Co(NH3)63+, and binding of Co(NH3)63+ and Mg(H2O)x2+ both induce deprotonation of C75. Thus, both of these ions are bound within the HDV ribozyme active site in proximity to C75. In addition, there is evidence that the inner sphere Mg2+ displaced by Co(NH3)63+is bound to a guanosine N7, and we have previously demonstrated that an Mg(H2O)x2+ ion is bound to the N7 of G1. Finally, solution biochemical data have demonstrated that Co(NH3)63+ inhibits the HDV ribozyme by binding competitively with Mg2+ (3). Thus, solution and Raman crystallographic data are both consistent with a model in which Co(NH3)63+ inhibits the HDV ribozyme by displacing the active site Mg(H2O)x2+ bound to G1. It is noteworthy that Been et al. (46) also suggested the possibility of the replacement of the inner sphere coordinated catalytic Mg2+ by Co(NH3)63+ due to the flexibility of HDV ribozyme active-site.
Nevertheless, there is also support for Model 2, in which Co(NH3)63+ inhibits the HDV ribozyme by inducing a conformational change upon displacing Mg2+ ions and stabilizing an inactive conformation of the ribozyme. Small conformational changes could play a critical role in positioning functional groups for the in-line attack during catalysis. Such a mechanism has been observed in the hammerhead ribozyme wherein lanthanides stabilize a ground-state, nonfunctional conformation of the ribozyme (47). Indeed, we have demonstrated that addition of Mg2+ or Co(NH3)63+ to the HDV ribozyme induces unique minor conformational changes of opposite effect and of unknown origin.
Of course, model 1 and model 2 are not mutually exclusive. Changes in activity may arise both from the ability of Co(NH3)63+ to displace inner sphere bound catalytic Mg2+ in HDV and from different conformations caused by Mg2+ and Co(NH3)63+ binding.
Finally, there is the observation that monovalent cations such as Na+ support catalysis, albeit less efficiently than Mg2+, while Co(NH3)63+ is inhibitory. If Model 1 best describes the mechanism of Co(NH3)63+ inhibition, then activity with small monovalent ions may be related to their ability to directly coordinate to the N7 and provide an ordered water from their hydration shell for general base catalysis, as seen crystallographically with Tl+ (7). However, if Model 2 is correct, then monovalent ions may simply support an active fold similar to that seen in the presence of Mg2+.
Clearly, it will take additional studies to fully understand roles of metal ions in the HDV ribozyme. Further, X-ray crystallographic analysis of the HDV ribozyme active site with a substrate-analog bound could provide deeper understanding of how metal ions bind within the active site. In addition, isotopic substitutions at the N7 of G1 and at specific phosphodiester bonds would allow Raman spectroscopy to follow these changes at the site-specific level.
Supplementary Material
Acknowledgments
We thank Mike Harris and Eric Christian for sharing unpublished data.
Abbreviations
- HDV
hepatitis delta virus
- Mg(H2O)x2+(x≤5)
inner sphere Mg hydrate
- -O-P-O-
phosphodiester backbond
- PO2−
phosphodioxy group
- Ki,app
apparent binding constant of Co(NH3)63+
- KiCo(NH3)63+
apparent inhibition constant of Co(NH3)63+
- KDMg2+
apparent dissociation constant of Mg2+
- n or nHill
Hill coefficient
- C75
cytosine residue at position 75 of the genomic HDV ribozyme
- C75U
cytosine-to-uracil mutation at position 75 of the genomic HDV ribozyme
Footnotes
Differential bands are of special interest in the Raman because they generally mark where molecular features of metal-free RNA have been converted to features of metal-bound RNA.
The fraction of deprotonated cytosine, assuming a simple single protonation event, can be calculated as , which gives a value of 0.41 using a pH of 6.0 and a pKa of 6.15. Given that the concentration of RNA in the crystal is 26 mM, approximately 10.7 mM would deprotonate upon raising the Mg2+ concentration, consistent with the data herein. These calculations assume that the pKa for C75 in the absence of Mg2+ is significantly elevated, consistent with an experimental pKa of 7.25 extrapolated to no bound Mg2+ from pH-dependent Mg2+ binding experiments in solution (42).
For pH titrations, we previously used the PO2− stretch at 1101 cm−1 to standardize the 1528 cm−1 stretch (24), but that was not used here because the 1101 cm−1 feature is sensitive to Mg2+ binding (Figure 2A).
Protonation of C41 is likely invisible to the Raman features observed for CMP and C75. This is because C41, but not C75 or CMP, engages in extensive base pairing when it protonates
This project was supported by NSF Grant MCB-0527102 (P.C.B.), the Purdue University Department of Biochemistry and Cancer Center (B.L.G), and NIH Grant GM-54072 (P.R.C.).
SUPPORTING INFORMATION AVAILABLE
Derivations of equation 1, 2 and 3, and analysis of competitive binding of Co(NH3)63+ and Mg2+ in the HDV ribozyme when Hill coefficient is held to 2 (Figure S1). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Suh YA, Kumar PK, Taira K, Nishikawa S. Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res. 1993;21:3277–3280. doi: 10.1093/nar/21.14.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih IH, Been MD. Ribozyme cleavage of a 2,5-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA. 1999;5:1140–1148. doi: 10.1017/s1355838299990763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 4.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 5.Nakano S, Cerrone AL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 6.Perrotta AT, Been MD. HDV ribozyme activity in monovalent cations. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- 7.Ke A, Ding F, Batchelor JD, Doudna JA. Structural roles of monovalent cations in the HDV ribozyme. Structure. 2007;15:281–287. doi: 10.1016/j.str.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Gong B, Chen YY, Christian EL, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Detection of Inner-sphere Interactions between Magnesium Hydrate and the Phosphate Backbone of the HDV Ribozyme Using Raman Crystallography. J Am Chem Soc. 2008;130:9670–9672. doi: 10.1021/ja801861s. [DOI] [PubMed] [Google Scholar]
- 9.Chen JH, Gong B, Bevilacqua PC, Carey PR, Golden BL. A catalytic metal ion interacts with the cleavage Site G.U wobble in the HDV ribozyme. Biochemistry. 2009;48:1498–1507. doi: 10.1021/bi8020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadkins TS, Shih I, Perrotta AT, Been MD. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J Mol Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 11.Cerrone-Szakal AL, Siegfried NA, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: solvent isotope effects and proton inventories in the absence of divalent metal ions support C75 as the general acid. J Am Chem Soc. 2008;130:14504–14520. doi: 10.1021/ja801816k. [DOI] [PubMed] [Google Scholar]
- 12.Hampel A, Cowan JA. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 13.Nesbitt S, Hegg LA, Fedor MJ. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 14.Young KJ, Gill F, Grasby JA. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duguid J, Bloomfield VA, Benevides J, Thomas GJ., Jr Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys J. 1993;65:1916–1928. doi: 10.1016/S0006-3495(93)81263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duguid JG, Bloomfield VA, Benevides JM, Thomas GJ., Jr Raman spectroscopy of DNA-metal complexes. II. The thermal denaturation of DNA in the presence of Sr2+, Ba2+, Mg2+, Ca2+, Mn2+, Co2+, Ni2+, and Cd2+ Biophys J. 1995;69:2623–2641. doi: 10.1016/S0006-3495(95)80133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JH, Xiao DG, Deng H, Callender R, Webb MR. Vibrational study of phosphate modes in GDP and GTP and their interaction with magnesium in aqueous solution. Biospectroscopy. 1998;4:219–227. doi: 10.1002/(SICI)1520-6343(1998)4:4%3C219::AID-BSPY1%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Langlais M, Tajmir-Riahi HA, Savoie R. Raman spectroscopic study of the effects of Ca2+, Mg2+, Zn2+, and Cd2+ ions on calf thymus DNA: binding sites and conformational changes. Biopolymers. 1990;30:743–752. doi: 10.1002/bip.360300709. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad R, Arakawa H, Tajmir-Riahi HA. A comparative study of DNA complexation with Mg(II) and Ca(II) in aqueous solution: major and minor grooves bindings. Biophys J. 2003;84:2460–2466. doi: 10.1016/S0006-3495(03)75050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller MR, Bruck MA, O’Connor T, Armatis FJ, Knolinski EA, Kottmair N, Tobias RS. Heavy metal -nucleotide interactions. 14. Raman difference spectrophotometric studies of competitive reactions in mixtures of four nucleotides with electrophiles. Factors governing selectivity in the binding reactions. Journal of the American Chemical Society. 1980;102:4589–4598. [Google Scholar]
- 21.O’Connor T, Bina M, McMillin DR, Haley MR, Tobias RS. Heavy metal-nucleotide interactions. 15. Reactions of calf thymus DNA with the electrophiles methylmercury(II) nitrate, cis-dichlorodiammineplatinum(II), and trans-dichlorodiammineplatinum(II) studied using Raman difference spectroscopy. Evidence for the formation of C-DNA upon metalation. Biophys Chem. 1982;15:223–234. [PubMed] [Google Scholar]
- 22.Callender R, Deng H. Nonresonance Raman difference spectroscopy: a general probe of protein structure, ligand binding, enzymatic catalysis, and the structures of other biomacromolecules. Annu Rev Biophys Biomol Struct. 1994;23:215–245. doi: 10.1146/annurev.bb.23.060194.001243. [DOI] [PubMed] [Google Scholar]
- 23.Carey PR. Raman crystallography and other biochemical applications of Raman microscopy. Annu Rev Phys Chem. 2006;57:527–554. doi: 10.1146/annurev.physchem.57.032905.104521. [DOI] [PubMed] [Google Scholar]
- 24.Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Direct measurement of a pK(a) near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J Am Chem Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 25.Gong B, Chen JH, Yajima R, Chen YY, Chase E, Chadalavada DM, Golden BL, Carey PR, Bevilacqua PC. Raman Crystallography of RNA. Methods. 2009 doi: 10.1016/j.ymeth.2009.1004.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. Direct measurement of a pK(a) near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J Am Chem Soc. 2007;129:13335–13342. doi: 10.1021/ja0743893. [DOI] [PubMed] [Google Scholar]
- 27.Thomas GJ, Jr, Tsuboi M. Raman Spectroscopy of Nucleic Acids and Their Complexes. Advances in Biophysical Chemistry. 1993:1–70. [Google Scholar]
- 28.Carey PR. Biochemical Application of Raman and Resonance Raman Spectroscopies. Academic Press; New York: 1982. [Google Scholar]
- 29.Lafleur L, Rice J, Thomas GJ., Jr Raman studies of nucleic acids. VII. Poly A-poly U and poly G-poly C. Biopolymers. 1972;11:2423–2437. doi: 10.1002/bip.1972.360111205. [DOI] [PubMed] [Google Scholar]
- 30.Fish SR, Hartman KA, Stubbs GJ, Thomas GJ., Jr Structural studies of tobacco mosaic virus and its components by laser Raman spectroscopy. Biochemistry. 1981;20:7449–7457. doi: 10.1021/bi00529a019. [DOI] [PubMed] [Google Scholar]
- 31.Small EW, Peticolas WL. Conformational dependence of the Raman scattering intensities from polynucleotides. 3. Order-disorder changes in helical structures. Biopolymers. 1971;10:1377–1418. doi: 10.1002/bip.360100811. [DOI] [PubMed] [Google Scholar]
- 32.Majoube M. Vibrational spectra of guanine. A noral coordinate analysis. J Chim Phys. 1984;81:303–315. [Google Scholar]
- 33.Moller MR, Bruck MA, O’Connor T, Armatis FJ, Jr, Knolinski EA, Kottmair N, Tobias RS. Heavy metal-nucleotide interactions. 14. Raman difference spectrophotometric studies of competitive reactions in mixtures of four nucleotides with the electrophiles methylmercury(II) perchlorate, cis dimethylgold(III) perchlorate, dichloroethylene-diamine-palladium(II), trans-dichlorodiaminepalladium(II), and aquopentaaminecobalt(III) perchlorate. Factors governing selectivity in the binding reactions. J Am Chem Soc. 1980;102:4589–4598. [Google Scholar]
- 34.Rodriguez-Casado A, Bartolome J, Carreno V, Molina M, Carmona P. Structural characterization of the 5′ untranslated RNA of hepatitis C virus by vibrational spectroscopy. Biophys Chem. 2006;124:73–79. doi: 10.1016/j.bpc.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Hobro AJ, Rouhi M, Blanch EW, Conn GL. Raman and Raman optical activity (ROA) analysis of RNA structural motifs in Domain I of the EMCV IRES. Nucleic Acids Res. 2007;35:1169–1177. doi: 10.1093/nar/gkm012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 37.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 38.Jou R, Cowan JA. Ribonuclease H activation by inert transition-metal complexes. Mechanistic probes for metallocofactors: insights on the metallobiochemistry of divalent magnesium ion. J Am Chem Soc. 1991;113:6685–6686. [Google Scholar]
- 39.Duffy TH, Nowak T. 1H and 31P relaxation rate studies of the interaction of phosphoenolpyruvate and its analogues with avian phosphoenolpyruvate carboxykinase. Biochemistry. 1985;24:1152–1160. doi: 10.1021/bi00326a014. [DOI] [PubMed] [Google Scholar]
- 40.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draper DE. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.http://www-lbit.iro.umontreal.ca/mcannotate-simple.
- 43.Nakano S, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: a mutant of the C41 motif provides insight into the positioning and thermodynamic linkage of metal ions and protons. Biochemistry. 2007;46:3001–3012. doi: 10.1021/bi061732s. [DOI] [PubMed] [Google Scholar]
- 44.Ferre-D’Amare AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 45.Weiss JN. The Hill equation revisited: uses and misuses. Faseb J. 1997;11:835–841. [PubMed] [Google Scholar]
- 46.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu Rev Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 47.Feig AL, Scott WG, Uhlenbeck OC. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb. Science. 1998;279:81–84. doi: 10.1126/science.279.5347.81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.